Abstract
The elevation of [cAMP]i is an important mechanism of platelet inhibition and is regulated by the opposing activity of adenylyl cyclase and phosphodiesterase (PDE). In this study, we demonstrate that a variety of platelet agonists, including thrombin, significantly enhance the activity of PDE3A in a phosphorylation-dependent manner. Stimulation of platelets with the PAR-1 agonist SFLLRN resulted in rapid and transient phosphorylation of PDE3A on Ser312, Ser428, Ser438, Ser465, and Ser492, in parallel with the PKC (protein kinase C) substrate, pleckstrin. Furthermore, phosphorylation and activation of PDE3A required the activation of PKC, but not of PI3K/PKB, mTOR/p70S6K, or ERK/RSK. Activation of PKC by phorbol esters also resulted in phosphorylation of the same PDE3A sites in a PKC-dependent, PKB-independent manner. This was further supported by the finding that IGF-1, which strongly activates PI3K/PKB, but not PKC, did not regulate PDE3A. Platelet activation also led to a PKC-dependent association between PDE3A and 14-3-3 proteins. In contrast, cAMP-elevating agents such as PGE1 and forskolin-induced phosphorylation of Ser312 and increased PDE3A activity, but did not stimulate 14-3-3 binding. Finally, complete antagonism of PGE1-evoked cAMP accumulation by thrombin required both Gi and PKC activation. Together, these results demonstrate that platelet activation stimulates PKC-dependent phosphorylation of PDE3A on Ser312, Ser428, Ser438, Ser465, and Ser492 leading to a subsequent increase in cAMP hydrolysis and 14-3-3 binding.
Upon vascular injury, platelets adhere to the newly exposed subintimal collagen and undergo activation leading to platelet spreading to cover the damaged region and release of thrombogenic factors such as ADP and thromboxane A2. In addition, platelets are activated by thrombin, which is generated as a result of activation of the coagulation pathway, and stimulates platelets by cleaving the protease-activated receptors (PAR),2 PAR-1 and PAR-4. The final common pathway is the exposure of fibrinogen binding sites on integrin αIIbβ3 resulting in platelet aggregation and thrombus formation.
Thrombin-mediated cleavage of PARs leads to activation of phospholipase C β (PLC), hydrolysis of phosphatidylinositol (PI) 4,5-bisphosphate and a subsequent increase in [Ca2+]i and activation of protein kinase C (PKC). Protein kinase C contributes to platelet activation both directly, through affinity regulation of the fibrinogen receptor, integrin αIIbβ3 (1), and indirectly by enhancing degranulation (2). Thrombin also stimulates activation of PI 3-kinases and subsequent generation of PI (3, 4, 5) trisphosphate and PI (3, 4) bisphosphate (3), which recruit protein kinase B (PKB) to the plasma membrane where it becomes phosphorylated and activated.
Platelet activation is opposed by agents that raise intracellular 3′-5′-cyclic adenosine monophosphate ([cAMP]i). cAMP is a powerful inhibitory second messenger that down-regulates platelet function by interfering with Ca2+ homeostasis, degranulation and integrin activation (4). Synthesis of cAMP is stimulated by mediators such as prostaglandin I2 (PGI2), which bind to Gs-coupled receptors leading to activation of adenylate cyclase (AC). This inhibitory pathway is opposed by thrombin, which inhibits the elevation of cAMP indirectly via autocrine activation of the Gi-coupled ADP receptor P2Y12. cAMP signaling is terminated by hydrolysis to biologically inert 5′-AMP by 3′-phosphodiesterases. Platelets express two cAMP phosphodiesterase isoforms, cGMP-stimulated PDE2 and cGMP-inhibited PDE3A. PDE3A is the most abundant isoform in platelets and has a ∼250-fold lower Km for cAMP than PDE2 (4). As a consequence of these properties, PDE3A exerts a greater influence on cAMP homeostasis, particularly at resting levels. The importance of PDE3A in platelet function is further emphasized by the finding that the PDE3A inhibitors cilostamide and milrinone raise basal cAMP levels and strongly inhibit thrombin-induced platelet activation (5). Furthermore, PDE3A-/- mice demonstrate increased resting levels of platelet cAMP and are protected against a model of pulmonary thrombosis (6). In contrast, the PDE2 inhibitor EHNA has no significant effect on cAMP levels and platelet aggregation (7, 8). The activity of PDE3A is therefore essential to maintain low equilibrium levels of cAMP and determine the threshold for platelet activation (7).
Like its paralogue PDE3B, it has recently become clear that PDE3A activity can be positively regulated by phosphorylation in platelets and human oocytes (9, 10). There is some evidence that PKB may be involved in this regulation, although the phosphorylation sites are poorly characterized. In contrast, phosphorylation of PDE3A in HeLa cells was stimulated by phorbol esters and blocked by inhibitors of PKC (11). In this study, we aimed to identify the signaling pathways and phosphorylation sites that are involved in regulation of platelet PDE3A. Here, we show strong evidence that PKC, and not PKB, is involved in agonist-stimulated PDE3A phosphorylation on Ser312, Ser428, Ser438, Ser465, and Ser492, leading to an increase in PDE3A activity, 14-3-3 binding and modulation of intracellular cAMP levels.
EXPERIMENTAL PROCEDURES
Materials—Total and site-specific phospho-PDE3A antibodies were generated as reported previously (11). pSer473 PKB, PKC phospho-motif (used for analysis of pleckstrin phosphorylation, pThr202/Tyr204 ERK, pSer65 4EBP1, and pSer239 VASP antibodies were from Cell Signaling Technologies (New England Biolabs, Hitchin, UK). PKBα and 14-3-3 (K-19), antibodies were from Santa Cruz (Insight Biotechnology, Wembley, UK)). ERK antibody and recombinant PKBα and PKCα were from Upstate Biotechnology (Millipore, Watford, UK). VASP (M4) antibody, U46619 and microcystin-LR were from Axxora (Nottingham, UK). Pleckstrin antibody was from Abcam (Cambridge, UK). 4EBP1 antiserum was provided by Professor Dick Denton (University of Bristol, UK). U73122, GF109203X, wortmannin, rapamycin, and U0126 were from Tocris (Avonmouth, UK). Cilostamide, forskolin, H89, and rottlerin were from Merck Chemicals (Nottingham, UK). PAR-1-activating peptide (SFLLRN-NH2) was from Bachem (Weil am Rhein, Germany). Cross-linked collagen-related peptide (CRP) was provided by Professor Richard Farndale (University of Cambridge). 8-[3H]adenosine 3′5′-cyclic monophosphate, ammonium salt, and enhanced chemiluminescent detection reagents were from GE Healthcare (Bucks, UK). Peroxidase-conjugated secondary antibodies were from Jackson (Stratech, Newmarket, UK). NuPAGE SDS-PAGE sample buffer was from Invitrogen (Paisley, UK). The P2Y12 antagonist AR-C69931-MX (Cangrelor) was a gift from AstraZeneca (Alderley Park, UK). All peptides were synthesized by Professor Graham Bloomberg (University of Bristol). All other reagents were sourced from Sigma unless otherwise indicated.
