Abstract
T-cell migration is a complex highly coordinated process that involves cell adhesion to the high endothelial venules or to the extracellular matrix by surface receptor/ligand interactions, cytoskeletal rearrangements, and phosphorylation-dependent signaling cascades. The mechanism(s) that regulates T-cell migration is of considerable relevance for understanding the pathogenesis of various diseases, such as chronic inflammatory diseases and cancer metastasis. This study was designed to identify potential involvement of STAT3, a latent transcription factor, in mediating integrin-induced T-cell migration. Using our previously characterized in vitro model for lymphocyte migration, we demonstrate that STAT3 is activated and translocated to the nucleus during the process of active motility of Hut78 T-lymphoma cells triggered via LFA-1. Blocking STAT3 signaling by multiple approaches inhibited LFA-1-induced T-cell locomotion via destabilization of microtubules and post-translational modification of tubulin. Here, we show that STAT3 physically interacts with stathmin to regulate microtubule dynamics in migrating T-cells. These observations strongly indicate that STAT3 is critically important for T-cell migration and associated signaling events.
Efficient operation of the adaptive immune system requires migration of T-lymphocytes from the vascular compartment across tissue barriers and through the extracellular matrix. This process involves a series of integrin ligand-receptor interactions (1) that initially retards lymphocyte flow and ultimately leads to arrest and diapedesis across the endothelium (2, 3). T-cells utilize the integrin, lymphocyte function-associated antigen-1 (LFA-1),3 when migrating in response to chemoattractants across the vasculature into lymph nodes or inflamed tissues (1, 4, 5). By engagement with ligands from the intercellular adhesion molecule group (ICAMs), in particular ICAM-1, LFA-1 also provides a strong adhesive force to promote and stabilize T-cell and antigen-presenting cell conjugate formation. We have demonstrated that LFA-1 transduces a variety of transmembrane signals in crawling T-cells involving protein kinase C activation and cytoskeletal rearrangement (4, 6-9). However, the exact sequence of downstream integrin-mediated signaling events resulting in cytoskeletal rearrangements and cell locomotion is not fully understood.
T-cell migration involves cross-talk between integrins and the cytoskeleton, coordinated changes in the cytoskeleton, and the controlled formation and dispersal of adhesion sites (10). Motile lymphocytes develop trailing extensions, which contain cytoskeletal and signaling elements (11). Microtubules (MTs) are essential components of the cytoskeleton and are important for many aspects of mammalian cell responses, including cell division, growth, migration, and signaling (12-14). Whereas the actin cytoskeleton provides the driving force at the cell front, the MT network assumes a regulatory function in coordinating rear retraction (15). MT retraction into the cellular uropod is an important step in T-cell motility (4, 8). MTs are necessary for directed migration of multiple cells, and there are several possible mechanisms by which disruption or interference of MTs could block cell motility. These include impairment of the repositioning of the microtubule organizing center (MTOC), changes in MT interaction with focal adhesions, inhibition of the MT polymerization and depolymerization cycle, inhibition of intracellular protein trafficking and vesicle transport, and interference with MT-mediated integrin clustering and increased avidity (16). The reorganization of the MT cytoskeleton depends on the global and local activity of several proteins that affect nucleation dynamics and arrangement of the filament systems. Tubulins, the building block of MTs, are subject to specific post-translational modifications, including acetylation, detyrosination, and tyrosination (13, 17), which potentially modulate the functions and localization of MTs within the cell.
The signal transducers and activators of transcription (STATs) are a family of latent cytoplasmic transcription factors that are activated by many cytokines and growth factors (18). The STAT family is composed of seven members in mammals, of which STAT3 is the most pleiotropic member (18-20), and appears to have important and unique functions. Cell stimulation can activate STAT family members by tyrosine phosphorylation to induce their dimerization; activated STAT3 translocates from the cytosol to the cell nucleus to mediate transcription of a number of STAT3-responsive genes (21). STAT3 was originally identified as a mediator of the acute phase of inflammatory response triggered by interleukin-6 (22). However, it is now known that STAT3 is implicated in several biological processes, including cell proliferation, differentiation, and survival (18-20, 23, 24). Although STAT3 knock-out in mice caused early embryonic lethality (25), conditional gene targeting using the Cre-loxP strategy has revealed a critical role for STAT3 in cell migration in keratinocytes (23, 26).
The 18-kDa phosphoprotein stathmin (also known as oncoprotein 18) is a tubulin-binding protein involved in the control of MT assembly and dynamics (27-30). Originally identified as a key factor in cell proliferation, it also plays roles as a relay protein and integrating protein within intracellular signaling networks (31). The inhibitory effect of stathmin on MT growth is believed to derive from its ability to sequester tubulin by binding tubulin dimers, which decreases the concentration of free heterodimers available to polymerization (20, 30). The MT-destabilizing activity of stathmin is turned off by cell surface receptor kinase cascades and cyclin-dependent kinases (32). By changing MT polymerization/depolymerization dynamics, stathmin may help the cell react to external stimuli. A recent study has suggested a functional link between stathmin and STAT3, whereby STAT3 interacts with stathmin and antagonizes its MT depolymerization activity (33).
Here, we report a role for STAT3 in LFA-1-induced T-cell migration. Upon LFA-1 cross-linking, STAT3 is activated by tyrosine phosphorylation that translocates to the nucleus and interacts with stathmin to regulate MT dynamics in migrating T-cells.
EXPERIMENTAL PROCEDURES
Antibodies and Reagents—Antibody used for induction of T-cell motility as previously described (8) was of the motility-inducing clone SPVL7 (monoclonal antibody to the α-chain of LFA-1 from Monosan, Sanbio, Uden, The Netherlands). Rabbit monoclonal anti-STAT3, anti-phospho-STAT3 (Tyr705), anti-stathmin, anti-phosphostathmin(Ser16), anti-glyceraldehyde-3-phosphate dehydrogenase, and horseradish peroxidase-conjugated anti-rabbit and anti-mouse antibodies were from Cell Signaling Technology (Danvers, MA). Mouse monoclonal anti-α-tubulin, anti-acetylated tubulin (clone 6-11B-1), anti-tyrosinated tubulin (TUB-1A2), and goat anti-human IgG (Fc-specific) antibodies were from Sigma. Goat anti-mouse IgG was from Dako A/S (Denmark). Human rICAM-1 was from R&D Systems (Minneapolis, MN). Alexa fluor-488-conjugated anti-mouse antibody and Alexa fluor-546 Phalloidin were from Invitrogen. Cell-permeable AG490 was obtained from Calbiochem.
Cell Culture—The human cutaneous T-lymphoma cell line Hut78 (American Type Culture Collection (ATCC), Manassas, VA) was used and cultured as described previously (4). Briefly, cells were cultured in RPMI 1640 medium (Invitrogen) containing 10% (v/v) heat-inactivated fetal bovine serum, 2 mm l-glutamine, and antibiotics (penicillin (100 IU/ml) and streptomycin (100 μg/ml)) in a humidified chamber at 37 °C containing 5% CO2.
Induction of Cell Motility—The ability of T-cells to display integrin-induced locomotory behavior was determined using our well characterized migration-triggering model system, as previously described (4). Briefly, 6- or 96-well tissue culture plates (flat bottom; Nunc™), depending on the particular assay type, were precoated with goat anti-mouse IgG or mouse anti-human Fc and subsequently incubated with cross-linking monoclonal motility-inducing anti-LFA-1 antibody or human rICAM-1. As a control (resting cells), wells were coated with poly-l-lysine (cells adhered to this surface without activation). Hut78 cells were loaded into the coated wells (60 × 104 cells/well in a 6-well plate or 1.0 × 104 cells/well in a 96-well plate) and incubated in 5% CO2 at 37 °C for the indicated time periods, depending on the particular experiments.
Deformation Index (DI)—In order to quantify migratory phenotype of T-cells, DI was calculated as described previously (3). DI provides a stronger measure of the degree of cell polarization in comparison with elongation index and circularity alone and therefore more accurately reflects the phenotype of polarized locomotory cells. Typically, values of 1-3 are assigned to cells that were nonmigratory (i.e. cell shape nearing circular). Those cells that display a polarized phenotype (cell body and trailing uropod) and therefore a higher degree of deformation will have higher DI values (>5) (17). On average, >40 randomly chosen cells were scored for each set of conditions. Semiautomated analysis was performed using the Scion Image software (Scion Corp., Frederick, MD).
