Abstract
TrkA receptor signaling is essential for nerve growth factor (NGF)-induced survival and differentiation of sensory neurons. To identify possible effectors or regulators of TrkA signaling, yeast two-hybrid screening was performed using the intracellular domain of TrkA as bait. We identified muc18-1-interacting protein 2 (Mint2) as a novel TrkA-binding protein and found that the phosphotyrosine binding domain of Mint2 interacted with TrkA in a phosphorylation- and ligand-independent fashion. Coimmunoprecipitation assays showed that endogenous TrkA interacted with Mint2 in rat tissue homogenates, and immunohistochemical evidence revealed that Mint2 and TrkA colocalized in rat dorsal root ganglion neurons. Furthermore, Mint2 overexpression inhibited NGF-induced neurite outgrowth in both PC12 and cultured dorsal root ganglion neurons, whereas inhibition of Mint2 expression by RNA interference facilitated NGF-induced neurite outgrowth. Moreover, Mint2 was found to promote the retention of TrkA in the Golgi apparatus and inhibit its surface sorting. Taken together, our data provide evidence that Mint2 is a novel TrkA-regulating protein that affects NGF-induced neurite outgrowth, possibly through a mechanism involving retention of TrkA in the Golgi apparatus.
The neurotrophin family member nerve growth factor (NGF)3 is essential for proper development, patterning, and maintenance of nervous systems (1, 2). NGF has two known receptors; TrkA, a single-pass transmembrane receptor-tyrosine kinase that binds selectively to NGF, and p75, a transmembrane glycoprotein that binds all members of the neurotrophin family (3, 4). NGF binding activates the kinase domain of TrkA, leading to autophosphorylation (5). The resulting phosphotyrosines become docking sites for adaptor proteins involved in signal transduction pathways that lead to the activation of Ras, Rac, phosphatidylinositol 3-kinase, phospholipase Cγ, and other effectors (2, 6). Many of these TrkA-interacting adaptor proteins have been identified and include, Grb2, APS, SH2B, fibroblast growth factor receptor substrate 2 (FRS-2), Shc, and human tumor imaginal disc 1 (TID1) (7-10). The identification of these binding partners has contributed greatly to our understanding of the mechanisms underlying the functional diversity of NGF-TrkA signaling.
Studies have indicated that the transmission of NGF signaling in neurons involves retrograde transport of NGF-TrkA complexes from the neurite tip to the cell body (11-14). TrkA associates with components of cytoplasmic dynein, and it is thought that vesicular trafficking of neurotrophins occurs via direct interaction of Trk receptors with the dynein motor machinery (14). Furthermore, the atypical protein kinase C-interacting protein, p62, associates with TrkA and plays a novel role in connecting receptor signals with the endosomal signaling network required for mediating TrkA-induced differentiation (15). Recently, the membrane-trafficking protein Pincher has been found to mediate macroendocytosis underlying retrograde signaling by TrkA (16). Despite the progress made to date in understanding Trk complex internalization and trafficking, the mechanisms remain poorly understood.
Mint2 (muc18-1-interacting protein 2) belongs to the Mint protein family, which consists of three members, Mint1, Mint2, and Mint3. Mint proteins were first identified as interacting proteins of the synaptic vesicle-docking protein Munc18-1 (17, 18). Mint1 is also sometimes referred to as mLIN-10, as it is the mammalian orthologue of the Caenorhabditis elegans LIN-10 (19). Additionally, Mint1, Mint2, and Mint3 are also referred to as X11α or X11, X11β or X11L (X11-like), and X11γ or X11L2 (X11-like 2), respectively (20). All Mint proteins contain a conserved central phosphotyrosine binding (PTB) domain and two contiguous C-terminal PDZ domains (repeated sequences in the brain-specific protein PSD-95, the Drosophila septate junction protein Discs large, and the epithelial tight junction protein ZO-1) (17, 18, 21). Mint1 and Mint2 are expressed only in neuronal tissue (17), whereas Mint3 is ubiquitously expressed (18). Although the function of Mints proteins is not fully clear, their interactions with the docking and exocytosis factors Mun18 -1 and CASK, ADP-ribosylation factor (Arf) GTPases involved in vesicle budding (22), and other synaptic adaptor proteins, such as neurabin-II/spinophilin (23), tamalin (24), and kalirin-7 (25), all suggest possible roles for Mints in synaptic vesicle docking and exocytosis. Mint proteins have also been implicated in the trafficking and/or processing of β-amyloid precursor protein (β-APP). Through their PTB domains, all three Mints bind to a motif within the cytoplasmic domain of β-APP (21, 26-29), and Mint1 and Mint2 can stabilize β-APP, affect β-APP processing, and inhibit the production and secretion of Aβ (28, 30-32). Although the mechanisms by which Mints inhibit β-APP processing are not yet well known, Mints and their binding partners have emerged as potential therapeutic targets for the treatment of Alzheimer disease.
To uncover new TrkA-interacting factors and gain insight into the mechanisms that guide TrkA intracellular trafficking and other aspects of TrkA signaling, we conducted a yeast two-hybrid screen of a brain cDNA library using the intracellular domain of TrkA as bait. The screen identified several candidate TrkA-interacting proteins, one of which was Mint2. Follow-up binding assays showed that the PTB domain of Mint2 alone was necessary and sufficient for mediating the interaction with TrkA. Endogenous Mint2 was also coimmunoprecipitated and colocalized with TrkA in rat DRG tissue. Overexpression and knockdown studies showed that Mint2 could significantly inhibit NGF-induced neurite outgrowth in both TrkA-expressing PC12 cells and DRG neurons. Moreover, Mint2 was found to induce the retention of TrkA in the Golgi apparatus and inhibit its surface sorting. Our results suggest that Mint2 is a novel regulator of TrkA receptor signaling.