Isolation of Human Platelets—Platelets were isolated from whole blood as previously described (12) and resuspended at 4 × 108/ml in modified HEPES-tyrode buffer (145 mm NaCl, 3 mm KCl, 0.5 mm Na2HPO4, 1 mm MgSO4, 10 mm HEPES pH 7.2, 0.1% (w/v) d-glucose, 0.02 units/ml apyrase, and 10 μm indomethacin).
Platelet Extraction and Immunoprecipitation—Washed platelets were incubated with vehicle (0.2% DMSO) or compound at the indicated concentration for 10 min at 37 °C prior to addition of stimulus. At the required time point, platelets were extracted by the addition of 1 volume of ice-cold Nonidet P-40 extraction buffer (50 mm HEPES pH 7.4, 2% Nonidet P-40, 120 mm NaCl, 40 mm sodium β-glycerophosphate, 20 mm tetrasodium pyrophosphate, 2 mm benzamidine, 2 mm EDTA, 10 mm sodium orthovanadate 2 μm microcystin-LR, and 2 μg/ml each pepstatin, antipain, and leupeptin). Whole cell lysate was prepared by direct lysis in NuPAGE sample buffer. PDE3A was immunoprecipitated from platelet lysates by incubation with 1 μg anti-PDE3A for 3 h. Immune complexes were captured with 10 μl of protein G-Sepharose for 1 h and washed with extraction buffer prior to elution with NuPAGE sample buffer.
14-3-3 Co-immunoprecipitation—For co-immunoprecipitation experiments, platelet lysates were incubated with 2 μg of anti-PDE3A covalently coupled to cyanogen bromide-activated Sepharose 4B for 4 h. Briefly, anti-PDE3A was exchanged into bicarbonate buffer (0.1 m NaHCO3 pH 8.3, 0.5 m NaCl) by ultrafiltration. CnBr-activated agarose was washed extensively with ice-cold 1 mm HCl to remove stabilizing agents. Swelled resin was equilibrated with bicarbonate buffer and incubated with anti-PDE3A for 90 min at room temp. Remaining active residues were quenched with 0.1 m Tris, pH 8, for a further 90 min. Uncoupled antibody was removed by sequential washes with bicarbonate and acetate buffer (0.1 m acetate, pH 4.0, 0.5 m NaCl). In some instances, 14-3-3 proteins were eluted from PDE3A immunoprecipitates by competition with 1 mm of the indicated synthetic peptide (in the presence of 5% glycerol) for 20 min at room temperature. Beads were subsequent washed twice in Nonidet P-40 extraction buffer including 1 mm peptide, followed by two washes in Nonidet P-40 extraction buffer without peptide.
In Vitro Phosphorylation—PDE3A immunoprecipitates were washed with kinase buffer (20 mm MOPS pH 7.2, 20 mm sodium β-glycerophosphate, 15 mm MgCl2, 1 mm sodium orthovanadate, 1 mm dithiothreitol) Additionally, 1 mm EGTA was added for PKB and 0.5 mm CaCl2, 50 μg/ml phosphatidylserine, 5 μg/ml diacylglycerol was added for optimal PKCα activity. Samples were incubated with 0.1 mm ATP and 0.5 units of kinase for 30 min at 30 °C prior to phosphodiesterase assay or elution with NuPAGE sample buffer and analysis by Western blotting.
In Vitro Dephosphorylation—Immunoprecipitated PDE3A was washed three times with calf intestinal phosphatase buffer (50 mm Tris-HCl, pH 8.0, 100 mm NaCl, 10 mm MgCl2, 1 mm dithiothreitol) and incubated with 5 units of calf intestinal phosphatase at 37 °C for 30 min. Reactions were transferred to ice and assayed for phosphodiesterase activity. As a control, duplicate samples were incubated with heat-inactivated phosphatase.
Electrophoresis and Immunoblotting—Proteins were separated on 8% bis-Tris gels (357 mm bis-Tris pH 6.5, 8% acrylamide 1.5% bis-acrylamide) using MOPS running buffer (50 mm MOPS, 50 mm Tris, 1 mm EDTA, 0.1% SDS, 5 mm sodium bisulfite) and transferred to polyvinylidene difluoride membranes. Membranes were blocked using 5% nonfat milk in Tris-buffered saline (20 mm Tris pH 7.5, 137 mm NaCl, 0.1% Tween-20) for 1 h at room temp and incubated with primary and secondary antibodies prepared in 5% (w/v) bovine serum albumin. Membranes were developed using an enhanced chemiluminescence detection system. Membranes were stripped in 62.5 mm Tris, pH 6.8, 2% SDS, 100 mm 2-mercaptoethanol, and reprobed for total protein to confirm equal loading.
Phosphodiesterase Assay—Platelets were treated as indicated and lysed with 1 volume of ice-cold extraction buffer (50 mm HEPES 7.4, 2% Triton X-100, 100 mm NaCl, 2 mm EDTA, 40 mm sodium β-glycerophosphate, 20 mm tetra-sodium pyrophosphate, 2 mm benzamidine, 2 μg/ml pepstatin/antipain/leupeptin, 10 mm sodium orthovanadate, 2 μm microcystin-LR, 20% glycerol). Extracts were snap-frozen in liquid nitrogen and stored at -80 °C prior to assay. Samples were rapidly thawed in a water bath at room temperature and 150 μl of lysate incubated with 1 μg of anti-PDE3A antibody and 10 μl of protein G-Sepharose for 3 h at 4 °C. Immune complexes were washed twice with lysis buffer and three times with assay buffer (20 mm HEPES pH 7.4, 10 mm MgCl2 and 5 mm 2-mercaptoethanol). Phosphodiesterase activity was measured using a modification of the two-step radiometric assay of Thompson and Appleman (13). Briefly, PDE3A immunoprecipitates (1/5th per reaction) were incubated for 10 min at 30 °C with 1 μm cAMP and 0.15 μCi of [3H]cAMP in a total volume of 100 μl. Reactions were terminated in a boiling water bath (2 min). Then, 25 μg of Crotalus atrox venom (a source of 5-nucleotidase) was added and incubated for 15 min at 30 °C. Charged species were extracted with 0.4 ml of Dowex 1 × 8 resin (1:1:1 resin:water:ethanol) for 15 min and pelleted at 14,000 × g for 3 min. [3H]Adenosine in the supernatant was determined by scintillation counting. The rate of hydrolysis was expressed as pmol of cAMP·min-1·108 platelets-1. Phosphodiesterase activity of PDE3A immunoprecipitates was blocked in the presence of 10 μm cilostamide.
cAMP Assay—Washed platelets (2 × 108/ml) were treated as indicated and extracted with 2.5 volumes of ice-cold ethanol at the desired time point. Samples were clarified at 20,000 × g for 15 min, and the supernatant evaporated under vacuum. The residue was reconstituted with assay buffer and cAMP concentration determined by enzyme immunoassay per the manufacturer's instructions. Results are expressed as percentage of control (containing all additions except for thrombin).
Statistics—All data are presented as mean ± S.E. of at least three independent observations. Data presented with statistical analysis were analyzed using a 2-tailed Students t test (paired) with Microsoft Excel 2000 and Data Analysis Toolpak.
