Abstract
Frontotemporal lobar degeneration (FTLD) with inclusion body myopathy and Paget disease of bone is a rare, autosomal dominant disorder caused by mutations in the VCP (valosin-containing protein) gene. The disease is characterized neuropathologically by frontal and temporal lobar atrophy, neuron loss and gliosis, and ubiquitin-positive inclusions (FTLD-U), which are distinct from those seen in other sporadic and familial FTLD-U entities. The major component of the ubiquitinated inclusions of FTLD with VCP mutation is TDP-43 (TAR DNA-binding protein of 43 kDa). TDP-43 proteinopathy links sporadic amyotrophic lateral sclerosis, sporadic FTLD-U, and most familial forms of FTLD-U. Understanding the relationship between individual gene defects and pathologic TDP-43 will facilitate the characterization of the mechanisms leading to neurodegeneration. Using cell culture models, we have investigated the role of mutant VCP in intracellular trafficking, proteasomal function, and cell death and demonstrate that mutations in the VCP gene 1) alter localization of TDP-43 between the nucleus and cytosol, 2) decrease proteasome activity, 3) induce endoplasmic reticulum stress, 4) increase markers of apoptosis, and 5) impair cell viability. These results suggest that VCP mutation-induced neurodegeneration is mediated by several mechanisms.
Frontotemporal lobar degeneration (FTLD)2 accounts for 10% of all late onset dementias and is the third most frequent neurodegenerative disease after Alzheimer disease and dementia with Lewy bodies (1). FTLD with ubiquitin-immunoreactive inclusions is genetically, clinically, and neuropathologically heterogeneous (2, 3). FTLD-U comprises several distinct entities, including sporadic forms and familial cases caused by mutations in the genes encoding VCP (valosin-containing protein), GRN (progranulin), CHMP2B (charged multivesicular body protein 2B), TDP-43 (TAR DNA-binding protein of 43 kDa) and an unknown gene linked to chromosome 9 (2, 3). Frontotemporal dementia with inclusion body myopathy and Paget disease of bone is a rare, autosomal dominant disorder caused by mutations in the VCP gene located on chromosome 9p13-p12 (4-10) (Fig. 1). This multisystem disease is characterized by progressive muscle weakness and atrophy, increased osteoclastic bone resorption, and early onset frontotemporal dementia, also called FTLD (9, 11). Mutations in VCP are also associated with dilatative cardiomyopathy with ubiquitin-positive inclusions (12). Neuropathologic features of FTLD with VCP mutation include frontal and temporal lobar atrophy, neuron loss and gliosis, and ubiquitin-positive inclusions (FTLD-U). The majority of aggregates are ubiquitin- and TDP-43-positive neuronal intranuclear inclusions (NIIs); a smaller proportion is made up of TDP-43-immunoreactive dystrophic neurites (DNs) and neuronal cytoplasmic inclusions (NCIs). A small number of inclusions are VCP-immunoreactive (5, 13). Pathologic TDP-43 in inclusions links a spectrum of diseases in which TDP-43 pathology is a primary feature, including FTLD-U, motor neuron disease, including amyotrophic lateral sclerosis, FTLD with motor neuron disease, and inclusion body myopathy and Paget disease of bone, as well as an expanding spectrum of other disorders in which TDP-43 pathology is secondary (14, 15).
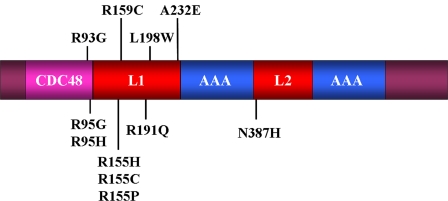
FIGURE 1.
Model of pathogenic mutations and domains in valosin-containing protein. CDC48 (magenta), located within the N terminus (residues 22-108), binds the following cofactors: p47, gp78, and Npl4-Ufd1 (23-25, 28). There are two AAA-ATPase domains (AAA; blue) at residues 240-283 and 516-569, which are joined by two linker regions (L1 and L2; red).
TDP-43 proteinopathy in FTLD with VCP mutation has a biochemical signature similar to that seen in other sporadic and familial cases of FTLD-U, including sporadic amyotrophic lateral sclerosis, FTLD-motor neuron disease, FTLD with progranulin (GRN) mutation, and FTLD linked to chromosome 9p (3, 16). TDP-43 proteinopathy in these disorders is characterized by hyperphosphorylation of TDP-43, ubiquitination, and cleavage to form C-terminal fragments detected only in insoluble brain extracts from affected brain regions (16). Identification of TDP-43 as the major component of the ubiquitin-immunoreactive inclusions of FTLD with VCP mutation supports the hypothesis that VCP gene mutations cause an alteration of VCP function, leading to TDP-43 proteinopathy.
VCP/p97 (valosin-containing protein) is a member of the AAA (ATPase associated with diverse cellular activities) superfamily. The N-terminal domain of VCP has been shown to be involved in cofactor binding (CDC48 (cell division cycle protein 48)) and two AAA-ATPase domains that form a hexameric complex (Fig. 1) (17). Recently, it has been shown that the N-terminal domain of VCP binds phosphoinositides (18, 19). AKT (activated serine-threonine protein kinase) phosphorylates VCP and is required for constitutive VCP function (20, 21). AKT is activated through phospholipid binding and phosphorylation via the phosphoinositide 3-kinase signaling pathway, which is involved in cell survival (22). The lipid binding domain may recruit VCP to the cell membrane where it is phosphorylated by AKT (19).
The diversity of VCP functions is modulated, in part, by a variety of intracellular cofactors, including p47, gp78, and Npl4-Ufd1 (23). Cofactor p47 has been shown to play a role in the maintenance and biogenesis of both the endoplasmic reticulum (ER) and Golgi apparatus (24). The structure of p47 contains a ubiquitin regulatory X domain that binds the N-terminus of VCP, and together they act as a chaperone to deliver membrane fusion machinery to the site of adjacent membranes (25). The function of the p47-VCP complex is dependent upon cell division cycle 2 (CDC2) serine-threonine kinase phosphorylation of p47 (26, 27). Also, VCP has been found to interact with the cytosolic tail of gp78, an ER membrane-spanning E3 ubiquitin ligase that exclusively binds VCP and enhances ER-associated degradation (ERAD) (28). The Npl4-Ufd1-VCP complex is involved in nuclear envelope assembly and targeting of proteins through the ubiquitin-proteasome system (29, 30). The cell survival response of this complex has been found to be important in DNA damage repair though activation by phosphorylation and its recruitment to double-stranded breaks (20, 31). The Npl4-Ufd1-VCP cytosolic complex is also recruited to the ER membrane, interacting with Derlin 1, VCP-interacting membrane proteins (VIMP), and other complexes. At the ER membrane, these misfolded proteins are targeted to the proteasome via ERAD (32-34). VCP also targets IKKβ for ubiquitination to the ubiquitin-proteasome system, implicating VCP in the cell survival pathway and neuroprotection (21, 35-37).
To investigate the mechanism of neurodegeneration caused by VCP mutations, we first tested the hypothesis that VCP mutations decrease cell viability in vitro using a neuroblastoma SHSY-5Y cell line and then investigated cellular pathways that are known to lead to neurodegeneration, including decrease in proteasome activity, caspase-mediated degeneration, and a change in cellular localization of TDP-43.
EXPERIMENTAL PROCEDURES
Tissue Collection and Processing—Human post-mortem brain tissues from clinically and/or neuropathologically well characterized cases were obtained from the Alzheimer's Disease Research Center, Washington University School of Medicine (St. Louis, MO). Brain tissue from the following cases was used: FTLD with the VCP R155H mutation, as described previously (13); FTLD with the GRN A9D mutation; sporadic FTLD-U; Alzheimer disease (n = 2); and two normal aged-matched control cases (see supplemental Table 1 for case descriptions) (7, 13, 16, 38).
Histology and Immunohistochemistry—Paraffin sections from all cases included the frontal lobe (middle frontal gyrus and Brodmann areas 8, 9, and 46), temporal lobe (middle/superior temporal gyrus and Brodmann areas 21 and 22), parahippocampal gyrus, hippocampus, striatum, pons, medulla oblongata, and, when available, spinal cord. Ubiquitin, VCP, and TDP-43 immunohistochemistry was also performed on sections with disease-representative pathology from cases with other neurodegenerative conditions and normal aged control cases (see supplemental Table 1). Antigen retrieval was performed by microwaving tissue sections in a solution of 0.1 mol/liter citrate buffer, pH 6.0, at 100 °C for 10 min. Immunohistochemistry was undertaken on 7-μm-thick sections prepared from formalin-fixed, paraffin-embedded tissue blocks using the avidin-biotin complex detection system (Vector Laboratories) and the chromagen 3,3-diaminobenzidine; sections were counterstained with hematoxylin, as previously described (3, 13, 16). Antibodies used included those that recognize epitopes of ubiquitin (rabbit polyclonal, 1:1,000; Dako), TDP-43 (rabbit polyclonal, 1:2,000; Proteintech Group, Inc.), and VCP (1:1,000; Abcam). The classification of FTLD-U proposed by the Consortium for Frontotemporal Lobar Degeneration was used because this nosology demonstrates a close correspondence between FTLD-U phenotype and genotype (2, 16, 39). FTLD with VCP mutation is distinguished from other FTLD-U subtypes by numerous NIIs and infrequent NCIs and DNs in neocortical areas with relative sparing of the hippocampus, consistent with the pathology previously described in cases with FTLD with VCP mutation (13, 16, 40).
