Abstract
Intersectin-short (intersectin-s) is a multimodule scaffolding protein functioning in constitutive and regulated forms of endocytosis in non-neuronal cells and in synaptic vesicle (SV) recycling at the neuromuscular junction of Drosophila and Caenorhabditis elegans. In vertebrates, alternative splicing generates a second isoform, intersectin-long (intersectin-l), that contains additional modular domains providing a guanine nucleotide exchange factor activity for Cdc42. In mammals, intersectin-s is expressed in multiple tissues and cells, including glia, but excluded from neurons, whereas intersectin-l is a neuron-specific isoform. Thus, intersectin-I may regulate multiple forms of endocytosis in mammalian neurons, including SV endocytosis. We now report, however, that intersectin-l is localized to somatodendritic regions of cultured hippocampal neurons, with some juxtanuclear accumulation, but is excluded from synaptophysin-labeled axon terminals. Consistently, intersectin-l knockdown (KD) does not affect SV recycling. Instead intersectin-l co-localizes with clathrin heavy chain and adaptor protein 2 in the somatodendritic region of neurons, and its KD reduces the rate of transferrin endocytosis. The protein also co-localizes with F-actin at dendritic spines, and intersectin-l KD disrupts spine maturation during development. Our data indicate that intersectin-l is indeed an important regulator of constitutive endocytosis and neuronal development but that it is not a prominent player in the regulated endocytosis of SVs.
Clathrin-mediated endocytosis (CME)4 is a major mechanism by which cells take up nutrients, control the surface levels of multiple proteins, including ion channels and transporters, and regulate the coupling of signaling receptors to downstream signaling cascades (1-5). In neurons, CME takes on additional specialized roles; it is an important process regulating synaptic vesicle (SV) availability through endocytosis and recycling of SV membranes (6, 7), it shapes synaptic plasticity (8-10), and it is crucial in maintaining synaptic membranes and membrane structure (11).
Numerous endocytic accessory proteins participate in CME, interacting with each other and with core components of the endocytic machinery such as clathrin heavy chain (CHC) and adaptor protein-2 (AP-2) through specific modules and peptide motifs (12). One such module is the Eps15 homology domain that binds to proteins bearing NPF motifs (13, 14). Another is the Src homology 3 (SH3) domain, which binds to proline-rich domains in protein partners (15). Intersectin is a multimodule scaffolding protein that interacts with a wide range of proteins, including several involved in CME (16). Intersectin has two N-terminal Eps15 homology domains that are responsible for binding to epsin, SCAMP1, and numb (17-19), a central coil-coiled domain that interacts with Eps15 and SNAP-23 and -25 (17, 20, 21), and five SH3 domains in its C-terminal region that interact with multiple proline-rich domain proteins, including synaptojanin, dynamin, N-WASP, CdGAP, and mSOS (16, 22-25). The rich binding capability of intersectin has linked it to various functions from CME (17, 26, 27) and signaling (22, 28, 29) to mitogenesis (30, 31) and regulation of the actin cytoskeleton (23).
Intersectin functions in SV recycling at the neuromuscular junction of Drosophila and C. elegans where it acts as a scaffold, regulating the synaptic levels of endocytic accessory proteins (21, 32-34). In vertebrates, the intersectin gene is subject to alternative splicing, and a longer isoform (intersectin-l) is generated that is expressed exclusively in neurons (26, 28, 35, 36). This isoform has all the binding modules of its short (intersectin-s) counterpart but also has additional domains: a DH and a PH domain that provide guanine nucleotide exchange factor (GEF) activity specific for Cdc42 (23, 37) and a C2 domain at the C terminus. Through its GEF activity and binding to actin regulatory proteins, including N-WASP, intersectin-l has been implicated in actin regulation and the development of dendritic spines (19, 23, 24). In addition, because the rest of the binding modules are shared between intersectin-s and -l, it is generally thought that the two intersectin isoforms have the same endocytic functions. In particular, given the well defined role for the invertebrate orthologs of intersectin-s in SV endocytosis, it is thought that intersectin-l performs this role in mammalian neurons, which lack intersectin-s. Defining the complement of intersectin functional activities in mammalian neurons is particularly relevant given that the protein is involved in the pathophysiology of Down syndrome (DS). Specifically, the intersectin gene is localized on chromosome 21q22.2 and is overexpressed in DS brains (38). Interestingly, alterations in endosomal pathways are a hallmark of DS neurons and neurons from the partial trisomy 16 mouse, Ts65Dn, a model for DS (39, 40). Thus, an endocytic trafficking defect may contribute to the DS disease process.
Here, the functional roles of intersectin-l were studied in cultured hippocampal neurons. We find that intersectin-l is localized to the somatodendritic regions of neurons, where it co-localizes with CHC and AP-2 and regulates the uptake of transferrin. Intersectin-l also co-localizes with actin at dendritic spines and disrupting intersectin-l function alters dendritic spine development. In contrast, intersectin-l is absent from presynaptic terminals and has little or no role in SV recycling.