RESULTS
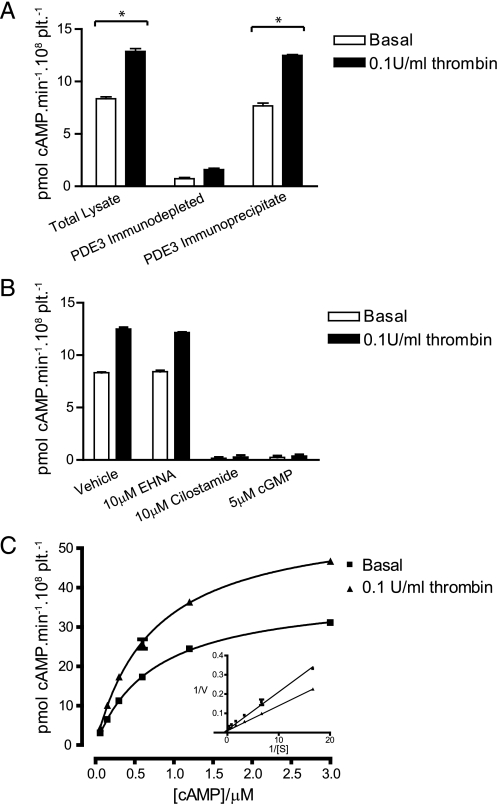
Phosphodiesterase 3A Activity Increases upon Platelet Stimulation—cAMP elevation is the most potent inhibitory mechanism in platelets and is tightly regulated through the coordinated activities of adenylate cyclase and a network of phosphodiesterases. We found a 40-50% increase in cAMP phosphodiesterase activity in lysates from thrombin-stimulated platelets (Fig. 1A). To determine whether this increase in activity is due to PDE3A, the most abundant cAMP-PDE in platelets (14), PDE3A was quantitatively depleted from platelet lysates using an isoform-specific antibody. This resulted in almost complete ablation of thrombin-stimulated activity, whereas ∼90% of PDE activity in crude lysate could be recovered in PDE3A immunoprecipitates (Fig. 1A). In contrast, a sheep control antibody had no effect (data not shown). Consistent with a role for PDE3A, the thrombin-stimulated increase in PDE3A activity in the immunoprecipitates was unaffected by the PDE2A inhibitor EHNA, but completely blocked by the PDE3A inhibitors cilostamide and cGMP (Fig. 1B). Kinetic analysis of PDE3A from basal and thrombin-stimulated platelets revealed that while the Km was unchanged (0.74 ± 0.2 μm versus 0.73 ± 0.03 μm) there was a 49% increase in Vmax (38.9 ± 0.5 pmol cAMP·min-1·108 plt-1 versus 58.0 ± 1.0 pmol cAMP·min-1·108 plt-1, Fig. 1C). Together, these results demonstrate that platelet activation is associated with an increase in cAMP PDE activity that is mediated specifically by PDE3A.
FIGURE 1.
PDE3A activity increases upon stimulation of platelets with thrombin. Washed platelets were stimulated with 0.1 unit/ml thrombin for 5 min, and 3′-cAMP phosphodiesterase activity was measured in whole cell lysates. Samples were depleted of PDE3A by immunoprecipitation with a PDE3A specific antibody and phosphodiesterase activity measured in depleted lysates and immunoprecipitates. *, p < 0.05 (A). PDE3A immunoprecipitates were assayed for 3′-cAMP phosphodiesterase activity in the presence of the indicated compounds (B). Km and Vmax were determined from PDE3A immunoprecipitates from basal and thrombin-stimulated platelets by non-linear regression of substrate saturation curves. A Lineweaver-Burk linear transformation is in-set into the plot (C).
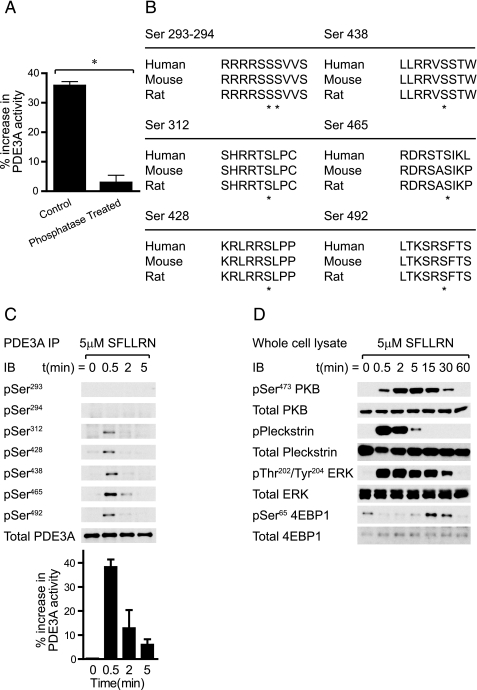
PDE3A Is Phosphorylated and Activated upon Platelet Activation—The paradigm of PDE3 regulation is N-terminal phosphorylation by members of the AGC kinase family (protein kinase A, protein kinase G, and protein kinase C) in adipocytes, hepatocytes, and oocytes, which increases the activity of the enzyme. Phosphatase treatment of PDE3A immunoprecipitates ablated the thrombin-stimulated increase in PDE activity confirming that platelet PDE3A is indeed regulated by phosphorylation (Fig. 2A). An in silico analysis of predicted S/T phosphorylation sites in PDE3A using the Scansite algorithm (15) identified seven serine residues as likely candidates, which are highly conserved among different species (see Fig. 2B). All sites loosely adhere to the consensus motif (K/R)X(K/R)XX(S/T)* preferred by the AGC family kinases PKA, PKB, PKC, p70S6K, and RSK.
FIGURE 2.
Thrombin-stimulated PDE3A activity is mediated by phosphorylation. PDE3A immunoprecipitates from basal and thrombin-stimulated platelets were incubated with calf-intestinal phosphatase (CIP) or heat-inactivated enzyme (control) for 30 min prior to assay for 3′-cAMP phosphodiesterase activity. Results are expressed as the percentage increase in PDE3A activity from basal (n = 3, A). Predicted S/T phosphorylation sites in PDE3A from human, mouse, and rat. Conserved phosphorylation sites are indicated with asterisks (B). Platelets were stimulated with 5 μm SFLLRN for the indicated time and extracted. PDE3A immunoprecipitates (C) or whole cell lysates (D) were analyzed by Western blotting or assayed for 3′-cAMP phosphodiesterase activity (C, bar graph). Results are representative for three independent experiments.
To investigate the phosphorylation status of PDE3A upon platelet activation we used affinity-purified phosphospecific antibodies generated against the candidate S/T phosphorylation sites on PDE3A. The specificity of these antibodies was verified by dot blots of synthetic peptides corresponding to the phosphorylated and unphosphorylated sites (see supplemental Fig. S1). Fig. 2C shows that PDE3A was rapidly phosphorylated on Ser312, Ser428, Ser438, Ser465, and Ser492 in response to the PAR-1 agonist, SFLLRN. Phosphorylation was markedly transient with maximal phosphorylation at 30 s and no detectable phosphorylation after 5 min. PDE activity followed a similar transient trend (Fig. 2C, bar graph). In contrast, phosphorylation at Ser293-294 was not detected.