Cell Culture and Constructs—Human neuroblastoma cell line SHSY-5Y (ATCC; CRL-226) was plated at ∼50% confluency on coverslips in 24-well plates in Iscove's modified Dulbecco's medium and 10% fetal calf serum supplemented with 100 units of penicillin/streptomycin. Retinoic acid (10 μm; Sigma) was used to induce differentiation, activate phosphatidylinositol 3-kinase signaling, and induce phosphorylation of AKT. The proteasome inhibitor MG132 (Calbiochem) was used as a negative control in the proteasome assay, and phosphatidylinositol 3-kinase inhibitor LY294002 (Cell Signaling) inhibits phosphorylation of AKT. The human cytomegalovirus promoter driving N-terminal fusion constructs pDsRed Monomer-C1 (Clontech) (VCP wild type, R155H, R155C, R95G, R191Q, A232E, and the dominant negative E305Q/E578Q mutation) were kindly provided by Dr. Mark Forman (University of Pennsylvania School of Medicine, Philadelphia, PA).
Immunoblotting—Cells were lysed in lysis buffer containing 50 mm Tris-HCl, pH 7.5, 150 mm sodium chloride, 1% Nonidet P-40, 0.5% sodium deoxycholate, 100 μm sodium orthovanadate, and a protease inhibitor mixture (Sigma). Sample concentrations were determined to ensure equal loading, placed in Laemmli sample buffer containing β-mercaptoethanol, and denatured at 100 °C for 5 min. Proteins were separated on a 4-20% SDS-PAGE Tris-glycine gel (Invitrogen), transferred to polyvinylidene difluoride membrane (Millipore), and blocked in 5% instant milk Tris-buffered saline with 0.1% Tween for 1 h. For immunoblotting, primary antibodies were incubated overnight at 4 °C, 1:1000 anti-actin (Abcam), 1:1500 anti-TDP-43 (Proteintech Group, Inc. and Abnova), 1:1000 anti-VCP, 1:1000 BiP/GRP78, 1:1000 AKT, and 1:1000 anti-AKTSer473 (Cell Signaling). Secondary antibodies were incubated for 1 h at room temperature, washed, and exposed to film as previously described (41).
Immunoprecipitation—Cells were lysed in immunoprecipitation buffer containing 50 mm Tris-HCl, pH 7.5, 150 mm sodium chloride, 1% Nonidet P40, 0.5% sodium deoxycholate, 100 μm sodium orthovanadate, and a protease inhibitor mixture (Sigma) and then sonicated briefly on ice. The remaining steps were performed at 4 °C. Lysates from cells were then centrifuged at 10,000 × g for 10 min. Supernatant from SHSY-5Y cells, high salt fractions from human brain were incubated on an orbital shaker with 50 μl of Protein A-agarose beads (Roche Applied Science) for 2-3 h to reduce nonspecific protein binding. Beads were pelleted for 20 s at 10,000 × g, and supernatant was placed in a fresh tube with 4 μl of anti-TDP-43- (Proteintech Group or Abnova) or anti-VCP-antibodies (Cell Signaling) overnight. 50 μl of Protein A-agarose beads were then added and allowed to incubate on a rocker for 2 h. Protein A-agarose antibody complexes were pelleted at 10,000 × g for 20 s, and the supernatant was discarded. Beads were washed twice in 500 μl of high salt buffer, 50 mm Tris-HCl, pH 7.5, 500 mm sodium chloride, 0.1% Nonidet P-40, 0.05% sodium deoxycholate, and then washed twice in low salt buffer, 50 mm Tris-HCl, pH 7.5, 0.1% Nonidet P40, 0.05% sodium deoxycholate. Protein A-agarose antibody complexes were resuspended in loading buffer. SDS-PAGE was performed, and proteins were transferred to polyvinylidene difluoride membrane for immunoblotting (see above).
Immunofluorescence Microscopy—Neuroblastoma SHSY-5Y cells were plated at ∼50-75% confluency in media on 0.1% gelatin- or poly-d-lysine-pretreated coverslips. After cells adhered, all constructs were transfected following the manufacturer's protocol (Lipofectamine 2000; Invitrogen). At 24, 48, and/or 72 h post-transfection, cells were treated as described, washed in PBS, pH 7.4, three times, and fixed in a 4% paraformaldehyde PBS solution for 20 min. After fixation, cells were washed three times in PBS and blocked for 30 min at room temperature in blocking buffer, 1× PBS, 5% bovine serum albumin, and 0.3% Triton X-100. Primary antibodies were diluted in blocking buffer: Golgi marker, anti-giantin (GOLGB1) (1:2000) (Covance Research Products); anti-TDP-43 (1:500) (Proteintech Group); protein-disulfide isomerase (1:500) (Stressgen), and FK2, which recognizes anti-Lys29-, Lys48-, and Lys63-linked polyubiquitinylated and monoubiquitinylated protein epitopes) at 1:1000 (BioMol) for 1 h. Cells were washed three times in PBS and incubated with Alexa Fluor (λex = 488 nm, 543 nm, and/or 633 nm; Invitrogen) fluorescently labeled secondary antibodies in blocking buffer for 1 h, washed three times in PBS, and either mounted directly or incubated with the nuclear marker TOPRO-3 (1:1000) (Invitrogen) for 5 min and then washed three times in PBS. Cells were mounted on slides for confocal microscopy (Prolong Gold; Invitrogen). Images were acquired using a LSM510 Meta laser-scanning confocal microscope (Carl Zeiss Inc., Thornwood, NY) equipped with argon (λex = 488 nm), HeNe1 (λex = 543 nm), and HeNe2 (λex = 633 nm) lasers. A ×63, 1.4 numerical aperture Zeiss Plan Apochromat oil objective was used. Confocal Z slices of 0.5 μm were obtained using the Zeiss LSM510 software (courtesy of Wandy Beatty; Molecular Microbiology Imaging Facility, Washington University).
Cell Death Assay—In order to evaluate cell viability, a trypan blue exclusion assay was performed as previously described (42). Cell counts were determined in triplicate from three separate experiments in four separate fields for transiently transfected SHSY-5Y cells over a 4-day period starting 24 h after transfection.
Caspase Activity Assays—Colorimetric substrates Ac-Asp-Glu-Val-Asp-p-nitroaniline (catalog number P-412; Biomol) and Ac-Leu-Glu-His-Asp-p-nitroaniline (catalog number PNA154; MP Biomedicals) were used to determine caspase-3/7 activity and caspase-9/4 activity, respectively. Cells were lysed 24 h after transfection in 50 mm HEPES, 100 mm NaCl, 0.1% CHAPS, 0.1 mm EDTA, pH 7.4, lysis buffer, and 50 μg of protein was incubated with 100 μl of 200 μm substrate in 50 mm HEPES, 100 mm NaCl, 0.1% CHAPS, 10 mm dithiothreitol, 0.1 mm EDTA, pH 7.4, in 96-well plates at 37 °C for 24 h. The absorbance of the cleaved product was measured at 405 nm in a microplate reader (Bio-Rad).
Proteasome Activity Assay—Cells were plated at ∼50% confluence and transfected with Lipofectamine 2000 (Invitrogen) as described above. After 24 h, cells were rinsed in PBS twice; suspended in 300 μl of lysis buffer containing 10 mm Tris-HCl, pH 7.5, 1 mm EDTA, 2 mm ATP, 4 mm dithiothreitol, 20% glycerol; and sonicated on ice. Lysates were centrifuged at 13,000 × g for 10 min at 4 °C, supernatant was removed, and protein concentration was assayed (Coomassie Plus; Pierce). A total of 2.5 μg of protein was loaded per well at a concentration of 0.1 μg/μl with 25 μl of 2× proteasome assay buffer containing 50 mm Tris-HCl, pH 8.8, 0.5 mm EDTA, 40 μm N-succinyl-Leu-Leu-Val-Tyr-7-amido-4-methyl-coumain (catalog number S-6510; Sigma), and 40 μm 7-amino-4-methylcoumarin (A989; Sigma). Standards (total of 50 μl in 1× proteasome assay buffer) and a negative control containing 100 μm MG-132 were incubated for 1 h at 37 °C. The reaction was stopped by the addition of 250 μl of ice-cold distilled H2O and assayed immediately on a fluorescent plate reader (λex = 380 nm and λem = 440 nm) (43-45).
Quantitative Real Time RT-PCR and cDNA Synthesis—RNA was isolated from SHSY-5Y cells (RNA Easy; Qiagen) and cDNA synthesis (High Capacity cDNA archive kit; Applied Biosystems). RT-PCR was performed using a 20-μl total reaction with SYBR Green I 2× master mix (Applied Biosystems) with 20 μm gene-specific primers and 2 μl of a 1:40 and 1:80 dilution of cDNA template. An ABI Prism 7900HT real time RT-PCR system (Applied Biosystems) was used for PCR amplification. The efficiency of each PCR was determined by six serial dilutions undertaken in duplicate and repeated three times for SHSY-5Y cDNA, and each expression ratio was based upon normalization with glyceraldehyde-3-phosphate dehydrogenase (GAPDH) cDNA (46). Sequences of real time primers used were as follows: GAPDH-F, TGGAAGGACTCATGACCACAGT; GAPDH-R, GGGCCATCCACAGTCTTCTG; GRP78-F, ACCAATTATCAGCAAACTCTATGGAA, GRP78-R, CATCTTTTTCTGCTGTATCCTCTTCA; CALR-F, TTTGACAACTTCCTCATCACCAA; CALR-R, CCACGTCTCGTTGCCAAAC. The use of GAPDH and β-actin in differentiating neurons has been previously evaluated (47-49).
Statistical Analysis—Data analyses were performed using a repeated measure one-way analysis of variance. For each experiment, the significance levels were determined and data expressed as mean ± S.D.
