Abstract
Clathrin-dependent endocytosis is mediated by a tightly regulated network of molecular interactions that provides essential protein-protein and protein-lipid binding activities. Here we report the hydrolysis of the α- and β2-subunits of the tetrameric adaptor protein complex 2 by calpain. Calcium-dependent α- and β2-adaptin hydrolysis was observed in several rat tissues, including brain and primary neuronal cultures. Neuronal α- and β2-adaptin cleavage was inducible by glutamate stimulation and was accompanied by the decreased endocytosis of transferrin. Heterologous expression of truncated forms of the β2-adaptin subunit significantly decreased the membrane recruitment of clathrin and inhibited clathrin-mediated receptor endocytosis. Moreover, the presence of truncated β2-adaptin sensitized neurons to glutamate receptor-mediated excitotoxicity. Proteolysis of α- and β2-adaptins, as well as the accessory clathrin adaptors epsin 1, adaptor protein 180, and the clathrin assembly lymphoid myeloid leukemia protein, was detected in brain tissues after experimentally induced ischemia and in cases of human Alzheimer disease. The present study further clarifies the central role of calpain in regulating clathrin-dependent endocytosis and provides evidence for a novel mechanism through which calpain activation may promote neurodegeneration: the sensitization of cells to glutamate-mediated excitotoxicity via the decreased internalization of surface receptors.
Cells must internalize necessary macromolecules from their surfaces and the extracellular space; this trafficking into inner compartments achieves transmembrane signaling functions as well as maintaining cellular homeostasis. Clathrin-dependent endocytosis is an important cargo internalization mechanism involved in a variety of cellular signaling and transport pathways, such as those involving epidermal growth factor or transferrin, respectively. Clathrin adaptor proteins play a central role in this process by recognizing specific cargo and facilitating the formation of clathrin-coated vesicles. The adaptor protein complex 2 (AP-2)3 is considered to be the central hub in this interaction network that links clathrin, accessory clathrin adaptors, other endocytic accessory factors, membrane lipids, and membrane-associated cargo proteins (1, 2).
The tetrameric AP-2 complex is composed of α-, β2-, μ2-, and σ-subunits (3, 4). Its structure is characterized by a trunk domain and two appendages that are connected to the trunk by flexible hinges. The trunk domain interacts with phosphatidylinositol phosphates and cargo (5), whereas the appendage domains recruit alternate adaptors and accessory proteins (6, 7). AP-2 has two clathrin binding sites, one each on the hinge and appendage domains of the β2-subunit; both binding sites contribute to clathrin recruitment (8).
The AP-2 core adaptor complex is important for the overall capacity of clathrin-dependent endocytosis and is crucial for the internalization of particular cargoes such as the transferrin receptor (9). In neurons, AP-2-dependent trafficking of N-methyl-d-aspartic acid- and α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid-type ionotropic glutamate receptor proteins has been reported to be an important determinant of synaptic strength and plasticity (10-13).
Calpains comprise a widely expressed family of calcium-dependent cysteine proteases. Strict determinants of calpain substrate utilization have been elusive, due to the fact that they are governed by structural constraints rather than primary sequence motifs (14). Known substrates for calpains include proteins of widely diverse functions, including membrane receptors, ion channels and transporters, enzymes, cytoskeletal proteins, and transcription factors. Thus, calpains are implicated in a diverse variety of normal and pathologic cellular processes, including signal transduction, cell motility, gene expression, and apoptotic and necrotic cell death (reviewed in Ref. 15). In the nervous system, calpain activation has been observed in both acute and chronic neurodegenerative conditions (16-20). Abnormal calpain activation can occur under these circumstances through increases in cytosolic calcium due to various causes, including changes in calcium homeostasis, the accumulation of disease-causing proteins, or the increased activation of NMDA receptors (16, 21). In many of these conditions, inhibition of calpain is neuroprotective (16, 20, 22).
Here we report that the α- and β2-subunits of the AP-2 clathrin adaptor complex are subject to calpain proteolysis. This cleavage separates the appendage domains from the adaptor trunk and results in decreased clathrin-dependent endocytosis. A significant fraction of this decrease can be attributed specifically to truncation of the β2-subunit, and incorporation of truncated β2-adaptin into AP-2 complexes can both decrease endocytosis and sensitize neurons to excitotoxicity. We also demonstrate calpain-mediated cleavage of the accessory clathrin adaptors clathrin assembly lymphoid myeloid leukemia protein (CALM), epsin 1, and adaptor protein 180 (AP180). Although the present studies are focused primarily on calpain proteolysis of clathrin adaptors in the nervous system, these findings are likely to be representative of a more widespread phenomenon that also occurs in other cell types.
EXPERIMENTAL PROCEDURES
Clathrin Adaptor Expression Vectors—cDNAs encoding full-length rat β2-adaptin (with C-terminal FLAG-, 6×-histidine-, or hemagglutinin (HA) tags), N-terminal or C-terminal rat β2-adaptin fragments (amino acids 1-691, with no tag or C-terminal HA tag; amino acids 692-951, with C-terminal 6×-histidine or HA tags), and human AP180 (with an N-terminal HA tag) were created using PCRs with specific primers (β2-adaptin: GenBank™ NM_001030006, nucleotides 190-3045; AP180: GenBank™ NM_014841, nucleotides 244-2967) to which the protein tag-encoding sequences were added. The resultant cDNAs were then cloned into self-inactivating lentiviral transfer vector plasmids under the control of the mouse phosphoglycerate kinase 1 (PGK) promoter (SIN-W-PGK) (23) or the tetracycline response element (TRE) promoter (SIN-W-TRE) using the pENTR/D-TOPO Cloning Kit (Invitrogen) and the LR recombination reaction (Gateway™ system, Invitrogen), and lentiviral vectors were produced as described previously (24).
To generate Δ692-694 and Δ688-697 deletion mutants of β2-adaptin with 6×-histidine tags, we used site-directed mutagenesis with complementary internal primers and two-step PCR. The first step consisted of producing a partial 5′ cDNA using a forward wild-type primer and a mutagenic reverse primer and a partial 3′ cDNA using a mutagenic forward primer and a wild-type reverse primer in two separate PCR reactions. The second step consisted of combining the obtained 5′ and 3′ cDNA fragments (which overlapped in sequence where the mutagenic primers were introduced) and amplifying the mutated full-length cDNA by PCR with wild-type primers.
Primary Cortical and Striatal Neuron Cultures—Primary neuronal cultures were prepared and maintained as described previously (postnatal day 2 cortical cultures according to (25) and embryonic day 16 striatal cultures per (26)). For lentiviral-mediated protein expression, cultures were infected on DIV 1 (50 ng of p24/ml of culture medium for each vector), half the medium was replaced on DIV 4, and half of the medium was replaced weekly thereafter. SIN-W-TRE expression vectors were applied at a concentration of 25 ng of p24/ml of culture medium together with the tetracycline transactivator in the SIN-W-PGK vector (50 ng of p24/ml of culture medium). Neuronal expression of the adaptin transproteins was ∼1× and 3× that of endogenous levels under the control of the PGK and TRE promoters, respectively.
Glutamate Stimulation of Cultured Neurons—At DIV 11-13, striatal and cortical neurons were exposed for 15 min to 100 μm glutamate in medium supplemented with 50 μm glycine and 2.7 mm CaCl2. Following glutamate treatment, cells were washed twice and incubated in Neurobasal medium for the indicated times. Where specified, cells were exposed to the NMDA-receptor antagonist MK801, calpain inhibitors ALLN, calpain inhibitor IV (benzyloxycarbonyl-LLY-fluoromethyl ketone (Z-LLY-FMK)), or PD 151746 (all purchased from Calbiochem/Merck) starting 1 h prior to glutamate stimulation, and these inhibitors were included in all subsequent manipulations.