Cell Lysis, Immunoprecipitation, and Immunoblotting—The cell lysis was performed as described previously (34). The protein content of the cell lysates was determined by the Bio-Rad protein assay kit according to the manufacturer's instructions. Equal amounts of precleared lysates were subjected to immunoprecipitation with antibodies to STAT3, α-tubulin, or stathmin, as described previously (35). The cell lysates or immunoprecipitated samples were boiled with Laemmli sample buffer for 5 min and resolved by SDS-PAGE. The separated proteins were electrophoretically transferred to polyvinylidene fluoride membranes by semidry blotting for 1 h. The polyvinylidene fluoride membranes were blocked in 5% nonfat dry milk in PBST (0.1% (v/v) Tween 20 in phosphate-buffered saline (PBS)) for 1 h at room temperature. After washing, the blots were incubated with the indicated primary antibodies (diluted according to the manufacturer's instructions) overnight at 4 °C with gentle rocking. After three washes in PBST, the membranes were incubated with appropriate horseradish peroxidase-conjugated secondary antibodies for 1 h at room temperature. The immunoreactive bands were visualized using the enhanced chemiluminescence detection system (Amersham Biosciences) and subsequent exposure to Kodak light-sensitive film.
STAT Activation Assay Using High Content Analysis (HCA)—Serum-starved Hut78 cells (10,000 cells/well in 100 μl of medium) were seeded into 96-well tissue culture plates precoated with anti-LFA-1, ICAM-1, or poly-l-lysine. Cells were incubated for different time periods, depending on particular experiments, fixed, and stained with the Cellomics® HCS reagent kit for STAT activation as per the manufacturer's instructions (Thermo Fisher Scientific Inc.). Plates were scanned (three randomly selected fields/well containing at least 300 cells) using an automated microscope (IN Cell Analyzer 1000 HCA platform; GE Healthcare). Images were acquired using a ×20 objective. STAT activation was measured and analyzed by IN Cell Investigator software using nuclear trafficking analysis module (GE Healthcare) that quantifies nuclear to cytoplasmic fluorescence intensity.
Peptide Synthesis—Peptides were synthesized on an Applied Biosystems (model 433A) peptide synthesizer (Foster City, CA) following standard Fmoc amino acid chemistry (36), using O-(benzotriazol-1-yl)-N,N,N′,N′-tetramethyluronium hexa-fluorophosphate 1-hydroxybenzotriazole diisopropylethylamine as coupling reagents. All Fmoc-protected amino acids and coupling reagents were purchased from Novabiochem (Darmstadt, Germany) except for Fmoc-protected phosphotyrosine derivative, which was from Bachem (Bachem Distribution Services GmbH, Weil am Rhein, Germany), with phosphate diprotected by (methyldiphenylsilyl)ethyl moiety (37). Peptides were cleaved from the polymer support and freed from side chain protections in trifluoroacetic acid/triisopropylsilane/water (95/2.5/2.5 in volume) for 4 h. After removing the resin polymer, the peptides were precipitated in cold ether and purified by semipreparative reversed-phase high pressure liquid chromatography on a C18 column (5 μm, 10 × 250 mm; Vydac). The molecular weights of peptides were confirmed by mass spectrometry (electrospray ionization m/z M + H+: 2246.4 for Antennapedia peptide (Antp) and 4186.2 for STAT3-inhibitory peptide (pY-pept)).
Immunofluorescent Staining and Microscopy—Immunostaining was performed as previously described (4). Conventional phase-contrast microscopy was performed on a Nikon TE3000 inverted microscope (Nikon, Tokyo, Japan) equipped with a Leica DC 100 digital camera. Fluorescence microscopy was performed by a Zeiss confocal work station attached to a Zeiss LSM 510 laser module using a ×63 objective (Carl Zeiss, Thornwood, NY). Images were acquired and processed using Zeiss LSM 510 (release 4.0) software (Carl Zeiss). At least 20 different microscopic fields were observed for each sample. Semiautomated analysis for MT fiber length was performed using the Zeiss LSM Image Examiner software (Carl Zeiss).
Cell Viability Assay—Cell viability was measured by Cell-Titer 96® AQueous One Solution cell proliferation assay kit according to the manufacturer's instructions (Promega, Madison, WI). Hut78 cells were distributed in 96-well microplates (1.5 × 104 cells/well) in 100 μl of medium and pretreated with the indicated inhibitors. Cells were then incubated with 20 μl of MTS tetrazolium solution (provided by the manufacturer) for an additional 4 h. The relative cell viability was calculated by determining the absorbance at 490 nm using a microplate reader (Tecan, Mannedorf, Switzerland).
Cell Adhesion Assay—The cell adhesion assay was performed as described previously (38) with minor modifications. Briefly, untreated or pretreated Hut78 cells (1.5 × 104 cells in 100 μl of medium) were added to the wells of an anti-LFA-1-coated 96-well microtiter plate (as described above). The cells were then incubated for 1 h at 37 °C. Nonadherent cells were removed by gentle washing three times with PBS and fixed with 1% glutaraldehyde, 0.5% sucrose in PBS for 15 min at room temperature. After two washes with PBS, the cells were stained with 0.5% crystal violet in 20% methanol plus PBS for 30 min at room temperature. The cells were then washed five times with PBS, and the cell-bound stain was resolubilized in methanol. Cell adhesion was quantified by measuring the absorbance at 590 nm using a microplate reader (Tecan).
Densitometric Analysis—Densitometric analyses of the Western blots were performed as described previously (34) using Gene-Tools software (Syngene, Cambridge, UK). The relative values of the samples were determined by giving an arbitrary value of 1.0 to the respective control samples of each experiment.
Statistical Analysis—The data are expressed as mean ± S.E. For comparison of two groups, p values were calculated by a two-tailed unpaired Student's t test. In all cases, p < 0.05 was considered to be statistically significant.
RESULTS
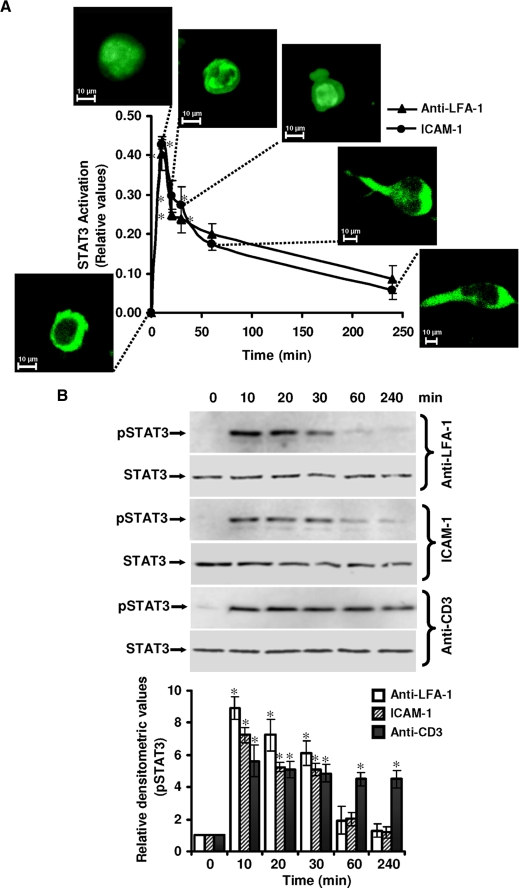
T-cell Motility Induced by LFA-1 Cross-linking Involves Activation of STAT3—In resting Hut78 cells, STAT3 was present mainly in the cytoplasm (Fig. 1A). Following LFA-1 cross-linking via immobilized anti-LFA-1, cytoplasmic STAT3 translocated into the nucleus within 10 min (Fig. 1A). Quantitation of STAT3 nuclear translocation showed an up to 42.8% increase in 10 min (Fig. 1A). This increase in STAT3 nuclear translocation was transitory, returning to basal level after 30 min (Fig. 1A). A similar STAT3 activation profile was observed when cells were incubated on immobilized ICAM-1 (Fig. 1A). No significant change in the activation of other STAT isoforms STAT1 or STAT2 was observed following LFA-1 cross-linking either via immobilized anti-LFA-1 or ICAM-1 to the Hut78 cells (supplemental Fig. 1).
FIGURE 1.
