Abstract
Complement component C5a is a potent pro-inflammatory agent inducing chemotaxis of leukocytes toward sites of infection and injury. C5a mediates its effects via its G protein-coupled C5a receptor (C5aR). Although under normal conditions highly beneficial, excessive levels of C5a can be deleterious to the host and have been related to numerous inflammatory diseases. A natural inhibitor of the C5aR is chemotaxis inhibitory protein of Staphylococcus aureus (CHIPS). CHIPS is a 121-residue protein excreted by S. aureus. It binds the N terminus of the C5aR (residues 1-35) with nanomolar affinity and thereby potently inhibits C5a-mediated responses in human leukocytes. Therefore, CHIPS provides a starting point for the development of new anti-inflammatory agents. Two O-sulfated tyrosine residues located at positions 11 and 14 within the C5aR N terminus play a critical role in recognition of C5a, but their role in CHIPS binding has not been established so far. By isothermal titration calorimetry, using synthetic Tyr-11- and Tyr-14-sulfated and non-sulfated C5aR N-terminal peptides, we demonstrate that the sulfate groups are essential for tight binding between the C5aR and CHIPS. In addition, the NMR structure of the complex of CHIPS and a sulfated C5aR N-terminal peptide reveals the precise binding motif as well as the distinct roles of sulfated tyrosine residues sY11 and sY14. These results provide a molecular framework for the design of novel CHIPS-based C5aR inhibitors.
The human complement system is a key component of the innate host defense directed against invading pathogens. Complement component C5a is a 74-residue glycoprotein generated via complement activation by cleavage of the α-chain of its precursor C5. C5a is a strong chemoattractant involved in the recruitment of neutrophils and monocytes, activation of phagocytes, release of granule-based enzymes, and in the generation of oxidants (1, 2). C5a exerts its effect by activating the C5a receptor (C5aR).3 Although this is a highly efficient process, excessive or erroneous activation of the C5aR can have deleterious effects on host tissues. C5a has been implicated in the pathogenesis of many inflammatory and immunological diseases, including rheumatoid arthritis, inflammatory bowel disease, immune complex disease, and reperfusion injury (3, 4). Consequently, there is an active ongoing search for compounds that suppress C5a-mediated inflammatory responses.
Chemotaxis inhibitory protein of Staphylococcus aureus (CHIPS) is a 121-residue protein excreted by S. aureus, which efficiently inhibits the activation of neutrophils and monocytes by formylated peptides and C5a (5, 6). CHIPS specifically binds to the formylated peptide receptor (FPR) and the C5aR with nanomolar affinity (Kd = 35.4 ± 7.7 nm and 1.1 ± 0.2 nm, respectively) (7), thereby suppressing the inflammatory response of the host. A CHIPS fragment lacking residues 1-30 (designated CHIPS31-121) has the same activity in blocking the C5aR compared with wild-type CHIPS (8). CHIPS31-121 is a compact protein comprising an α-helix packed onto a four-stranded anti-parallel β-sheet (8). C5a has an entirely different fold (PDB ID code 1KJS) and is comprised of an anti-parallel bundle of four α-helices stabilized by three disulfide bonds (9, 10). Preliminary experiments indicated that CHIPS binds exclusively to the extracellular N-terminal portion of the C5aR (7). In contrast, the binding of C5a by its receptor involves two separate binding sites: C5a residues located in the region between 12-46 (11, 12) bind to a primary binding site partly coinciding with the binding site of CHIPS, while the C terminus of C5a (residues 69-74) binds to the activation domain of the C5aR located in the receptor core (13). Because of their dissimilarity in sequence and structure, the binding sites of CHIPS and C5a are not identical (11). The present working model is that CHIPS interferes with the primary binding site of C5a located at the N terminus of the C5aR, thereby preventing the C-terminal tail of C5a from contacting the activation domain of the C5aR and blocking downstream signaling. Currently, the development of C5aR inhibitors has been focused primarily on mimicking C5a in order to directly interrupt C5a-mediated C5aR signaling (3, 4, 14). Understanding the interactions between CHIPS and the C5aR may provide valuable insights toward the development of new C5aR antagonists.
Postma et al. (15) proposed that residues involved in CHIPS binding are located between residues 10-18 of the C5aR. Specifically, the acidic residues Asp-10, Asp-15, and Asp-18 and residue Gly-12 appear to be critical for binding. High affinity binding was observed between 125I-labeled CHIPS and the N-terminal portion of the C5aR (residues 1-38) expressed on the cell surface of HEK293 cells (Kd = 29.7 ± 4.4 nm). In contrast, very moderate affinity between CHIPS and a synthetic C5aR N-terminal peptide (residues 1-37; Kd = 40 ± 19 μm), measured by isothermal titration calorimetry (ITC), was recently reported by Wright et al. (16). The discrepancy in the magnitude of these dissociation constants may be explained by the presence of two sulfate groups on tyrosine 11 and 14 of the C5aR N terminus expressed on the cell surface of HEK293 cells, which are absent in the synthetic C5aR peptide utilized by Wright et al. (16). Farzan et al. (17) stressed the critical role of these sulfate groups in activation of the C5aR by C5a. Previous mutational studies employing FITC-labeled CHIPS, however, suggested that the sulfate groups had only a limited effect on the binding affinity (15).
To resolve these discrepancies, we set out to chemically synthesize several sulfated and unsulfated peptides representing the N terminus of the human C5aR. We have measured the binding affinities of these peptides to CHIPS31-121 by ITC and used the C5aR peptide with the highest affinity to determine the structure of the complex between CHIPS31-121 and the C5aR N terminus by NMR spectroscopy.