Protein Extraction and Immunoblotting—Except where otherwise noted, protein samples for proteolytic analyses were harvested in lysis buffer (comprised of 20 mm HEPES, pH 7.4, 1% Triton X-100, 100 mm KCl, 0.1 mm dithiothreitol, and protease inhibitor mixture (Sigma-Aldrich catalog no. P8340), and supplemented with either 2 mm EDTA plus 2 mm EGTA or 2 mm CaCl2, to prevent or allow calpain digestion, respectively. Lysates were incubated on ice for 30 min and vortexed every 10 min. Protein extracts were cleared of cell debris by centrifugation at 20,000 × g for 15 min at 4 °C, separated by SDS-PAGE on a 12% polyacrylamide gel, and analyzed by immunoblotting.
Recombinant calpain treatments of rat brain or HEK 293T cell extracts consisted of exposure to recombinant rat calpain 2 (11 μg/ml, Calbiochem/Merck) and 2 mm CaCl2 for 1 h at room temperature. Calpain was inactivated by addition of 4 mm EDTA plus 4 mm EGTA.
The following primary antibodies were used for protein immunodetection: anti-β2-adaptin (sc-6425), anti-α-adaptin (immunoreactive to αA-adaptin; sc-6421), anti-CALM (sc-5395), and anti-epsin 1 (sc-8673) goat polyclonal antibodies from Santa Cruz Biotechnology; mouse monoclonal anti-α-adaptin (immunoreactive to αA- and αC-adaptin; NB300-721) from Novus Biologicals; anti-clathrin heavy chain (610499) and anti-dynamin 2 (610263) mouse monoclonal antibodies from BD Transduction Laboratories; anti-AP180 (A4825), anti-α-tubulin (T5168), and anti-FLAG M2 (F1804) mouse monoclonal antibodies from Sigma-Aldrich; anti-hemagglutinin mouse monoclonal antibody from Covance; rabbit polyclonal anti-histidine antibody (PA1-23022) from Affinity BioReagents; rabbit polyclonal anti-α-tubulin antibody (ab4074) from Abcam; and anti-Na+/K+ ATPase mouse monoclonal antibody from the Developmental Studies Hybridoma Bank. Secondary antibodies consisted of Alexa Fluor 680-conjugated polyclonal goat anti-mouse or goat anti-rabbit IgG or rabbit anti-goat (Invitrogen) or IRDye 800CW-conjugated donkey anti-mouse IgG (LI-COR). Protein bands were visualized using the Odyssey Infrared Imaging System (LI-COR).
Identification of the Calpain Cleavage Site in β2-Adaptin by Edman Degradation—HEK 293T cells were transfected with SIN-W-PGK plasmid vector encoding FLAG-tagged β2-adaptin using CaPO4 transfection. Cells were harvested 24 h post-transfection in chelator-free lysis buffer, and lysates were treated with rat recombinant calpain 2 plus 2 mm CaCl2 for 1 h at room temperature. The FLAG-tagged C-terminal proteolytic fragment of β2-adaptin was then immunoprecipitated using anti-FLAG M2 Affinity Gel (Sigma-Aldrich) according to the manufacturer's instructions. Precipitated proteins were then separated by SDS-PAGE and transferred to an Invitrolon polyvinylidene difluoride membrane and stained with Ponceau S. The C-terminal β2-adaptin fragment band (25 kDa) was cut from the membrane and subjected to sequencing by Edman degradation at the Functional Genomics Center Zurich (Zurich, Switzerland) on a commercial basis.
Transferrin and Dextran Internalization Assays—Neuronal cultures grown on poly-l-lysine-coated glass coverslips were used for transferrin internalization assay on DIV 11-13. Where indicated, cells were pretreated with 25 μm ALLN for 1 h, and the same concentration of inhibitor was added to all subsequent reagents. Cells were stimulated with glutamate (as described above) and incubated in B27-supplement-free Neurobasal medium for 1 h. Cells were then exposed to Alexa Fluor 488-conjugated transferrin (Invitrogen) at a concentration of 5 μg/ml. To allow transferrin internalization, cells were incubated at 37C for 1 h. Samples were then placed on ice, and uninternalized transferrin was stripped from the cell surface by incubation in 0.2 m acetic acid (pH 2.8) plus 0.5 m NaCl for 5 min. Cells were then fixed with 4% paraformaldehyde in PBS (pH 7.4) on ice for 15 min. After washing, coverslips were mounted with Immunomount (Thermo Shandon) and analyzed on a TCS-SP2 AOBS confocal laser-scanning microscope (Leica Microsystems). In some experiments, uptake of Rhodamine B isothiocyanate-dextran (average molecular mass 10 kDa, Sigma-Aldrich, 5 μg/ml) was assayed in parallel using the same procedure. Signals were analyzed in 52-210 neurons per condition from five independent cultures.
Immunoprecipitation of α-Adaptin—Mouse monoclonal antibodies to α-adaptin (Novus Biologicals) were conjugated to Protein G-Sepharose 4 Fast Flow beads (Amersham Biosciences) and used as described previously (27) to immunoprecipitate α-adaptin-containing protein complexes from HEK 293T cells transfected with SIN-W-TRE plasmid vectors encoding either full-length or N-terminal and C-terminal fragments of rat β2-adaptin, all with C-terminal HA tags. Immunoprecipitated protein complexes were analyzed by immunoblotting.
Cellular Fractionation Studies—Monolayer cultures of HEK 293T cells overexpressing full-length rat β2-adaptin or a combination of N-terminal and C-terminal rat β2-adaptin fragments were washed twice with PBS (pH 7.4) and harvested in 50 mm HEPES (pH 7.4). Cells were lysed by 15 passages through a 25-gauge needle, and precleared by a 15-min centrifugation at 3,000 × g. After removal of an aliquot for total protein analysis, lysates were subjected to ultracentrifugation at 100,000 × g for 1 h at 4 °C. The pellet of the 100,000 × g centrifugation (comprising the membrane fraction) was resuspended in 50 mm HEPES (pH 7.4) and analyzed by immunoblotting together with its corresponding total protein extract.
Immunostaining of Primary Cortical Neurons—Primary cortical neurons cultured on glass coverslips were fixed on DIV 12 with ice-cold 4% paraformaldehyde in PBS (pH 7.4) for 10 min and blocked with 0.1% bovine serum albumin in PBS for 20 min. A rabbit polyclonal antiserum against clathrin heavy chain (Cat. 2419, Cell Signaling) was diluted in PBS with 0.5 mg/ml saponin and applied to the cells for 1 h at room temperature. Alexa Fluor 488-conjugated polyclonal donkey anti-rabbit IgG (Invitrogen) was used as a secondary antibody. Coverslips were mounted with FluoroSafe (Calbiochem) and analyzed on a TCS-SP2 AOBS confocal laser-scanning microscope (Leica Microsystems).
Lactate Dehydrogenase Assay—Measurements of lactate dehydrogenase release into the culture medium were performed using the CytoTox 96 Non-Radioactive Cytotoxicity Assay (Promega) as described in a previous study (25).
Neuronal Survival Assessed by NeuN-positive Cell Counting—DIV 11 striatal neurons were exposed for 15 min to 50 μm glutamate in culture medium supplemented with 50 μm glycine and 2.7 mm CaCl2. Following glutamate treatment, cells were washed twice and incubated in fresh Neurobasal medium for 48 h. Cell cultures were then fixed and immunostained with anti-NeuN antibody (1/400, Chemicon) as described previously (26) followed by analysis with a (BD Pathway 855 Bioimager, BD Biosciences).