STAT3 activation during LFA-1-induced T-cell migration. A, serum-starved Hut78 cells were incubated on a poly-l-lysine-coated (0 min) or anti-LFA-1- or ICAM-1-coated 96-well plate for 10, 20, 30, 60, or 240 min and fixed. Cells were labeled with the Cellomics® HCS reagent kit for STAT3 activation. Cellular images were acquired using an IN Cell Analyzer 1000 automated microscope and analyzed by Investigator image analysis software that quantifies nuclear to cytoplasmic fluorescence intensity. Data are normalized mean ± S.E. of three independent experiments in triplicate from three randomly selected fields per well containing at least 300 cells. The insets show representative cell images obtained at the indicated time points. B, serum-starved Hut78 cells were incubated on anti-LFA-1-, ICAM-1-, or anti-CD3-coated 6-well plates for the indicated times and lysed. Cell lysates (20 μg each) were resolved by SDS-PAGE and after Western blotting were probed with anti-phospho-STAT3 (Tyr705; pSTAT3) or anti-STAT3 (as a loading control) antibody. Relative densitometric analysis of the individual pSTAT3 band was performed and presented. Data are mean ± S.E. of three independent experiments. *, p < 0.05 with respect to corresponding control.
Next, we sought to determine whether LFA-1 signaling induced tyrosine phosphorylation of STAT3 in Hut78 cells. Following LFA-1 cross-linking in Hut78 cells via immobilized anti-LFA-1 or ICAM-1, a greater than 7-fold increase in tyrosine phosphorylation of STAT3 (Tyr705) was observed in 10 min, declining toward base line level after 30 min (Fig. 1B). In contrast, T-cell receptor (TCR) activation by anti-CD3-induced tyrosine phosphorylation of STAT3 showed a distinctly different activation pattern detected at 10 min that remained constant for up to 4 h (Fig. 1B). However, CD3 ligation via immobilized anti-CD3 did not induce migration of Hut78 cells up to 4 h (data not shown). Therefore, for further experiments on migrating T-cells, LFA-1 cross-linking of Hut78 cells was employed. No change in the expression of STAT3 protein was observed during these time periods (Fig. 1B). Together, these results demonstrate that signaling through LFA-1 in Hut78 cells induces changes associated with STAT3 functional activation, including nuclear translocation and tyrosine phosphorylation.
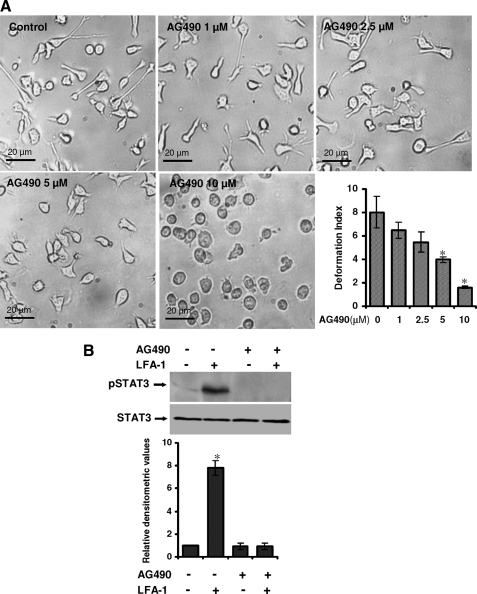
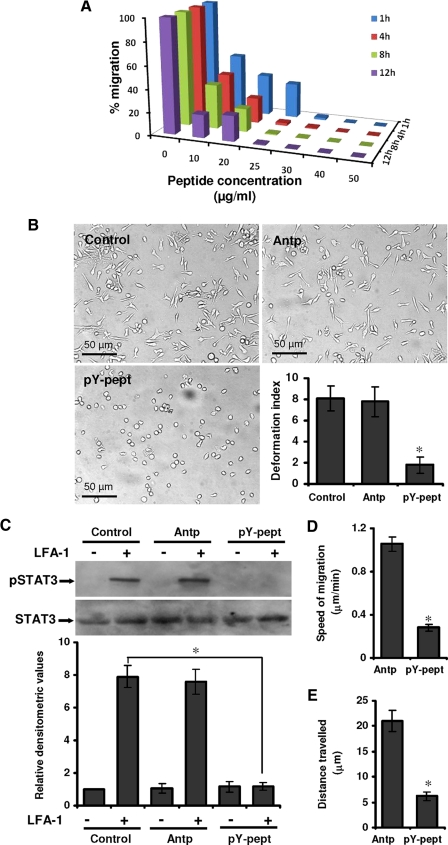
Suppression of STAT3 Activity Attenuates LFA-1-induced T-Cell Polarization and Migration—Hut78 cells triggered via LFA-1 receptor/ligand interaction rapidly polarize and develop a locomotion-associated phenotype (4, 8). To further investigate the role of STAT3, selective inhibitory strategies were utilized. Initially, we used AG490, a Janus kinase-specific inhibitor that subsequently reduces STAT3 tyrosine phosphorylation. Hut78 cells were pretreated with different concentrations of AG490 ranging from 1 to 10 μm for 1 h and incubated on anti-LFA-1-coated 96-well plates for 4 h (Fig. 2A). Pretreatment of Hut78 cells with AG490 resulted in dose-dependent inhibition of the LFA-1-induced locomotion-associated phenotype (Fig. 2A). Quantitation of the observed inhibitory effect of AG490 on motile phenotype of Hut78 cells demonstrated that DI reduced from 7.9 in controls cells to 1.8 when cells were treated with 10 μm AG490 (Fig. 2A). Pretreatment of Hut78 cells with 10 μm AG490 also inhibited LFA-1-induced tyrosine phosphorylation of STAT3 (Fig. 2B). AG490 treatment had no significant affect on Hut78 cell survival (supplemental Fig. 2A) or adhesion to anti-LFA-1-coated plates (supplemental Fig. 2B). We extended these studies using cell-permeable isoform-specific inhibitory peptide specifically targeted against STAT3. This 12-mer phosphopeptide (AYRNRpYRRQYRY, where pY represents phosphotyrosine), identified through combinatorial chemistry (39), binds with high affinity and specificity to STAT3 in a BiaCore competition assay. This peptide was conjugated, with a spacer 6-aminohexanoic, to a cell-permeable peptide sequence called penetratine or Antennapedia (RQIKIWFQNRRMKWKK), a vector that has been shown to be highly efficient for delivering bioactive molecules (40, 41). Antp alone was used as a control peptide. Hut78 cells were pretreated with a range of concentrations of pY-pept for varying times, ranging from 1 to 12 h. Pretreatment of cells with pY-pept inhibited LFA-1-induced cell migration in a time- and dose-dependent manner (Fig. 3A). Complete loss of locomotion-associated phenotype was observed at 25 μg/ml concentration of pY-pept treated for at least 4 h (Fig. 3A). Control untreated cells or cells treated with nonspecific peptide Antp displayed a polarized and elongated morphology when triggered via LFA-1 with DI 8.1 or 7.8, respectively, whereas cells treated with 25 μg/ml pY-pept for 4 h were almost round with DI 1.78 (Fig. 3B). However, the attachment of cells and formation of associated filopodia remained intact in these cells (data not shown). Pretreatment of Hut78 cells with pY-pept treatment had no significant effect on cell survival (supplemental Fig. 3A) or adhesion to anti-LFA-1-coated plates (supplemental Fig. 3B). Therefore, for further experimentations, short pretreatment of the cells for 4 h with a25 μg/ml concentration of pY-pept was employed. Hut78 cells pretreated with pY-pept did not show LFA-1-induced STAT3 tyrosine phosphorylation (Fig. 3C). The use of small interfering RNA-mediated approaches was constrained by the capacity of STAT-3-specific small interfering RNA to induce apoptosis in transfected cells.4
FIGURE 2.
Effect of AG490 on LFA-1-induced locomotory phenotype of T-cells. A, Hut78 cells were pretreated with either vehicle (0.01% (v/v) DMSO; Control) or AG490 (1, 2.5, 5, or 10 μm) for 1 h and incubated on an anti-LFA-1-coated 96-well plate for 4 h. At least 20 microscopic fields were photographed, and a representative image is shown. Dose response migration inhibition by AG490 in Hut78 cells stimulated via immobilized anti-LFA-1 was quantified by measurement of DI and presented. B, untreated or AG490 (10 μm)-treated serum-starved Hut78 cells were stimulated with or without anti-LFA-1 for 10 min and lysed. Cell lysates (20 μg each) were resolved by SDS-PAGE and after Western blotting were probed with anti-phospho-STAT3 (Tyr705; pSTAT3) or anti-STAT3 antibody. Relative densitometric analysis of the individual pSTAT3 band was performed and presented. Data are means ± S.E. of three independent experiments. *, p < 0.05 with respect to control.