EXPERIMENTAL PROCEDURES
Peptide Synthesis—The C5aR peptides (supplemental Table S1) were synthesized on a 433A peptide synthesizer (Applied Biosystems) applying Fmoc/tBu chemistry according to a method described elsewhere.4 The final peptides were checked for purity (>98%) and composition by HPLC and mass spectrometry (supplemental Table S1). Peptide concentrations were determined by weight.
Protein Production and Purification—Wild type CHIPS31-121 and mutants were cloned and expressed as described before (8). Inclusion bodies were isolated using CelLytic B Bacterial Cell Lysis/Extraction Reagent (Sigma-Aldrich) and lysozyme, according to the manufacturer's description. Inclusion bodies were intensively washed with 0.5% N,N-dimethyldodecylamine N-oxide (Sigma-Aldrich) in 20 mm sodium phosphate buffer with 0.5 m NaCl, pH 7.8, pelleted, and solubilized in 50 mm Tris, pH 8.0, containing 6 m guanidine. After dialysis in 20 mm sodium phosphate buffer, pH 8.0 the protein was loaded on a HiTrap SP XL cation exchange column (GE Healthcare BioSciences) and eluted with a 1 m NaCl gradient. Protein-containing fractions were pooled, analyzed on SDS-PAGE for purity, and dialyzed in phosphate-buffered saline. Protein concentrations were determined by measuring the absorbance at 280 nm in the presence of 8 m urea and using the extinction coefficients obtained from the ExPASy ProtParam tool.
ITC Measurements—All ITC measurements were performed at 298 K on a MCS Isothermal Titration Calorimeter (MicroCal). The measuring cell was filled with 1.345 ml of a 26 μm solution of CHIPS31-121 in a 20 mm sodium phosphate buffer at pH 6.5. The syringe was loaded with 250 μl of a 0.5 mm solution of one of the peptides in the same buffer. After each addition of 7.5-15 μl of peptide solution, the heat change due to binding of the added peptide was measured. The data were analyzed using Microcal Origin software and fitted by non-linear regression analysis.
Calcium Mobilization—Calcium mobilization studies were performed in Fluo-3-labeled U937/C5aR cells as described (8). The potency of the peptides C5aR7-28 and C5aR7-28S2 to block the CHIPS-inhibiting effect on the C5a-induced calcium mobilization was investigated. Therefore, 10 nm CHIPS31-121 was preincubated with peptides C5aR7-28 and C5aR7-28S2 (3 nm to 10 μm) for 30 min at room temperature. Next, the CHIPS/peptide mixture was added to Fluo-3-labeled U937/C5aR cells for 30 s before 0.3 nm C5a was used as a stimulus for calcium mobilization in the FACSCalibur flow cytometer (Becton Dickinson). In separate experiments, testing the inhibiting potency of peptides C5aR7-28 and C5aR7-28S2 on the C5a-induced calcium mobilization, 0.3 nm C5a (Sigma-Aldrich) was preincubated with peptides C5aR7-28 and C5aR7-28S2 (3 μm to 300 μm) for 30 min at room temperature before using this C5a/peptide mixture as a stimulus in the flow cytometer.
CD Spectroscopy—CD spectra (190-260 nm) were recorded on an Olis RSM1000 spectrophotometer operating at 0.3 nm spectral resolution (slit size 0.2 mm). Samples of wild-type CHIPS31-121 (47 μm) and mutants (46-51 μm) in 20 mm sodium phosphate buffer (pH 7.4) were measured at 298 K using a 0.5-mm cuvette. To gain sufficient S/N ratio multiple scans were summed, with data points averaged by three-point smoothing.
NMR Spectroscopy—NMR data were collected on a Varian Inova 500, a Varian Inova 600 MHz, and a Bruker Avance 900 MHz spectrometer. NMR assignments of all the sulfated peptides and unsulfated peptide C5aR10-18, free in solution, are provided in supplemental Tables S2-S5. Assignments of unsulfated peptide C5aR7-28 are similar to the values previously reported by Chen et al. (11) for the C5aR fragment 1-35. NMR experiments of the protein-peptide complexes were recorded on samples containing 20 mm sodium phosphate (pH 6.5), 0.1% (w/v) NaN3 in 5-10% (v/v) D2O/H2O solution at 298 K. A titration study was performed by titrating unlabeled C5aR peptide C5aR7-28S2 to uniformly 15N-labeled CHIPS31-121. The peptide/protein molar ratio ranged between 0-1.31 with a protein concentration of 0.5 mm. Backbone 15N relaxation experiments and experiments for spectral assignments were carried out as described previously (8) on a sample containing a 1:1 complex of peptide C5aR7-28S2 and uniformly 13C/15N-labeled CHIPS31-121 at a concentration of 1 mm. Chemical shift perturbations observed in this sample were calculated as a weighted sum of 1H, 15N, 13Cα/β chemical shifts changes. Three-dimensional 13C-edited-ω3-filtered NOESY spectra, two-dimensional 13C-15N-filtered TOCSY, and two-dimensional 13C-15N-filtered NOESY spectra were recorded on a sample containing a 1:1 complex of peptide C5aR7-28S2 and uniformly 13C/15N-labeled CHIPS31-121 to assign intermolecular NOEs (see supplemental Experimental Procedures and supplemental Fig. S1).
Structure Determination of the CHIPS31-121:C5aR7-28S2 Complex—The structures of both components of the CHIPS31-121: C5aR7-28S2 complex were first calculated separately using the standard Aria1.2/CNS 1.1 simulated annealing protocol (18). Subsequently, distance restraint-docking between the experimentally derived peptide structures and CHIPS was performed using the Yasara Structure/Whatif 8.3.3 Twinset software. The flexible docking was based on a set of 568 intermolecular NOE distance restraints derived from various isotope-filtered experiments. The average RMS deviations to the mean structure of the well-defined regions of CHIPS (residues 36-113) and C5aR7-28S2 (residues 10-23) of the final ensemble of 25 refined structures are 0.058 nm for the backbone atoms and 0.089 nm for all heavy atoms. For further details concerning the structure calculations and the final ensemble of structures see supplemental Experimental Procedures and supplemental Table S6.