Rat Tissues and Rat Ischemia Model—All animal experiments were carried out in accordance with the European Community directive for the care and use of laboratory animals (86/-609/-EEC) and the Swiss Academy of Medical Science and were authorized by the veterinary office of the Canton of Vaud. For multitissue analyses of AP-2 subunit proteolysis, adult female Wistar rats (180-200 g, Iffa Credo/Charles River) were sacrificed by sodium pentobarbital overdose and transcardially perfused with phosphate buffer. Organ tissues were removed and homogenized in lysis buffer plus 2 mm CaCl2 or 2 mm EGTA by mechanical disruption.
Focal ischemia was induced in 12-day-old male Sprague-Dawley rats according to (28). Animals were decapitated 24 h after the end of ischemia, and the infarcted and contralateral control tissues were dissected in PBS with 1 mm MgCl2 on ice. Tissues were then homogenized rapidly in ice-cold lysis buffer (20 mm HEPES, pH 7.4, 10 mm NaCl, 3 mm MgCl2, 2.5 mm EGTA, 0.1 mm dithiothreitol, 50 mm NaF, 1 mm Na3VO4, 1% Triton X-100, and a protease inhibitor mixture (Roche Applied Science)) using a manual potter apparatus. Homogenates were subsequently sonicated and then centrifuged at 750 × g for 5 min at 4 °C. Supernatants were collected, and protein concentrations were determined by the Bradford method (Bio-Rad Protein Assay). Samples were stored at -80 °C until use.
Human Brain Samples—Neurodegenerative changes were staged using biochemical means. Tau staging on a scale from 0 to 10 was determined by immunoblot quantification of Tau pathology as described in a previous study (29); controls were limited to Tau stages 3 and 4 (tauopathy restricted to the hippocampal area), whereas AD samples were stage 10 (reflecting global tauopathy of all neocortical areas). Amyloidopathy staging was assessed in the neocortex on a scale from 0 to 10 as described before (30); amyloid stage was 0 in controls (no trace of amyloid deposition), whereas AD samples exhibited stages 7-9 (reflecting a substantial quantity of Aβ aggregates). Frozen samples from two control brains and five AD brains were homogenized in SDS-PAGE loading buffer.
Statistical Analysis—Numerical data represent the mean values ± S.E. The one-way analysis of variance was used to compare multiple conditions, and the two-tailed Student's t test was used for two-group comparisons. p < 0.05 was set as the threshold for statistical significance.
RESULTS
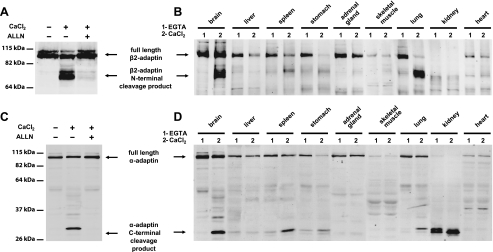
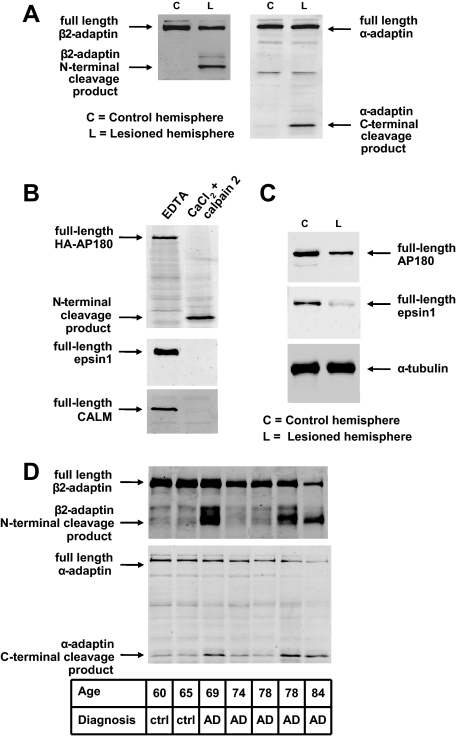
Calcium-dependent Proteolysis of α- and β2-Adaptins in Brain and Other Rat Tissues—In the context of assessing potential calcium-dependent protein interactions that modulate synaptic plasticity (12), we noted a calcium-dependent change in the electrophoretic mobility of the major anti-N-terminal β2-adaptin-immunoreactive protein species in rat brain extracts. In the presence of EDTA or EGTA, the major β2-adaptin protein species was visualized at the position expected for this 100-kDa protein, whereas a more rapidly migrating band (corresponding to an approximate molecular mass of 75 kDa) was observed in the presence of exogenous calcium (2 mm) (Fig. 1A). A similar phenomenon was observed in immunoblots for anti-C-terminal α-adaptin immunoreactive species, whose apparent migration changed from a position corresponding to a 100-kDa protein to that of a 25-kDa protein (also corresponding to the expected and a faster-migrating species, respectively (Fig. 1C)). To explore whether this phenomenon also occurred in non-brain tissues, we subjected samples of peripheral organs to a similar analysis. This examination revealed the same calcium-dependent effects in spleen, stomach, and lung (Fig. 1, B and D). Upon noting these results, we reasoned that the strong calcium dependence and the large apparent change in electrophoretic migration of these proteins might be explained by calpain-mediated proteolysis. We therefore examined whether a broad spectrum calpain inhibitor (ALLN) would prevent this apparent modification of AP-2 proteins. The change in the electrophoretic migration of AP-2 proteins was indeed prevented by ALLN, indicating calpain cleavage of both α- and β2-adaptins (Fig. 1, A and C).
FIGURE 1.
Cleavage of the β2- and α-subunits of the AP-2 complex in rat tissues. A, immunoblot analysis of rat brain homogenates with an antiserum that recognizes an N-terminal epitope of β2-adaptin. A novel molecular species consistent with a molecular mass of 75 kDa appears in rat brain extracts in the presence of CaCl2, the appearance of this band is blocked by the broad spectrum calpain inhibitor ALLN. B, immunoblot analysis of various rat tissue homogenates shows a calcium-dependent change in migration of the β2-adaptin immunoreactive species in brain, spleen, lung, and stomach. C, immunoblot analysis of rat brain homogenates with antiserum recognizing a C-terminal epitope of α-adaptin. A molecular species consistent with a molecular mass of 30 kDa appears in the rat brain protein extract when CaCl2 is present in the lysis buffer. This change in migration is blocked by the broad spectrum calpain inhibitor ALLN. D, immunoblot analysis of rat tissue homogenates using anti-C-terminal-α-adaptin antiserum shows a calcium-dependent change in migration of the immunoreactive species in samples of brain, spleen, lung, and stomach.
Proteolysis of Adaptins by Calpain 2—To establish the relevance of this proteolytic event in living cells, we tested whether activation of calpains by neuronal stimulation would also result in adaptin hydrolysis. 15-min exposures of primary striatal neurons to glutamate (Fig. 2, A and B) or the selective ionotropic glutamate receptor agonist NMDA (data not shown) also resulted in calcium-dependent and ALLN-sensitive α- and β2-adaptin hydrolysis. Glutamate-stimulated hydrolysis was sensitive to the NMDA receptor antagonist MK801 and EGTA, showing a major dependence on NMDA receptors and extracellular calcium (Fig. 2, A and B).
FIGURE 2.
Cleavage of β2- and α-subunits of AP-2 in cultured striatal neurons. A and B, C-terminally 6×-histidine-tagged β2-adaptin-expressing primary neurons were stimulated with 100 μm glutamate for 15 min in the presence of 2.7 mm CaCl2 (or 3 mm EGTA) in combination with calpain 1 inhibitor PD 151746 (25 μm), calpain 2 inhibitor Z-LLY-FMK (10 μm), or NMDA-receptor antagonist MK801 (10 μm). Protein extracts were collected 1 h after the end of the glutamate application and subjected to immunoblotting with antibodies against the 6×-histidine tag (A) or α-adaptin (B). C, C-terminally 6×-histidine-tagged β2-adaptin expressed in HEK 293T cells is degraded in the same manner when exposed to purified calpain 1 and calpain 2, but not caspase 3. D, α-adaptin is degraded in the same manner when exposed to purified calpain 2.