RESULTS
Inclusions of FTLD with VCP R155H Mutation Contain Ubiquitinated TDP-43 as a Major Component and VCP as a Minor Component—Immunohistochemistry using anti-ubiquitin and anti-TDP-43 antibodies produced similar findings, including a spectrum of neuronal inclusions (NCIs, DNs, and NIIs) (Fig. 2). All NCIs, DNs, and NIIs that were ubiquitin-immunoreactive were also TDP-43-positive; a minority of inclusions was VCP-immunoreactive. Unlike cases of FTLD with GRN mutation, where NCIs and DNs are numerous and NIIs much less frequent, in FTLD-U with VCP mutation, the most frequent pathological inclusion was the NII, a morphological feature that distinguishes this FTLD-U from other subtypes, as previously described (13, 40). These and our previous immunohistochemical and biochemical data indicate that TDP-43 proteinopathy is a signature feature of FTLD with VCP mutation (13, 17).
FIGURE 2.
Neuropathology of the brain of a 47-year-old man with frontotemporal lobar degeneration and VCP mutation R155H. A, low power micrograph of the superficial laminae of the frontal lobe showing two neurons (arrows) containing intranuclear aggregates of VCP. B, high power micrograph showing a VCP-immunoreactive intranuclear inclusion. C, a TDP-43-immunoreactive intranuclear aggregate. D, a ubiquitin-immunoreactive neuronal intranuclear inclusion. E, a TDP-43 NCI. F, a ubiquitin-immunoreactive NCI. A and B, VCP immunohistochemistry (IHC); C and E, TDP-43 immunohistochemistry; D and F, ubiquitin immunohistochemistry. Scale bar, 20 μm (A) and 5 μm (B-F).
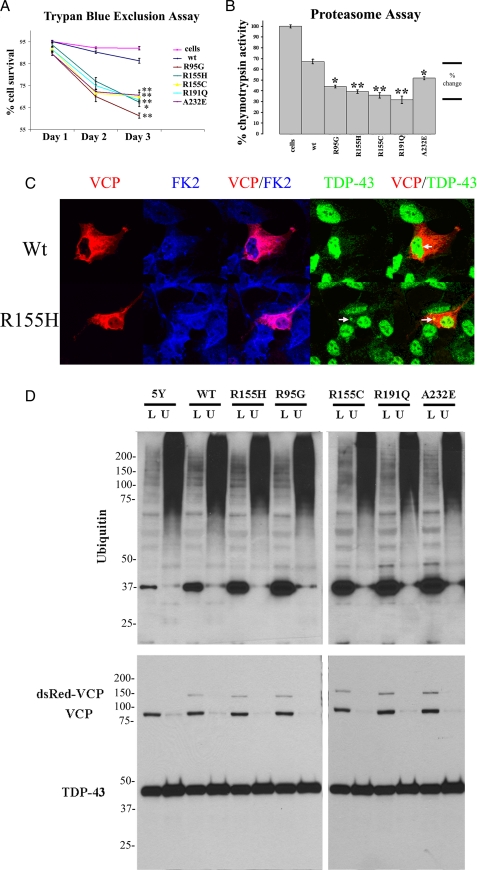
Mutations in VCP Induce Cell Death—To determine the effects on cell viability of overexpressed wild-type (WT) and mutant VCP in neuroblastoma cells, we used the trypan blue exclusion assay for evaluating cell viability starting 24 h post-transfection (Fig. 3A). All disease-related VCP mutations caused a significant decrease in cell viability at 48 and 72 h post-transfection (R95G, 20.2 ± 2.0% (p < 0.001); R155H, 13.0 ± 1.2% (p < 0.05); R155C, 18.8 ± 1.0% (p < 0.001); R191Q, 15.3 ± 1.4% (p < 0.001); A232E, 18.2 ± 2.3% (p < 0.001) when compared with WT; means were calculated from three separate experiments).
FIGURE 3.
Mutations in VCP decrease cell viability and proteasome activity and increase the amount of soluble ubiquitin in neuroblastoma, SHSY-5Y, cells. A, trypan blue exclusion assay measured cell viability over a 3-day time course after transfection in three separate experiments. The graph shows viability of WT compared with mutant VCP transiently transfected SHSY-5Y cells (n = 3; *, p < 0.05; **, p < 0.001). B, proteasome chymotrypsin cleavage activity in retinoic acid induced differentiated SHSY-5Y cells transiently transfected with VCP constructs assessed 24 h post-transfection (n = 6; *, p < 0.05; **, p < 0.001). C, SHSY-5Y cells transiently transfected with monomeric dsRED-VCP constructs; arrows indicate colocalization of TDP-43/VCP (yellow) and visualization by confocal microscopy (VCP dsRED fusion constructs (red), anti-FK2 (blue), and anti-TDP-43 (green)). D (top), immunoblotting of low salt and urea-soluble cell fractions reveals a distinct ubiquitin-positive band of ∼37 kDa. Soluble low salt (L) and soluble urea (U) fractions are shown. 5Y, control SHSY-5Y cells not transfected. Top, blot probed with anti-ubiquitin antibodies. D (bottom), fractions were probed with anti-VCP and anti-TDP-43 antibodies. A single band of ∼43 kDa of TDP-43 was identified; no other bands and no cleavage products were detected in the cellular protein fractions. VCP was detected using anti-VCP antibodies, and two bands were identified: full-length VCP of ∼97 kDa and VCP-dsRED fusion protein of ∼150 kDa.
Proteasome Activity Is Impaired by Mutations in VCP—Since cell viability was reduced with mutant VCP, and because VCP has previously been shown to be involved in the transport of misfolded proteins to the proteasome, we next tested whether mutations in VCP affected proteasome activity (32). Proteasome activity was measured 24 h post-transfection by cleavage of N-succinyl-Leu-Leu-Val-Tyr-7-amido-4-methyl-coumain for chymotrypsin-like activity (Fig. 3B). We observed a 32.8 ± 1.5% (p < 0.001) decrease in proteasome activity in cells overexpressing WT VCP compared with nontransfected cells or vehicle. There was also a significant decrease in proteasome activity in VCP mutants compared with WT: R95G, 23.2 ± 2.6% (p < 0.05); R155H, 27.7 ± 1.3% (p < 0.001); R155C, 31.4 ± 1.6% (p < 0.001); R191Q, 35.6 ± 3.5% (p < 0.001); A232E, 15.4 ± 1.5% (p < 0.05) (means were calculated from three separate experiments). Since ubiquitin-positive inclusions were observed in post-mortem brain (Fig. 2) and mutations in VCP with defects in ERAD have been shown to cause an increase in polyubiquitinated aggregates in muscle, we tested whether these aggregates were present in our tissue culture model (50). There was no apparent increase in the relative amount of FK2 (specific for mono- and polyubiquitin protein conjugates) in the R155H mutant compared with WT cells (Fig. 3C), but we did observe that VCP and FK2 had more robust colocalization in R155H than WT-transfected cells (Fig. 3C; for mutant VCP and TDP-43 and FK2 colocalization, see supplemental Fig. 1). Interestingly, in WT-expressing cells, TDP-43 and VCP were observed in proximity to the ER/nuclear envelope (Fig. 3C) in both mutant and WT VCP in 60% of transfected cells (Fig. 3C). In R155H and other mutant VCP-expressing cells, there were punctate cytosolic TDP-43/VCP-positive aggregates (Fig. 3C). This colocalization in the neuroblastoma cell line is reminiscent of that observed in the pathological inclusions of FTLD-U with VCP mutation (Fig. 2, C-F).
Sequentially fractionated post-mortem brain tissue from amyotrophic lateral sclerosis and FTLD with TDP-43 proteinopathy displays a unique biochemical signature: hyperphosphorylated TDP-43, an increase in ubiquitinated protein aggregates, and cleaved C-terminal fragments (15). To determine whether our tissue culture system displayed this biochemical signature, we took soluble low salt and urea fractions of SHSY-5Y cells and analyzed these fractions by Western blot (Fig. 3D). In the soluble fractions, there was a relative increase in the amount of ubiquitinated proteins compared with WT and untransfected cells (Fig. 3, D and F). Although we did not detect a 47 kDa band or a 25-kDa cleavage product, as has been reported in FTLD-U and amyotrophic lateral sclerosis brain, there was a distinct ubiquitin-positive band of ∼37 kDa that is most likely a covalently linked ubiquitin tetramer that increased in the mutant-expressing cells, consequent to a decrease in proteasome activity (Fig. 3B). We found no increase in the quantity of ubiquitinated proteins in the insoluble fractions of SHSY-5Y cells expressing mutant VCP (Fig. 3D). Unlike in human disease, we were unable to detect any hyperphosphorylated or cleaved TDP-43 in either fraction in the transfected overexpressed mutant VCP (Fig. 3D) in neuroblastoma cells.
Since VCP contributes to ERAD function and a decrease in proteasome activity has been shown to increase ER stress and the unfolded protein response, we investigated whether mutations in VCP cause an increase in ER stress (50-52).
ER Stress and Unfolded Protein Response Play a Role in VCP Pathogenesis—Because AKT phosphorylation of VCP has been shown to play a role in activation of VCP, we investigated whether induction and/or inhibition of phosphatidylinositol 3-kinase signaling plays a role in the post-translational modification of TDP-43 (21, 53). AKT is induced during differentiation in neuroblastoma cells by the addition of retinoic acid and is inhibited by the phosphoinositide 3-kinase inhibitor LY294002 (LY) and untreated SHSY-5Y cells (5Y) (Fig. 4A; see supplemental Fig. 2) (54). Endogenous VCP (lower band) has been shown previously to be up-regulated in differentiating U937 leukemia and pancreatic ductal adenocarcinoma, but we observed no change in endogenous VCP in retinoic acid-induced differentiated SHSY-5Y cells (Fig. 4A, lower band) (55, 56). There appeared to be no posttranslational modifications of TDP-43 in mutant VCP-expressing cells compared with WT (Fig. 4A).
FIGURE 4.