SFLLRN is an ideal tool for dissecting platelet phosphorylation as its activation of several S/T kinases is kinetically distinct (Fig. 2D). The phosphorylation of the major PKC substrate, pleckstrin was rapid and short-lived, and closely paralleled PDE3A phosphorylation (compare Fig. 2, C and D). PKB activation was much slower and peak activity coincided with the waning of PKC. ERK2 phosphorylation was also rapid and transient. The activation of mTOR as assessed by 4EBP1 phosphorylation exhibited significant lag and was detectable 15 min of post-stimulation. Although all five PDE3A phosphorylation sites are potential substrates for several AGC kinases, the kinetic profile is most consistent with PKC.
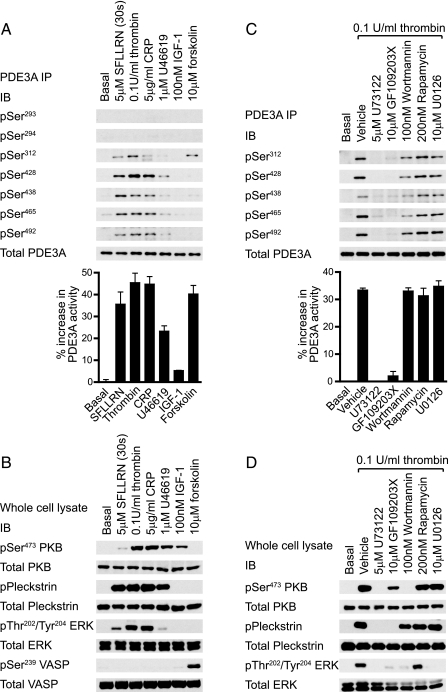
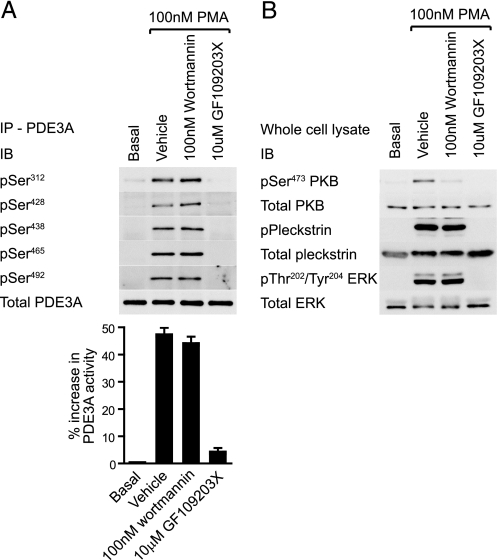
PDE3A Phosphorylation Is PKC-dependent—PDE3A phosphorylation and PDE activity were assessed in response to a range of stimuli and compared with the profile of S/T kinase activation to identify critical pathways (Fig. 3). SFLLRN, thrombin and collagen-related peptide (CRP) induced strong phosphorylation of Ser312, Ser428, Ser438, Ser465, and Ser492, whereas the thromboxane A2 receptor agonist, U46619 was less efficacious (Fig. 3A). PDE3A phosphorylation paralleled an increase in PDE activity (Fig. 3A, bar graph) and the phosphorylation of the PKC substrate pleckstrin (Fig. 3B). Interestingly, IGF-1 did not stimulate significant PDE3A phosphorylation or PDE3A activity (Fig. 3A), under conditions where PKB was the sole AGC kinase activated (Fig. 3B). Elevation of cAMP in response to the adenylate cyclase activator, forskolin, promoted the selective phosphorylation of Ser312 and a 40% increase in PDE3A catalytic activity (Fig. 3A) in parallel with phosphorylation of the PKA marker, vasodilator-stimulated phosphoprotein (VASP) (Fig. 3B). Phosphorylation of Ser293-294 was not detected in response to any of the tested reagents. Multisite phosphorylation of PDE3A is therefore associated with the ability of agonists to activate PKC. Inhibition of PKC and downstream pleckstrin phosphorylation by targeting PLC (U73122) or PKC itself (GF109203X) indeed correlated with significant inhibition of thrombin-stimulated PDE3A phosphorylation (Fig. 3C) and PDE activity (Fig. 3C, bar graph). In contrast, the PI 3-kinase inhibitor wortmannin only marginally decreased PDE3A phosphorylation and had no effect on PDE3A activity (Fig. 3C), under conditions where downstream activation of PKB was completely blocked (Fig. 3D). The partial effect of wortmannin correlates with its effect on pleckstrin phosphorylation (compare Fig. 3, C and D). Selective inhibition of mTOR > p70S6K with rapamycin or blocking ERK2 > RSK using the MEK inhibitor U0126 had no effect on PDE3A phosphorylation or activity (Fig. 3C). These results strongly implicate PKC, and not PKB, in the regulation of PDE3A. Indeed, direct activation of PKC by phorbol esters (phorbol 12-myristate 13-acetate, PMA) stimulated strong phosphorylation and activation of PDE3A, which was blocked by GF109203X but unaffected by wortmannin (Fig. 4A). Furthermore, the PKB inhibitor Akti-1/2 (16, 17) had no effect on thrombin-stimulated PDE3A phosphorylation and catalytic activity, under conditions where PKB phosphorylation was blocked (data not shown).
FIGURE 3.
PDE3A is phosphorylated upon platelet activation in a PKC-dependent manner. Platelets were stimulated with the indicated agonists for 5 min (A, B, SFLLRN for 30 s) or incubated with the indicated compounds for 10 min prior to stimulation with 0.1 unit/ml thrombin for 5 min (C, D). Platelets were subsequently extracted and PDE3A immunoprecipitates (A, C, top) were analyzed by immunoblotting with the indicated antibodies or analyzed for 3′-cAMP phosphodiesterase activity (bar graphs). PDE3A activity is expressed as the % increase in PDE3A activity from basal (n = 3). The whole cell lysate was analyzed by immunoblotting with the indicated antibodies (B, D).
FIGURE 4.
PMA stimulates PKC-dependent phosphorylation of PDE3A. Platelets were incubated with the indicated compounds for 10 min prior to stimulation with 100 nm PMA for 5 min and subsequently extracted. PDE3A immunoprecipitates (A, top) were analyzed by immunoblotting with the indicated antibodies or analyzed for 3′-cAMP phosphodiesterase activity (A, bar graph). PDE3A activity is expressed as % increase in PDE3A activity from basal (n = 3). The whole cell lysate was analyzed by immunoblotting with the indicated antibodies (B).