EXPERIMENTAL PROCEDURES
Neuronal Cultures and Transfections—Experiments were performed on primary hippocampal cultures from E18 rats, prepared as described (41) and maintained in culture from between 10 and 21 days in vitro (DIV). Neurons were grown in Neurobasal medium supplemented with penicillin/streptomycin, l-glutamine, B-27, and N-2 supplements (Invitrogen). At 4-7 DIV, neurons were transduced with lentivirus (see below) or transfected using Lipofectamine 2000 (Invitrogen), following the manufacturer's guidelines.
miRNA Vectors and Viruses—Three separate microRNA (miRNA) sequences (intersectin #1, #2, and #3) were generated to target rat intersectin-l. Intersectin #1 and #2 are against regions that are also found in intersectin-s, whereas intersectin #3 is specific to intersectin-l. The sequences target intersectin-l mRNA starting at nucleotides 2085 (intersectin #1), 2205 (intersectin #2), and 5337 (intersectin #3) and are CAAGCCGGAAGTGCAAGACAA, GAGTGCCAAAGTGGTGTATTA, and GGTCTTGTCATGGCTTCTAGA, respectively. A non-targeting miRNA was used for controls, and its sequence is AATTCTCCGAACGTGTCACGT. Oligonucleotides encoding the miRNA sequences were cloned into pcDNA 6.2-GW/EmGFP-miR plasmid (Invitrogen). The EmGFP-miR cassette was then amplified by PCR and subcloned into the pRRLsinPPT vector, and VSVG pseudotyped virus was produced in HEK-293T cells. Virus particles were purified by ultracentrifugation and titered in HEK-293T cells.
Antibodies for Immunofluorescence and Western Blots—Mouse monoclonal antibodies against the following proteins were from the indicated source: intersectin (ESE-1) and AP-1 (BD Bioscience, Franklin Lakes, NJ); PSD95 (mab-045, ABR); synaptophysin (clone SVP38, Sigma); CHC (X22 produced from hybridomas purchased from ATCC); and α-adaptin (AP.6, ABR). F-actin was stained with TRITC-phalloidin (Sigma). Rabbit polyclonal antibodies for intersectin (27) and clathrin light chain (42) have been described previously. For immunofluorescence, fluorophore-conjugated secondary antibodies were from Jackson ImmunoResearch laboratories and Invitrogen. Immunofluorescence images were acquired with a Zeiss LSM 510 Meta confocal inverted microscope using Plan-Apochromat 63× (numerical aperture, 1.4) or 100× (numerical aperture, 1.4) lenses.
Endocytosis Assays—FM4-64 uptake and analysis were performed as previously described (42). For CME of transferrin, neuronal cultures were starved with unsupplemented Neurobasal media overnight prior to the experiment. Alexa-Fluor 546-transferrin (Invitrogen) was diluted to 12.5 μg/ml in Hanks' balanced salt solution (Invitrogen) and was continuously perfused in the imaging chamber during the entire length of live cell imaging. For quantification, regions of interest were drawn to outline the somatodendritic regions of neurons using ImageJ software (NIH). Total fluorescence in the regions of interest was measured and plotted over time. A linear regression was performed on the fluorescence time courses to calculate the rate of uptake. Uptake rates were averaged over multiple experiments as indicated in the legend to Fig. 6.
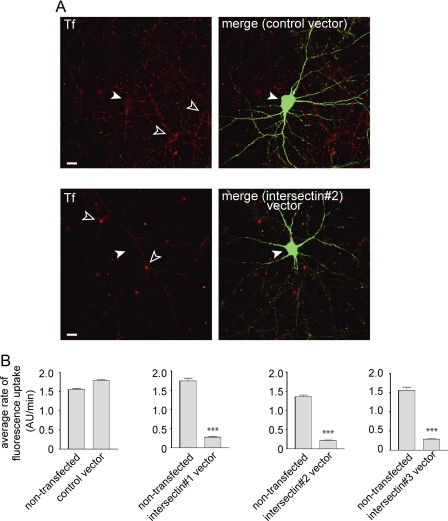
FIGURE 6.
Intersectin KD delays CME of transferrin in hippocampal neurons. Hippocampal neurons were transfected with control vector or intersectin miRNA vectors as indicated. A, transfected cells (green) were identified due to EmGFP expression from the vectors. The neurons were then processed for transferrin (Tf) uptake (red), and images at the end of 30 min of uptake are displayed. Filled arrowheads indicated transfected neurons, and open arrowheads indicate surrounding non-transfected neurons. Movies corresponding to these images are in supplemental Fig. S1. B, the bar graphs indicate the average rate of fluorescent Tf uptake ± S.E. (arbitrary fluorescence units (AU)/min) from a population of neurons coming from at least three separate transfections/cultures. ***, p < 0.001 F-test.
Analysis of Dendritic Spines—Primary hippocampal cultures at 9-11 DIV were transferred to a recording chamber mounted on an upright confocal microscope (DMLFSA, Leica Microsystems, Heidelberg, Germany) with a Leica SP2 scanhead, and cultures were continuously perfused with ∼30 °C Hanks' balanced salt solution. EmGFP-labeled tertiary dendrites were imaged with a Leica 63× (numerical aperture, 0.9) water immersion lens in three-dimensional stacks with voxel dimensions of 61 × 61 × 250-300 nm. Areas of interest were cropped and further processed for deconvolution using Huygens Pro Software (Scientific Volume Imaging, Hilversum, The Netherlands) through a full maximum likelihood extrapolation algorithm. Volume rendering and quantification was carried out using Imaris software (Bitplane, Zurich). Dendritic spines were quantified as previously described (43). Briefly, spines that have short necks but a large, mushroom-shaped head correspond to the “mushroom” class. Spines that have an elongated neck (>1 μm in length) but only a small head correspond to the “long-thin” class. In addition, “filopodia”-type protrusions were typically >3 μm in length, lacking a distinct head, similar to those found during synaptogenesis (44).