FIGURE 3.
Effect of STAT3-inhibitory peptide on LFA-1-induced locomotory phenotype of T-cells. A, Hut78 cells were pretreated with different concentrations of pY-pept (ranging from 1 to 50 μg/ml) for varying time periods (ranging from 1 to 12 h) and incubated on an anti-LFA-1-coated 96-well plate for 4 h. Quantitative evaluation of the number of Hut78 cells developing a polarized migratory phenotype was performed using an IN Cell Analyzer 1000 microscope and Investigator image analysis software. Data are normalized mean values of three independent experiments in triplicate. B, Hut78 cells untreated (control) or pretreated with 25 μg/ml nonspecific peptide (Antp) or 25 μg/ml STAT3-inhibitory peptide (pY-pept) for 4 h were incubated on an anti-LFA-1-coated 96-well plate for another 4 h. At least 20 microscopic fields were photographed, and a representative figure is shown. Migratory phenotypes were quantified by measurement of DI and presented. C, untreated or 25 μg/ml Antp- or pY-pept-treated serum-starved cells were stimulated with or without anti-LFA-1 for 10 min and lysed. Cell lysates (20 μg each) were resolved by SDS-PAGE and after Western blotting were probed with anti-phospho-STAT3 (pSTAT3) or anti-STAT3 antibody. Relative densitometric analysis of the individual pSTAT3 band was performed and presented. D and E, Hut78 cells were pretreated with 25 μg/ml Antp or pY-pept, and cell nuclei were stained with Hoechst. Cells were incubated on an anti-LFA-1-coated 96-well plate, and time lapse images at 2-min intervals were recorded for 20 min using an IN Cell Analyzer 1000 automated microscope. Data were analyzed using Image-Pro Plus version 6.1 analysis software for speed of migration and distance traveled. Data are means ± S.E. of three independent experiments. *, p < 0.05 with respect to corresponding control.
Further, to determine if the failure of STAT3-inhibited cells to develop a migratory phenotype was also accompanied by the loss of active locomotion, HCA experiments utilizing time lapse video imaging by an IN Cell Analyzer 1000 automated microscope were performed. A slow and apparently random mode of locomotion was observed in control Hut78 cells triggered via cross-linking of LFA-1. Measuring live cell migration over a period of 20 min gave clear evidence that pY-pept treatment significantly decreased LFA-1-stimulated Hut78 cell migration (Fig. 3, D and E). The average speed of Hut78 cell migration was recorded to be 1.055 μm/min, which was decreased by 73.1% (0.283 μm/min) when cells were pretreated with pY-pept (Fig. 3D). The average distance traveled by control cells was 21.1 μm (ranging from 8.3 to 69.5 μm), which was decreased to 6.2 μm (ranging from 0.1 to 22 μm) when cells were pretreated with pY-pept (Fig. 3E). These findings clearly demonstrate the requirement of active STAT3 function for T-cell locomotion in a physiological microenvironment.
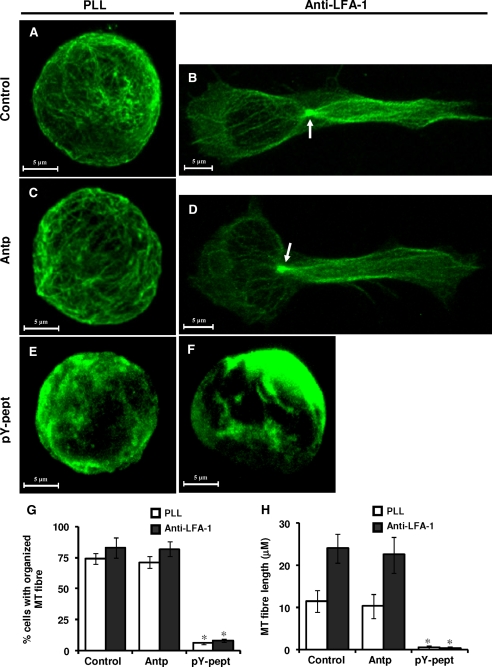
Inhibition of STAT3 Affects MTs of T-cells—We have recently demonstrated that MT targeting agents, such as paclitaxael (a tubulin stabilizer), nocodazole, or PBOX-15 (tubulin depolymerizers) inhibited T-cell migration (34). To determine whether inhibition of STAT3 by pY-pept affects MTs of T-lymphocytes, causing inhibition of cell migration, intracellular distribution and functional involvement of the MTs were analyzed. Control untreated Hut78 cells or cells treated with nonspecific peptide Antp showed radial arrays of organized tubulin distribution (Fig. 4, A and C). LFA-1-induced migrating T-cells displayed the characteristic array of MT in that MTOC was located at the side of the nucleus opposing the direction of cell migration and long MT extended to the uropods from here (Fig. 4, B and D). In contrast, exposure of Hut78 cells to STAT3-inhibitory peptide pY-pept dramatically impaired the tubulin network (Fig. 4E), resulting in complete loss of the typical motile phenotype upon LFA-1 cross-linking (Fig. 4F). Quantitative analysis of the α-tubulin cytoskeletal phenotypes in the population of cells by IN Cell Investigator HCA software (GE Healthcare) indicated that inhibition of STAT3 activity by pY-pept significantly reduced the number of cells (up to less than 8%) with organized MTs (Fig. 4G). Semiautomated analysis for MT fiber length of 10 randomly selected cells by Zeiss LSM Image Examiner (Carl Zeiss) indicated that pY-pept treatment significantly reduced the average length of tubulin fiber from greater than 11.5 μm to less than 0.5 μm (Fig. 4H). STAT3-inhibitory peptide did not significantly affect the actin cytoskeletal network (supplemental Fig. 4).
FIGURE 4.
Effect of STAT3 inhibition on the organization of MT network in resting and LFA-1-stimulated T-cells. Hut78 cells untreated (Control; A and B) or pretreated with 25 μg/ml nonspecific peptide (Antp; C and D) or 25 μg/ml STAT3-inhibitory peptide (pY-pept; E and F) for 4 h were incubated on poly-l-lysine-coated (PLL; A, C, and E) or anti-LFA-1-coated (B, D, and F) Permanox® chamber slides for 4 h. After this time, the medium was carefully removed, and cells were fixed in 3% paraformaldehyde. Cells were immunostained for α-tubulin (green). The organization of the MT network was visualized by confocal microscopy using a ×63 oil immersion lens. The arrow indicates clearly distinguishable MTOC. G, organization of MT network in the population of cells. A minimum of 100 cells was scored by IN Cell Investigator HCA analysis software. H, semiautomated analysis of MT fiber length of 10 randomly selected cells was performed using the Zeiss LSM Image Examiner software. Results shown are representative of three independent experiments. *, p < 0.05 with respect to corresponding control.
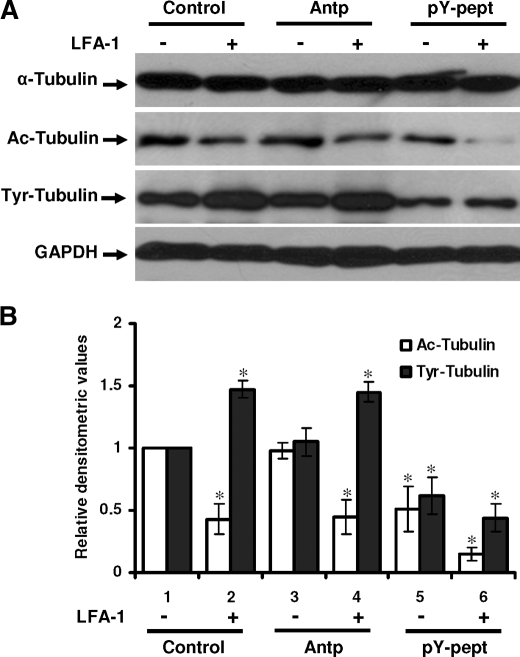
Inhibition of STAT3 Activity Affects Post-translational Modifications of Tubulin in T-cells—α-Tubulin undergoes post-translational modifications, including acetylation and tyrosination, that are thought to modulate MT stability and dynamics within the cell (13). To further explore the effect of STAT3 inhibition on post-translational modification of tubulin in migrating T-cells, we examined the total expression of α-tubulin, acetylated α-tubulin, and tyrosinated α-tubulin by Western immunoblotting and with densitometric quantitation normalized for glyceraldehyde-3-phosphate dehydrogenase expression (Fig. 5). Acetylated α-tubulin level in LFA-1-stimulated Hut78 cells was 57% lower as compared with resting cells (Fig. 5B, open bars, lane 2 versus lane 1). Pretreatment of cells with pY-pept reduced acetylated tubulin level to 51% (Fig. 5B, open bars, lane 5 versus lane 1), which was further reduced to 11% after LFA-1 cross-linking as compared with control untreated resting cells (Fig. 5B, open bars, lane 6 versus lane 1). No significant change in the level of acetylated tubulin was observed when Hut78 cells were pretreated with nonspecific peptide Antp (Fig. 5B, open bars, lane 3 versus lane 1 and lane 4 versus lane 2). The tyrosinated tubulin level in migrating Hut78 cells was increased up to 47% compared with the resting cells (Fig. 5B, closed bars, lane 2 versus lane 1). Treatment of these cells with pY-pept reduced the level of tyrosinated tubulin by 51% in resting (Fig. 5B, closed bars, lane 5 versus lane 1) and by 56% upon LFA-1 cross-linking as compared with control (untreated) cells (Fig. 5B, closed bars, lane 6 versus lane 1). No significant change in the level of tyrosinated tubulin was observed when Hut78 cells were pretreated with nonspecific peptide Antp (Fig. 5B, closed bars, lane 3 versus lane 1 and lane 4 versus lane 2).