Mutations in VCP increase markers of ER stress. A, Western blot analysis of VCP constructs in SHSY-5Y cells under different conditions: untreated SHSY-5Y cells (5Y), retinoic acid (RA)-induced differentiation, and cells in the presence of phosphatidylinositol 3-kinase inhibitor, LY294002 (LY). B, there was an increase, but not statistically significant, in GRP78 levels in mutants compared with controls. Values represent relative optical density values of GRP78 in untreated cells in arbitrary units normalized to β-actin (ACTB) in three separate experiments. C, GRP78 mRNA normalized to GAPDH was significantly increased in mutants, as determined by quantitative RT-PCR (qRT-PCR). D, quantitative RT-PCR of calreticulin mRNA levels were significantly reduced in mutants compared with WT control when normalized to GAPDH 24 h after transfection and 24 h after retinoic acid-induced differentiation in SHSY-5Y cells. Data are expressed as mean ± S.D.; Student's t test; *, p < 0.05; **, p < 0.001.
Recently, VCP was shown to have a role in the autophagic clearance of protein aggregates (57), and the loss of endosomal sorting complexes required for transport has been shown to be implicated in the accumulation of TDP-43 (58). In order to investigate the role of aggregation in our overexpression system, we induced autophagy through the phosphatidylinositol 3-kinase inhibitor LY294002 (59). This showed altered localization of TDP-43; there was no difference between the untransfected cells and WT and mutant VCP (see supplemental Fig. 2).
GRP78 (BiP), GRP94, CHOP, and XBP1 are activated in response to ER stress (60, 61). GRP78 protein level in untreated cells (5Y) showed no significant difference between nontransfected cells, WT, and all the mutant VCP cells, but there was a small increase in protein level when WT transfected cells were compared with R155C, R191Q, and A232E cells (Fig. 4, A and B) (60). In order to investigate this trend in GRP78 expression, we looked at transcriptional levels by quantitative RT-PCR of GRP78, calreticulin, GRP94, CHOP, and XBP1. We found no significant change in the level of transcription of GRP94, CHOP, or alternative splicing of XBP1 (data not shown). In contrast, the GRP78 transcription level in untreated cells was significantly lower than in WT transfected cells 57.7 ± 2.0 (p < 0.01). Compared with WT cells, all VCP mutants expressed higher levels of GRP78 mRNA than cells overexpressing WT VCP: R95G, 42.2 ± 1.1% (p < 0.05); R155H, 25.5 ± 8.1% (p < 0.05); R155C, 12.7 ± 3.1% (p < 0.05); R191Q, 70.6 ± 8.4% (p < 0.05); A232E, 74.8 ± 5.6% (p < 0.001). The data were obtained from two independent experiments with four replicates. Interestingly, calreticulin displayed no significant change in transcription when WT was compared with mutant VCP in untreated cells (data not shown). However, 24 h after the addition of retinoic acid, wild-type VCP-overexpressing cells showed a 237.5 ± 5.3% (p < 0.001) increase in calreticulin transcription compared with retinoic acid-treated cells. Wild-type expressing cells compared with mutant VCP showed a smaller increase in calreticulin transcription: R95G, 56.5 ± 7.0% (p < 0.05); R155H, 141.2 ± 10.3% (p < 0.001); R155C, 170.1 ± 1.8% (p < 0.001); R191Q, 172.4 ± 7.1% (p < 0.001); A232E, 156.6 ± 9.0% (p < 0.001). The data were obtained from two independent experiments with four replicates (Fig. 4D).
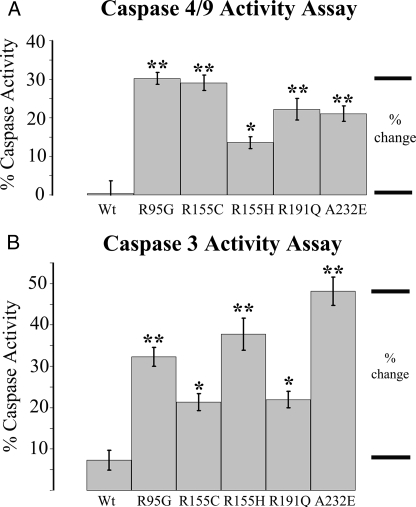
Mutations in VCP Increase Cleaved Caspase Activity—Previously, it has been shown that caspase activity is responsible for post-translational cleavage associated with TDP-43 proteinopathy in FTLD with GRN mutation (62). To investigate this pathway in FTLD with VCP mutation, we used a colorimetric substrate that can be cleaved by caspase-4 and -9. We demonstrated that cleaved caspase-4 and/or -9 increased significantly in cells overexpressing mutant VCP (R95G, 29.0 ± 2.0% (p < 0.001); R155H, 30.2 ± 1.5% (p < 0.001); R155C, 13.6 ± 1.5% (p < 0.05); R191Q, 22.2 ± 2.8% (p < 0.001); A232E, 21.0 ± 2.0% (p < 0.001; n = 10) (Fig. 5A) (63). Cleaved caspase-3 activity was also increased in cells expressing mutant VCP: R95G, 25.0 ± 2.3% (p < 0.001); R155H, 13.7 ± 2.0% (p < 0.05); R155C, 30.5 ± 3.9% (p < 0.001); R191Q, 14.7 ± 2.0% (p < 0.05); A232E, 40.8 ± 3.5% (p < 0.001; n = 10) (Fig. 5B) (64).
FIGURE 5.
Apoptotic markers are elevated in mutant cell lines compared with controls. A, caspase-4 and/or -9 activity was significantly increased in all mutants. B, caspase-3 activity was also elevated in all mutants compared with WT control cells. Transiently transfected overexpressed WT VCP was compared with mutant VCP in SHSY-5Y cells (n = 10; *, p < 0.05; **, p < 0.001).
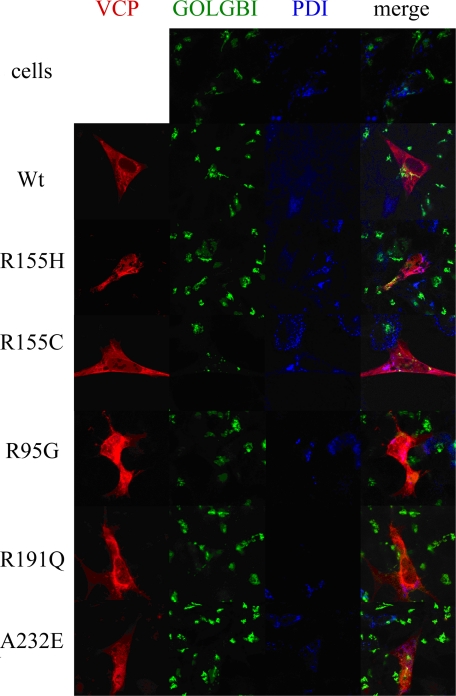
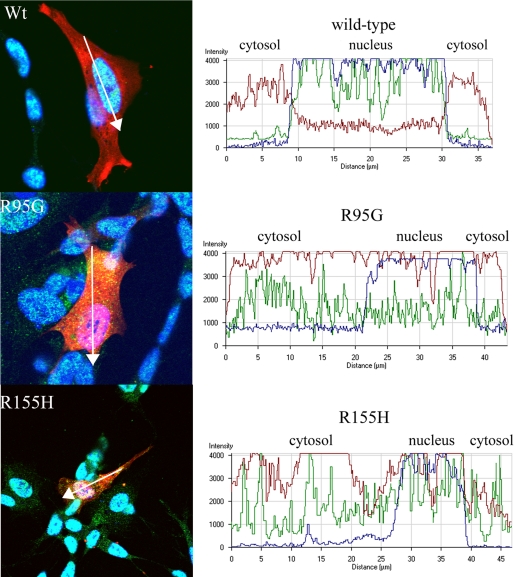
Mutations in VCP Alter Endogenous TDP-43 Localization—To investigate localization and normal distribution of VCP in SHSY-5Y cells, we used confocal microscopy. Distribution of WT VCP was primarily cytosolic, with both ER and Golgi colocalization (Fig. 6). There was no apparent change in the distribution of giantin, a trans-Golgi marker, or protein-disulfide isomerase, an ER marker, in cells transfected with mutant VCP, indicating that the Golgi and ER do not appear to be fragmented, a feature of some neurodegenerative diseases (Fig. 6) (65). There was no increase in accumulation or change in aggregation of overexpressed WT compared with mutant VCP. However, mutant VCP was distributed throughout the entire cell compared with WT, which was predominantly localized to the nucleus (Fig. 6).
FIGURE 6.
VCP colocalizes with the ER-Golgi network. Overexpressed dsRED-VCP fusion protein (red), GOLGB1 (anti-giantin, trans-Golgi) (green), and protein-disulfide isomerase (PDI; ER marker, secondary; blue) were visualized by confocal microscopy.