RESULTS
Affinity of C5aR Model Peptides to CHIPS Determined by ITC—In total six sulfated and non-sulfated peptides, serving as models of the N terminus of the human C5aR (Fig. 1), were synthesized and their affinity to CHIPS31-121 was measured by ITC (Table 1). The sequences of these peptides were selected based upon previous studies (15) and preliminary NMR experiments. The influence of the two sulfate groups at positions 11 and 14 was tested by comparing the non-sulfated peptides C5aR10-18 and C5aR7-28 with their sulfated counterparts C5aR10-18S2 and C5aR7-28S2. Clearly, the sulfate groups have a substantial influence on binding affinity. Peptide C5aR10-18 did not show any detectable binding, while affinity in the micromolar range was observed for sulfated peptide C5aR10-18S2 (Kd = 15.9 ± 1.4 μm). Sulfation of peptide C5aR7-28 enhanced the affinity to CHIPS more than two orders of magnitude (Table 1). The minimal sequence required for high affinity binding was further investigated by comparing the affinities of sulfated sequences 10-18, 10-24, 7-28, and 1-35, respectively (Table 1). Extension of peptide C5aR10-18S2 with six additional amino acids at the C-terminal side, peptide C5aR10-24S2, resulted in nanomolar binding to CHIPS (Kd = 24.7 ± 0.4 nm). Even stronger binding was observed for peptide C5aR7-28S2 (Kd = 8.4 ± 1.1 nm). The thermodynamic binding parameters of peptide C5aR1-35S2 indicated that the sequence beyond residues 7-28 is not involved in favorable interactions with CHIPS (Table 1). In summary, the ITC data generated for the various peptides revealed that tight binding to CHIPS requires at least residues 10-24 of the C5aR as well as the presence of O-sulfated tyrosine residues. The doubly sulfated peptide C5aR7-28S2 comprises all residues involved in CHIPS binding.
FIGURE 1.
Amino acid sequence of residues 1-35 of the human C5aR (numbering according to Swiss-Prot entry P21730). The sequence of peptide C5aR7-28S2 is indicated in gray. The positions of tyrosine residues 11 and 14 are in black.
TABLE 1.
Thermodynamic binding parameters of CHIPS31-121:C5aR peptide complexes determined by ITC
C5aR peptides are numbered according to Swiss-Prot entry P21730. The presence of sulfate groups on tyrosine residues 11 and 14 is indicated by S2. The data are averages from at least three independent experiments unless indicated otherwise (mean ± S.E.). The errors in the thermodynamic parameters were estimated by Monte Carlo simulations using the standard deviations of the individual experiments. The average stoichiometric value n = 1.13 ± 0.04 (the error bound represents ± S.E.; n = 14).
| Peptide | Kd | ΔG | ΔH | ΔS |
|---|---|---|---|---|
| nm | kJ mol−1 | kJ mol−1 | J K−1 mol−1 | |
| C5aR10-18 | −a | − | − | − |
| C5aR7-28 | (3.2 ± 0.1) × 103 | −31.3 ± 0.1 | −78.0 ± 2.5 | −157 ± 8 |
| C5aR10-18S2 | (15.9 ± 1.4) × 103 | −27.4 ± 0.2 | −41.9 ± 3.1 | −49 ± 10 |
| C5aR10-24S2b | 24.7 ± 0.4 | −43.4 ± 0.1 | −85.4 ± 0.3 | −141 ± 1 |
| C5aR7-28S2 | 8.4 ± 1.1 | −46.1 ± 0.3 | −94.5 ± 2.2 | −162 ± 7 |
| C5aR1-35S2 | 27.8 ± 5.0 | −43.1 ± 0.4 | −93.8 ± 2.2 | −170 ± 7 |
No detectable binding observed in two independent experiments.
Data from one experiment. Error bounds represent ± S.D.
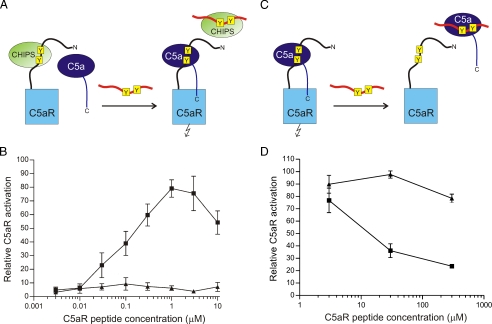
Inhibitory Potency of the Peptides C5aR7-28 and C5aR7-28S2—The role of the sulfate groups on bioactivity was investigated by comparing the model peptides C5aR7-28 and C5aR7-28S2 for their ability to compete with the C5aR for binding either CHIPS or C5a. C5a-induced calcium mobilization in a U937/C5aR cell line, stably expressing the C5aR, was used as a functional C5a-dependent activation assay. First, the potency of the C5aR7-28 and C5aR7-28S2 peptides to compete with the binding between CHIPS and the native C5aR was tested. In this experiment 10 nm of CHIPS31-121 was added to the activation assay representing the minimal amount of CHIPS required to completely suppress C5aR activation by C5a. Subsequently, either peptide C5aR7-28 or C5aR7-28S2 was titrated to interfere with CHIPS inhibition, as schematically represented in Fig. 2A. In Fig. 2B it is shown that at concentrations >10 nm, peptide C5aR7-28S2 competes with the native C5aR in binding CHIPS, releasing the C5aR for C5a-induced activation.
FIGURE 2.