FIGURE 5.
Effect of STAT3 inhibition on post-translational modifications of tubulin in resting and LFA-1-stimulated T-cells. A, Hut78 cells untreated (Control), pretreated with 25 μg/ml nonspecific peptide (Antp) or 25 μg/ml STAT3-inhibitory peptide (pY-pept) for 4 h were stimulated with or without anti-LFA-1 for 4 h and lysed. Cell lysates (20 μg each) were resolved by SDS-PAGE and after Western blotting were probed with anti-α-tubulin, anti-acetylated tubulin (Ac-Tubulin), anti-tyrosinated tubulin (Tyr-Tubulin), or glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (as a loading control) antibody. B, relative densitometric analysis of the individual band was performed and presented. Data are mean ± S.E. of three independent experiments. *, p < 0.05 with respect to corresponding controls.
Next, to explore the possibility (if any) that STAT3 exerts its effect on MT assembly and post-translational modification of tubulin by direct physical association with tubulin, cell lysates of resting or LFA-1-stimulated Hut78 cells were immunoprecipitated with tubulin and probed with anti-STAT3 antibody. However, we did not detect STAT3 protein in tubulin-immunoprecipitated samples (data not shown).
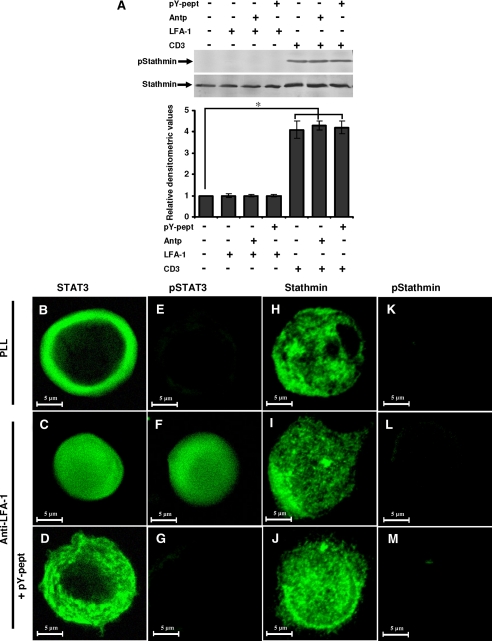
STAT3 Exerts Its Effect on MT via Stathmin—The spatial organization of the MT cytoskeleton is thought to be directed by several regulatory molecules. Stathmin-tubulin interaction is known to regulate MT dynamics in motile and mitotic cells (28). Stathmin is known to undergo serine phosphorylation at Ser16 upon cell stimulation (27). However, we did not detect stathmin phosphorylation (Ser16) due to LFA-1 cross-linking (Fig. 6A, lane 2 versus lane 1). In contrast, CD3 cross-linking induced stathmin phosphorylation on Ser16 (Fig. 6A, lane 5 versus lane 1). Moreover, CD3-induced stathmin phosphorylation was not affected by pY-pept (Fig. 6A, lane 7 versus lane 5). Microscopic examination of stathmin indicated its cytoplasmic distribution in the resting Hut78 cells (Fig. 6, H-J). Upon LFA-1 stimulation, STAT3 underwent tyrosine phosphorylation and nuclear translocation (Fig. 6, B-G); however, stathmin remained in the cytoplasm but displayed polarized staining, which was not affected by pY-pept treatment (Fig. 6, H-J). Phosphorylation (Ser16) of stathmin was not detected in resting or LFA-1-stimulated Hut78 cells (Fig. 6, K-M) but was detected in response to stimulation with anti-CD3 (Fig. 6A).
FIGURE 6.
Stathmin phosphorylation and localization during LFA-1-induced T-cell migration. A, serum-starved Hut78 cells were pretreated with 25 μg/ml nonspecific peptide Antp or 25 μg/ml STAT3-inhibitory peptide pY-pept for 4 h and incubated on anti-LFA-1- or anti-CD3-coated 6-well plates for 10 min and lysed. Cell lysates (20 μg each) were resolved by SDS-PAGE and after Western blotting were probed with anti-phosphostathmin (Ser16; pStathmin) or anti-stathmin antibody. Relative densitometric analysis of the individual pStathmin band was performed and presented. Data are the mean ± S.E. of three independent experiments. *, p < 0.05 with respect to control. B-M, Hut78 cells untreated or pretreated with 25 μg/ml STAT3-inhibitory peptide (pY-pept; D, G, J, and M) were incubated on poly-l-lysine (PLL) or anti-LFA-1-coated Permanox® chamber slides for 10 min. After this time, the medium was carefully removed, and cells were fixed in 3% paraformaldehyde. Cells were immunostained for STAT3, phospho-STAT3 (Tyr705; pSTAT3), stathmin, or phosphostathmin (Ser16; pStathmin) and visualized by confocal microscopy using a ×63 oil immersion lens.
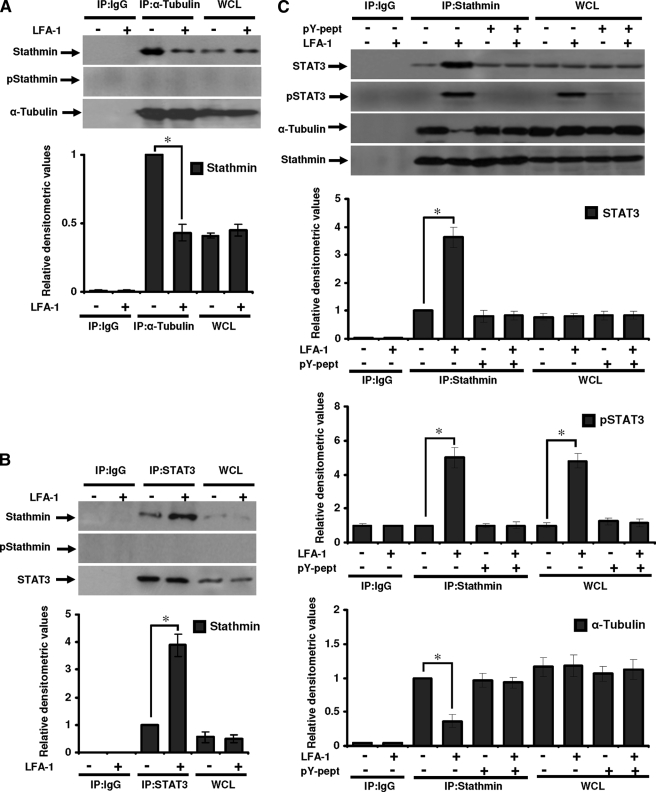
Next, we examined whether stathmin physically interacts with α-tubulin in T-cells during migration. Resting and LFA-1-stimulated migrating Hut78 cells were immunoprecipitated with anti-α-tubulin or anti-IgG (isotype control) and probed with anti-stathmin (Fig. 7A). Results showed that stathmin binding to tubulin was reduced to 43% in migrating cells as compared with nonmigrating cells (Fig. 7A). There was no change in the expression of total stathmin protein in resting and LFA-1-stimulated cells as detected by immunoblotting for whole cell lysates (Fig. 7A). Phosphostathmin was not detected either in α-tubulin-immunoprecipitated samples or in whole cell lysates (Fig. 7A).