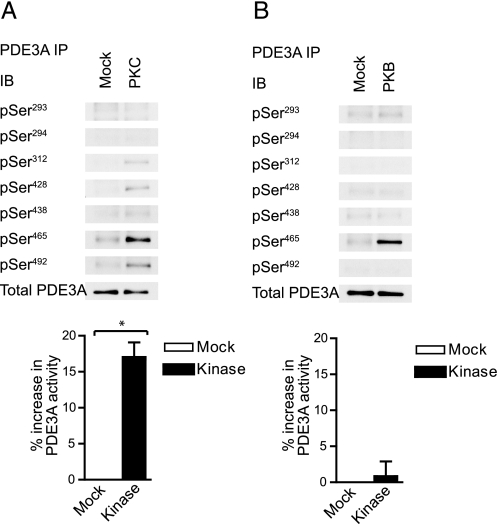
PDE3A Is an in Vitro Substrate for PKC—To establish whether PKC is able to phosphorylate PDE3A directly, recombinant PKCα was incubated with immunoprecipitated PDE3A in the presence of ATP. Fig. 5A demonstrates that phosphorylation was detectable on all sites identified in vivo (Ser312, Ser428, Ser438, Ser465, and Ser492) and correlated with a significant increase in PDE activity (see bar graph). Recent studies have indicated a role for PKB in the regulation of PDE3A (9, 10). However, parallel experiments using recombinant PKBα showed that only one of the phosphorylation sites identified in vivo (the classical “RXRXXS*” PKB site Ser465) was phosphorylated by PKBα in vitro (Fig. 5B). Furthermore, no phosphorylation of the candidate PKB sites Ser293-294 (RRRRSS*S*VVS) or a significant increase in PDE3A activity could be detected upon incubation with PKBα (Fig. 5B). The latter implies that phosphorylation of Ser465 alone is unable to regulate PDE3A catalytic activity. Together, these results demonstrate that PKC, but not PKB, is able to (i) directly phosphorylate PDE3A on the same sites as identified in vivo, and (ii) increase PDE3A activity.
FIGURE 5.
PDE3A is phosphorylated in vitro by PKC and PKB. Platelets were extracted and immunoprecipitated PDE3A was incubated with buffer (Mock) or recombinant PKCα (A) for 30 min in the presence of 0.5 mm CaCl2, 50 μg/ml phosphatidylserine, 5 μg/ml diacylglycerol, and 0.1 mm ATP. Alternatively, PDE3A immunoprecipitates were incubated with buffer (Mock) or recombinant PKBα in the presence of 1 mm EGTA and 0.1 mm ATP. Samples were analyzed by immunoblotting with the indicated antibodies (top) or analyzed for 3′-cAMP phosphodiesterase activity (bar graphs). Results are expressed as % increase from basal (n = 3, *, p < 0.05).
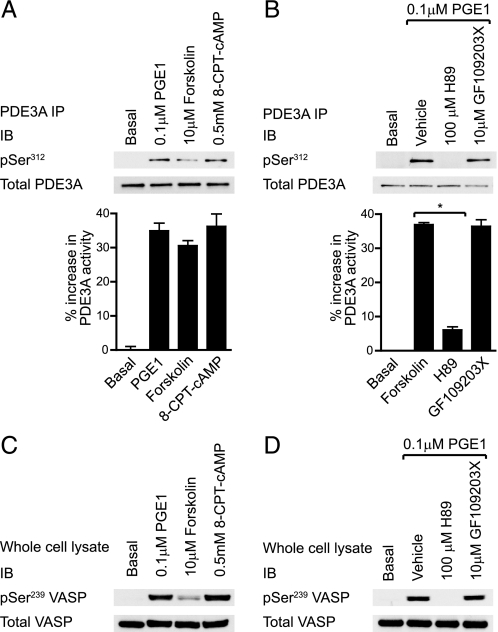
PDE3A Regulation by PKA—As described earlier, Ser312 phosphorylation and PDE3A activation was observed in response to cAMP elevation by the AC activator forskolin (Fig. 3A). Similar results were obtained by activation of the IP/EP receptors by PGE1 and direct activation of PKA using a cell permeable cAMP analogue, 8-(4-chlorophenylthio)-cAMP (8-CPT-cAMP) (Fig. 6A). Ser312 phosphorylation increased in parallel with the PKA substrate VASP (Fig. 6C) and correlated with increased hydrolysis of cAMP (Fig. 6A, bar graph). In contrast to thrombin-stimulated PDE3A phosphorylation and activation (Fig. 3C), these effects were blocked by the PKA inhibitor H89, but unaffected by PKC inhibition with GF109203X (Fig. 6B).
FIGURE 6.
PDE3A regulation by protein kinase A. Platelets were stimulated with the indicated PKA activators for 3 min (A, C) or with the indicated compounds for 30 min before treatment with 0.1 μm PGE1 for 3 min (B, D). Platelets were subsequently extracted and PDE3A immunoprecipitates (A, B, top) were analyzed by immunoblotting with the indicated antibodies or analyzed for 3′-cAMP phosphodiesterase activity (bar graphs). PDE3A activity is expressed as the % increase in PDE3A activity from basal (n = 3, *, p < 0.05). Whole cell lysate was analyzed by immunoblotting with the indicated antibodies (C and D).
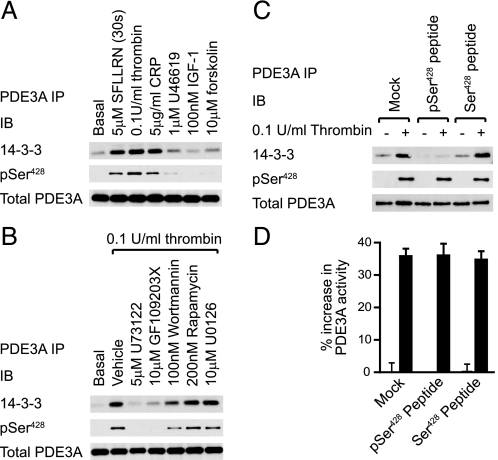
PDE3A Phosphorylation on Ser428 Increases 14-3-3 Binding—The phosphorylation of adipocyte PDE3B is associated with increased binding to 14-3-3 proteins, a group of evolutionarily conserved eukaryotic proteins that regulate protein function by binding to specific phosphoserine motifs (18). Co-immunoprecipitation studies showed that a low level of 14-3-3 was constitutively associated with PDE3A isolated from resting platelets (Fig. 7A), which was significantly enhanced upon stimulation with SFLLRN, thrombin, CRP, but not IGF-1. This correlated with the ability of primary agonists to activate PKC (compare Fig. 7A to Fig. 3B). Equally, thrombin-stimulated PKC activity (Fig. 3D) and 14-3-3 binding to PDE3A was blocked by U73122 and GF109203X and partially affected by wortmannin (Fig. 7B). Rapamycin and U0126 had no effect. Similar results were obtained in 14-3-3 overlay assays using digoxygenin-labeled 14-3-3 probes (results not shown). Sequence analysis identified two candidate 14-3-3 binding sites, Ser312 (mode 1 R(S/X)XS*XP) and Ser428 (mode 2 RXXXS*XP). PKA-dependent phosphorylation of Ser312 by cAMP elevation did not cause a significant increase in 14-3-3 binding (Fig. 7A, last lane). However, there was a good correlation between Ser428 phosphorylation and 14-3-3 binding (Fig. 7, A and B) and an excess of the synthetic peptide 423KRLRRpS428LPPGL433 blocked the interaction (Fig. 7C). It is therefore likely that the interaction between PDE3A and 14-3-3 is mediated by Ser428, which is in agreement with a previous report (11). 14-3-3 proteins are known to regulate the localization, degradation, and activity of their substrates (19). However, we found that PDE3A remained cytosolic upon thrombin stimulation, as determined by subcellular fractionation (data not shown). Furthermore, elution of bound 14-3-3 with excess phosphopeptide had no effect on the thrombin-stimulated increase in PDE3A activity in vitro (Fig. 7D). Together, these results demonstrate that 14-3-3 binding to PDE3A had no obvious effect on the localization or activity of PDE3A.