RESULTS
Intersectin Knockdown Does Not Affect SV Recycling in Hippocampal Neurons—Intersectin has been shown to be present and to regulate SV recycling at invertebrate neuromuscular synapses (21, 32-34). Because invertebrates only express orthologs of intersectin-s, and intersectin-s is not expressed in mammalian neurons (26, 28, 35, 36), it is possible that intersectin-l fulfills this function in vertebrates including mammals. In fact, in lamprey, which expresses intersectin-l, disruption of intersectin function through injection of antibodies or overexpression of SH3 domains disrupts CME of SVs, although these reagents do not distinguish intersectin-s from intersectin-l (45). To test for a possible role for intersectin-l in SV recycling in mammalian neurons, we first analyzed its localization in hippocampal cultures. When neurons expressing monomeric red fluorescent protein to reveal cell morphology were stained for intersectin, the protein was found in puncta that were distributed in neuronal processes and throughout the cell body (Fig. 1A). Double labeling immunofluorescence with an antibody against synaptophysin, a marker of axonal boutons revealed little or no co-localization of intersectin with synaptophysin (Fig. 1, A and B). This result is in fact entirely consistent with the recent observation of Nishimura et al. (19) that GFP-intersectin-l transfected in hippocampal neurons does not co-localize with synaptophysin. Thus, intersectin does not appear to be a major component of pre-synaptic boutons but is instead localized to somatodendritic regions.
FIGURE 1.
Intersectin is absent from presynaptic nerve terminals. A: top panels, confocal fluorescent images of a representative cultured hippocampal neuron transfected with monomeric red fluorescent protein (mRFP) to reveal its morphology (red) and immunostained for intersectin (green) and synaptophysin (blue). Scale bar is 15 μm. Bottom panels, higher magnification images from the fields indicated by white boxes in the upper panels. Note that intersectin immunoreactivity is in dendritic branches (yellow) and is largely absent from synaptic boutons labeled by synaptophysin (arrowheads). Scale bars are 5 μm. B: top panels, confocal fluorescent images of representative dendrites immunostained for intersectin (green) and synaptophysin (red). Bottom panel, higher magnification image from the field indicated by the white box in the upper panels. Scale bars are 5 μm.
Recently, Yu et al. (36) demonstrated that intersectin null mice display a subtle but significant slowing of SV endocytosis. Thus, despite the fact there is little or no intersectin expression in the presynaptic compartment of hippocampal neurons, we sought to examine for possible defects in CME of SVs using a loss-of-function approach. To this end, we took advantage of a lentiviral system that we have developed to deliver inhibitory miRNA into post-mitotic cells, including neurons.5 We used three different miRNA sequences to KD intersectin-l in neurons. Two of the miRNAs (intersectin #1 and #2) target sequences that are common with intersectin-s, whereas a third miRNA (intersectin #3) targets a sequence specific to intersectin-l. Neuronal cultures transduced with the various intersectin targeted viruses or a control virus were lysed and processed for Western blot analysis. Intersectin #1- and #2-encoding viruses caused a decrease in the levels of both intersectin-s (∼140 kDa) and intersectin-l (∼190 kDa) as detected with two different antibodies (Fig. 2A). Intersectin #3 miRNA caused a selective decrease in the level of intersectin-l with no affect on the level of intersectin-s coming from glia (Fig. 2A). The levels of the multiple variants of clathrin light chains serve as loading controls and are equivalent between samples.
FIGURE 2.
Intersectin KD in hippocampal neurons. A, lysates of hippocampal neuron cultures transduced with control lentivirus or with virus producing miRNAs targeting both intersectin-s and intersectin-l (#1 and #2) or intersectin-l specifically (#3) were blotted with two different antibodies (polyclonal and monoclonal as indicated) recognizing the two intersectin variants. An antibody that recognizes clathrin light chains (CLC) a and b and their respective splice variants reveals equal loading. Similar results were obtained from four different cultures. B, confocal fluorescent images of representative fields of cultured hippocampal neurons transduced with different lentiviruses as indicated. All viruses express EmGFP as a reporter. The cells were stained with antibody specific for intersectin (red). Scale bar is 15 μm. C, efficient KD of intersectin expression was also achieved through transfection of miRNA-encoding plasmid vectors. Note the weak intersectin immunoreactivity (red) of transfected neurons (green, the vectors express EmGFP as a reporter) when compared with surrounding, non-transfected cells. Scale bar is 15 μm.
Intersectin KD was also readily detectable by immunofluorescence where images from cultures treated with different miRNA-encoding viruses and control virus were taken with the same microscope settings (Fig. 2B). Alternatively, we used transfection of miRNA-encoding plasmids. Neurons transfected with the various intersectin miRNAs showed reduced intersectin immunofluorescence staining (Fig. 2C). These data demonstrate the effectiveness of the intersectin KD, and, importantly, they indicate that the immunofluorescence signal seen with the intersectin antibody is specific for intersectin. We also observed that intersectin miRNAs targeting the region of intersectin-l common with intersectin-s were able to eliminate intersectin staining in glia, whereas the intersectin-l-specific miRNA did not affect the levels of intersectin in glial cells (data not shown). Thus, the intersectin miRNA-encoding reagents are effective for intersectin KD.
We measured SV endocytosis with the membrane dye FM4-64 as previously described (42). When comparing FM4-64 uptake in axons of transfected neurons with their non-transfected neighbors, we found no difference in axonal bouton fluorescence intensity upon intersectin KD (Fig. 3A). Averaging the amount of FM4-64 uptake in hundreds of boutons from multiple cultures/transfections revealed no significant effect of intersectin KD, regardless of which intersectin miRNA was used (Fig. 3B). Our data demonstrate that intersectin is not present in axonal boutons and that it does not appear to function in CME of SVs in mammalian hippocampal neurons.
FIGURE 3.
SV retrieval is normal following intersectin KD. A, confocal images of axonal processes of cultured hippocampal neurons transfected with control vector or vector expressing intersectin #1 miRNA. Both vectors express EmGFP as a reporter. The neurons were processed for FM4-64 uptake (red). Scale bars are 5 μm. B, bar graph summarizing FM4-64 uptake following transfection with the indicated vectors. The data were accumulated from three separate cultures/transfections (N = 3) and the total number of boutons analyzed (n) is indicated.