FIGURE 7.
STAT3 interaction with tubulin or stathmin in migrating T-cells. Serum-starved Hut78 cells were stimulated with or without anti-LFA-1 for 10 min and lysed. A, cell lysates (500 μg each) were immunoprecipitated with anti-α-tubulin (IP:α-Tubulin) or IgG (as isotype control; IP:IgG). Immunoprecipitates and whole cell lysates (used as control, 20 μg each; WCL) were Western blotted and probed with anti-stathmin, anti-phosphostathmin (Ser16; pStathmin), or anti-α-tubulin. B, cell lysates (500 μg each) were immunoprecipitated with anti-STAT3 (IP:STAT3) or IgG (IP:IgG). Immunoprecipates and whole cell lysates (20 μg each; WCL) were Western blotted and probed with anti-stathmin, anti-phospho-stathmin (Ser16; pStathmin), or anti-STAT3. C, untreated or pY-pept-treated Hut78 cells were stimulated with or without anti-LFA-1 for 10 min and lysed. Cell lysates (500 μg each) were immunoprecipitated with anti-stathmin (IP: Stathmin) or IgG (IP:IgG). Immunoprecipates and whole cell lysates (20 μg each; WCL) were Western blotted and probed with anti-STAT3, anti-phospho-STAT3 (Tyr705; pSTAT3), anti-α-tubulin, or anti-stathmin. Relative densitometric analysis of the individual band was performed and presented. Data are mean ± S.E. of three independent experiments. *, p < 0.05 with respect to corresponding controls.
To determine whether STAT3 exerts its effect on MT via stathmin in migrating T-cells, we immunoprecipitated resting and LFA-1-stimulated Hut78 cells with anti-STAT3 or isotype control IgG and probed with anti-stathmin (Fig. 7B). Interestingly, stathmin co-immunoprecipitated with STAT3, with a substantially greater amount (3.6-fold) of stathmin co-immunoprecipitated in migrating cells as compared with resting cells (Fig. 7B). Phosphostathmin was not detected either in STAT3 immunoprecipitated samples or in whole cells lysates (Fig. 7B).
Similarly, untreated or pY-pept-treated resting and LFA-1-stimulated Hut78 cells were immunoprecipitated with anti-stathmin or isotype control IgG and probed for the presence of STAT3, pSTAT3, or α-tubulin. A 3.8-fold higher association between stathmin and STAT3 was observed in LFA-1-induced samples as compared with resting cells (Fig. 7C). Pretreatment of Hut78 cells with pY-pept inhibited STAT3-stathmin association (Fig. 7C). Stathmin association with pSTAT3 was increased to 5-fold in LFA-1-stimulated Hut78 cells as compared with resting cells (Fig. 7C). pSTAT3 was not detected in pY-pept-treated samples (Fig. 7C). α-Tubulin co-precipitated with stathmin in both the resting and LFA-1-induced samples. Association of α-tubulin with stathmin reduced to 0.57-fold in LFA-1-stimulated Hut78 cells as compared with resting cells (Fig. 7C). This decrease in α-tubulin-stathmin association in LFA-1-stimulated cells was not detected when these cells were pretreated with pY-pept (Fig. 7C). These data suggest that STAT3 competitively binds to stathmin and thereby inhibits its MT depolymerization activity during T-cell migration.
Taken together, these results demonstrate that STAT3 regulates T-cell migration. Inhibition of STAT3 activity disrupts the microtubule network, interferes with post-translational modifications of tubulin, and suppresses microtubule dynamics via its physical association with stathmin, thus affecting the function of this cytoskeletal system in the migratory process.
DISCUSSION
T-cell migration in response to integrin signaling is a complex, highly coordinated, multistep process involving 1) extension of membrane protrusions at the leading edge, 2) formation of attachment sites to the extracellular matrix at the newly formed cell periphery, 3) cell traction, which might be driven by contraction of the cytoskeleton, and 4) the release of adhesions at the rear portions of the cell (detachment) (42). These events are regulated by multiple signaling mechanisms, such as kinase and/or phosphatase signaling, small GTPase signaling (i.e. Rho and Rac), and cytoskeletal reorganization (42). Previously, we have demonstrated that T-cells activated through LFA-1 undergo reorganization of the tubulin cytoskeleton during migration (4, 8). Further, we demonstrated that disruption of tubulin polymerization process or MT dynamics by known tubulin-targeting agents, such as paclitaxael (MT stabilizer), nocodazole (MT depolymerizer), or PBOX-15 (a novel tubulin depolymerizing compound), inhibits T-cell migration (34). Here, we demonstrate that suppression of STAT3 activity impaired the MT network, resulting in the inhibition of T-cell migration. In addition to the multifunctional role of STAT3 in mediating several biological processes, including cell proliferation, differentiation, survival, and motility and promotion of tumor growth (20, 43, 44), a role of STAT3 is emerging in cell migration, as described in both keratinocytes (23, 26) and mouse embryonic fibroblasts (33). In this study, we demonstrate involvement of STAT3 in T-cell migration, suggesting possible wider roles for this molecule in the migratory processes.
STAT3 is known to be activated by various cytokines and growth factors, such as interleukin-6, epidermal growth factor, and hepatocyte growth factor (22). In response to external stimuli, STAT3 is recruited from the cytosol to phosphorylated receptors via its Src homology 2 domain. STAT3 molecules then become tyrosine-phosphorylated (Tyr705) within receptor complexes, form dimers, and translocate to the nucleus to effect expression of responsive genes (18, 24, 44). Taking advantage of the STAT3 intracellular localization and phosphorylation status associated with its activation state, we demonstrate that LFA-1 cross-linking can activate STAT3 in T-cells by virtue of nuclear translocation as well as induction of Tyr705 phosphorylation. Pharmacological inhibition of STAT3 by both AG490 and specific peptide blocked LFA-1-induced migration of Hut78 cells. These observations lead to a consistent conclusion that STAT3 activation is required for T-cell migration either by altering expression and/or activation of proteins involving T-cell migration or through its function as an adaptor molecule. Oligonucleotide gene arrays and quantitative real time PCR analysis have revealed that STAT3 regulates a common set of genes involved in wound healing and cancer (24) that are implicated in cell migration, metastasis, and remodeling of extracellular matrix, including, for example, ICAM-1, CCL2, CXCL2, EPAS-1, proteases in the cathepsin and SERPIN families, such as uPA and its receptor uPAR, PAI-1, etc. (24). Many of these genes also play a role in the process of T-cell migration and trafficking. Further functional genomic and proteomic studies are required to determine the role of STAT3 in the regulation of genes and proteins involved in T-cell migration.
The adaptor function of STAT3 has also been implicated in the cell migration process. Intracellular signaling of STAT3 in keratinocytes modulates tyrosine phosphorylation of p130cas (a focal adhesion molecule), affecting cell adhesiveness to the substratum and cell migration in response to growth factor (45). In endothelial cells, phosphorylation of STAT3 in response to VEGFR stimulation resulted in its nuclear translocation and induction of migration, and a dominant negative mutant of STAT3 completely inhibited cell migration (46). Last, an association has been reported between STAT3 and Rho GTPases (47), the later being critically involved in the selective stabilization of MTs in the lamella of crawling cells.
Here, we observed that the suppression of STAT3 activity significantly reduced both the acetylated and the tyrosinated form of α-tubulin. These reversible post-translational modifications of α-tubulin have been implicated in regulating MT stability and functions (48, 49) that influence important biological processes, such as cell motility, signaling, and homeostasis (13, 48), in a cell- or context-dependent manner (50). We have recently demonstrated that a tubulin-targeting compound PBOX-15 altered tubulin post-translational modifications and thus inhibited T-cell migration (34). Acetylated α-tubulin is most abundant in stable MTs but is absent from dynamic cellular structures (13, 48). Acetylation of α-tubulin may be required for coordination and differential interaction of MTs with MT-associated proteins and motor proteins (such as kinesin and dynein) for regulating their activity or localization (51); kinesin is known to regulate distribution of intermediate filaments. Change in tubulin acetylation affects the conformation of the MT surface to which MT-associated proteins bind (50), which in turn could affect processes involved in T-cell motility. In addition, it regulates dynamics of cell adhesion and motility by alteration in Rac and Rho signaling, targeting of MTs to focal adhesions, and localization of adhesion assembly and disassembly signals (48, 50). Tubulin tyrosination acts as a signal for the interaction of MT with other signaling proteins (49) that may affect cell morphology and locomotory potential. It regulates the interactions of intermediate filaments with MT via a kinesin-dependent mechanism (51). Moreover, tubulin tyrosination is important for the coordination of different cytoskeletal elements, such as vimentin (49), which in turn may provide structural support for the extensive microtentacles (extensive and motile MT-enriched membrane protrusions) (52) in migrating T-cells. Both acetylation and tyrosination of tubulin regulate localized recruitment of complexes composed of CLIP-170 (a cytoplasmic linker protein) and CLIP-associating proteins and/or components forming adhesions to the plus end of dynamic MTs (50, 53). A role of tubulin post-translational modification in MT-dependent organelle polarization (Golgi, MTOC) typically found in migrating cells has recently been proposed (50).