To examine whether the hydrolysis of adaptins was mediated directly by calpains, we exposed adaptins to recombinant enzymes. Whereas caspase-3 was unable to proteolyze calpain, proteolytic fragments of β2-adaptin were produced after exposure to either calpain-1 or calpain-2 (Fig. 2C). To assess the relevant calpain(s) activated by stimulation in neuronal cells, we determined whether specific calpain inhibitors would prevent the calcium-dependent adaptor cleavage in vivo. Whereas the calpain 1 inhibitor PD 151746 had no effect on glutamate-stimulated neuronal adaptin proteolysis (Fig. 2, A and B), the calpain 2 inhibitor Z-LLY-FMK prevented this hydrolysis. Moreover, the protein species produced by neuronal stimulation show identical SDS-PAGE migration characteristics to those observed when α- and β2-adaptins were subjected to recombinant calpain 2 treatment in vitro (Fig. 2, C and D). Although these results do not rule out the possibility of adaptin hydrolysis by other calpain isoforms, the data indicate that calpain 2 activation mediates the direct in vivo hydrolysis of α- and β2-adaptins in neuronal cells.
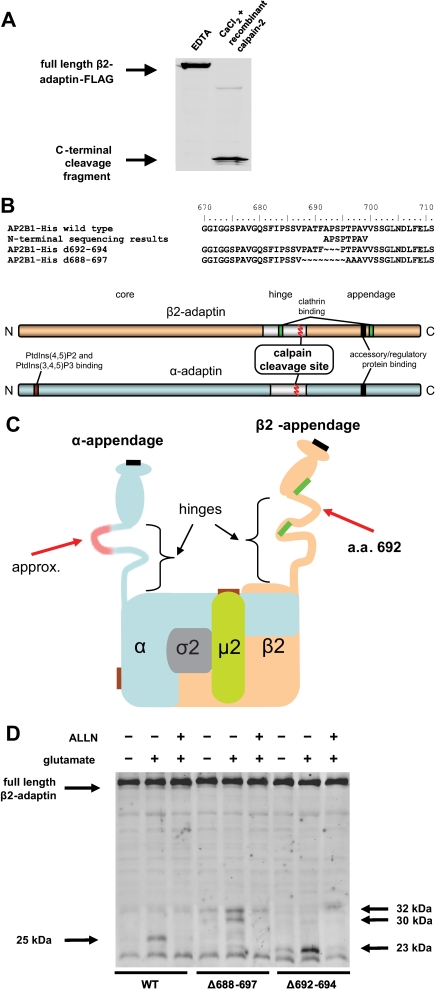
Identification of the Calpain Cleavage Sites in the Hinge Regions of the Adaptor Subunits—To facilitate the determination of its endogenous calpain cleavage site in β2-adaptin, a C-terminally FLAG-tagged expression construct was produced. Both glutamate-stimulated neuronal proteolysis and in vitro calpain 2 proteolysis of the tagged protein resulted in the formation of the expected 25-kDa product (complementary to the 75-kDa N-terminal fragment previously observed in immunoblot experiments) (Fig. 3A). This C-terminal β2-adaptin fragment was subsequently purified and subjected to Edman degradation. Results of this N-terminal sequencing identified the calpain 2 cleavage site as the peptide bond between phenylalanine 691 and alanine 692 (Fig. 3B). In confirmation of this result, deletion mutations of β2-adaptin lacking amino acids 692-694 or 688-697 abrogated the formation of the 25-kDa cleavage product of calpain proteolysis of wild-type β2-adaptin (Fig. 3D). However, ALLN-sensitive cleavage of β2-adaptin was nonetheless observed at alternate sites. Both the endogenous and alternate cleavage-susceptible sites are positioned in the hinge region of the AP-2 β2-subunit, with the endogenous cleavage position occurring between two previously defined clathrin binding sites (8) (Fig. 3C). Although parallel experiments were unable to determine the precise cleavage site in the AP-2 α-subunit because of a cellular contaminant in the recombinant fragment purification, the location predicted by the N- and C-terminal proteolysis fragments (Figs. 1C,1D,2B, and 2D) together with deletion mutagenesis studies (data not shown) also map the calpain cleavage site in α-adaptin to its hinge region (Fig. 3). Thus, cleavage of the β2- and α-subunits of the AP-2 complex by calpain occurs such that a clathrin binding site and two accessory adaptor-binding domains become separated from the AP-2 core.
FIGURE 3.
Sites of calpain cleavage in AP-2. A, protein extracts from HEK 293T cells expressing β2-adaptin with C-terminal FLAG tag were treated with recombinant rat calpain 2 in presence of 2 mm CaCl2 and subjected to immunoblotting with an anti-FLAG tag antibody. B, text shows the alignment of amino acid sequences of wild-type β2-adaptin, protein sequencing results for the N terminus of its C-terminal calpain proteolytic fragment, and Δ692-694 and experimental Δ688-697 deletion mutants. The scheme represents the linearized domain structures of β2-adaptin (orange) and α-adaptin (blue). C, schematic representation of the heterotetrameric AP-2 complex, depicting binding sites for clathrin (bright green), clathrin accessory factors (black), and phosphatidylinositol phosphates (brown), as determined in previous studies (5-8). Red arrows indicate the approximate positions of calpain cleavage sites. D, cleavage of β2-adaptin at Ala-692 is abrogated by deletion mutations. Cultured striatal neurons were infected with lentiviral vectors encoding wild-type β2-adaptin, Δ692-694 or Δ688-697 deletion mutants, all with C-terminal 6×-histidine tags. Neurons were stimulated with 100 μm glutamate plus 2.7 mm CaCl2 for 15 min. Protein was extracted 1 h after the end of the glutamate stimulation, and the extracts were subjected to immunoblotting with an anti-6×-histidine antibody. The expected 25-kDa cleavage product of wild-type β2-adaptin is not observed with the deletion mutants (although both deletion mutants are partially cleaved by a calpain-like activity at alternate sites, as indicated by arrows at right); cleavage of both wild-type and mutant adaptors is blocked by the addition of the calpain inhibitor ALLN. These data are consistent with the presence of at least three potential calpain cleavage sites: the physiological one (described in Fig. 2) and two alternate sites that are utilized only after mutation of the endogenous proteolytic site. These findings are not unexpected given that calpain hydrolysis is governed largely by structural rather than primary sequence determinants (14).
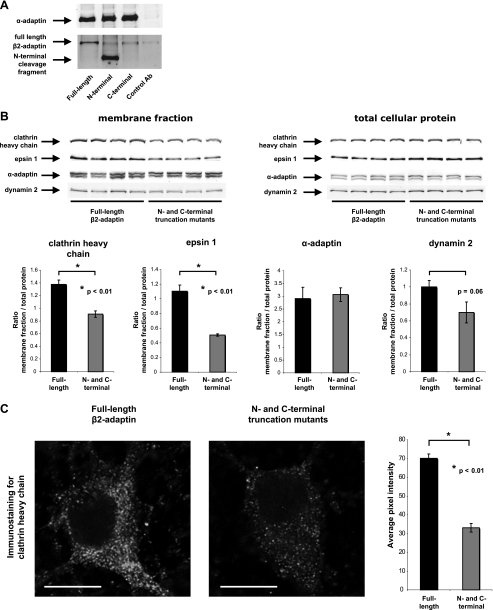
β2-Adaptin Cleavage Decreases Clathrin Recruitment to Membranes—Because we were unable to create a cleavage-resistant form of β2-adaptin through conservative mutations in the endogenous cleavage site (Fig. 3D), we assessed the consequences of calpain proteolysis by examining the effects of cleavage-mimicking β2-adaptin mutants. Heterologously expressed N-terminal fragment and wild-type β2-adaptin proteins were incorporated normally into AP-2 complexes (Fig. 4A).