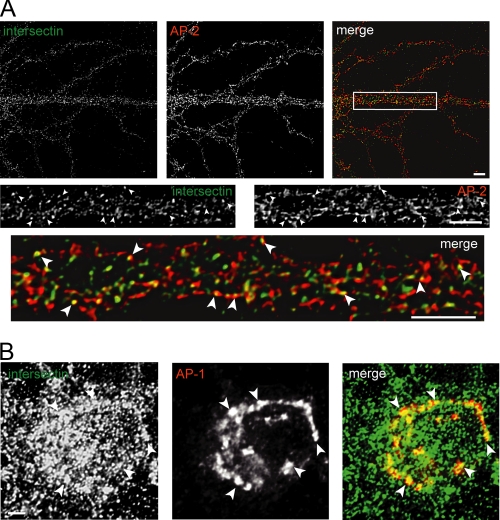
Intersectin Co-localizes with Markers of Clathrin-coated Structures in the Soma and Dendrites—Despite its seeming lack of function in SV endocytosis, intersectin could still participate in endocytic events in neurons. To examine this possibility, we studied the localization of intersectin in relationship to CHC, the central component of clathrin-coated pits and clathrin-coated vesicles. In neurons, CHC is localized to clathrin-coated structures that form at the trans-Golgi network (TGN) and the plasma membrane in somatodendritic regions. CHC is also found in synaptic boutons, were it co-localizes with synaptophysin and other pre-synaptic markers. Intersectin was found to partially co-localize with CHC in dendrites (Fig. 4) suggesting that a pool of the protein is present on endocytic clathrin-coated pits. Consistent with this notion we detected co-localization of intersectin with AP-2 in dendrites (Fig. 5A). Because, like CHC, a significant pool of AP-2 in neurons is found in pre-synaptic boutons, the CHC and AP-2 puncta that are negative for intersectin likely represent, to a large part, the pre-synaptic component. We also noted a pool of intersectin in the juxtanuclear area (Figs. 4 and 5). This pool of intersectin was also found to co-localize with CHC (Fig. 4), suggesting that intersectin is present on clathrin-coated structures at the TGN. Consistent with this notion, the juxtanuclear pool of intersectin was seen to co-localize in part with adaptor protein-1 (AP-1), a component and marker of TGN-derived clathrin-coated pits (Fig. 5B). Little co-localization was seen with GM130, a marker of the cis-Golgi (data not shown). Therefore, intersectin localizes to multiple clathrin-coated membranes in the somatodendritic region of neurons.
FIGURE 4.
Intersectin co-localizes with CHC in somatodendritic regions. Top panels, confocal fluorescent images of a representative cultured hippocampal neuron immunostained for intersectin (green) and CHC (red). Scale bar is 15 μm. Bottom six panels, higher magnification images from the fields indicated by the white boxes in the upper panels. Arrowheads indicate co-localizing structures in dendritic branches (a) and in the cell soma (b). Scale bars are 5 μm.
FIGURE 5.
Intersectin co-localizes with clathrin adaptor proteins. A: top panels, confocal fluorescent images of a dendritic region of a representative cultured hippocampal neuron immunostained for intersectin (green) and AP-2 (red). Middle and lower panels, higher magnification images from the field indicated by the white box in the upper panels. Examples of co-localizing structures are indicated by arrowheads. Scale bars are 5 μm. B: confocal fluorescent images of the cell body of a representative cultured hippocampal neuron immunostained for intersectin (green) and AP-1 (red). Scale bar is 10 μm.
Intersectin Functions in CME of Transferrin—Given the somatodendritic localization of intersectin-l to clathrin/AP-2-positive clathrin-coated pits, we sought to examine if it contributes to constitutive endocytosis in neurons. Constitutive endocytosis of transferrin occurs exclusively in dendrites and the soma of cultured hippocampal neurons (46). We thus used live-cell time-lapse monitoring of Alexa-Fluor 546-labeled transferrin to examine this CME pathway in cultures transfected with control and intersectin miRNA-encoding plasmids. Intersectin KD caused a marked reduction in the rate of transferrin uptake when monitored over a period of 30-50 min, and similar results were seen using all of the intersectin miRNAs (Fig. 6, supplemental Fig. S1, and supplemental movies S1 and S2). A reduction in CME of transferrin was also seen when neurons were transduced with lentivirus encoding the miRNAs (supplemental Fig. 2). Thus, intersectin functions in constitutive CME of transferrin in neurons.
Intersectin Co-localizes with F-actin in Dendritic Spines—Intersectin-l is a GEF for Cdc42. In vitro, intersectin-l stimulates the formation of branched actin filaments by activating Cdc42 with subsequent downstream activation of N-WASP (23). F-actin is enriched in dendritic spines, and in many cases, the intersectin staining that we observe in dendritic shafts appears to progress into dendritic spines. We thus sought to determine if intersectin co-localizes with actin. Interestingly, intersectin immunoreactivity was found to co-localize with assembled actin as determined by staining with phalloidin (Fig. 7A). Co-localization was seen both in dendrites and in a juxtanuclear pool (Fig. 7A). It is unclear if the juxtanuclear corresponds to intersectin at the TGN. To further examine the potential localization of intersectin to spines, we performed co-staining with the post-synaptic marker PSD95, an important scaffold protein that organizes the different components of the post-synaptic machinery (47). Neurons were transfected with EmGFP to reveal neuronal morphology and then co-stained for intersectin and PSD95. As expected, PSD95 was generally found at the distal ends of the spines (Fig. 7B). In many cases, intersectin was also localized within the spines, and in other cases, it was found in the dendritic shaft at the base of spines (Fig. 7B). Triple-labeling experiments further highlighted this observation. F-actin was found throughout the spines, PSD95 was co-localized with F-actin primarily in the distal part of the spine, and intersectin was co-localized with F-actin at the more proximal part of the spine (Fig. 7C). Thus, in addition to its role in CME, intersectin is likely to have a role in the regulation of actin assembly in spines.