Inhibition of the C5a-induced calcium mobilization. A, schematic representation of the C5a/C5aR7-28S2 inhibition experiment in the presence of CHIPS. The sulfated tyrosines in both the C5aR and C5aR7-28S2 are marked in yellow. Activation of the C5aR is indicated by the flash symbol. B, C5a inhibition experiment in the presence of CHIPS. CHIPS31-121 (10 nm) was preincubated with different concentrations of the C5aR N-terminal peptides C5aR7-28 (triangles) and C5aR7-28S2 (squares) for 30 min at room temperature. Fluo-3-labeled U937/C5aR cells were incubated with these CHIPS31-121/peptide mixtures for 30 s, before stimulation of the cells with 0.3 nm C5a. The increase in fluorescence was measured by flow cytometry. Activation of cells by C5a incubated in the absence of CHIPS and peptide is set to 100%. C, schematic representation of the C5a/C5aR7-28S2 inhibition experiment in the absence of CHIPS. D, C5a inhibition experiment in the absence of CHIPS. C5a (0.3 nm) was preincubated with different concentrations of the C5aR N-terminal peptides C5aR7-28 (triangles) and C5aR7-28S2 (squares) for 30 min at room temperature. These C5a/peptide mixtures were then used for stimulation of Fluo-3-labeled U937/C5aR cells.
Next, the inhibitory potency of the C5aR peptides toward C5a was tested, as schematically represented in Fig. 2C. In Fig. 2D it is shown that >10 μm of peptide C5aR7-28S2 is needed to inhibit the C5a-induced calcium mobilization. It is important to note that peptide C5aR7-28S2 does not completely recapitulate the dual binding mode of C5a to its receptor. As a result, the affinity of C5a for C5aR7-28S2 is four orders of magnitude lower compared with its affinity for the native C5aR (Kd = 1 nm) (17, 19). The decrease in relative C5aR activation at C5aR7-28S2 concentrations > 1 μm, observed in the experiment in the presence of CHIPS (Fig. 2B), is due to C5a:C5aR7-28S2 complex formation, in a similar way as observed in the experiment without CHIPS (Fig. 2D). In both experiments, the unsulfated C5aR7-28 peptide was inactive (Fig. 2, B and D). These experiments clearly demonstrate the crucial role of the sulfate moieties in inhibition of the C5aR by CHIPS.
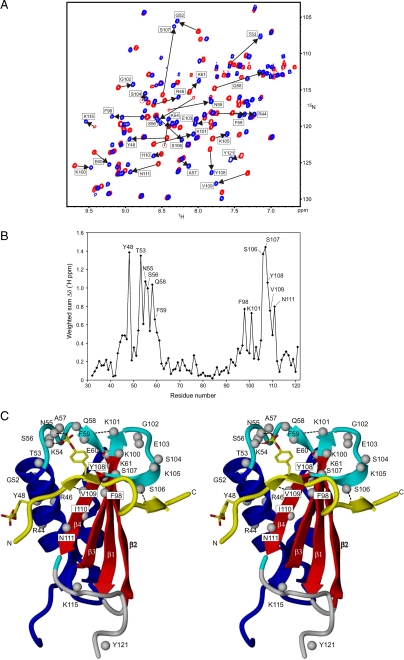
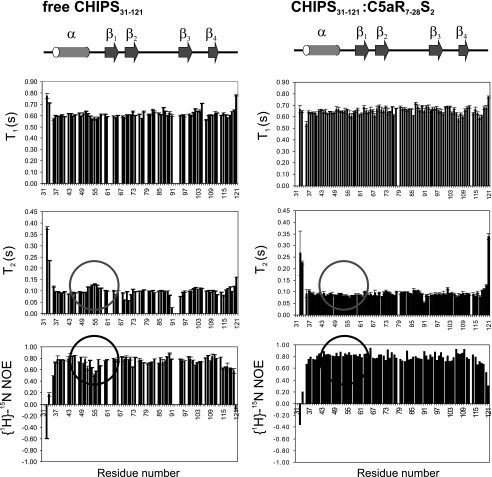
NMR Titration and Relaxation Studies—Residue specific information concerning the interactions between the C5aR and CHIPS was derived from NMR experiments. Titration of the unlabeled model peptide C5aR7-28S2, which is unstructured free in solution, to 15N-labeled CHIPS31-121 resulted in perturbation of numerous peaks in the 15N heteronuclear single quantum correlation (HSQC) NMR spectrum of CHIPS31-121. Increasing concentrations of C5aR7-28S2 resulted in the disappearance of free CHIPS31-121 peaks and the concomitant appearance of CHIPS31-121:C5aR7-28S2 peaks, compatible with a slow-exchange binding regime (20) (Fig. 3A). We reassigned the NMR spectra of 13C/15N-labeled CHIPS31-121 in complex with unlabeled C5aR7-28S2 and compared the protein amide 15N-1H and 13Cα/β chemical shifts with those of free CHIPS31-121. A weighted sum of these chemical shift changes was plotted versus residue number (Fig. 3B). Two regions show a relatively large perturbation in backbone and Cβ chemical shifts upon binding: (i) Residues 43-61, which include part of the α-helix and subsequent loop 52-59 and (ii) residues 95-111, which include strands β3 and β4 (Fig. 3C). Previously, we concluded from backbone 15N relaxation measurements (8) that residues residing in the 52-59 loop region of free CHIPS31-121 are less ordered compared with the rest of the protein backbone (without considering the N and C termini). In the CHIPS31-121: C5aR7-28S2 complex, this loop adopts an ordered conformation as can be concluded from backbone 15N relaxation data (Fig. 4).
FIGURE 3.