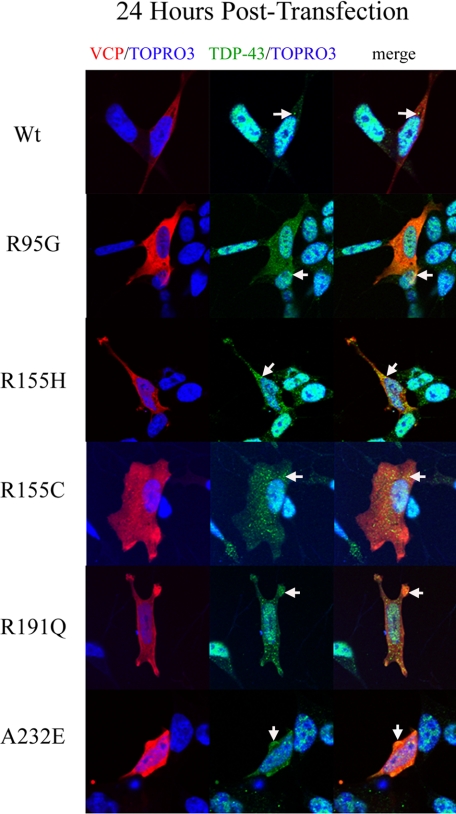
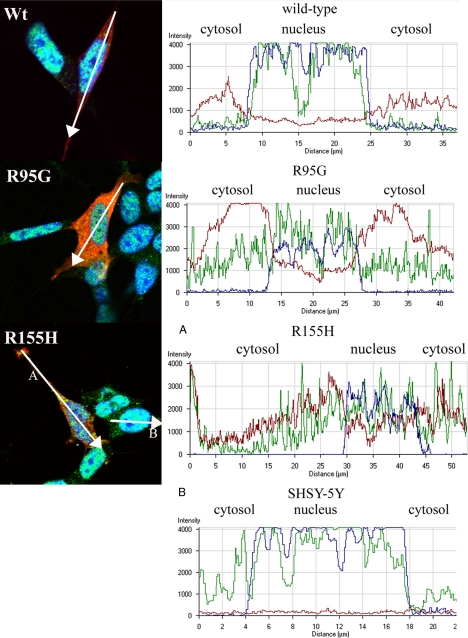
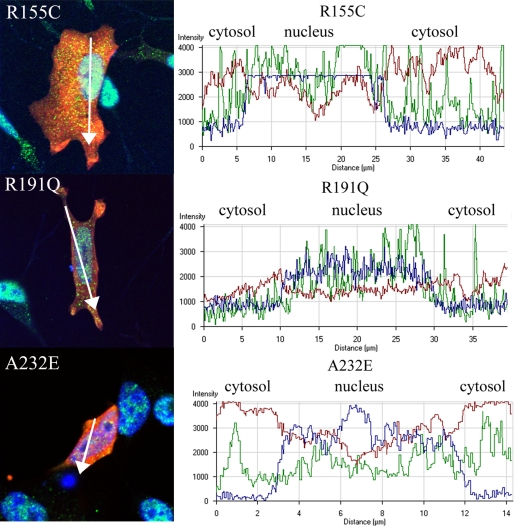
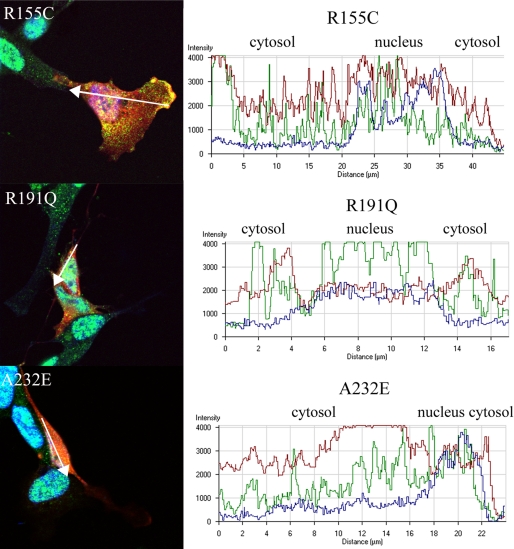
Since TDP-43 is a predominantly nuclear protein and translocation from the nucleus to the cytoplasm is a signature feature of TDP-43 proteinopathies, including FTLD with VCP mutation, we investigated whether overexpression of mutant VCP causes a change in localization of endogenous TDP-43 (5, 40, 66). Observations were made at 24 and 48 h time points after transfection. At 24 h after transfection, the WT VCP-expressing cells showed no apparent change in distribution or accumulation of TDP-43; there were punctate TDP-43-positive speckles or granules within the cytoplasm in the WT-expressing cells that were less abundant compared with the cells that were not transfected (Fig. 7). VCP and TDP-43 colocalized in both the WT and mutant VCP-expressing cells. This TDP-43/VCP colocalization was observed in 65% of the mutant-expressing cells (Fig. 7). Also, in 75% of cells transfected, mutant VCP was observed in both the cytoplasm and nucleus. In contrast, in the WT-expressing cells, VCP was predominantly found within the cytoplasm with some punctuate expression in the nucleus (Fig. 7). In order to investigate this distribution, we looked at cytoplasmic and nuclear intensity distribution within the cell using a graphical method (Figs. 8 and 9) (67). As indicated in the intensity line profile, in the WT-transfected cells, TDP-43 was predominantly nuclear, with some cytosolic staining, and where VCP was more abundant. In R95G cells, there was an increase in nuclear localization of VCP that was even more pronounced in R155H, R155C, R191Q, and A232E mutants (Figs. 7, 8, 9). TDP-43 distribution within the nucleus was more granular, with some areas being more punctate, most prominently in the R155C-expressing cells (Fig. 9). In untransfected SHSY-5Y cells, the distribution of intensity indicated a normal cytoplasmic distribution of TDP-43 (Fig. 8B).
FIGURE 7.
Mutant VCP alters endogenous TDP-43 localization 24 h post-transfection. At 24 h post-transfection, dsRED-VCP fusion protein (red), TDP-43 (green), and TOPRO-3 (nuclear marker; blue) were visualized by confocal microscopy (arrows indicate TDP-43 and VCP colocalization and accumulation (merge/yellow)).
FIGURE 8.
Mutant VCP localizes to the nucleus from cytoplasm 24 h post-transfection. Wild-type VCP shows no change in cytosolic distribution. R95G and R155H showed a similar spatial pattern of protein localization; the graphs show the intensity distribution profile of dsRED-VCP (red line), TDP-43 (green line), and TOPRO-3, nuclear marker (blue line) of the merged image. Intensity profile of R155H (A) and control, nontransfected cell (SHSY-5Y) profile are represented in the graph (B) and show predominantly nuclear and subtle TDP-43 cytoplasmic staining.
FIGURE 9.
Mutant VCP translocates to the nucleus from the cytoplasm. VCP translocates to the nucleus and TDP-43 appears more abundant in the cytoplasm of R155C-, R191Q-, and A232E-transfected cells. The graphs show the intensity distribution profile of dsRED-VCP (red line), TDP-43 (green line), and TOPRO-3 (nuclear marker; blue line) in the analyzed cells.
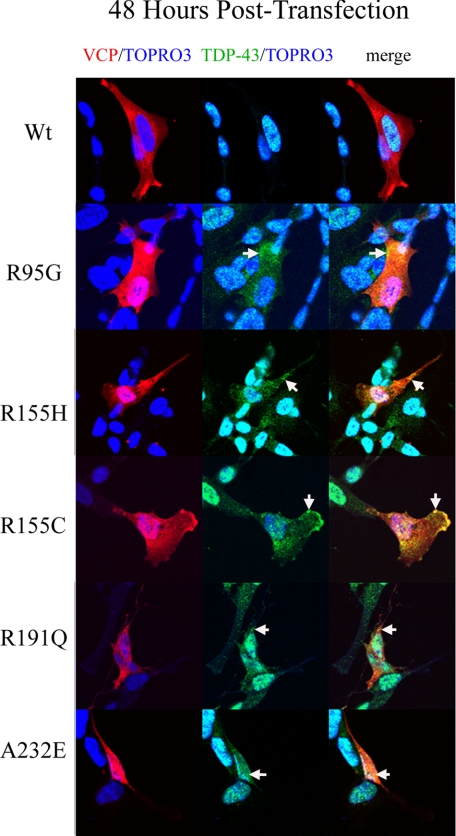
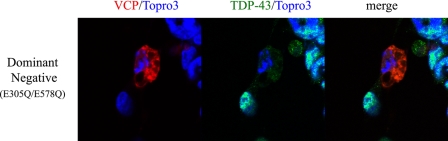
In the viability experiments described above, we observed that cell death occurred 48 h post-transfection (Fig. 3A). In order to look at downstream morphological effects and redistribution of TDP-43, we observed these cells 48 h after transfection (Fig. 10). At 48 h post-transfection, WT VCP remained predominantly cytosolic in over 85% of all transfected cells, whereas over 85% of the mutant expressing cells showed a nuclear distribution (Fig. 10). Colocalization of TDP-43 and VCP was apparent in the mutant-expressing cells (Fig. 10). In R95G, R155H, and R155C cells, there was an increase in nuclear colocalization of VCP/TDP-43 as large punctate inclusions (Figs. 11 and 12). Interestingly, A232E mutant-expressing cells displayed a morphologically condensed nucleus, which has been shown previously to be associated with apoptosis (Fig. 12) (68). There was also a punctate cytosolic distribution of TDP-43 that colocalized with VCP (Fig. 11). Nuclear TDP-43 was reduced in mutant VCP cells in comparison with WT cells, as demonstrated by immunoblotting and confocal immunocytochemistry (see supplemental Fig. 3, A and B; see Figs. 11 and 12). VCP was present at a relatively higher intensity within the nucleus of R95G-, R155H-, R155C-, R191Q-, and A234E-expressing cells with very low intensity of wild-type VCP localized to the nucleus (Fig. 12) (see supplemental material for video files showing complete Z-stack distribution of TDP-43 at 48 h post-transfection comparing WT with mutant VCP R155H). Overexpression of E305Q/E578Q, a dominant-negative VCP mutant, led to nuclear fragmentation consistent with neuronal cell death at 24 h after transfection with little to no TDP-43 present either in the nucleus or cytoplasm (Fig. 13).
FIGURE 10.
Mutant VCP alters endogenous TDP-43 localization 48 h post-transfection. dsRED-VCP fusion protein (red), TDP-43 (green), and TOPRO-3 (nuclear marker; blue) were visualized by confocal microscopy At 48 h post-transfection. The arrows indicate TDP-43 and VCP colocalization in the cytosol (merge/yellow).
FIGURE 11.