FIGURE 7.
Intersectin co-localizes with F-actin in dendritic spines. A, confocal fluorescent image of a representative cultured hippocampal neuron immunostained for intersectin (green) with F-actin detected by phalloidin (red). Scale bar is 15 μm. The panels on the right and below are magnified images from the fields indicated by the white boxes in the main panel. Scale bars on the magnified images are 10 μm. Examples of co-localizing structures are indicated by arrowheads. B, confocal fluorescent images of a representative cultured hippocampal neuron transfected with EmGFP to reveal its morphology (green) and immunostained for intersectin (red) and PSD95 (blue). Scale bar is 15 μm. The lower panel is a magnified image from the field indicated by the white box in the upper panels. Arrowheads indicate examples of PSD95 immunoreactivity in dendritic spines. In several cases, the spines also contain intersectin immunoreactivity although intersectin is often found at the base of the spines. Scale bar is 5 μm. C, confocal fluorescent images of a dendritic region immunostained for intersectin (green) and PSD95 (blue) with F-actin detected by phalloidin (red). Scale bar is 1 μm.
Intersectin Regulates Spine Maturation—Overexpression and dominant-negative studies in neurons have implicated intersectin-l in the regulation of spine development (19, 24). We thus sought to directly examine for a role for intersectin in spine maturation using our loss-of-function approach. Neurons transfected with control or intersectin miRNA encoding vectors were imaged in three-dimensional stacks. Resulting stacks were deconvolved using Huygens Pro Software and were subjected to surface rendering using Imaris software. When compared with neurons transfected with control vector, neurons in which intersectin had been knocked down appeared to have an increased number of long, thin, and immature looking filopodia-type protrusions and fewer spines that appeared morphologically mature (Fig. 8A). Quantification of numerous images from multiple cultures/transductions revealed a significant increase in filopodia-type protrusions and a significant decrease in mushroom spines in intersectin KD neurons (Fig. 8B). There was also a slight decrease in the number of long-thin type spines, but this was not statistically significant (Fig. 8B). Thus, it appears that loss of intersectin function influences the maturation of dendritic spines.
FIGURE 8.
Intersectin KD disrupts the development of dendritic spines. A, surface rendering of confocal z-stack images of representative dendritic regions of hippocampal neurons transfected with control or intersectin #3 miRNA vectors. Both vectors express EmGFP as a reporter. The images were surfaced rendered with Imaris software, and examples of filopodia-type protrusions (filled arrowheads), mushroom spines (open arrowheads), and long-thin spines (arrows), as defined under “Experimental Procedures” are indicated. Scale bars are 2 μm. B, bar graph summarizing the number of processes per 10-μm length of dendrite, separated into the indicated classes from control versus intersectin #3 transfected neurons. The data were accumulated from 18 dendritic branches from 5 separate cultures/transfections. *, p < 0.05 and **, p < 0.01 Student's one-tailed t test.
DISCUSSION
In Drosophila, dynamin-associated protein of 160 kDa (Dap160) is a four SH3-domain containing ortholog of intersectin-s that localizes to the neuromuscular junction (32, 33). Both temperature-sensitive (32) and loss-of-function mutants (33) have been used to examine its function. Dap160 mutants cause alterations in synaptic morphology, decreased synaptic levels of other endocytic accessory proteins such as dynamin and synaptojanin, and reductions in SV endocytosis measured with FM dyes (32, 33). In C. elegans, intersectin-s is expressed throughout the nervous system and at the neuromuscular junction, and intersectin-s mutants exhibit reduced frequency of miniature end-plate synaptic currents (21). Mutants also exhibit synaptic structure defects that are typically caused by disruptions in SV recycling (21). These data strongly support a role for intersectin-s in SV recycling in invertebrates. It is thus interesting that intersectin-s is not expressed in mammalian neurons (26, 28, 35, 36), and as reported here, intersectin-l KD has no influence on SV endocytosis. Although this result may seem surprising, it is in agreement with our observation that there is little or no intersectin in synaptic boutons of hippocampal neurons and the observation of Nishimura et al. (19) that overexpressed GFP-intersectin-l fails to localize to pre-synaptic boutons in mammalian neurons but instead shows somatodendritic localization. Moreover, such a situation is not without precedent. For example, in mammalian neurons, amphiphysin is enriched in pre-synaptic nerve terminals where it functions in SV endocytosis, in part through recruiting dynamin and synaptojanin to synaptic sites (48, 49). In contrast, in Drosophila, amphiphysin variants lack CHC- and AP-2-binding motifs, the protein is predominantly post-synaptic, and it does not function in SV endocytosis (50-52). It is thus interesting to speculate that the role played by intersectin as a pre-synaptic scaffold for SV endocytosis in invertebrates is played by amphiphysin in mammalian systems. However, it must be noted that while intersectin does not function in SV endocytosis in hippocampal neurons, we have not formally ruled out a pre-synaptic trafficking role for the protein in mammalian neuromuscular junctions.
Yu et al. (36) recently reported studies on intersectin null mice. Using the pH-sensitive SV marker synaptophluorin (53) in cultured neurons from the mice, they reported a small decrease in SV endocytosis in a narrow time window between 10 and 20 s of stimulation in the nulls when compared with the wild type (36). It is possible that this effect is caused by the loss of a pool of intersectin-l that is present at low levels in synapses and is thus undetectable in our studies. The relatively subtle alterations recorded by Yu et al. (36) would likely be below the level of detection of FM4-64 assays. However, given that intersectin-l is present in the post-synaptic compartment and is required for normal development of dendritic spines (Refs. 19,24 and our current results), it is also possible that the effects on SV endocytosis reflect indirect changes in pre-synaptic function occurring from developmental defects occurring post-synaptically. Further studies will be required to address these issues.