Chemical shift perturbations of CHIPS31-121 upon complex formation with the C5aR peptide C5aR7-28S2. A, overlay of the 15N-1H HSQC spectrum of free uniformly 15N-labeled CHIPS31-121 (red) and of the titrated stoichiometric [15N]CHIPS31-121:C5aR7-28S2 complex (blue). Amide protons of CHIPS31-121 that experience substantial chemical shift perturbations (shown by the arrows) upon complex formation with C5aR peptide C5aR7-28S2 are labeled as such. B, weighted sum of CHIPS amide 15N-1H and 13Cα/β chemical shift changes, Δδ(1H ppm) = [(ΔδNH)2 + (0.25ΔδCα)2 + (0.25ΔδCβ)2 + (0.1ΔδN)2]1/2, upon binding the C5aR7-28S2 peptide, plotted versus residue number. Residues that were most affected are indicated by labels. Values for proline residues are based exclusively on carbon Cα/β shifts. C, side-by-side stereo cartoon representation of mapped positions within the CHIPS31-121 structure that display perturbed chemical shifts after formation of the CHIPS31-121:C5aR7-28S2 complex. Perturbed amide protons are indicated by gray balls. The peptide strand (colored yellow), running from residue 9-24 and including both sulfated tyrosines sY11 and sY14, is displayed as a visual reference of the binding interface. Broken lines indicate newly formed amide hydrogen bonds.
FIGURE 4.
Backbone 15N relaxation parameters T1, T2, and heteronuclear {1H}-15N-NOE of free CHIPS31-121 (left) and of the CHIPS31-121:C5aR7-28S2 complex (right) plotted against the residue numbers. The location of the secondary structure elements was derived from the structure of free CHIPS31-121, and is shown at the top of the graph. The data points of T2 and heteronuclear {1H}-15N-NOE concerning the loop between the helix and strand β1 (residues 52-59) are indicated by circles.
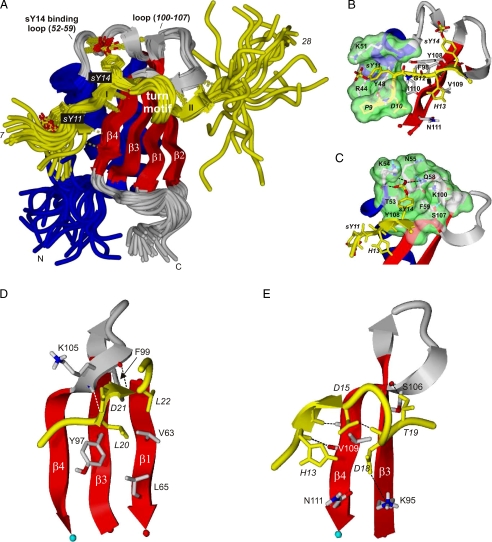
Structure of the CHIPS31-121:C5aR7-28S2 Complex—We have determined the structure of the stoichiometric complex of 13C/15N-labeled CHIPS31-121 and unlabeled C5aR7-28S2 by NMR. An overlay of 25 selected low-energy structures of the CHIPS31-121:C5aR7-28S2 complex is presented in Fig. 5A. The structure of CHIPS31-121 in the complex is largely similar to free CHIPS31-121 (Fig. 6). Subtle changes in the structure of CHIPS upon binding include ordering of the helix-strand loop between residues 52-59. The C5aR7-28S2 peptide is wrapped around a large portion of CHIPS31-121 covering a substantial part of its solvent accessible surface area (buried ∼18 nm2 versus total ∼92 nm2). The C5aR7-28S2 peptide, which is a random coil free in solution, clearly adopts a well-defined conformation upon binding pre-organized CHIPS. Essentially, two amino acid stretches of equal size define the binding interface with CHIPS: Residues 10-14 and 19-24 of C5aR7-28S2 form two short β-strands (labeled I and II in Fig. 5A) running in an antiparallel fashion with respect to strand β4 and residues 104-107 of CHIPS31-121. These two stretches are connected by a single turn comprised of residues 15-18. Residues 25-28 of C5aR7-28S2 are disordered in the structural ensemble (Fig. 5A) and do not interact with CHIPS31-121. Residues 7-9 are less well defined compared with the 10-24 core region, probably because of increased conformational freedom.
FIGURE 5.
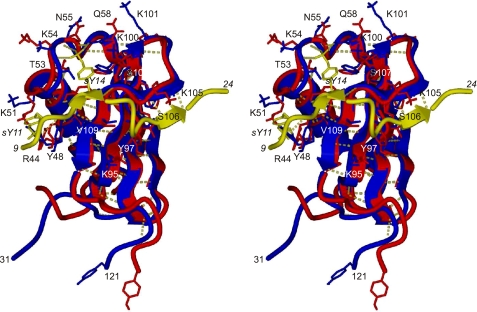
Structure of the CHIPS31-121:C5aR7-28S2 complex. Specific residues belonging to C5aR7-28S2 are annotated by italics. A, overlay of the 25 selected low-energy NMR structures determined of the complex between CHIPS31-121 and C5aR7-28S2. Newly formed β-strands in the peptide are labeled I and II. B, binding pocket of sY11. Residues lining the pocket are labeled. C, binding pocket of sY14. D, view of the binding strand II and the hydrophobic interactions of peptide residues Leu-20 and Leu-22, positioned toward the β-sheet surface of CHIPS. E, view of the binding strand I and turn-motif of the complex. Residues Asp-16 and Lys-17 of the peptide stick out into solution and are omitted from the plot for clarity. Intermolecular interactions are indicated by broken lines. Coloring scheme: C5aR7-28S2 in yellow; the side chains of sY11 and sY14 in yellow sticks and the sulfate oxygens in red; the CHIPS31-121 β-strands in red; the CHIPS31-121 binding loops, the β1-β2 loop, and the C terminus (113-121) in gray; the CHIPS31-121 α-helix and remaining backbone in blue. The solvent-accessible surface surrounding the sulfated tyrosine is colored green.