Mutant VCP induces TDP-43 distribution to the cytosol 48 h post-transfection. Wild-type VCP-expressing cells show no change in nuclear distribution of TDP-43. In contrast, VCP R95G and R155H mutants display a relative increase in cytosolic TDP-43. The graphs show the intensity distribution profile of dsRED-VCP (red line), TDP-43 (green line), and TOPRO-3 (nuclear marker; blue line) of the merged image.
FIGURE 12.
Mutant VCP induces TDP-43 distribution from the nucleus to the cytosol 48 h post-transfection. R155C, R191Q, and A232E mutants display a relative increase in cytosolic TDP-43. The graphs show the intensity distribution profile of dsRED-VCP (red line), TDP-43 (green line), and TOPRO-3 (nuclear marker; blue line) of the merged image.
FIGURE 13.
Overexpression of dsRED-VCP E305Q/E578Q (dominant negative) leads to nuclear fragmentation consistent with neuronal cell death at 24 h post-transfection. There is little to no TDP-43 present either in the nucleus or cytoplasm. dsRED-VCP E305Q/E578Q (dominant negative) (red), TDP-43 (green) and TOPRO-3 (nuclear marker; blue) and visualized by confocal microscopy.
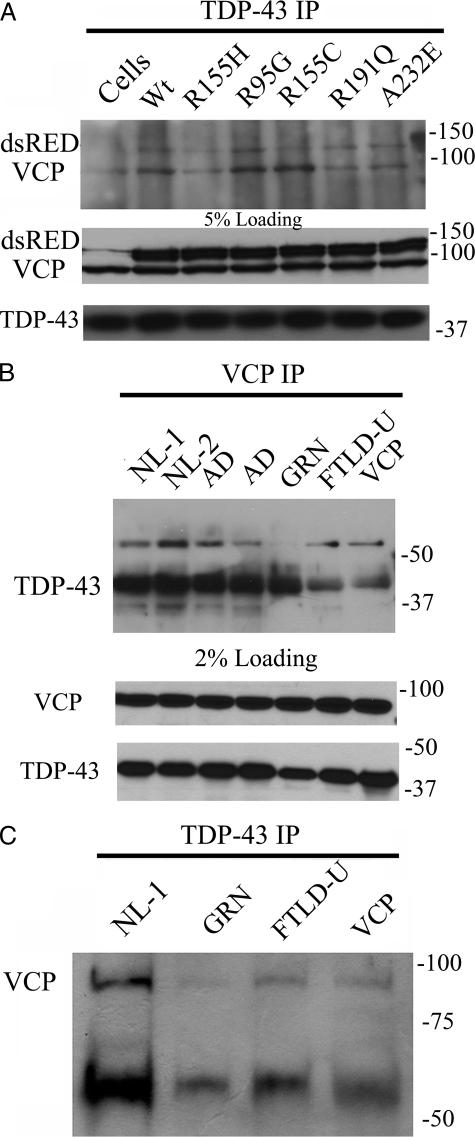
TDP-43 and VCP Are Coimmunoprecipitated from SHSY-5Y Cells and Brain—In order to expand our understanding of the colocalization observed in brain and in tissue culture models (Figs. 2 and 8A), we investigated whether TDP-43 interacts with VCP using immunoprecipitation methods. We found that TDP-43 and VCP immunoprecipitate together in SHSY-5Y cells and that the VCP mutations did not affect the formation of these complexes (Fig. 14A). We confirmed that VCP and TDP-43 also coimmunoprecipitate from brain homogenates (Fig 14, B and C). Interestingly, we found that there was less TDP-43 immunoprecipitated by VCP in a spectrum of TDP-43 proteinopathies (Fig. 14C).
FIGURE 14.
TDP-43 complexes with VCP in co-immunoprecipitation assays in both SHSY-5Y cells and human brain. A, TDP-43 immunoprecipitation (IP) in VCP-transfected SHSY-5Y cells probed with anti-VCP (top) and 5% loading control (bottom). B, VCP immunoprecipitation in high salt (HS) fraction of human brain probed with anti-TDP-43. Samples were immunoprecipitated with VCP: normal adult control cases (NL-1 and NL-2); Alzheimer disease (AD); familial FTLD-U with GRN A9D mutation (GRN); sporadic (FTLD-U); and FTLD-U with the VCP R155H mutation (VCP) (top) and 2% loading control (bottom). C, TDP-43 immunoprecipitation in HS of human brain probed with anti-VCP: age-matched control (NL-1); familial FTLD-U with GRN A9D mutation (GRN); sporadic (FTLD-U); and FTLD-U with VCP R155H mutation (VCP).
DISCUSSION
Loss of VCP function using knockdown experiments results in a decrease in proteasome activity and an increase in ubiquitinated proteins (37). We describe a tissue culture model of FTLD with VCP mutation in which mutant transfectants cause a loss of function. Several mechanisms contribute to this loss of function. Mutant VCP lowers chymotrypsin cleavage activity of the proteasome and increases soluble ubiquitin, which resembles the accumulation of ubiquitinated proteins, aggregation, change in solubility, and inclusion formation present in the human disease (13, 40). Proteasome dysfunction is implicated in the pathogenesis of Huntington disease, Parkinson disease, motor neuron disease, and other neurodegenerative diseases, but the mode of activation has yet to be elucidated (62, 69, 70). A decrease in proteasome activity not only increases ubiquitinated proteins targeted for degradation but may also affect activation of signaling pathways that are regulated by ubiquitination (43, 71). VCP has been shown to be phosphorylated by AKT and also has a role in ubiquitination of IκBα activating the NF-κB cell survival pathway (21, 35, 53). Modulation of VCP expression has been implicated in cancer and apoptotic pathways (56, 72). Proteasome malfunction has been shown to activate the caspase cascade and, eventually, apoptotic cell death (69). Activation of ER stress and the unfolded protein response has been shown previously to induce the caspase cascade (63, 73, 74). We describe a specific induction of ER stress involving an increase in the molecular chaperone GRP78. A unique function of VCP in retinoic acid-induced differentiation of neuroblastoma cells is to affect the transcriptional level of calreticulin. This protein monitors unfolded glycoproteins, which may lead to an accumulation of these substrates not targeted correctly through ERAD to the proteasome (Fig. 4C) (75-78). Although mutations in VCP have been shown previously to be defective in targeting specific substrates through ERAD, the specific mechanism leading to this defect has not been elucidated, and the relationship between VCP and calreticulin needs further investigation (50).
In the SHSY-5Y model system, the spatial relocalization of TDP-43 in cells expressing mutant VCP resembles the altered localization of TDP-43 with KCl-induced neuronal stress (79). Although the predominant pathology in FTLD with VCP mutation is TDP-43 proteinopathy in the form of NIIs, we demonstrate in this tissue culture model that this change in localization of TDP-43 is present before cell death. Also, the mutant VCP localizes to the nucleus and cytosol, whereas WT VCP was predominantly cytosolic, which recapitulates features of the human disease (2).
In summary, we describe an in vitro model of FTLD with VCP mutation. Transfected mutant VCP in neuroblastoma cells induced cell death and caused relocation of TDP-43. To determine potential mechanisms of neurodegeneration, we report a novel interaction between TDP-43 and VCP in tissue culture that is also detected in brain. This interaction requires further investigation, but it suggests that TDP-43 may be a substrate targeted to the proteasome or may modulate VCP function as a cofactor. The ability of TDP-43 to bind single-stranded DNA may indicate a role for VCP targeted to sites of DNA repair (17, 20). Elucidating the relationship of the interaction between TDP-43 and VCP will provide further insights into the pathogenesis of this TDP-43 proteinopathy (21, 35, 53, 66, 80) and provide novel targets for therapeutic intervention.
Supplementary Material
Acknowledgments
We thank Dr. John C. Morris and the clinical, genetic, pathology, and technical staff of the Washington University Alzheimer's Disease Research Center for making information and tissue samples available for this study, and we thank the families of patients whose generosity made this research possible.
This work was supported, in whole or in part, by National Institutes of Health, NIA, Grants P50-AG05681, P01-AG03991, and P30-NS057105. This work was also supported by the Buchanan Fund, the Charles and Joanne Knight Alzheimer Research Initiative, and the Barnes-Jewish Hospital Foundation (St. Louis, MO). A preliminary report of these data was presented (81).
The on-line version of this article (available at http://www.jbc.org) contains supplemental Table 1, Figs. 1-3, and Videos 1 and 2.