We further demonstrate a direct interaction between STAT3 and stathmin. Moreover, stathmin associates with tubulin, although no direct association of STAT3 with α-tubulin was observed. Stathmin family proteins are important for appropriate cell cycle progression in many types of eukaryotic cells and promote neurite outgrowth through regulation of MT dynamics in growth cones (30). A reduced level of stathmin-tubulin interaction has been described at the leading edge of migrating Xenopus A6 cells (28). Interestingly, stathmin is highly expressed in many types of cancers, and overexpression correlates with abnormal motility and tissue invasion of human sarcomas in vivo (54). Experimental evidence suggests that stathmin MT-depolymerizing activity is negatively regulated by phosphorylation on serine residues, of which Ser16 plays a major role(s) (27). We and others (55) observed that TCR activation by CD3 cross-linking of T-cells induced stathmin phosphorylation (Ser16). However, we did not detect stathmin phosphorylation (Ser16) due to LFA-1 cross-linking to Hut78 cells. These findings suggest that LFA-1 regulation of stathmin is different from that of TCR. Further, we show that TCR induced phosphorylation of stathmin is independent on STAT3, since inhibition of STAT3 activity by pY-pept did not affect anti-CD3-stimulated stathmin phosphorylation (Ser16).
Our results are in agreement with a recent report that STAT3 interacts with stathmin and regulates MT dynamics by antagonizing its depolymerization activity (33). However, these authors utilized exogenous expression of wild-type or mutant STAT3 and stathmin proteins. Moreover, they demonstrated by cell-free in vitro experiments that STAT3 attenuated the MT-destabilizing activity of stathmin (33). Here, we demonstrate a functional involvement of endogenous STAT3 by direct interaction with stathmin to control MT dynamics in migrating T-cells. Specific inhibition of STAT3 activity by pY-pept inhibited stathmin interaction with STAT3 and α-tubulin, thus disrupting its regulation of MT dynamics. It remains to be determined whether STAT3 interacts with other signal-transducing molecules involved in T-cell migration.
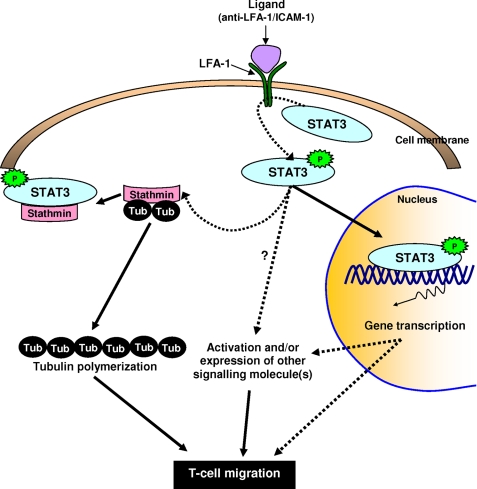
Based on our data, we propose a model for STAT3 involvement in regulating T-cell migration (Fig. 8). LFA-1 cross-linking activates STAT3 by inducing tyrosine phosphorylation and nuclear translocation. The interaction of activated STAT3 with stathmin plays a key role(s) in inhibiting the tubulin depolymerization activity of stathmin. This interaction makes tubulin heterodimers available for polymerization during active cytoskeleton reorganization process in migrating T-cells. This presents a novel mechanism through which STAT3 mediates T-cell migration possibly also in combination with its nuclear transcriptional activity.
FIGURE 8.
A model for LFA-1-induced STAT3 activation and regulation of MT dynamics. The figure illustrates STAT3 activation by tyrosine phosphorylation, its nuclear translocation, and its predicted interaction with stathmin. Stathmin-bound tubulin heterodimers are unable to polymerize. Activated STAT3 binding to stathmin inhibits its tubulin depolymerization activity, making tubulin available for polymerization in actively migrating T-cells. Dashed lines with arrows indicate that the interactions either are putative or involve multiple steps. P in the solid star, Tyr705 phosphorylation.
In conclusion, we have demonstrated that STAT3 is involved in mediating T-cell migration by regulating MT dynamics. The expectation that activated STAT3 would turn on specific transcriptional events and target genes that are integral parts of migrating T-cells forms the basis for further experiments. These studies will greatly enhance the understanding of fundamental intracellular mechanisms involved in T-cell migration. Our results suggest that STAT3 may be potentially used as a therapeutic target for various pathological conditions involving T-cell responses, including chronic inflammatory disease, such as inflammatory bowel disease and the inflammatory arthropathies.
This work was supported by a Grant from the Higher Education Authority of Ireland under the department of Education and Science's Program for Research in Third Level Institutions and the Health Research Board of Ireland.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1-4.
Footnotes
The abbreviations used are: LFA-1, lymphocyte function-associated antigen-1; ICAM, intercellular adhesion molecule; STAT, signal transducers and activators of transcription; MT, microtubule; MTOC, microtubule organizing center; TCR, T-cell receptor; Fmoc, 9-fluorenylmethoxycarbonyl; DI, deformation index; PBS, phosphate-buffered saline; HCA, high content analysis; Antp, Antennapedia peptide; pY-pept, STAT3-inhibitory peptide.