FIGURE 6.
Side-by-side stereo plot displaying the difference between a representative structure of free CHIPS31-121 (red) (PDB ID code 1XEE.pdb) and the corresponding structure of the CHIPS31-121 protein as part of the CHIPS31-121:C5aR7-28S2 complex (blue). Amino acid side chains of CHIPS31-121 that are important for binding C5aR7-28S2 in the core region between residues 9-24, and that have different orientations in the free and unbound form, are displayed in stick representation. The backbone trace of residues 9-24 of C5aR7-28S2 is displayed in yellow with residues sY11 and sY14 in stick representation. The two models mutually overlay with an RMS deviation of 0.089 nm for the well-defined backbone atoms between residues 36-113. The ensemble average backbone RMS deviation is 0.13 ± 0.02 nm.
Details concerning specific interactions between C5aR7-28S2 and CHIPS31-121 are presented in Fig. 5, B-E. The first stretch of C5aR7-28S2 involved in CHIPS binding includes the two sulfated tyrosine residues Tyr-11 and Tyr-14 (subsequently referred to as sY11 and sY14). sY11 is stacked on top of Tyr-48, with the sulfate group of sY11 neutralized by the positive charge of residues Arg-44 and Lys-51 located in the CHIPS helix (Fig. 5B). sY14 is bound by the loop region 52-59 of CHIPS, while residues Tyr-108 and Lys-100 surround the binding pocket (Fig. 5C). The oxygen atoms of the sulfate group of sY14 are primarily coordinated to the backbone amide protons of Lys-54 and Asn-55 and the side chain amide of Gln-58. Surprisingly, the positively charged side chains of Lys-54 and Lys-100 are only partially positioned within the coordination sphere of the sulfate group of sY14. Instead, these side chains are mostly involved in cationic-π stacking with the aromatic ring of sY14 or in long-range electrostatic interactions.
The second stretch of C5aR7-28S2 (residues 19-24) involved in binding is located on the opposite face of the CHIPS molecule near residues 104-107 between strand β3 and β4. This short β-strand is stabilized by backbone hydrogen bonds between residues Leu-20 and Leu-22 of C5aR7-28S2 and Ser-104 and Ser-106 of CHIPS31-121. The side chains of Leu-20 and Leu-22 are in contact with the hydrophobic β-sheet surface of CHIPS (Fig. 5D). Finally, oppositely charged residues Asp-21 and Lys-105 are in close proximity to each other while the side chains of Thr-19 and Ser-106 are within hydrogen bonding distance (Fig. 5E). Experimentally, we observed the hydroxyl proton of Thr-19 in the NMR spectra, implying it is protected from solvent exchange.
The turn-motif comprising residues 15-18 of C5aR7-28S2 is stabilized by intra-molecular interactions between the negatively charged carboxyl group of Asp-15 and the backbone amide protons of Asp-16, Lys-17, and Asp-18 comprising the turn (Fig. 5E). The side chains of Asp-16 and Lys-17 are on average not in close contact to CHIPS. Finally, a salt-bridge between the side chains of Asp-18 and Lys-95 of CHIPS31-121 provides additional stability (Fig. 5E).
CHIPS Mutation Data—To assess the role of specific molecular interactions in the CHIPS31-121:C5aR7-28S2 complex, several mutants of CHIPS31-121 were prepared to verify their inhibitory propensities in a biological assay with intact C5aR. We quantified this as the concentration of C5a needed to achieve 50% activation of the C5aR (AC50) in the presence of 100 nm wild type CHIPS31-121 or mutant. We selected a number of mutants based upon their position in the structure. Only mutants Tyr-97 and Ser-106 show substantial decreased AC50 values (supplemental Fig. S2). CD spectroscopy was used to check the structural integrity of selected CHIPS mutants (supplemental Fig. S3). No differences in secondary structure were observed between wild type CHIPS, and mutants Y97A and S106A. The same holds for mutants R44A and K95A studied previously (8). This indicates that the loss of inhibitory potency of these mutants is not caused by loss of structural integrity, but by altered specific binding.
DISCUSSION
The important role of O-sulfated tyrosine residues in a wide range of biological processes is becoming increasingly recognized (21). Several G protein-coupled receptors, including the C5aR, have been demonstrated to contain O-sulfated tyrosine residues within their N termini. This post-translational modification is required for proper ligand binding and receptor activation (17, 22-25). Here, we assessed the role the O-sulfated tyrosine residues in the interaction of the C5aR with the immune evasion protein CHIPS. The data obtained demonstrate that the O-sulfated tyrosine residues 11 and 14 of the C5aR are essential for tight binding to CHIPS. However, residues beyond the smallest C5aR fragment we have investigated (residues 10-18) also add substantially to the binding affinity (Table 1). Among the fragments measured by ITC, peptide C5aR7-28S2 exhibited the highest affinity for CHIPS (Kd = 8.4 ± 1.1 nm). The observed Kd value is comparable to the Kd values of 125I-labeled CHIPS for the naturally sulfated intact C5aR (Kd = 1.1 ± 0.2 nm) and C5aR N terminus (residues 1-38; Kd = 29.7 ± 4.4 nm), as previously reported by Postma et al. (7, 15). Peptide C5aR7-28S2 competed with the C5aR N terminus in binding CHIPS in a calcium mobilization assay at concentrations >10 nm (Fig. 2). In this bioactivity assay, the unsulfated peptide C5aR7-28 did not show any effect. We conclude that peptide C5aR7-28S2 comprises all moieties of the C5aR essential for tight binding to the CHIPS protein and is therefore a representative model of the N-terminal portion of the human C5aR.