Footnotes
The abbreviations used are: FTLD, frontotemporal lobar degeneration; FTLD-U, FTLD with ubiquitin-positive inclusions; NII, neuronal intranuclear inclusion; DN, dystrophic neurite; NCI, neuronal cytoplasmic inclusion; ER, endoplasmic reticulum; ERAD, ER-associated degradation; PBS, phosphate-buffered saline; CHAPS, 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonic acid; RT, reverse transcription; GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
References
- 1.Bird, T., Knopman, D., VanSwieten, J., Rosso, S., Feldman, H., Tanabe, H., Graff-Raford, N., Geschwind, D., Verpillat, P., and Hutton, M. (2003) Ann. Neurol. 54 Suppl. 5, 29-31 [DOI] [PubMed] [Google Scholar]
- 2.Cairns, N. J., Bigio, E. H., Mackenzie, I. R., Neumann, M., Lee, V. M., Hatanpaa, K. J., White, C. L., III, Schneider, J. A., Grinberg, L. T., Halliday, G., Duyckaerts, C., Lowe, J. S., Holm, I. E., Tolnay, M., Okamoto, K., Yokoo, H., Murayama, S., Woulfe, J., Munoz, D. G., Dickson, D. W., Ince, P. G., Trojanowski, J. Q., and Mann, D. M. (2007) Acta Neuropathol. 114 5-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cairns, N. J., Neumann, M., Bigio, E. H., Holm, I. E., Troost, D., Hatanpaa, K. J., Foong, C., White, C. L., III, Schneider, J. A., Kretzschmar, H. A., Carter, D., Taylor-Reinwald, L., Paulsmeyer, K., Strider, J., Gitcho, M., Goate, A. M., Morris, J. C., Mishra, M., Kwong, L. K., Stieber, A., Xu, Y., Forman, M. S., Trojanowski, J. Q., Lee, V. M., and Mackenzie, I. R. (2007) Am. J. Pathol. 171 227-240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bersano, A., Del Bo, R., Lamperti, C., Ghezzi, S., Fagiolari, G., Fortunato, F., Ballabio, E., Moggio, M., Candelise, L., Galimberti, D., Virgilio, R., Lanfranconi, S., Torrente, Y., Carpo, M., Bresolin, N., Comi, G. P., and Corti, S. (2007) Neurobiol. Aging 10.1016/jneurobiolaging.2007.08.009 [DOI] [PubMed]
- 5.Guyant-Maréchal, L., Laquerriére, A., Duyckaerts, C., Dumanchin, C., Bou, J., Dugny, F., Le, B. I., Frébourg, T., Hannequin, D., and Campion, D. (2006) Neurology 67 644-651 [DOI] [PubMed] [Google Scholar]
- 6.Haubenberger, D., Bittner, R. E., Rauch-Shorny, S., Zimprich, F., Mannhalter, C., Wagner, L., Mineva, I., Vass, K., Auff, E., and Zimprich, A. (2005) Neurology 65 1304-1305 [DOI] [PubMed] [Google Scholar]
- 7.Kimonis, V. E., and Watts, G. D. (2005) Alzheimer Dis. Assoc. Disord. 19 Suppl. 1, 44-47 [DOI] [PubMed] [Google Scholar]
- 8.Kovach, M. J., Waggoner, B., Leal, S. M., Gelber, D., Khardori, R., Levenstien, M. A., Shanks, C. A., Gregg, G., Al Lozi, M. T., Miller, T., Rakowicz, W., Lopate, G., Florence, J., Glosser, G., Simmons, Z., Morris, J. C., Whyte, M. P., Pestronk, A., and Kimonis, V. E. (2001) Mol. Genet. Metab. 74 458-475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Watts, G. D., Wymer, J., Kovach, M. J., Mehta, S. G., Mumm, S., Darvish, D., Pestronk, A., Whyte, M. P., and Kimonis, V. E. (2004) Nat. Genet. 36 377-381 [DOI] [PubMed] [Google Scholar]
- 10.Watts, G. D., Thomasova, D., Ramdeen, S. K., Fulchiero, E. C., Mehta, S. G., Drachman, D. A., Weihl, C. C., Jamrozik, Z., Kwiecinski, H., Kaminska, A., and Kimonis, V. E. (2007) Clin. Genet. 72 420-426 [DOI] [PubMed] [Google Scholar]
- 11.Daroszewska, A., and Ralston, S. H. (2006) Nat. Clin. Pract. Rheumatol. 2 270-277 [DOI] [PubMed] [Google Scholar]
- 12.Hubbers, C. U., Clemen, C. S., Kesper, K., Boddrich, A., Hofmann, A., Kamarainen, O., Tolksdorf, K., Stumpf, M., Reichelt, J., Roth, U., Krause, S., Watts, G., Kimonis, V., Wattjes, M. P., Reimann, J., Thal, D. R., Biermann, K., Evert, B. O., Lochmuller, H., Wanker, E. E., Schoser, B. G., Noegel, A. A., and Schroder, R. (2007) Brain 130 381-393 [DOI] [PubMed] [Google Scholar]
- 13.Forman, M. S., Mackenzie, I. R., Cairns, N. J., Swanson, E., Boyer, P. J., Drachman, D. A., Jhaveri, B. S., Karlawish, J. H., Pestronk, A., Smith, T. W., Tu, P. H., Watts, G. D., Markesbery, W. R., Smith, C. D., and Kimonis, V. E. (2006) J. Neuropathol. Exp. Neurol. 65 571-581 [DOI] [PubMed] [Google Scholar]
- 14.Davidson, Y., Kelley, T., Mackenzie, I. R., Pickering-Brown, S., Du, P. D., Neary, D., Snowden, J. S., and Mann, D. M. (2007) Acta Neuropathol. 113 521-533 [DOI] [PubMed] [Google Scholar]
- 15.Neumann, M., Sampathu, D. M., Kwong, L. K., Truax, A. C., Micsenyi, M. C., Chou, T. T., Bruce, J., Schuck, T., Grossman, M., Clark, C. M., McCluskey, L. F., Miller, B. L., Masliah, E., Mackenzie, I. R., Feldman, H., Feiden, W., Kretzschmar, H. A., Trojanowski, J. Q., and Lee, V. M. (2006) Science 314 130-133 [DOI] [PubMed] [Google Scholar]
- 16.Neumann, M., Mackenzie, I. R., Cairns, N. J., Boyer, P. J., Markesbery, W. R., Smith, C. D., Taylor, J. P., Kretzschmar, H. A., Kimonis, V. E., and Forman, M. S. (2007) J. Neuropathol. Exp. Neurol. 66 152-157 [DOI] [PubMed] [Google Scholar]
- 17.Zhang, H., Wang, Q., Kajino, K., and Greene, M. I. (2000) DNA Cell Biol. 19 253-263 [DOI] [PubMed] [Google Scholar]
- 18.Shiozawa, K., Maita, N., Tomii, K., Seto, A., Goda, N., Akiyama, Y., Shimizu, T., Shirakawa, M., and Hiroaki, H. (2004) J. Biol. Chem. 279 50060-50068 [DOI] [PubMed] [Google Scholar]
- 19.Shiozawa, K., Goda, N., Shimizu, T., Mizuguchi, K., Kondo, N., Shimozawa, N., Shirakawa, M., and Hiroaki, H. (2006) FEBS J. 273 4959-4971 [DOI] [PubMed] [Google Scholar]
- 20.Livingstone, M., Ruan, H., Weiner, J., Clauser, K. R., Strack, P., Jin, S., Williams, A., Greulich, H., Gardner, J., Venere, M., Mochan, T. A., DiTullio, R. A., Jr., Moravcevic, K., Gorgoulis, V. G., Burkhardt, A., and Halazonetis, T. D. (2005) Cancer Res. 65 7533-7540 [DOI] [PubMed] [Google Scholar]
- 21.Vandermoere, F., Yazidi-Belkoura, I., Slomianny, C., Demont, Y., Bidaux, G., Adriaenssens, E., Lemoine, J., and Hondermarck, H. (2006) J. Biol. Chem. 281 14307-14313 [DOI] [PubMed] [Google Scholar]
- 22.Hui, L., Pei, D. S., Zhang, Q. G., Guan, Q. H., and Zhang, G. Y. (2005) Brain Res. 1052 1-9 [DOI] [PubMed] [Google Scholar]
- 23.Alzayady, K. J., Panning, M. M., Kelley, G. G., and Wojcikiewicz, R. J. (2005) J. Biol. Chem. 280 34530-34537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lavoie, C., Chevet, E., Roy, L., Tonks, N. K., Fazel, A., Posner, B. I., Paiement, J., and Bergeron, J. J. (2000) Proc. Natl. Acad. Sci. U. S. A. 97 13637-13642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dreveny, I., Kondo, H., Uchiyama, K., Shaw, A., Zhang, X., and Freemont, P. S. (2004) EMBO J. 23 1030-1039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kano, F., Tanaka, A. R., Yamauchi, S., Kondo, H., and Murata, M. (2004) Mol. Biol. Cell 15 4289-4298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kano, F., Kondo, H., Yamamoto, A., Tanaka, A. R., Hosokawa, N., Nagata, K., and Murata, M. (2005) Genes Cells 10 333-344 [DOI] [PubMed] [Google Scholar]
- 28.Ballar, P., Shen, Y., Yang, H., and Fang, S. (2006) J. Biol. Chem. 281 35359-35368 [DOI] [PubMed] [Google Scholar]
- 29.Miyachi, K., Hirano, Y., Horigome, T., Mimori, T., Miyakawa, H., Onozuka, Y., Shibata, M., Hirakata, M., Suwa, A., Hosaka, H., Matsushima, S., Komatsu, T., Matsushima, H., Hankins, R. W., and Fritzler, M. J. (2004) Clin. Exp. Immunol. 136 568-573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pye, V. E., Beuron, F., Keetch, C. A., McKeown, C., Robinson, C. V., Meyer, H. H., Zhang, X., and Freemont, P. S. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 467-472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Partridge, J. J., Lopreiato, J. O., Jr., Latterich, M., and Indig, F. E. (2003) Mol. Biol. Cell 14 4221-4229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.DeLaBarre, B., Christianson, J. C., Kopito, R. R., and Brunger, A. T. (2006) Mol. Cell 22 451-462 [DOI] [PubMed] [Google Scholar]
- 33.McCracken, A. A., and Brodsky, J. L. (1996) J. Cell Biol. 132 291-298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schmitz, A., and Herzog, V. (2004) Eur. J. Cell Biol. 83 501-509 [DOI] [PubMed] [Google Scholar]