FIGURE 7.
Phosphorylation of PDE3A increases binding to 14-3-3 proteins. Platelets were stimulated with the indicated agonists for 5 min (SFLLRN for 30 s, A), incubated with the indicated compounds for 30 min before treatment with 0.1 units/ml thrombin for 5 min (B) or stimulated with 0.1 units/ml thrombin for 5 min (C, D) prior to extraction. PDE3A was immunoprecipitated using anti-PDE3A covalently coupled to Sepharose (A-D). Immunoprecipitates were untreated (A, B) or incubated with 1 mm free peptide for 30 min at room temperature to compete off bound 14-3-3 (C, D). PDE3A immunoprecipitates were subsequently analyzed by immunoblotting with the indicated antibodies (A-C) or assayed for 3′-cAMP phosphodiesterase activity (D).
PDE3A Activation Contributes to cAMP Depression upon Platelet Activation—Thrombin potently reduces the intracellular cAMP concentration upon platelet activation (20). This is largely mediated by indirect coupling to Gi protein, via the ADP receptor P2Y12, leading to inhibition of AC and suppression of cAMP synthesis (21). However, thrombin still retains the ability to reduce cAMP in the absence of Gi signaling. We hypothesize that this is the consequence of a PKC-dependent increase in PDE3A Vmax.
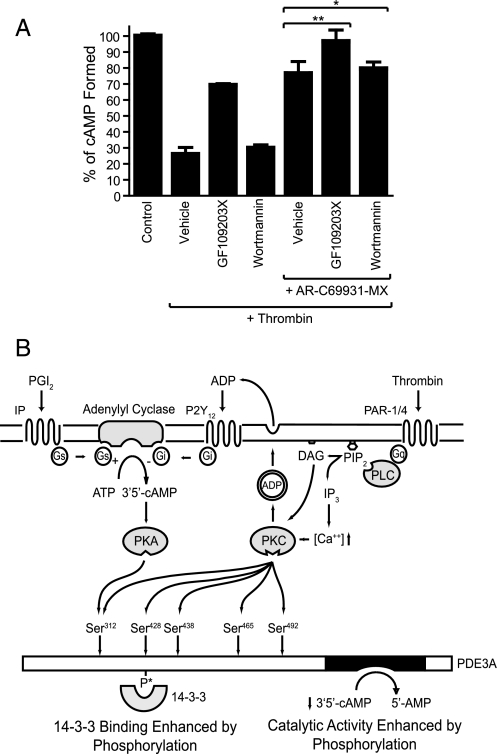
To investigate the contribution of PDE3A, we elevated the cAMP concentration with PGE1 and subsequently stimulated platelets with thrombin. As expected, the increase in cAMP concentration in response to PGE1 was largely reversed upon stimulation with thrombin (Fig. 8A, bar 2). The ability of thrombin to reduce cAMP was significantly inhibited by the PKC inhibitor GF109203X, (Fig. 8A, bar 3), demonstrating a major role for PKC in thrombin-mediated cAMP regulation. In contrast, targeting PKB using wortmannin had no effect (Fig. 8A, bar 4). In agreement with earlier reports, antagonism of the Gi pathway with AR-C69931MX significantly reduced the ability of thrombin to inhibit cAMP accumulation (Fig. 8A, bar 5). The residual effect of thrombin was fully blocked by PKC inhibition (GF109203X), whereas wortmannin had no effect (Fig. 8A, bar 6 and 7, respectively). These results strongly suggest that PKC-mediated activation of PDE3A contributes to intracellular cAMP regulation in human platelets.
FIGURE 8.
PDE3A activation plays a role in cAMP depression in stimulated platelets. Platelets were treated with the indicated compounds/vehicle (0.2% DMSO) for 10 min prior to sequential stimulation with 0.1 μm PGE1 for 3 min and 0.1 units/ml thrombin or vehicle (control) for 3 min. [cAMP] was measured as described under “Experimental Procedures.” Results are expressed as % of controls (containing all additions except for thrombin) ± S.E. (n = 3, *, p > 0.1, **, p < 0.05) (A). Proposed model of cAMP regulation in platelets is shown. Elevation of cAMP in response to prostacyclin activates PDE3A via PKA-mediated phosphorylation of Ser312. This negative feedback loop limits cAMP accumulation. Upon vascular injury and thrombin stimulation, PAR activation leads to Ca2+ release and PKC activation, which promotes dense granule secretion and inhibition of adenylate cyclase by P2Y12 agonism. The concomitant phosphorylation and activation of PDE3A by PKC synergize with Gi signaling to remove cAMP below an inhibitory threshold for platelet activation (B).
DISCUSSION
The intracellular cAMP concentration in human platelets is tightly regulated by the rate of synthesis by adenylate cyclase and hydrolysis by 3′-phosphodiesterases. Under resting conditions, a non-zero level of cAMP is maintained by tonic production and hydrolysis of cAMP. Indeed, PDE3A inhibitors such as cilostamide significantly increase basal cAMP levels and potently inhibit platelet function. PDE3A therefore regulates platelet responsiveness even in the absence of exogenous agents that raise cAMP through Gs-coupled receptors (7).
It has recently been demonstrated that thrombin enhances PDE3A activity in human platelets through a phosphorylation-dependent mechanism, which may involve the PI3K/PKB signaling pathway (9). In this study we aimed to better characterize the signaling pathways and phosphorylation sites that are involved in PDE3A regulation and cAMP homeostasis in platelets. We provide strong evidence that PKC, and not PKB, is involved in agonist-stimulated up-regulation of PDE3A activity. We demonstrate that: (i) the PAR-1 agonist SFLLRN stimulates rapid and transient phosphorylation of PDE3A on Ser312, Ser428, Ser438, Ser465, and Ser492. Phosphorylation of these sites closely follows phosphorylation of the PKC substrate pleckstrin and is associated with an increase in PDE3A activity. (ii) PKC, but not PKB, is able to directly phosphorylate these same sites in vitro and increase PDE3A activity. (iii) Thrombin-mediated PDE3A phosphorylation and activation are blocked by the PKC inhibitor GF109203X but is largely unaffected by wortmannin, a PI3 kinase inhibitor that blocks activation of downstream PKB, and the direct PKB inhibitor Akti-1/2. (iv) Direct activation of PKC using PMA stimulates phosphorylation and activation of PDE3A, which is unaffected by wortmannin and blocked by GF109203X, and (v) IGF-1, which strongly activates PKB (12), but not PKC, did not phosphorylate or increase the activity of PDE3A. We furthermore show that thrombin-mediated PDE3A phosphorylation resulted in a PKC-dependent association of PDE3A and 14-3-3. In addition, cAMP elevating agents induced phosphorylation of Ser312 and increased PDE3A activity, but did not induce 14-3-3 binding. Importantly, PKC-mediated activation of PDE3A likely contributes to elevated [cAMP]i by thrombin, as the Gi-independent reduction of [cAMP]i by thrombin was blocked by the PKC inhibitor GF109203X.