The structure of the CHIPS31-121:C5aR7-28S2 complex defines residues 10-24 as the core region of the C5aR N terminus interacting with CHIPS. The binding motif is comprised of two short β-strands interconnected by a turn. These two strands, labeled I and II in Fig. 5A, are hydrogen bonded in an antiparallel fashion to strand β4 and the residues 104-107 of CHIPS, respectively. The sulfated tyrosine residues sY11 and sY14 indeed play a key role in stabilization of the complex. Sulfated tyrosine sY11 is located near the α-helix of CHIPS and interacts primarily with the aromatic ring of tyrosine Tyr-48, and with the end groups of Arg-44 and Lys-51 closely placed around the negatively charged sulfate group. sY14 is tightly bound by residues of the loop region 52-59 of CHIPS. The charged sulfate group of sY14 is coordinated by the backbone amide protons of Lys-54 and Asn-55, and by the side chain amide protons of Gln-58. Unexpectedly, the positively charged side chains of nearby lysine residues Lys-54 and Lys-100 are not involved in specific interactions with the sulfate group of sY14 (Fig. 5C). The slightly higher affinity to CHIPS of peptide C5aR7-28S2 compared with peptide C5aR10-24S2 (Table 1) indicates that residues outside the core region 10-24 of C5aR also contribute to binding. This contribution is assumed to involve nonspecific interactions of the residues at the N-terminal side of C5aR7-28S2 (T7-P9).
Postma et al. (15) did not observe major differences in the binding of CHIPS-FITC to a sulfated or non-sulfated C5aR N terminus (residues 1-38) expressed on HEK293 cells. The FITC label, which was employed to detect binding by means of fluorescence flow cytometry, was introduced covalently at random positions to surface-exposed side chains of lysine residues. The structure of the CHIPS31-121:C5aR7-28S2 complex reveals that lysine Lys-51 is near tyrosine sY11 while Lys-54 and Lys-100 are in close proximity to tyrosine sY14. It is likely that modification of these particular lysine residues in CHIPS by the large FITC group interferes with the binding of C5aR and decreases the overall affinity for steric reasons.
Recently, we have investigated the activity of 16 lysine and arginine to alanine single-point mutants of full length CHIPS1-121 with respect to activation of the C5aR and FPR (8). Of these mutants, R44A, R46A, and K95A were most affected. Although the side chain of Arg-44 is not well-defined in the NMR ensemble, the guanidinium group of Arg-44 is on average in close proximity to the sulfated tyrosine sY11 of C5aR7-28 S2. The role of mutant R46A is related to the structural integrity of CHIPS as was accounted for previously (8). Residue Lys-95 is located at the start of strand β3 of CHIPS and its side chain is in direct contact with the side chain of residue Asp-18 of C5aR7-28S2 (Fig. 5E). This interaction is evidently important for maintaining the turn-motif, which places the two short β-strands of C5aR7-28S2 in an optimal position to interact with CHIPS (Fig. 5A). Among the CHIPS mutants reported in this work, Y97A and S106A have the lowest inhibitory potency with respect to C5a-mediated C5aR activation. The aromatic ring of Tyr-97 supports the backbone of Asp-18 and Thr-19 and is in contact with residue Leu-20 of C5aR7-28S2 (Fig. 5D). The sensitivity of the S106A mutant is explained by the fact that the hydroxyl group of Ser-106 is involved in a hydrogen-bond network together with Thr-19 of C5aR7-28S2.
Previous mutational studies showed that residues Asp-10, Gly-12, Asp-15, and Asp-18 of the C5aR are essential for binding of CHIPS (15). The structure of the complex suggests that Asp-15 and Asp-18 are important for maintaining the turn motif between the two short β-strands of C5aR7-28S2. The side chain carboxyl group of residue Asp-10 is within hydrogenbond distance with residues residing in β-strand β4 of CHIPS. Finally, residue Gly-12 is favored in the complex, because the side chain of any other l-amino acid would lead to clashes with the adjacent strand β4 of CHIPS. The side chains of residues Asp-16 and Lys-17 are not involved in key interactions with CHIPS and are therefore not critical for binding.
In conclusion, all moieties of the C5aR required for high affinity binding to the immune evasion protein CHIPS are present in the synthetic peptide C5aR7-28S2. In particular, two O-sulfated tyrosine residues at positions 11 and 14 of the C5aR are crucial for tight binding to CHIPS. The key C5aR binding element of CHIPS is the loop located between the α-helix and strand β1 (residues 52-59). This loop accommodates sulfated tyrosine sY14 and represents a potential lead sequence for the development of novel C5aR antagonists.
Acknowledgments
We thank Nathaniel Martin and Ruud Scheek for helpful discussions; Eefjan Breukink for assistance with the ITC experiments; Klaas Dijkstra for recording the 600 MHz NMR spectra; Hugo van Ingen for help with the 900 MHz filtered NOESY experiments; and Cees Versluis for the analysis by mass spectrometry. 900 MHz NMR spectra were recorded at the SON NMR Large Scale Facility in Utrecht (FP-2005-RII3-Contract-no. 026145).
The atomic coordinates and structure factors (code 2K3U) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
Chemical shift assignments have been deposited in the BioMagResBank, www.bmrb.wisc.edu (Accession No. 15778).
This work was supported in part by the Dutch Technology Foundation STW, Applied Science Division of NWO, and the Technology Program of the Ministry of Economic Affairs (UKG.06609).
The on-line version of this article (available at http://www.jbc.org) contains supplemental Experimental Procedures, Tables S1-S6, and Figs. S1-S3.
Footnotes
The abbreviations used are: C5aR, C5a receptor; CHIPS, chemotaxis inhibitory protein of Staphylococcus aureus; RMS, root mean square; ITC, isothermal titration calorimetry.