FIGURE 4.
Effects of β2-adaptin cleavage products on the membrane distribution of endocytosis-related proteins. A, an antibody against α-adaptin was used to immunoprecipitate AP-2-containing protein complexes from HEK 293T cells overexpressing wild-type β2-adaptin or β2-adaptin mutants comprising the N- or C-terminal products of calpain cleavage. Immunoblotting of the precipitates with an antibody against the N-terminal portion of β2-adaptin shows that the heterologously expressed N-terminal truncation mutant is present in the precipitated AP-2 complexes. B, membrane fractions and total lysates of HEK 293T cells transfected with plasmid vectors encoding wild-type β2-adaptin or N- and C-terminal truncation mutants were subjected to immunoblotting with antibodies against the clathrin heavy chain, dynamin 2, α-adaptin, and epsin 1. Fluorescence intensities of protein immunodetection in the cellular membrane fractions were quantified and normalized to the intensity of the corresponding bands in parallel samples of total cellular protein. Four samples per condition were analyzed. The amounts of pelletable clathrin heavy chain and epsin 1 were significantly lower in the cells overexpressing N- and C-terminal truncation mutants of rat β2-adaptin than in the cells overexpressing wild-type rat β2-adaptin. Dynamin 2 also showed a non-significant trend toward a decrease in the membrane fractions of the truncation mutant-expressing cells. The amount of membrane-associated α-adaptin was not changed by coexpression of N- and C-terminal β2-adaptin truncation mutants. C, primary cultured cortical neurons were transduced with a lentiviral encoding wild-type β2-adaptin or a combination of vectors encoding N- and C-terminal truncation mutants and immunostained with an antibody against clathrin heavy chain. For each cell, the immunofluorescence intensity was quantified in a 2-μm2 area at the visible limit of the cell border. β2-Adaptin truncation mutant-expressing cells demonstrated a significant decrease in clathrin immunostaining as compared with controls expressing wild-type β2-adaptin. Average pixel intensities were quantified from seven wild-type- and six truncation mutant-expressing cells. Bar = 10 μm.
To determine the specific contribution of β2-subunit proteolysis to the plasma membrane recruitment of clathrin and other endocytic proteins, we examined the effects of cleavage-mimicking mutants on their association with cellular membranes by biochemical fractionation. Expression of N- and C-terminal truncation products resulted in a significant decrease in membrane recruitment of clathrin heavy chain and epsin 1 (Fig. 4B). In addition, a non-significant trend toward decreased membrane recruitment was also observed for dynamin 2 (Fig. 4B). In contrast, the plasma membrane recruitment of α-adaptin was unchanged, consistent with the predominance of the direct lipid binding activity of AP-2.
We also used immunofluorescence to analyze the ability of the calpain-derived fragments of AP-2 to decrease the association of clathrin with the plasma membrane (Fig. 4C). As expected, plasma membrane compartmentalization of clathrin was decreased in neurons expressing N- and C-terminal truncation mutants compared with neurons expressing full-length β2-adaptin. These data indicate that β2-adaptin cleavage decreases the ability of AP-2 to recruit clathrin and other endocytic proteins to cellular membranes.
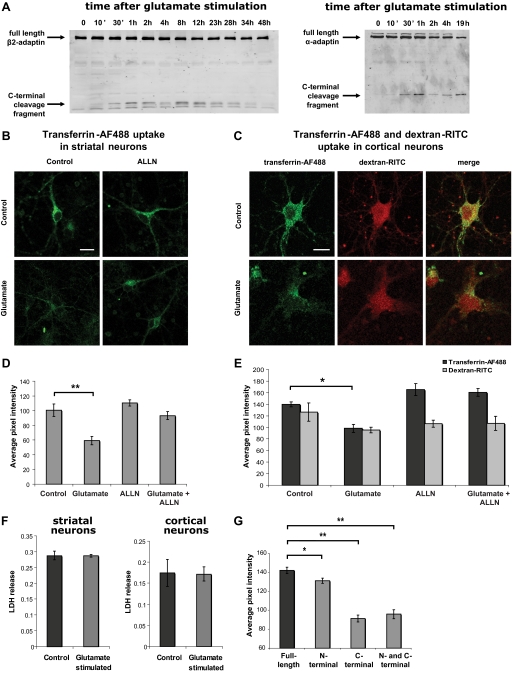
Cleavage of Adaptins Is Paralleled by a Decrease in Receptor-mediated Endocytosis—To determine the functional consequences of adaptin cleavage by calpain, we assessed the effects of adaptor hydrolysis in primary neuronal cultures. Calpain-proteolyzed adaptin fragments were visible within 30 min after glutamate stimulation and persisted for at least 48 h (Fig. 5A). Adaptin cleavage in glutamate-stimulated striatal or cortical primary neuron cultures was paralleled by a significant calpain-dependent decrease in transferrin endocytosis (Fig. 5, B-E). In contrast, dextran uptake, an indicator of total fluid-phase endocytosis, was not decreased under these conditions (Fig. 5, C and E). Moreover, lactate dehydrogenase release was also unchanged, solidly ruling out neuronal cell death as an alternative explanation for decreased transferrin uptake (Fig. 5F). These results show that AP-2-dependent endocytosis is diminished in conjunction with adaptin hydrolysis by calpain 2 in living neurons.
FIGURE 5.
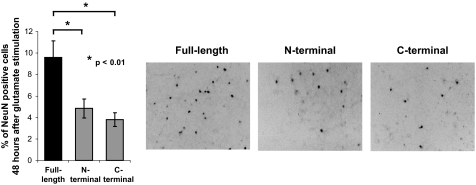
Transferrin uptake in neurons after AP-2 subunit truncation or proteolysis. A, time course of β2- and α-adaptin cleavage after glutamate stimulation of striatal neurons. Striatal neurons were monitored for cleavage of C-terminal 6×-histidine-tagged β2-adaptin and wild-type α-adaptin cleavage after stimulation with 100 μm glutamate for 15 min in the presence of 2.7 mm CaCl2. Protein was extracted at the indicated time points after the end of the glutamate stimulation and the extracts were subjected to immunoblotting with anti-histidine tag or anti-α-adaptin antibodies, respectively. Cleavage of adaptins was observed within 30 min after neuronal stimulation. Cleavage, both β2- and α-adaptin, was also observed within 30 min in primary cultured cortical neurons (data not shown). B and D, uptake of Alexa Fluor 488-conjugated transferrin (transferrin-AF488) by striatal neurons was assessed 1 h after a 15-min stimulation with 100 μm glutamate as measured by confocal microscopy (representative images shown in the top panels). Transferrin-AF488 uptake was quantified from the confocal plane with the highest overall AF488 intensity by calculating the average pixel intensity for an area of ∼4 μm2 within the limits of the neuronal plasma membrane but excluding the neuronal nucleus. The following numbers of cells were analyzed for each condition: 210 control, 142 glutamate, 181 ALLN, 176 glutamate plus ALLN. Transferrin-AF488 uptake was significantly reduced in glutamate-treated samples, whereas the calpain inhibitor ALLN restored transferrin uptake to the basal level. Bar = 10 μm. C and E, decreased transferrin uptake was also observed in cortical neurons after glutamate-stimulated calpain activation and AP-2 proteolysis. Transferrin-AF488 and dextran-Rhodamine B isothiocyanate (dextran-RITC) uptake was quantified as for striatal cells with analysis of the following numbers of cells per group: 54 control, 54 glutamate, 52 ALLN, 61 glutamate plus ALLN. Uptake of the fluid-phase endocytosis indicator dextran-RITC (10 kDa) into the same compartment was not significantly altered by glutamate stimulation or ALLN treatment. Bar = 10 μm. F, no cell death was detected in striatal or cortical neurons within 1 h of glutamate stimulation, as analyzed by lactate dehydrogenase release into the culture medium. G, expression of N-terminal, C-terminal, or both truncation mutants of β2-adaptin via lentiviral vectors recapitulates the calpain-mediated effect on transferrin endocytosis. Transferrin uptake by neurons expressing mutants comprising the N- and C-terminal products of calpain cleavage were compared with the parallel lentiviral-induced expression of full-length wild-type β2-adaptin. The following numbers of cells were analyzed for each condition: 196 wild-type, 260 N-terminal alone, 79 C-terminal alone, and 105 N- and C-terminal combined. *, p < 0.01; **, p < 0.005.