N. K. Verma, J. Dourlat, A. M. Davies, A. Long, W.-Q. Liu, C. Garbay, D. Kelleher, and Y. Volkov, unpublished data.
References
- 1.Hogg, N., Laschinger, M., Giles, K., and McDowall, A. (2003) J. Cell Sci. 116 4695-4705 [DOI] [PubMed] [Google Scholar]
- 2.Springer, T. A. (1994) Cell 76 301-314 [DOI] [PubMed] [Google Scholar]
- 3.Fanning, A., Volkov, Y., Freely, M., Kelleher, D., and Long, A. (2005) Int. Immunol. 17 449-458 [DOI] [PubMed] [Google Scholar]
- 4.Volkov, Y., Long, A., and Kelleher, D. (1998) J. Immunol. 161 6487-6495 [PubMed] [Google Scholar]
- 5.Stanley, P., Smith, A., McDowall, A., Nicol, A., Zicha, D., and Hogg, N. (2008) EMBO J. 27 62-75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Long, A., Kelleher, D., Lynch, S., and Volkov, Y. (2001) J. Immunol. 167 636-640 [DOI] [PubMed] [Google Scholar]
- 7.Freely, S., Volkov, Y., Kelleher, D., and Long, A. (2005) Biochem. Biophys. Res. Commun. 334 619-630 [DOI] [PubMed] [Google Scholar]
- 8.Volkov, Y., Long, A., McGrath, S., Eidhin, D. N., and Kelleher, D. (2001) Nat. Immunol. 2 508-514 [DOI] [PubMed] [Google Scholar]
- 9.Volkov, Y., Long, A., Freeley, M., Golden-Mason, L., O'Farrelly, C., Murphy, A., and Kelleher, D. (2005) Gastroenterology 130 482-492 [DOI] [PubMed] [Google Scholar]
- 10.Kupfer, A., and Singer, S. J. (1989) J. Exp. Med. 170 1697-1713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hauzenberger, D., Klominek, J., and Sundqvist, K. G. (1995) Crit. Rev. Immunol. 15 285-316 [DOI] [PubMed] [Google Scholar]
- 12.Watanabe, T., Noritake, J., and Kaibuchi, K. (2005) Trends Cell Biol. 15 76-83 [DOI] [PubMed] [Google Scholar]
- 13.Westermann, S., and Weber, K. (2003) Nat. Rev. Mol. Cell Biol. 4 938-947 [DOI] [PubMed] [Google Scholar]
- 14.Gundersen, G. G., and Bulinski, J. C. (1986) J. Cell Biol. 102 1118-1126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wehrle-Haller, B., and Imhof, H. A. (2003) Int. J. Biochem. Cell Biol. 35 39-50 [DOI] [PubMed] [Google Scholar]
- 16.Waterman-Storer, C. M., and Salmon, E. D. (1999) Curr. Opin. Cell Biol. 11 61-67 [DOI] [PubMed] [Google Scholar]
- 17.Luduena, R. F. (1998) Int. Rev. Cytol. 178 207-275 [DOI] [PubMed] [Google Scholar]
- 18.Levy, D. E., and Darnell, J. E., Jr. (2002) Nat. Rev. Mol. Cell Biol. 3 651-662 [DOI] [PubMed] [Google Scholar]
- 19.Yamashita, S., Miyagi, C., Carmany-Rampey, A., Shimizu, T., Fujii, R., Schier, A. F., and Hirano, A. F. (2002) Dev. Cell 2 363-375 [DOI] [PubMed] [Google Scholar]
- 20.Yu, H., Kortylewski, M., and Pardoll, D. (2007) Nat. Rev. Immunol. 7 41-51 [DOI] [PubMed] [Google Scholar]
- 21.Brierley, M. M., and Fish, N. (2005) J. Inteferon Cytokine Res. 25 733-744 [DOI] [PubMed] [Google Scholar]
- 22.Ihle, J. N. (2001) Curr. Opin. Cell Biol. 13 211-217 [DOI] [PubMed] [Google Scholar]
- 23.Akira, S., Nishio, Y., Inoue, M., Wang, X. J., Wei, S., Matsusaka, T., Yoshida, K., Sudo, T., Naruto, M., and Kishimoto, T. (1994) Cell 77 63-71 [DOI] [PubMed] [Google Scholar]
- 24.Dauer, D. J., Ferraro, B., Song, L., Yu, B., Mora, L., Buettner, R., Enkemann, S., Jove, R., and Haura, E. B. (2005) Oncogene 24 3397-3408 [DOI] [PubMed] [Google Scholar]
- 25.Takeda, K., Noguchi, K., Shi, W., Tanaka, T., Matsumoto, M., Yoshida, N., Kishimoto, T., and Akira, S. (1997) Proc. Natl. Acad. Sci. U. S. A. 94 3801-3804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sano, S., Itami, S., Takeda, K., Tarutani, M., Yamaguchi, Y., Miura, H., Yoshikawa, K., Akira, S., and Takeda, J. (1999) EMBO J. 18 4657-4668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Andersen, S. S. (2000) Trends Cell Biol. 10 261-267 [DOI] [PubMed] [Google Scholar]
- 28.Niethammer, P., Bastiaens, P., and Karsenti, E. (2004) Science 303 1862-1866 [DOI] [PubMed] [Google Scholar]
- 29.Sellin, M. E., Holmfeldt, P., Stenmark, S., and Gullberg, M. (2008) Mol. Biol. Cell 19 2897-2906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Steinmetz, M. O. (2007) J. Struct. Biol. 158 137-147 [DOI] [PubMed] [Google Scholar]
- 31.Sobel, A. (1991) Trends Biochem. Sci. 16 301-305 [DOI] [PubMed] [Google Scholar]
- 32.Giampietro, C., Luzzati, F., Gambarotta, G., Giacobini, P., Boda, E., Fasolo, A., and Perroteau, I. (2005) Endocrinology 146 1825-1834 [DOI] [PubMed] [Google Scholar]
- 33.Ng, D. C. H., Lin, B. H., Lim, C. P., Huang, G., Zhang, T., Poli, V., and Cao, X. (2006) J. Cell Biol. 172 245-257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Verma, N. K., Dempsey, E., Conroy, J., Olwell, P., McElligott, A. M., Davies, A. M., Kelleher, D., Butini, S., Campiani, G., Williams, D. C., Zisterer, D. M., Lawler, M., and Volkov, Y. (2008) J. Mol. Med. 86 457-469 [DOI] [PubMed] [Google Scholar]
- 35.Verma, N. K., and Dey, C. S. (2006) Diabetologia 49 1656-1660 [DOI] [PubMed] [Google Scholar]
- 36.Fields, G. B., and Noble, R. L. (1990) Int. J. Pept. Protein Res. 35 161-241 [DOI] [PubMed] [Google Scholar]
- 37.Chao, H. G., Bernatowicz, M. S., Reiss, P. D., and Matsueda, G. R. (1994) J. Org. Chem. 59 6687-6691 [Google Scholar]
- 38.Aoudjit, F., and Vuori, K. (2000) Blood 95 2044-2051 [PubMed] [Google Scholar]
- 39.Wiederkehr-Adam, M., Ernst, P., Muller, K., Bieck, E., Gombert, F. O., Ottl, J., Graff, P., Grossmüller, F., and Heim, M. H. (2003) J. Biol. Chem. 278 16117-16128 [DOI] [PubMed] [Google Scholar]
- 40.Derossi, D., Joliot, A. H., Chassaing, G., and Prochiantz, A. (1994) J. Biol. Chem. 269 10444-10450 [PubMed] [Google Scholar]
- 41.Dunican, D. J., and Doherty, P. (2001) Biopolymers 60 45-60 [DOI] [PubMed] [Google Scholar]
- 42.Horwitz, A. R., and Parsons, J. T. (1999) Science 286 1102-1103 [DOI] [PubMed] [Google Scholar]
- 43.Silver, D. L., Naora, H., Liu, J., Cheng, W., and Montel, D. J. (2004) Cancer Res. 64 3550-3558 [DOI] [PubMed] [Google Scholar]
- 44.Bowman, T., Garcia, R., Turkson, J., and Jove, R. (2000) Oncogene 19 2474-2488 [DOI] [PubMed] [Google Scholar]
- 45.Kira, M., Sano, S., Takagi, S., Yoshikawa, K., Takada, T., and Itami, S. (2002) J. Biol. Chem. 277 12931-12936 [DOI] [PubMed] [Google Scholar]
- 46.Yahata, Y., Shirakata, Y., Tokumaru, S., Yamasaki, K., Sayama, K., Hanakawa, Y., Detmar, M., and Hashimoto, K. (2003) J. Biol. Chem. 278 40026-40031 [DOI] [PubMed] [Google Scholar]
- 47.Debidda, M., Wang, L., Zang, H., Ploi, V., and Zheng, Y. (2005) J. Biol. Chem. 280 17275-17285 [DOI] [PubMed] [Google Scholar]
- 48.Hubbert, C., Guardiola, A., Shao, R., Kawaguchi, Y., Ito, A., Nixon, A., Yoshida, M., Wang, X. M., and Yao, T. P. (2002) Nature 417 455-458 [DOI] [PubMed] [Google Scholar]
- 49.Bulinski, J. C., Gundersen, G. C., and Webster, D. R. (1987) Nature 328 676-676 [DOI] [PubMed] [Google Scholar]
- 50.Tran, A. D., Marmo, T. P., Salam, A. A., Che, S., Finkelstein, E., Kabarriti, R., Xenias, H. S., Mazitschek, R., Hubbert, C., Kawaguchi, Y., Sheetz, M. P., Yao, T. P., and Bulinski, J. C. (2007) J. Cell Sci. 120 1469-1479 [DOI] [PubMed] [Google Scholar]
- 51.Kreitzer, G., Liao, G., and Gundersen, G. C. (1999) Mol. Biol. Cell 10 1105-1118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Whipple, R. A., Balzer, E. M., Cho, E. H., Matrone, M. A., Yoon, J. R., and Martin, S. S. (2008) Cancer Res. 68 5678-5688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Peris, L., Thery, M., Fauré, J., Saoudi, Y., Lafanechère, L., Chilton, J. K., Gordon-Weeks, P., Galjart, N., Bornens, M., Wordeman, L., Wehland, J., Andrieux, A., and Job, D. (2006) J. Cell Biol. 174 839-849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Belletti, B., Nicoloso, M. S., Schiappacassi, M., Berton, S., Lovat, F., Wolf, K., Canzonieri, V., D'Andrea, S., Zucchetto, A., Friedl, P., Colombatti, A., and Baldassarre, G. (2008) Mol. Biol. Cell 19 2003-2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tanaka, Y., Hamano, S., Gotoh, K., Murata, Y., Kunisaki, Y., Nishikimi, A., takii, R., Kawaguchi, M., Inayoshi, A., Masuko, S., Himeno, K., Sasazuki, T., and Fukui, Y. (2007) Nat. Immunol. 8 1067-1075 [DOI] [PubMed] [Google Scholar]