Platelets express cGMP-stimulated PDE2 and cGMP-inhibited PDE3A that are both able to hydrolyze cAMP. In this study, we demonstrate that thrombin stimulates a 40-50% increase in cAMP phosphodiesterase activity that is mediated by an ∼50% increase in the Vmax of PDE3A. An increase in Vmax is indeed the most efficient mechanism to increase substrate turnover as PDE3A has a submicromolar Km and is therefore operating close to saturation and zero-order kinetics under resting conditions with [cAMP]i of around 4.4 μm (22). A similar change in Vmax was found for calcium/calmodulin-dependent PDE1 upon phosphorylation by PKC in hamster hearts (23).
Phosphatase treatment of immunoprecipitated PDE3A demonstrated that the thrombin-stimulated increase in PDE3A activity was dependent on phosphorylation, in agreement with a recent study in human platelets (9). Activated PDE3A was not detected by anti-phosphotyrosine antibody (4G10, data not shown) and therefore sequence analysis of PDE3A was focused on identification of putative S/T phosphorylation sites. Multiple serine residues were identified with basic residues upstream of the phosphoacceptor site and are therefore potential substrates of the AGC kinase family. To identify the signaling pathways involved in PDE3A phosphorylation, we used the PAR-1 agonist SFLLRN, as it stimulates various signaling pathways with different temporal kinetics. SFLLRN-stimulated phosphorylation of PDE3A on Ser312, Ser428, Ser438, Ser465, and Ser492 was rapid and transient, correlated with an increase in activity and closely followed phosphorylation of the PKC substrate pleckstrin. Indeed, PKC inhibition using GF109203X blocked PDE3A phosphorylation and the increase in cAMP hydrolysis.
Interestingly, we could not detect any phosphorylation at the consensus PKB sites, Ser293-294, in vivo, or in vitro with recombinant PKBα. These residues are analogous to the well characterized PKB site, Ser295 in PDE3B and are often attributed importance to PDE3A regulation on this basis. For example, it has been suggested that platelet modulators such as insulin (24) and leptin (25) can stimulate phosphorylation and activation of platelet PDE3A through the PI3K/PKB pathway. However, in our study we were unable to detect PDE3A phosphorylation in response to insulin or leptin (data not shown). Furthermore, IGF-1, which promotes strong activation of PKB, did not induce phosphorylation and activation of PDE3A. A recent study by Zhang and Colman (9) established that thrombin regulates PDE3A activity, but concluded this was PKB-dependent. Interestingly, the authors identified a 140-kDa-phosphorylated species in partially purified PDE3A using a generic PKB phospho-motif antibody (9). This corresponds to the full-length transcript found in cardiac myocytes which possesses N-terminal hydrophobic domains that target PDE3A to internal membranes (26). In contrast we identified a single truncated isoform in PDE3A immunoprecipitates (∼110 kDa) with a cytosolic localization consistent with several previous studies (27-29). This band was only weakly detected by the PKB substrate antibody but in a manner indicative of a PKC-induced signal (data not shown). The discrepancy between these studies may be explained by use of a specific PDE3A antibody in the present study, which allowed us to study PDE3A in isolation confirming that the 110 kDa is the major, if not only, isoform expressed in platelets. It also excluded phosphodiesterase activity from PDE3B derived from contaminating red cells (30). However, a synergy between P2Y12 antagonism and PKB inhibition in the ability of thrombin to lower cAMP, as described by Zhang and Colman (9), is difficult to justify, considering that inhibition of autocrine P2Y12 signaling alone ablates PAR-stimulated PKB activation (31), thereby ruling out a role for PKB in Gi-independent cAMP regulation. Furthermore, the lack of effect of the PKC inhibitor on Gi-independent cAMP regulation in Zhang and Colman's study (9) is surprising as PKC inhibition has a significant effect on PKB activation by reducing ADP secretion (see also Fig. 3D). Indeed, a role for PKC has previously been suggested by Giesberts et al. (32) and has been confirmed in the present study.
Our data clearly demonstrate that PKC regulates the phosphorylation and activation of PDE3A in human platelets. Indeed, in this context PKC appears to be the logical choice over related AGC kinases. PKC is a highly abundant kinase that is rapidly activated by allosteric binding of diacylglyercol and/or Ca2+. In contrast the activation of PKB, RSK, and p70S6K is much slower due to multiple phosphorylation events required downstream of the generation of second messenger. PDE3A activation would be a priming event in platelet activation and require rapid activation to assist the clearance of cAMP below an inhibitory threshold. Although, we cannot rule out the possibility that a PKC-regulated kinase phosphorylates PDE3A, we demonstrated that PKC is capable of directly phosphorylating PDE3A in vitro.
Previous studies showed that an increase in [cAMP]i stimulates a negative feedback pathway by activation of PKA and subsequent phosphorylation and activation of PDE3A (27). We show that the adenylate cyclase activator, forskolin, stimulated phosphorylation of a single site, namely Ser312, and increased PDE3A activity to a similar extent as thrombin and CRP. These effects were blocked by the PKA inhibitor H89, but unaffected by the PKC inhibitor GF109203X, highlighting that this mode of regulation is distinct from the PKC-dependent pathway triggered by major platelet agonists. Consequently, Ser312 appears to function as a node, integrating signals from both activating (via PKC) and inhibitory (via PKA) stimuli.
PDE3A phosphorylation in platelets not only increased its activity but also promoted binding to 14-3-3 proteins, which are highly conserved proteins that bind to phosphoserine residues within specific motifs. PDE3A has two putative 14-3-3 binding motifs, Ser312 and Ser428, of which Ser428 is likely to be the major binding site (11). This was confirmed in our studies by the finding that forskolin stimulated single phosphorylation of Ser312 but did not stimulate 14-3-3 binding and that pS428 phosphopeptide reduced the PDE3A/14-3-3 interaction. 14-3-3 binding to PDE3A may affect its localization by behaving as an adaptor, nucleating the formation of a signalosome (33). However, we found that PDE3A is an exclusively cytosolic protein independent of phosphorylation status (data not shown). Furthermore, although the cytoskeletal linker plectin was identified as a PDE3A binding partner (11) and may target PDE3A to signaling complexes assembled on actin scaffolds, no PDE3A was detected in platelet cytoskeletons extracted by Triton X-100 (data not shown). Interestingly, 14-3-3 binding distinguishes vasodilator-activated PDE3A from activation by a thrombotic stimuli. Perhaps, this observation is related to the function of 14-3-3.
An early observation in the study of cyclic nucleotides in platelets was that platelet activation is associated with a decrease in [cAMP]i (20). Seminal work revealed that the ADP receptor P2Y12 is an essential component, mediating the inhibition of adenylyl cyclase and cAMP synthesis (21). However P2Y12 antagonism does not ablate the effect of thrombin and prior work has suggested that complete inhibition of cAMP accrual requires activation of both Gi and PKC (32). Similarly, we have demonstrated that in platelets treated with exogenous PGE1, inhibiting PKC (GF109203X) or Gi signaling (AR-C69931MX) significantly reversed the reduction of cAMP by thrombin. Indeed, blocking both PKC and Gi signaling completely prevented the reduction in cAMP by thrombin, whereas the PI3K inhibitor wortmannin had no effect. Together, these results strongly suggest that the Gi-independent regulation of [cAMP]i is due to PKC-dependent phosphorylation of PDE3A and enhancement of cAMP hydrolysis.