Calpain Cleavage-mimicking Truncation Mutants of β2-Adaptin Decrease AP-2-dependent Endocytosis and Sensitize Neurons to Glutamate-mediated Excitotoxicity—To confirm the specific role of adaptin cleavage in regulating endocytosis, we examined the neuronal effects of the hydrolysis-mimicking truncation mutants of β2-adaptin. Lentiviral-mediated expression of N- and C-terminal fragments of β2-adaptin resulted in significant decreases in transferrin uptake in primary neurons as compared with parallel expression of wild-type β2-adaptin (Fig. 5G). Comparative analyses of the individual fragments showed that the effect of the C-terminal appendage domain predominated (Fig. 5G). These results indicate that adaptin cleavage contributes substantially to the effect of calpain on clathrin-dependent endocytosis.
Because calpain activation is known to occur in neurodegenerative diseases, we considered the possible functional consequences of a calpain-mediated decrease in AP-2 activity in this context. We hypothesized that one such effect might be to decrease the internalization of glutamate receptors, thereby sensitizing cells to glutamatergic excitotoxicity. We therefore tested this hypothesis by examining how cells expressing β2-adaptin truncation mutants responded to an excitotoxic stimulus. As shown in Fig. 6, these experiments demonstrated a significant effect on cell viability, with only half as many neurons overexpressing truncated β2-adaptin subunits surviving as compared with cells overexpressing full-length β2-adaptin. These data provide evidence that calpain-mediated hydrolysis of the AP-2 complex may contribute significantly to neuronal degeneration.
FIGURE 6.
β2-Adaptin truncation sensitizes neurons to excitotoxicity. Lentiviral expression of N- or C-terminal truncation mutants of β2-adaptin sensitizes striatal neurons to excitotoxicity, as compared with parallel expression of full-length wild-type β2-adaptin. The left bar graph shows % remaining NeuN-positive neuronal cells remaining 48 h after excitotoxic stimulation (compared with unstimulated cells). Right panels show representative fields of remaining NeuN-positive cells (appearing as dark profiles) in each of the three conditions.
Cleavage of Core and Accessory Clathrin Adaptors in Brain via Endogenous Activation of Calpains—To determine whether calpain cleavage of clathrin adaptors could also be detected in brain in situations where endogenous calpain activation is present, we examined adaptor hydrolysis in two such conditions. Because ischemia-reperfusion injury is known to cause increases in free intracellular calcium and to activate calpains (31), we examined α- and β2-adaptin hydrolysis in the ischemic rat brain, implementing a model developed by Renolleau et al. (28). Immunoblot analysis of ischemic and control brain samples indeed revealed specific ischemia-induced cleavage of β2- and α-adaptins (Fig. 7A). Moreover, protein extracts of ischemic brain and/or in vitro exposure to recombinant calpain 2 showed hydrolysis of additional clathrin adaptors: CALM, AP180, and epsin 1 (Fig. 7, B and C).
FIGURE 7.
Neurodegeneration-related cleavage of clathrin adaptors. A, immunoblotting shows that β2- and α-subunits of AP-2 are cleaved within 24 h after reperfusion in a rat transient cerebral ischemia model. B, accessory adaptors AP180, epsin 1, and CALM are also substrates of calpain 2. Lysates of HEK 293T cells overexpressing HA-tagged AP180 protein (top panel) or untransfected HEK293T cells (bottom two panels) were exposed to recombinant rat calpain 2 plus 2 mm CaCl2 and subjected to immunoblotting with antibodies against the HA tag, epsin 1, or CALM, respectively. C, immunoblotting with antibodies against AP180 and epsin 1 shows that these adaptors are proteolyzed in a rat transient cerebral ischemia model. D, immunoblots of human prefrontal cortex (Brodmann area 9) detect proteolysis of β2- and α-adaptins in three of five AD samples.
Chronic neurodegeneration has also been reported to activate calpains (16, 32) and to diminish clathrin adaptor abundance (33). Therefore, we examined the proteolysis of AP-2 and other adaptor proteins in extracts from postmortem human Alzheimer disease (AD) brain. Whereas α- and β2-adaptin fragments were minimal in control brain samples, abundant cleavage products consistent with the sizes of calpain hydrolysis fragments were observed in three of five AD brains (Fig. 7D). Taken together, the above data indicate that the endogenous activation of calpains can result in substantial hydrolysis of AP-2 and other clathrin adaptors in vivo. These data suggest important roles for calpain in regulating clathrin-dependent endocytosis of a multitude of cargoes.
DISCUSSION
Here we report that calpain activation leads to the proteolysis of the α- and β2-subunits of the core AP-2 clathrin adaptor complex and that this cleavage contributes significantly to the calpain-mediated inhibition of clathrin-dependent receptor endocytosis. Proteolysis of the AP-2 subunits by calpain may decrease receptor-dependent endocytosis of membrane cargoes by several non-exclusive molecular effects. Based on previous work (8) as well as data from the present study, the effect predicted to be of greatest functional significance is the separation of two clathrin-binding domains in the β2-subunit hinge, resulting in decreased recruitment of AP-2 core-bound proteins to clathrin-coated vesicles. This mechanism may largely account for the observed decrease in the internalization of the transferrin receptor, whose YXXØ sorting signal binds directly to the core of the AP-2 complex. This mechanism is also pertinent for the trafficking of ionotropic glutamate receptor subunits that utilize the same core AP-2 binding sites. A second independent consequence of AP-2 proteolysis may be the decreased promotion of clathrin assembly by cleaved adaptors. A potential third effect is that the AP-2 subunit cleavage may prevent the necessary recruitment of accessory clathrin adaptors or other accessory proteins whose binding sites are localized on their appendage domains. Accessory adaptors are thought to play significant roles in both the promotion of clathrin assembly and the targeting of specific cargoes to endocytotic vesicles. Although previous data show that an appendage-deficient α-adaptin mutant does not inhibit transferrin receptor uptake (as might be expected given that its sorting signal binds to the AP-2 core, as described above) (34), other potential cargo-specific effects of this domain still remain to be explored.
Earlier sources describe calpain association with clathrin-coated vesicles from bovine brain (35) and report the in vitro hydrolysis of CALM by calpain (36). Our study elucidates the specific calpain cleavage of α- and β2-adaptins, confirms the cleavage of CALM, and shows for the first time that the accessory adaptors AP180 and epsin 1 are susceptible to calpain proteolysis. Interestingly, calpain-mediated proteolysis has also been suggested to regulate trans-Golgi AP-1-dependent trafficking of growth hormone-containing secretory vesicles (37), and proteolysis of β1-adaptin (attributed to caspase-3) has been implicated in the maturation of mouse bone marrow-derived dendritic cells (38).