The first evidence of a role for intersectin in CME came from transferrin uptake in cells lines (17, 27). In the studies of Simpson et al. (27) CME of transferrin was inhibited following expression of individual SH3 domains and in Sengar et al. (17), inhibition of transferrin endocytosis was observed following overexpression of the full-length protein. Given that both dominant-negative and gain-of-function approaches lead to alterations in transferrin uptake, it is important to examine this crucial endocytic event in a loss-of-function paradigm. Moreover, a potential role for intersectin-l in CME of transferrin in neurons has not been previously examined. This is critical, because intersectin has been implicated in DS, and alterations in early endosomes have been reported as a hallmark feature of DS neurons (38-40). In fact, in the study of Yu et al. (36), which analyzed neurons from intersectin null animals, whereas there was a small decrease in SV endocytosis, the most striking phenotype observed was the enlargement (swelling) of endosomes in the cell body of neurons. Such a change could result from alterations in somatodendritic endocytosis. In the present study, we demonstrate that a pool of intersectin-l is co-localized with AP-2 and CHC. Because there is little or no intersectin-l detectable in the pre-synaptic compartment where AP-2 and CHC are enriched, the co-localization of intersectin-l with these proteins is modest. However, the pool that does co-localize most likely represents intersectin-l associated with plasma membrane-derived clathrin-coated structures in the somatodendritic region. Consistently, intersectin-l KD leads to a significant decrease in the rate of CME of transferrin. Coupled with previous studies demonstrating that KD of intersectin-s in COS-7 cells leads to defects in epidermal growth factor receptor endocytosis (54), these data clearly demonstrate an endocytic function for intersectin family members in constitutive and regulated forms of CME. Interestingly, we also detected a pool of intersectin co-localizing with CHC and AP-1 at the TGN of hippocampal neurons. Future studies will determine if intersectin has a role in clathrin-mediated budding at the TGN and will also examine the contribution of clathrin-mediated trafficking to the pathophysiology of DS.
An important binding partner for intersectin is N-WASP. We previously reported that N-WASP binds through its proline-rich domain to various SH3 domains of intersectin, notably SH3A and SH3E (23). We also demonstrated that, in the case of intersectin-l, N-WASP binding was required to activate the GEF activity of the full-length protein toward Cdc42 (23). This was subsequently confirmed, and it was shown that binding of proline-rich domains to the SH3 domains of intersectin disrupts an intramolecular interaction between the GEF-containing tail and the adjacent SH3 domain region, creating an open conformation in which the DH domain has access to Cdc42 (55). Furthermore, Irie and Yamaguchi (24) demonstrated that the ephrin receptor EphB2 binds to intersectin-l and stimulates its GEF activity in cooperation with N-WASP. Activation of Cdc42 by intersectin-l subsequently leads to N-WASP activation and stimulation of branched actin filaments (23). Importantly, in hippocampal neurons at 9 DIV, disruption of the interaction between intersectin-l and N-WASP, with a corresponding decrease in GEF activity, leads to disruption of spine development (24). Notably in the Irie and Yamaguchi study (24) there was a significant increase in immature protrusions >2 μm in length and a significant decrease in spines >0.5 μm in width. This is very similar to what we observe here following intersectin-l KD, specifically that there is an increase in long, thin filopodia-like protrusions and a decrease in more mature, mushroom spines. Dendritic spines are composed of a meshwork of branched actin filaments (56). Interestingly, a recent study demonstrates that N-WASP is a critical regulator of actin in the development of dendritic spines and that N-WASP KD leads to a decrease in the number of spines in rat hippocampal neurons (57). Thus, intersectin-l, through activation of Cdc42 and N-WASP appears critical for the development of dendritic spines. In this regard, it is interesting to note that abnormalities in dendritic spines are another hallmark feature of neurons form DS brains (58). Interesting avenues for the future will be to better understand the role of intersectin in the pathophysiology of DS.
Supplementary Material
Acknowledgments
We thank Jacynthe Philie for excellent technical assistance.
This work was supported by Grants MOP-15396 and MOP-86724 from the Canadian Institutes of Health Research (to P. S. M. and R. A. M.), respectively, and from Natural Sciences and Engineering Rgpin 341942-07 (to R. A. M.).
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2 and Movies S1 and S2.
Footnotes
The abbreviations used are: CME, clathrin-mediated endocytosis; SV, synaptic vesicle; CHC, clathrin heavy chain; AP-2, adaptor protein-2; SH3, Src homology 3; intersectin-l, intersectin-long; intersectin-s, intersectin-short; GEF, guanine nucleotide exchange factor; DS, Down syndrome; DIV, days in vitro; miRNA, microRNA; TRITC, tetramethylrhodamine isothiocyanate; KD, knockdown; TGN, trans-Golgi network; EmGFP, emerald green fluorescent protein.