- 35.Asai, T., Tomita, Y., Nakatsuka, S., Hoshida, Y., Myoui, A., Yoshikawa, H., and Aozasa, K. (2002) Jpn. J. Cancer Res. 93 296-304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nowis, D., McConnell, E., and Wojcik, C. (2006) Exp. Cell Res. 312 2921-2932 [DOI] [PubMed] [Google Scholar]
- 37.Wojcik, C., Rowicka, M., Kudlicki, A., Nowis, D., McConnell, E., Kujawa, M., and DeMartino, G. N. (2006) Mol. Biol. Cell 17 4606-4618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mukherjee, O., Pastor, P., Cairns, N. J., Chakraverty, S., Kauwe, J. S., Shears, S., Behrens, M. I., Budde, J., Hinrichs, A. L., Norton, J., Levitch, D., Taylor-Reinwald, L., Gitcho, M., Tu, P. H., Tenenholz, G. L., Liscic, R. M., Armendariz, J., Morris, J. C., and Goate, A. M. (2006) Ann. Neurol. 60 314-322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sampathu, D. M., Neumann, M., Kwong, L. K., Chou, T. T., Micsenyi, M., Truax, A., Bruce, J., Grossman, M., Trojanowski, J. Q., and Lee, V. M. (2006) Am. J. Pathol. 169 1343-1352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gitcho, M. A., Baloh, R. H., Chakraverty, S., Mayo, K., Norton, J. B., Levitch, D., Hatanpaa, K. J., White, C. L., III, Bigio, E. H., Caselli, R., Baker, M., Al Lozi, M. T., Morris, J. C., Pestronk, A., Rademakers, R., Goate, A. M., and Cairns, N. J. (2008) Ann. Neurol. 63 535-538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mukherjee, O., Wang, J., Gitcho, M., Chakraverty, S., Taylor-Reinwald, L., Shears, S., Kauwe, J. S., Norton, J., Levitch, D., Bigio, E. H., Hatanpaa, K. J., White, C. L., Morris, J. C., Cairns, N. J., and Goate, A. (2008) Hum. Mutat. 29 512-521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bursztajn, S., Feng, J. J., Nanda, A., and Berman, S. A. (2001) Brain Res. Mol. Brain Res. 91 57-72 [DOI] [PubMed] [Google Scholar]
- 43.Li, X., Yang, D., Li, L., Peng, C., Chen, S., and Le, W. (2007) Neurochem. Int. 50 959-965 [DOI] [PubMed] [Google Scholar]
- 44.Qiu, J. H., Asai, A., Chi, S., Saito, N., Hamada, H., and Kirino, T. (2000) J. Neurosci. 20 259-265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Snider, B. J., Tee, L. Y., Canzoniero, L. M., Babcock, D. J., and Choi, D. W. (2002) Eur. J. Neurosci. 15 419-428 [DOI] [PubMed] [Google Scholar]
- 46.Muller, P. Y., Janovjak, H., Miserez, A. R., and Dobbie, Z. (2002) Bio-Techniques 32 1372-1379 [PubMed] [Google Scholar]
- 47.Nilbratt, M., Friberg, L., Mousavi, M., Marutle, A., and Nordberg, A. (2007) J. Neurosci. Res. 85 504-514 [DOI] [PubMed] [Google Scholar]
- 48.Christophersen, N. S., Meijer, X., Jorgensen, J. R., Englund, U., Gronborg, M., Seiger, A., Brundin, P., and Wahlberg, L. U. (2006) Brain Res. Bull. 70 457-466 [DOI] [PubMed] [Google Scholar]
- 49.Aravindan, N., Madhusoodhanan, R., Natarajan, M., and Herman, T. S. (2008) Mol. Cell Biochem. 310 167-179 [DOI] [PubMed] [Google Scholar]
- 50.Weihl, C. C., Dalal, S., Pestronk, A., and Hanson, P. I. (2006) Hum. Mol. Genet. 15 189-199 [DOI] [PubMed] [Google Scholar]
- 51.Shenkman, M., Tolchinsky, S., and Lederkremer, G. Z. (2007) Cell Stress Chaperones 12 373-383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shenkman, M., Tolchinsky, S., Kondratyev, M., and Lederkremer, G. Z. (2007) Biochem. J. 404 509-516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Klein, J. B., Barati, M. T., Wu, R., Gozal, D., Sachleben, L. R., Jr., Kausar, H., Trent, J. O., Gozal, E., and Rane, M. J. (2005) J. Biol. Chem. 280 31870-31881 [DOI] [PubMed] [Google Scholar]
- 54.Lopez-Carballo, G., Moreno, L., Masia, S., Perez, P., and Barettino, D. (2002) J. Biol. Chem. 277 25297-25304 [DOI] [PubMed] [Google Scholar]
- 55.Bertram, C., von Neuhoff, N., Skawran, B., Steinemann, D., Schlegelberger, B., and Hass, R. (2008) BMC Cell Biol. 9 1-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yamamoto, S., Tomita, Y., Hoshida, Y., Nagano, H., Dono, K., Umeshita, K., Sakon, M., Ishikawa, O., Ohigashi, H., Nakamori, S., Monden, M., and Aozasa, K. (2004) Ann. Surg. Oncol. 11 165-172 [DOI] [PubMed] [Google Scholar]
- 57.Ju, J. S., Miller, S. E., Hanson, P. I., and Weihl, C. C. (2008) J. Biol. Chem. [DOI] [PMC free article] [PubMed]
- 58.Filimonenko, M., Stuffers, S., Raiborg, C., Yamamoto, A., Malerod, L., Fisher, E. M., Isaacs, A., Brech, A., Stenmark, H., and Simonsen, A. (2007) J. Cell Biol. 179 485-500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Xing, C., Zhu, B., Liu, H., Yao, H., and Zhang, L. (2008) Acta Biochim. Biophys. Sin. 40 194-201 [DOI] [PubMed] [Google Scholar]
- 60.Kondratyev, M., Avezov, E., Shenkman, M., Groisman, B., and Lederkremer, G. Z. (2007) Exp. Cell Res. 313 3395-3407 [DOI] [PubMed] [Google Scholar]
- 61.Lass, A., Kujawa, M., McConnell, E., Paton, A. W., Paton, J. C., and Wojcik, C. (2008) Int. J. Biochem. Cell Biol. 40 2865-2879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang, Y. J., Xu, Y. F., Dickey, C. A., Buratti, E., Baralle, F., Bailey, R., Pickering-Brown, S., Dickson, D., and Petrucelli, L. (2007) J. Neurosci. 27 10530-10534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kim, S. J., Zhang, Z., Hitomi, E., Lee, Y. C., and Mukherjee, A. B. (2006) Hum. Mol. Genet. 15 1826-1834 [DOI] [PubMed] [Google Scholar]
- 64.King, T. D., Bijur, G. N., and Jope, R. S. (2001) Brain Res. 919 106-114 [DOI] [PubMed] [Google Scholar]
- 65.Nakagomi, S., Barsoum, M. J., Bossy-Wetzel, E., Sutterlin, C., Malhotra, V., and Lipton, S. A. (2008) Neurobiol. Dis. 29 221-231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Neumann, M., Kwong, L. K., Truax, A. C., Vanmassenhove, B., Kretzschmar, H. A., Van Deerlin, V. M., Clark, C. M., Grossman, M., Miller, B. L., Trojanowski, J. Q., and Lee, V. M. (2007) J. Neuropathol. Exp. Neurol. 66 177-183 [DOI] [PubMed] [Google Scholar]
- 67.Beatty, W. L. (2008) Infect. Immun. 76 2872-2881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhivotosky, B., and Orrenius, S. (2001) Current Protocols in Cell Biology, Unit 18.3, John Wiley & Sons, Hoboken, NJ [DOI] [PubMed]
- 69.Mishra, M., Paunesku, T., Woloschak, G. E., Siddique, T., Zhu, L. J., Lin, S., Greco, K., and Bigio, E. H. (2007) Acta Neuropathol. 114 81-94 [DOI] [PubMed] [Google Scholar]
- 70.Wang, J., Wang, C. E., Orr, A., Tydlacka, S., Li, S. H., and Li, X. J. (2008) J. Cell Biol. 180 1177-1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Peng, Y., Liu, X., and Schoenberg, D. R. (2008) Mol. Biol. Cell 19 546-552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zucchini, C., Rocchi, A., Manara, M. C., De Sanctis, P., Capanni, C., Bianchini, M., Carinci, P., Scotlandi, K., and Valvassori, L. (2008) Int. J. Oncol. 32 17-31 [PubMed] [Google Scholar]
- 73.Meares, G. P., Zmijewska, A. A., and Jope, R. S. (2008) Cell. Signal. 20 347-358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Medigeshi, G. R., Lancaster, A. M., Hirsch, A. J., Briese, T., Lipkin, W. I., Defilippis, V., Fruh, K., Mason, P. W., Nikolich-Zugich, J., and Nelson, J. A. (2007) J. Virol. 81 10849-10860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ito, M., Onuki, R., Bando, Y., Tohyama, M., and Sugiyama, Y. (2007) Biochem. Biophys. Res. Commun. 360 615-620 [DOI] [PubMed] [Google Scholar]
- 76.Langer, R., Feith, M., Siewert, J. R., Wester, H. J., and Hoefler, H. (2008) BMC Cancer 8 1-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hebert, D. N., Foellmer, B., and Helenius, A. (1995) Cell 81 425-433 [DOI] [PubMed] [Google Scholar]
- 78.Okuda-Shimizu, Y., and Hendershot, L. M. (2007) Mol. Cell 28 544-554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wang, I. F., Wu, L. S., Chang, H. Y., and Shen, C. K. (2008) J. Neurochem. 105 797-806 [DOI] [PubMed] [Google Scholar]
- 80.Arai, T., Hasegawa, M., Akiyama, H., Ikeda, K., Nonaka, T., Mori, H., Mann, D., Tsuchiya, K., Yoshida, M., Hashizume, Y., and Oda, T. (2006) Biochem. Biophys. Res. Commun. 351 602-611 [DOI] [PubMed] [Google Scholar]
- 81.Gitcho, M. A., Strider, J., Carter, D., Taylor-Reinwald, L., Forman, M. S., Goate, A. M., and Cairns, N. J. (2008) 6th International Conference on Frontotemporal Dementias, Rotterdam, The Netherlands, September 6, 2008 (abstr.)
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.