In addition to its potential roles in regulating AP complexes, calpain's impact on clathrin-dependent vesicle trafficking has also been reported to involve the cleavage of dynamin 1 (39) and amphiphysin 1 (40). Our discovery of AP-2 cleavage in living cells shows that calpain-mediated regulation of endocytosis can be achieved upstream of dynamin 1 and amphiphysin 1, providing further evidence that calpain is a strong, multipartite regulator of clathrin-dependent endocytosis under conditions that allow its activity to become manifest. Although calpain has the potential to cleave and regulate the activities of multiple molecular components of this pathway, calpain-mediated hydrolysis of these proteins is highly cell type-specific, as evidenced by the selective cleavage of adaptors in rat tissues (Fig. 1) and the inability to induce calpain-mediated hydrolysis of adaptins, even after heterologous calpain expression or purified calpain addition to extracts of numerous transformed cell lines.4 Where calpain cleavage of clathrin adaptors and other accessory proteins reaches a threshold level, as appears to be the case in some cell and tissue subtypes, many significant physiological consequences can be envisaged. Although a challenging endeavor, a detailed exploration of these many effects will be an important contribution to learning how adaptor cleavage may regulate cell and organ system functioning.
Calpain activity in neurons has been previously proposed to mediate normal synaptic plasticity, and synaptic proteins such as glutamate receptor subunits (40-42) and synaptic scaffolding proteins (43) have also been shown to be calpain substrates. Although, at first consideration, it might seem that the hydrolysis of multiple endocytotic proteins does not have the requisite specificity to underlie such a delicate process, additional opportunities to selectively regulate protein internalization occur at the level of accessory clathrin adaptors. Therefore, it will be interesting to learn to what degree cargo specificity can be achieved by differential adaptor utilization and hydrolysis, and whether these might be major determinants of the regulation of synaptic strength. Because N-methyl-d-aspartic acid- and α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid-type-receptor subunits can bind to AP-2 (10, 11, 13), calpain-mediated AP-2 hydrolysis has obvious ramifications for plasticity-related changes in synaptic glutamate receptor populations via changes in surface receptor levels. The ways in which AP-2 hydrolysis relates to other previously identified calpain-mediated mechanisms for regulating receptor number, localization, and activity are crucial parameters that remain to be defined.
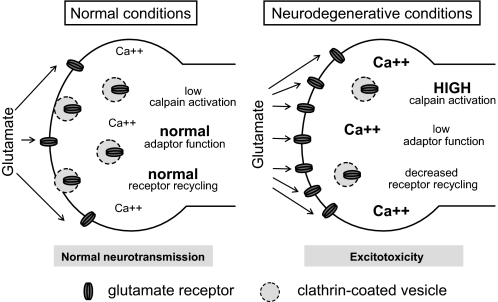
Whereas previous studies have largely focused on possible secretory and other presynaptic-related roles of calpain's regulation of clathrin adaptor complexes, we provide evidence for a substantial contribution to receptive cell (e.g. postsynaptic) effects. These effects seem to be highly plausible contributors to neurodegeneration, where glutamate receptor-expressing cells susceptible to calpain activation may be further sensitized to excitotoxicity resulting from decreased receptor internalization (Fig. 8). The data presented here indicate that the proteolysis of AP-2 alone is sufficient to enhance neuronal glutamate responses.
FIGURE 8.
Model for calpain-driven sensitization of neuronal cells to excitotoxicity via decreased endocytosis. The left panel represents normal (non-degenerative) conditions, where calcium homeostasis is normal and calpain activity is low. The right panel represents a potential disease state where primary abnormalities of protein aggregation, energy metabolism, endoplasmic reticular stress, or dysregulated neurotransmission could potentially sensitize cells to excitatory neurotransmitters such as glutamate. In this case, calpain cleavage of endocytic proteins is envisaged to decrease excitatory receptor endocytosis, which would result in their increased presence on the neuronal surface and thereby mediate excitotoxic damage or death. Not shown is the possibility that an abnormal cell surface turnover of receptors could also result in their being abnormally trafficked to extrasynaptic sites, which could further sensitize them to negative effects of excitatory signals.
Calpain activation is known to occur during acute neural insult and chronic neurodegenerative disease (16-20). Consistent with known calpain activation under these circumstances, we observed α- and β2-adaptin fragments in both a rodent ischemia model and postmortem AD brain. One mechanism underlying calpain activation during chronic neurodegeneration appears to be the accumulation of aggregate-prone proteins (39). In addition to the sensitization of neurons to excitotoxicity, anticipated decreases in endocytosis-dependent growth and trophic factor signaling could also have detrimental consequences in the degenerating brain. Taken together with previous results showing neurodegeneration-associated increases in calpain activation, our results suggest that endocytotic function might be a major cellular target of calpain inhibitors, which have already been proposed as candidate therapies for acute and chronic degenerative conditions (16, 18, 20).
Our detection of α- and β2-adaptin cleavage in extraneural tissues and previous data showing that calpain activation contributes to other normal and pathological processes (44-47) indicate additional roles for calpain-mediated inhibition of endocytosis outside of the nervous system. We therefore postulate that calpain's regulation of endocytosis in other physiological contexts will be an important area of further discovery.
Acknowledgments
We thank Maria Rey, Meredith Dixon, Liliane Glauser, and the Functional Genomics Center Zurich for excellent technical support, Gisou van der Goot, Ralf Schneggenburger, Olexiy Kochubey, Patrick Fraering, Graham Knott, and Michel Kropf for helpful discussions, and David Taylor for critical reading of the manuscript.
This work was supported by the Swiss National Science Foundation and the École Polytechnique Fédérale de Lausanne.
Footnotes
The abbreviations used are: AP-2, adaptor protein complex 2; NMDA, N-methyl-d-aspartic acid; CALM, clathrin assembly lymphoid myeloid leukemia protein; AP180, adaptor protein 180; HA, hemagglutinin; SIN, self-inactivating lentiviral vector; PGK, phosphoglycerate kinase 1; TRE, tetracycline response element; DIV, days in vitro; ALLN, N-acetyl-Leu-Leu-norleucinal; Z-LLY-FMK, benzyloxycarbonyl-LLY-fluoromethyl ketone; PBS, phosphate-buffered saline; AD, Alzheimer disease.