B. Ritter and P.S. McPherson, unpublished results.
References
- 1.Kobayashi, T., Gu, F., and Gruenberg, J. (1998) Semin. Cell Dev. Biol. 9 517-526 [DOI] [PubMed] [Google Scholar]
- 2.Conner, S. D., and Schmid, S. L. (2003) Nature 422 37-44 [DOI] [PubMed] [Google Scholar]
- 3.Moos, T., Rosengren Nielsen, T., Skjorringe, T., and Morgan, E. H. (2007) J. Neurochem. 103 1730-1740 [DOI] [PubMed] [Google Scholar]
- 4.von Zastrow, M., and Sorkin, A. (2007) Curr. Opin. Cell Biol. 19 436-445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hanyaloglu, A. C., and von Zastrow, M. (2008) Annu. Rev. Pharmacol. Toxicol. 48 537-568 [DOI] [PubMed] [Google Scholar]
- 6.Sara, Y., Mozhayeva, M. G., Liu, X., and Kavalali, E. T. (2002) J. Neurosci. 22 1608-1617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mani, M., Lee, S. Y., Lucast, L., Cremona, O., Di Paolo, G., De Camilli, P., and Ryan, T. A. (2007) Neuron 56 1004-1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu, L. G., and Betz, W. J. (1998) Biophys. J. 74 3003-3009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Evergren, E., Zotova, E., Brodin, L., and Shupliakov, O. (2006) Neuroscience 141 123-131 [DOI] [PubMed] [Google Scholar]
- 10.Kavalali, E. T. (2007) J. Physiol. 585 669-679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Poodry, C. A., and Edgar, L. (1979) J. Cell Biol. 81 520-527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McPherson, P. S., and Ritter, B. (2005) Mol. Neurobiol. 32 73-87 [DOI] [PubMed] [Google Scholar]
- 13.Salcini, A. E., Confalonieri, S., Doria, M., Santolini, E., Tassi, E., Minenkova, O., Cesareni, G., Pelicci, P. G., and Di Fiore, P. P. (1997) Genes Dev. 11 2239-2249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Montesinos, M. L., Castellano-Munoz, M., Garcia-Junco-Clemente, P., and Fernandez-Chacon, R. (2005) Brain Res. 49 416-428 [DOI] [PubMed] [Google Scholar]
- 15.McPherson, P. S. (1999) Cell Signal. 11 229-238 [DOI] [PubMed] [Google Scholar]
- 16.Yamabhai, M., Hoffman, N. G., Hardison, N. L., McPherson, P. S., Castagnoli, L., Cesareni, G., and Kay, B. K. (1998) J. Biol. Chem. 273 31401-31407 [DOI] [PubMed] [Google Scholar]
- 17.Sengar, A. S., Wang, W., Bishay, J., Cohen, S., and Egan, S. E. (1999) EMBO J. 18 1159-1171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fernandez-Chacon, R., Achiriloaie, M., Janz, R., Albanesi, J. P., and Sudhof, T. C. (2000) J. Biol. Chem. 275 12752-12756 [DOI] [PubMed] [Google Scholar]
- 19.Nishimura, T., Yamaguchi, T., Tokunaga, A., Hara, A., Hamaguchi, T., Kato, K., Iwamatsu, A., Okano, H., and Kaibuchi, K. (2006) Mol. Biol. Cell 17 1273-1285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Okamoto, M., Schoch, S., and Sudhof, T. C. (1999) J. Biol. Chem. 274 18446-18454 [DOI] [PubMed] [Google Scholar]
- 21.Wang, W., Bouhours, M., Gracheva, E. O., Liao, E. H., Xu, K., Sengar, A. S., Xin, X., Roder, J., Boone, C., Richmond, J. E., Zhen, M., and Egan, S. E. (2008) Traffic 9 742-754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tong, X. K., Hussain, N. K., de Heuvel, E., Kurakin, A., Abi-Jaoude, E., Quinn, C. C., Olson, M. F., Marais, R., Baranes, D., Kay, B. K., and McPherson, P. S. (2000) EMBO J. 19 1263-1271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hussain, N. K., Jenna, S., Glogauer, M., Quinn, C. C., Wasiak, S., Guipponi, M., Antonarakis, S. E., Kay, B. K., Stossel, T. P., Lamarche-Vane, N., and McPherson, P. S. (2001) Nat. Cell Biol. 3 927-932 [DOI] [PubMed] [Google Scholar]
- 24.Irie, F., and Yamaguchi, Y. (2002) Nat. Neurosci. 5 1117-1118 [DOI] [PubMed] [Google Scholar]
- 25.Jenna, S., Hussain, N. K., Danek, E. I., Triki, I., Wasiak, S., McPherson, P. S., and Lamarche-Vane, N. (2002) J. Biol. Chem. 277 6366-6373 [DOI] [PubMed] [Google Scholar]
- 26.Hussain, N. K., Yamabhai, M., Ramjaun, A. R., Guy, A. M., Baranes, D., O'Bryan, J. P., Der, C. J., Kay, B. K., and McPherson, P. S. (1999) J. Biol. Chem. 274 15671-15677 [DOI] [PubMed] [Google Scholar]
- 27.Simpson, F., Hussain, N. K., Qualmann, B., Kelly, R. B., Kay, B. K., McPherson, P. S., and Schmid, S. L. (1999) Nat. Cell Biol. 1 119-124 [DOI] [PubMed] [Google Scholar]
- 28.Ma, Y. J., Okamoto, M., Gu, F., Obata, K., Matsuyama, T., Desaki, J., Tanaka, J., and Sakanaka, M. (2003) J. Neurosci. Res. 71 468-477 [DOI] [PubMed] [Google Scholar]
- 29.Mohney, R. P., Das, M., Bivona, T. G., Hanes, R., Adams, A. G., Philips, M. R., and O'Bryan, J. P. (2003) J. Biol. Chem. 278 47038-47045 [DOI] [PubMed] [Google Scholar]
- 30.Adams, A., Thorn, J. M., Yamabhai, M., Kay, B. K., and O'Bryan, J. P. (2000) J. Biol. Chem. 275 27414-27420 [DOI] [PubMed] [Google Scholar]