In conclusion, this is the first study to demonstrate that PKC, and not PKB, is involved in regulation of PDE3A activity in platelets. We furthermore show that thrombin stimulation of platelets results in the phosphorylation of Ser312, Ser428, Ser438, Ser465, and Ser492 leading to a subsequent increase in cAMP hydrolysis and 14-3-3 binding (summarized in Fig. 8B).
Supplementary Material
Acknowledgments
We thank Prof. Miles Houslay (University of Glasgow) for his advice on the phosphodiesterases assays, Dr. Jean Harthill (University of Dundee) for performing the 14-3-3 overlay experiments, and the blood donors within the School of Medical Sciences for their generous donations.
This work was supported by the British Heart Foundation (Grants FS/05/047 and FS/04/027).
The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1.
Footnotes
The abbreviations used are: PAR, protease-activated receptors; PKC, protein kinase C; PLC, phospholipase C; PI, phosphatidylinositol; MOPS, 4-morpholinepropanesulfonic acid.
References
- 1.Hers, I., Donath, J., van Willigen, G., and Akkerman, J. W. (1998) Arterioscler. Thromb. Vasc. Biol. 18 404-414 [DOI] [PubMed] [Google Scholar]
- 2.Walker, T. R., and Watson, S. P. (1993) Biochem. J. 289, 277-282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Banfic, H., Downes, C. P., and Rittenhouse, S. E. (1998) J. Biol. Chem. 273 11630-11637 [DOI] [PubMed] [Google Scholar]
- 4.Schwarz, U. R., Walter, U., and Eigenthaler, M. (2001) Biochem. Pharmacol. 62 1153-1161 [DOI] [PubMed] [Google Scholar]
- 5.Manns, J. M., Brenna, K. J., Colman, R. W., and Sheth, S. B. (2002) Thromb. Haemost. 87 873-879 [PubMed] [Google Scholar]
- 6.Sun, B., Li, H., Shakur, Y., Hensley, J., Hockman, S., Kambayashi, J., Manganiello, V. C., and Liu, Y. (2007) Cell Signal. 19 1765-1771 [DOI] [PubMed] [Google Scholar]
- 7.Feijge, M. A., Ansink, K., Vanschoonbeek, K., and Heemskerk, J. W. (2004) Biochem. Pharmacol. 67 1559-1567 [DOI] [PubMed] [Google Scholar]
- 8.Dickinson, N. T., Jang, E. K., and Haslam, R. J. (1997) Biochem. J. 323, 371-377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang, W., and Colman, R. W. (2007) Blood 110 1475-1482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Han, S. J., Vaccari, S., Nedachi, T., Andersen, C. B., Kovacina, K. S., Roth, R. A., and Conti, M. (2006) EMBO J. 25 5716-5725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pozuelo Rubio, M., Campbell, D. G., Morrice, N. A., and Mackintosh, C. (2005) Biochem. J. 392, 163-172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hers, I. (2007) Blood 110 4243-4252 [DOI] [PubMed] [Google Scholar]
- 13.Thompson, W. J., and Appleman, M. M. (1971) Biochemistry 10 311-316 [PubMed] [Google Scholar]
- 14.Haslam, R. J., Dickinson, N. T., and Jang, E. K. (1999) Thromb. Haemost. 82 412-423 [PubMed] [Google Scholar]
- 15.Obenauer, J. C., Cantley, L. C., and Yaffe, M. B. (2003) Nucleic Acids Res. 31 3635-3641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Logie, L., Ruiz-Alcaraz, A. J., Keane, M., Woods, Y. L., Bain, J., Marquez, R., Alessi, D. R., and Sutherland, C. (2007) Diabetes 56 2218-2227 [DOI] [PubMed] [Google Scholar]
- 17.Hunter, R. W., Harper, M. T., and Hers, I. (2008) J. Thromb. Haemost. 6 1923-1932 [DOI] [PubMed] [Google Scholar]
- 18.Onuma, H., Osawa, H., Yamada, K., Ogura, T., Tanabe, F., Granner, D. K., and Makino, H. (2002) Diabetes 51 3362-3367 [DOI] [PubMed] [Google Scholar]
- 19.Mackintosh, C. (2004) Biochem. J. 381, 329-342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lagarde, M., and Dechavanne, M. (1977) Biomedicine 27 110-112 [PubMed] [Google Scholar]
- 21.Kim, S., Foster, C., Lecchi, A., Quinton, T. M., Prosser, D. M., Jin, J., Cattaneo, M., and Kunapuli, S. P. (2002) Blood 99 3629-3636 [DOI] [PubMed] [Google Scholar]
- 22.Eigenthaler, M., Nolte, C., Halbrugge, M., and Walter, U. (1992) Eur. J. Biochem. 205 471-481 [DOI] [PubMed] [Google Scholar]
- 23.Yu, H., Cai, J. J., and Lee, H. C. (1996) Mol. Pharmacol. 50 549-555 [PubMed] [Google Scholar]
- 24.Lopez-Aparicio, P., Belfrage, P., Manganiello, V. C., Kono, T., and Degerman, E. (1993) Biochem. Biophys. Res. Commun. 193 1137-1144 [DOI] [PubMed] [Google Scholar]
- 25.Elbatarny, H. S., and Maurice, D. H. (2005) Am. J. Physiol. Endocrinol. Metab. 289 E695-E702 [DOI] [PubMed] [Google Scholar]
- 26.Shakur, Y., Takeda, K., Kenan, Y., Yu, Z. X., Rena, G., Brandt, D., Houslay, M. D., Degerman, E., Ferrans, V. J., and Manganiello, V. C. (2000) J. Biol. Chem. 275 38749-38761 [DOI] [PubMed] [Google Scholar]
- 27.Macphee, C. H., Reifsnyder, D. H., Moore, T. A., Lerea, K. M., and Beavo, J. A. (1988) J. Biol. Chem. 263 10353-10358 [PubMed] [Google Scholar]
- 28.Sheth, S. B., Brennan, K. J., Biradavolu, R., and Colman, R. W. (1997) Thromb. Haemost. 77 155-162 [PubMed] [Google Scholar]
- 29.Degerman, E., Moos, M., Jr., Rascon, A., Vasta, V., Meacci, E., Smith, C. J., Lindgren, S., Andersson, K. E., Belfrage, P., and Manganiello, V. (1994) Biochim. Biophys. Acta 1205 189-198 [DOI] [PubMed] [Google Scholar]
- 30.Hanson, M. S., Stephenson, A. H., Bowles, E. A., Sridharan, M., Adderley, S., and Sprague, R. S. (2008) Am. J. Physiol. Heart Circ. Physiol. 295 H786-H793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim, S., Jin, J., and Kunapuli, S. P. (2004) J. Biol. Chem. 279 4186-4195 [DOI] [PubMed] [Google Scholar]
- 32.Giesberts, A. N., van Willigen, G., Lapetina, E. G., and Akkerman, J. W. (1995) Biochem. J. 309, 613-620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ahmad, F., Lindh, R., Tang, Y., Weston, M., Degerman, E., and Manganiello, V. C. (2007) Biochem. J. 404 257-268 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.