Y. Grishchuk, N. Rudinskiy, and R. Luthi-Carter, unpublished results.
References
- 1.Kirchhausen, T. (2000) Annu. Rev. Biochem. 69 699-727 [DOI] [PubMed] [Google Scholar]
- 2.Schmid, E. M., and McMahon, H. T. (2007) Nature 448 883-888 [DOI] [PubMed] [Google Scholar]
- 3.Collins, B. M., McCoy, A. J., Kent, H. M., Evans, P. R., and Owen, D. J. (2002) Cell 109 523-535 [DOI] [PubMed] [Google Scholar]
- 4.Robinson, M. S. (2004) Trends Cell Biol. 14 167-174 [DOI] [PubMed] [Google Scholar]
- 5.Honing, S., Ricotta, D., Krauss, M., Spate, K., Spolaore, B., Motley, A., Robinson, M., Robinson, C., Haucke, V., and Owen, D. J. (2005) Mol. Cell 18 519-531 [DOI] [PubMed] [Google Scholar]
- 6.Praefcke, G. J., Ford, M. G., Schmid, E. M., Olesen, L. E., Gallop, J. L., Peak-Chew, S. Y., Vallis, Y., Babu, M. M., Mills, I. G., and McMahon, H. T. (2004) EMBO J. 23 4371-4383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schmid, E. M., Ford, M. G., Burtey, A., Praefcke, G. J., Peak-Chew, S. Y., Mills, I. G., Benmerah, A., and McMahon, H. T. (2006) PLoS Biol. 4 e262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Owen, D. J., Vallis, Y., Pearse, B. M., McMahon, H. T., and Evans, P. R. (2000) EMBO J. 19 4216-4227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Motley, A., Bright, N. A., Seaman, M. N., and Robinson, M. S. (2003) J. Cell Biol. 162 909-918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kastning, K., Kukhtina, V., Kittler, J. T., Chen, G., Pechstein, A., Enders, S., Lee, S. H., Sheng, M., Yan, Z., and Haucke, V. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 2991-2996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lavezzari, G., McCallum, J., Lee, R., and Roche, K. W. (2003) Neuropharmacology 45 729-737 [DOI] [PubMed] [Google Scholar]
- 12.Palmer, C. L., Lim, W., Hastie, P. G., Toward, M., Korolchuk, V. I., Burbidge, S. A., Banting, G., Collingridge, G. L., Isaac, J. T., and Henley, J. M. (2005) Neuron 47 487-494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Prybylowski, K., Chang, K., Sans, N., Kan, L., Vicini, S., and Wenthold, R. J. (2005) Neuron 47 845-857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tompa, P., Buzder-Lantos, P., Tantos, A., Farkas, A., Szilagyi, A., Banoczi, Z., Hudecz, F., and Friedrich, P. (2004) J. Biol. Chem. 279 20775-20785 [DOI] [PubMed] [Google Scholar]
- 15.Goll, D. E., Thompson, V. F., Li, H., Wei, W., and Cong, J. (2003) Physiol. Rev. 83 731-801 [DOI] [PubMed] [Google Scholar]
- 16.Gafni, J., and Ellerby, L. M. (2002) J. Neurosci. 22 4842-4849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kampfl, A., Posmantur, R. M., Zhao, X., Schmutzhard, E., Clifton, G. L., and Hayes, R. L. (1997) J. Neurotrauma 14 121-134 [DOI] [PubMed] [Google Scholar]
- 18.Raynaud, F., and Marcilhac, A. (2006) FEBS J. 273 3437-3443 [DOI] [PubMed] [Google Scholar]
- 19.Yamashima, T. (2000) Prog. Neurobiol. 62 273-295 [DOI] [PubMed] [Google Scholar]
- 20.Crocker, S. J., Smith, P. D., Jackson-Lewis, V., Lamba, W. R., Hayley, S. P., Grimm, E., Callaghan, S. M., Slack, R. S., Melloni, E., Przedborski, S., Robertson, G. S., Anisman, H., Merali, Z., and Park, D. S. (2003) J. Neurosci. 23 4081-4091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lipton, P. (1999) Physiol. Rev. 79 1431-1568 [DOI] [PubMed] [Google Scholar]
- 22.Ray, S. K., Karmakar, S., Nowak, M. W., and Banik, N. L. (2006) Neuroscience 139 577-595 [DOI] [PubMed] [Google Scholar]
- 23.Deglon, N., Tseng, J. L., Bensadoun, J. C., Zurn, A. D., Arsenijevic, Y., Pereira de Almeida, L., Zufferey, R., Trono, D., and Aebischer, P. (2000) Hum. Gene Ther. 11 179-190 [DOI] [PubMed] [Google Scholar]
- 24.Perrin, V., Regulier, E., Abbas-Terki, T., Hassig, R., Brouillet, E., Aebischer, P., Luthi-Carter, R., and Deglon, N. (2007) Mol. Ther. 15 903-911 [DOI] [PubMed] [Google Scholar]
- 25.Vaslin, A., Puyal, J., Borsello, T., and Clarke, P. G. (2007) J. Neurochem. 102 789-800 [DOI] [PubMed] [Google Scholar]
- 26.Zala, D., Benchoua, A., Brouillet, E., Perrin, V., Gaillard, M. C., Zurn, A. D., Aebischer, P., and Deglon, N. (2005) Neurobiol. Dis. 20 785-798 [DOI] [PubMed] [Google Scholar]
- 27.Steiner, P., Sarria, J. C., Glauser, L., Magnin, S., Catsicas, S., and Hirling, H. (2002) J. Cell Biol. 157 1197-1209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Renolleau, S., Aggoun-Zouaoui, D., Ben-Ari, Y., and Charriaut-Marlangue, C. (1998) Stroke 29 1454-1460 [DOI] [PubMed] [Google Scholar]
- 29.Delacourte, A., David, J. P., Sergeant, N., Buee, L., Wattez, A., Vermersch, P., Ghozali, F., Fallet-Bianco, C., Pasquier, F., Lebert, F., Petit, H., and Di Menza, C. (1999) Neurology 52 1158-1165 [DOI] [PubMed] [Google Scholar]
- 30.Deramecourt, V., Bombois, S., Maurage, C. A., Ghestem, A., Drobecq, H., Vanmechelen, E., Lebert, F., Pasquier, F., and Delacourte, A. (2006) J. Neuropathol. Exp. Neurol. 65 278-288 [DOI] [PubMed] [Google Scholar]
- 31.Neumar, R. W., Meng, F. H., Mills, A. M., Xu, Y. A., Zhang, C., Welsh, F. A., and Siman, R. (2001) Exp. Neurol. 170 27-35 [DOI] [PubMed] [Google Scholar]
- 32.Tsuji, T., Shimohama, S., Kimura, J., and Shimizu, K. (1998) Neurosci. Lett. 248 109-112 [DOI] [PubMed] [Google Scholar]
- 33.Yao, P. J., Morsch, R., Callahan, L. M., and Coleman, P. D. (1999) Neuroscience 94 389-394 [DOI] [PubMed] [Google Scholar]
- 34.Motley, A. M., Berg, N., Taylor, M. J., Sahlender, D. A., Hirst, J., Owen, D. J., and Robinson, M. S. (2006) Mol. Biol. Cell 17 5298-5308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sato, K., Saito, Y., and Kawashima, S. (1995) Eur J. Biochem. 230 25-31 [DOI] [PubMed] [Google Scholar]
- 36.Kim, J. A., and Kim, H. L. (2001) Exp. Mol. Med. 33 245-250 [DOI] [PubMed] [Google Scholar]
- 37.Ohkawa, K., Takada, K., Asakura, T., Hashizume, Y., Okawa, Y., Tashiro, K., Ueda, J., Itoh, Y., and Hibi, N. (2000) Neuroreport 11 4007-4011 [DOI] [PubMed] [Google Scholar]
- 38.Santambrogio, L., Potolicchio, I., Fessler, S. P., Wong, S. H., Raposo, G., and Strominger, J. L. (2005) Nat. Immunol. 6 1020-1028 [DOI] [PubMed] [Google Scholar]
- 39.Kelly, B. L., and Ferreira, A. (2006) J. Biol. Chem. 281 28079-28089 [DOI] [PubMed] [Google Scholar]
- 40.Wu, Y., Liang, S., Oda, Y., Ohmori, I., Nishiki, T., Takei, K., Matsui, H., and Tomizawa, K. (2007) EMBO J. 26 2981-2990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xu, W., Wong, T. P., Chery, N., Gaertner, T., Wang, Y. T., and Baudry, M. (2007) Neuron 53 399-412 [DOI] [PubMed] [Google Scholar]
- 42.Yuen, E. Y., Gu, Z., and Yan, Z. (2007) J. Physiol. 580 241-254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lu, X., Rong, Y., and Baudry, M. (2000) Neurosci. Lett. 286 149-153 [DOI] [PubMed] [Google Scholar]
- 44.Glading, A., Bodnar, R. J., Reynolds, I. J., Shiraha, H., Satish, L., Potter, D. A., Blair, H. C., and Wells, A. (2004) Mol. Cell Biol. 24 2499-2512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pariat, M., Carillo, S., Molinari, M., Salvat, C., Debussche, L., Bracco, L., Milner, J., and Piechaczyk, M. (1997) Mol. Cell Biol. 17 2806-2815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rock, M. T., Dix, A. R., Brooks, W. H., and Roszman, T. L. (2000) Exp. Cell Res. 261 260-270 [DOI] [PubMed] [Google Scholar]
- 47.Squier, M. K., Sehnert, A. J., Sellins, K. S., Malkinson, A. M., Takano, E., and Cohen, J. J. (1999) J. Cell Physiol. 178 311-319 [DOI] [PubMed] [Google Scholar]