- 31.O'Bryan, J. P., Mohney, R. P., and Oldham, C. E. (2001) Oncogene 20 6300-6308 [DOI] [PubMed] [Google Scholar]
- 32.Koh, T. W., Verstreken, P., and Bellen, H. J. (2004) Neuron 43 193-205 [DOI] [PubMed] [Google Scholar]
- 33.Marie, B., Sweeney, S. T., Poskanzer, K. E., Roos, J., Kelly, R. B., and Davis, G. W. (2004) Neuron 43 207-219 [DOI] [PubMed] [Google Scholar]
- 34.Rose, S., Malabarba, M. G., Krag, C., Schultz, A., Tsushima, H., Di Fiore, P. P., and Salcini, A. E. (2007) Mol. Biol. Cell 18 5091-5099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tsyba, L., Skrypkina, I., Rynditch, A., Nikolaienko, O., Ferenets, G., Fortna, A., and Gardiner, K. (2004) Genomics 84 106-113 [DOI] [PubMed] [Google Scholar]
- 36.Yu, Y., Chu, P. Y., Bowser, D. N., Keating, D. J., Dubach, D., Harper, I., Tkalcevic, J., Finkelstein, D. I., and Pritchard, M. A. (2008) Hum. Mol. Genet. 17 3281-3290 [DOI] [PubMed] [Google Scholar]
- 37.Snyder, J. T., Worthylake, D. K., Rossman, K. L., Betts, L., Pruitt, W. M., Siderovski, D. P., Der, C. J., and Sondek, J. (2002) Nat. Struct. Biol. 9 468-475 [DOI] [PubMed] [Google Scholar]
- 38.Pucharcós, C., Fuentes, J. J., Casas, C., de la Luna, S., Alcántara, S., Arbonés, M. L., Soriano, E., Estivill, X., and Pritchard, M. (1999) Eur. J. Hum. Genet. 7 704-712 [DOI] [PubMed] [Google Scholar]
- 39.Cataldo, A. M., Peterhoff, C. M., Troncoso, J. C., Gomez-Isla, T., Hyman, B. T., and Nixon, R. A. (2000) Am. J. Pathol. 157 277-286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cataldo, A. M., Petanceska, S., Peterhoff, C. M., Terio, N. B., Epstein, C. J., Villar, A., Carlson, E. J., Staufenbiel, M., and Nixon, R. A. (2003) J. Neurosci. 23 6788-6792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Burman, J. L., Wasiak, S., Ritter, B., de Heuvel, E., and McPherson, P. S. (2005) FEBS Lett. 579 2177-2184 [DOI] [PubMed] [Google Scholar]
- 42.Allaire, P. D., Ritter, B., Thomas, S., Burman, J. L., Denisov, A. Y., Legendre-Guillemin, V., Harper, S. Q., Davidson, B. L., Gehring, K., and McPherson, P. S. (2006) J. Neurosci. 26 13202-13212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McKinney, R. A., Capogna, M., Dürr, R., Gähwiler, B. H., and Thompson, S. M. (1999) Nat. Neurosci. 2 44-49 [DOI] [PubMed] [Google Scholar]
- 44.Fiala, J. C., Feinberg, M., Popov, V., and Harris, K. M. (1998) J. Neurosci. 18 8900-8911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Evergren, E., Gad, H., Walther, K., Sundborger, A., Tomilin, N., and Shupliakov, O. (2007) J. Neurosci. 27 379-390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mundigl, O., Matteoli, M., Daniell, L., Thomas-Reetz, A., Metcalf, A., Jahn, R., and De Camilli, P. (1993) J. Cell Biol. 122 1207-1221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nagano, T., Jourdi, H., and Nawa, H. (1998) J. Biochem. 124 869-875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ramjaun, A. R., Micheva, K. D., Bouchelet, I., and McPherson, P. S. (1997) J. Biol. Chem. 272 16700-16706 [DOI] [PubMed] [Google Scholar]
- 49.Di Paolo, G., Sankaranarayanan, S., Wenk, M. R., Daniell, L., Perucco, E., Caldarone, B. J., Flavell, R., Picciotto, M. R., Ryan, T. A., Cremona, O., and De Camilli, P. (2002) Neuron 33 789-804 [DOI] [PubMed] [Google Scholar]
- 50.Leventis, P. A., Chow, B. M., Stewart, B. A., Iyengar, B., Campos, A. R., and Boulianne, G. L. (2001) Traffic 2 839-850 [DOI] [PubMed] [Google Scholar]
- 51.Zelhof, A. C., Bao, H., Hardy, R. W., Razzaq, A., Zhang, B., and Doe, C. Q. (2001) Development 128 5005-5015 [DOI] [PubMed] [Google Scholar]
- 52.Razzaq, A., Robinson, I. M., McMahon, H. T., Skepper, J. N., Su, Y., Zelhof, A. C., Jackson, A. P., Gay, N. J., and O'Kane, C. J. (2001) Genes Dev. 15 2967-2979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Burrone, J., Li, Z., and Murthy, V. N. (2006) Nat. Protoc. 1 2970-2978 [DOI] [PubMed] [Google Scholar]
- 54.Martin, N. P., Mohney, R. P., Dunn, S., Das, M., Scappini, E., and O'Bryan, J. P. (2006) Mol. Pharmacol. 70 1643-1653 [DOI] [PubMed] [Google Scholar]
- 55.Zamanian, J. L., and Kelly, R. B. (2003) Mol. Biol. Cell 14 1624-1637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Landis, D. M., and Reese, T. S. (1983) J. Cell Biol. 97 1169-1178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wegner, A. M., Nebhan, C. A., Hu, L., Majumdar, D., Meier, K. M., Weaver, A. M., and Webb, D. J. (2008) J. Biol. Chem. 283 15912-15920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fiala, J. C., Spacek, J., and Harris, K. M. (2002) Brain Res. Brain Res. Rev. 39 29-54 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.