Bunschoten, A., Kruijtzer, J. A. W., Ippel, J. H., de Haas, C. J. C., van Strijp, J. A. G., Kemmink, J., and Liskamp, R. M. J. (2009) Chem. Commun., in press.
References
- 1.Guo, R. F., and Ward, P. A. (2005) Annu. Rev. Immunol. 23 821-852 [DOI] [PubMed] [Google Scholar]
- 2.Haas, P. J., and van Strijp, J. A. G. (2007) Immunol. Res. 37 161-175 [DOI] [PubMed] [Google Scholar]
- 3.Heller, T., Hennecke, M., Baumann, U., Gessner, J. E., zu Vilsendorf, A. M., Baensch, M., Boulay, F., Kola, A., Klos, A., Bautsch, W., and Kohl, J. (1999) J. Immunol. 163 985-994 [PubMed] [Google Scholar]
- 4.Strachan, A. J., Woodruff, T. M., Haaima, G., Fairlie, D. P., and Taylor, S. M. (2000) J. Immunol. 164 6560-6565 [DOI] [PubMed] [Google Scholar]
- 5.de Haas, C. J. C., Veldkamp, K. E., Peschel, A., Weerkamp, F., Van Wamel, W. J., Heezius, E. C., Poppelier, M. J., Van Kessel, K. P., and van Strijp, J. A. (2004) J. Exp. Med. 199 687-695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Veldkamp, K. E., Heezius, H. C., Verhoef, J., van Strijp, J. A., and van Kessel, K. P. (2000) Infect. Immun. 68 5908-5913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Postma, B., Poppelier, M. J., van Galen, J. C., Prossnitz, E. R., van Strijp, J. A., de Haas, C. J., and van Kessel, K. P. (2004) J. Immunol. 172 6994-7001 [DOI] [PubMed] [Google Scholar]
- 8.Haas, P. J., de Haas, C. J. C., Poppelier, M. J. J. C., van Kessel, K. P. M., van Strijp, J. A. G., Dijkstra, K., Scheek, R. M., Fan, H., Kruijtzer, J. A. W., Liskamp, R. M. J., and Kemmink, J. (2005) J. Mol. Biol. 353 859-872 [DOI] [PubMed] [Google Scholar]
- 9.Zuiderweg, E. R. P., Nettesheim, D. G., Mollisson, K. W., and Carter, G. W. (1989) Biochemistry 28 172-185 [DOI] [PubMed] [Google Scholar]
- 10.Zhang, X., Boyar, W., Toth, M. J., Wennogle, L. P., and Gonnella, N. C. (1997) Proteins 28 261-267 [DOI] [PubMed] [Google Scholar]
- 11.Chen, Z., Zhang, X., Gonnella, N. C., Pellas, T. C., Boyar, W. C., and Ni, F. (1998) J. Biol. Chem. 273 10411-10419 [DOI] [PubMed] [Google Scholar]
- 12.Monk, P. N., Scola, A.-M., Madala, P., and Fairlie, D. P. (2007) Brit. J. Pharmacol. 152 429-448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gerard, C., and Gerard, N. P. (1994) Annu. Rev. Immunol. 12 775-808 [DOI] [PubMed] [Google Scholar]
- 14.March, D. R., Proctor, L. M., Stoermer, M. J., Sbaglia, R., Abbenante, G., Reid, R. C., Woodruff, T. M., Wadi, K., Paczkowski, N., Tyndall, J. D., Taylor, S. M., and Fairlie, D. P. (2004) Mol. Pharmacol. 65 868-879 [DOI] [PubMed] [Google Scholar]
- 15.Postma, B., Kleibeuker, W., Poppelier, M. J., Boonstra, M., van Kessel, K. P., van Strijp, J. A., and de Haas, C. J. (2005) J. Biol. Chem. 280 2020-2027 [DOI] [PubMed] [Google Scholar]
- 16.Wright, A. J., Higginbottom, A., Philippe, D., Upadhyay, A., Bagby, S., Read, R. C., Monk, P. N., and Partridge, L. J. (2007) Mol. Immunol. 44 2507-2517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Farzan, M., Schnitzler, C. E., Vasilieva, N., Leung, D., Kuhn, J., Gerard, C., Gerard, N. P., and Choe, H. (2001) J. Exp. Med. 193 1059-1065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Linge, J. P., Habeck, M., Rieping, W., and Nilges, M. (2003) Bioinformatics 19 315-316 [DOI] [PubMed] [Google Scholar]
- 19.Van Epps, D. E., Simpson, S. J., and Johnson, R. (1993) J. Immunol. 150 246-252 [PubMed] [Google Scholar]
- 20.Cavanagh, J., Fairbrother, W. J., Palmer, A. G., Rance, M., and Skelton, N. J. (2007) Protein NMR Spectroscopy: Principles and Practice (2nd edition), Elsevier Academic Press, San Diego
- 21.Moore, K. L. (2003) J. Biol. Chem. 278 24243-24246 [DOI] [PubMed] [Google Scholar]
- 22.Bannert, N., Craig, S., Farzan, M., Sogah, D., Santo, N. V., Choe, H., and Sodroski, J. (2001) J. Exp. Med. 194 1661-1673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Colvin, R. A., Campanella, G. S. V., Manice, L. A., and Luster, A. D. (2006) Mol. Cell. Biol. 26 5838-5849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Veldkamp, C. T., Seibert, C., Peterson, F. C., Sakmar, T. P., and Volkman, B. F. (2006) J. Mol. Biol. 359 1400-1409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu, J., Louie, S., Hsu, W., Yu, K. M., Nicholas, H. B., and Rosenquist, G. L. (2008) Am. J. Resp. Cell Mol. 38 738-743 [DOI] [PMC free article] [PubMed] [Google Scholar]