MATERIALS AND METHODS
Plasmid Constructs—Full-length TrkA constructs were a generous gifts from Dr. David D. Ginty, and full-length Mint2 constructs were provided by Dr. Thomas C. Südhof. The intracellular domain of human TrkA was generated by PCR and cloned in-frame into the LexA fusion vector pGilda (Clontech, Palo Alto, CA) as bait pGilda-TrkAIC. Full-length Mint2 and the PTB, PDZ1, PDZ2, and N-terminal domains of Mint2 were amplified by PCR and subcloned into pB42AD vectors. To construct the Myc-tagged Mint2 (Myc-Mint2) and GFP-tagged Mint2 (GFP-Mint2), we subcloned full-length Mint2 into pcDNA3 (Invitrogen) and pEGFP-N2 vector (Clontech) vectors, respectively. The K547N TrkA mutant was generated by applying the QuikChange site-directed mutagenesis method from Stratagene (La Jolla, CA). All constructs were fully sequenced before being used for transformation or transfection.
For inhibition of Mint2 expression by siRNA, three unique 21-bp nucleotide sequences were designed and synthesized by GeneChem (Shanghai, China). They were: No.1 (bp 948-969), 5′-GCCUGAGCAUGACCAGUAUtt-3′; No.2 (bp 1192-1213), 5′-GGUUUGCAAUGGCUUGGAAtt-3′, and No.3 (bp 1468-1489), 5′-GGCUGCUAAGAUCAAGAAAtt-3′. To determine the knockdown efficacy of the siRNAs, HEK293 cells in 12-well culture plates were cotransfected using Lipofectamine 2000 (Invitrogen) with Myc-Mint2 plasmid and nonsense sequence or Myc-Mint2 plasmid and siRNA. Cells were lysed 48 h after transfection, and lysates were subjected to immunoblotting analysis. The most efficient siRNA sequence was inserted into pSuper vector.
Yeast Two-hybrid Screening—Yeast two-hybrid screening was performed according to the manufacturer's protocols (Clontech). The Saccharomyces cerevisiae yeast strain EGY48 (MATa, his3, trp1, ura3, LEU2::plexAop6-LEU2), the LexA yeast two-hybrid system, and MatchMaker human brain cDNA library were all purchased from Clontech. The pGilda-TrkAIC construct and human brain cDNA library fused to pB42AD were cotransformed into the EGY48 strain with polyethylene glycol/LiAc solution. The cotransformants were plated on SD-Gal Ura-His-Trp-Leu drop-out galactose induction medium for 3-4 days at 30 °C to induce the expression of reporter proteins fused with the activation domain. Filter-lift color assays and leucine-deficient (Leu-) culture assays were performed as described before (33). During the analysis, pGilda-53 cotransformed with pB42AD-T was used as a positive control, whereas pGilda cotransformed with pB42AD was used as a negative control. Potential positive clones were selected, and prey-plasmid-containing library cDNA inserts were isolated and shuttled into Escherichia coli KC8 cells. Positive colonies were further confirmed by testing pB42AD-cDNA constructs against LexA-lamin to eliminate false positives and then sequenced.
GST Pulldown Assay—To generate glutathione S-transferase (GST) fusion proteins, full-length Mint2 and PTB, PDZ1, PDZ2 domain, and N-terminal domain cDNAs were amplified by PCR and subcloned into pGEX-4T-3 vector (Amersham Biosciences). All fusion proteins were expressed in E. coli BL21 cells, precipitated with glutathione-Sepharose beads, and eluted with 10 mm glutathione (in 50 mm Tris, pH 8.0) according to the Amersham Biosciences protocol. Binding assays were performed with PC12 cell extracts as described previously (33). Equal amounts of immobilized GST fusion proteins were mixed and incubated for 3 h at 4 °C with PC12 cell extract in GST binding buffer containing 40 mm HEPES, 50 mm sodium acetate, 200 mm NaCl, 2 mm EDTA, 5 mm dithiothreitol, 0.5% Nonidet P-40, and protease inhibitors. Glutathione beads were washed three times in binding buffer and boiled in SDS sample buffer to elute bound proteins, which were then resolved by SDS-PAGE and followed by Western blot analysis and visualized with enhanced chemiluminescence (ECL, Pierce).
Preparation of Tissue Homogenates and Immunoprecipitation—DRG, cerebellum, and basal forebrain tissue were dissected from adult male Sprague-Dawley rats and dissociated. Samples were homogenized in a pestle tissue grinder in solubilization buffer (25 mm HEPES-NaOH, pH 7.4, 125 mm potassium acetate, 5 mm MgCl2, 0.32 m sucrose, and 1% Triton X-100). Proteins solubilized from each homogenate were quantified using the BCA method. Equal amounts of protein were incubated with primary antibodies for 3 h at 4 °C under mild agitation. Protein G-agarose beads (Roche Applied Science) were added for 3 h, and immunoprecipitated samples were then washed 3 times with solubilization buffer, boiled 3-5 min in sample-loading buffer, and subjected to Western blotting analysis. Antibodies used in Western blot and immunoprecipitation experiments included rabbit anti-TrkA, phospho-Tyr-490 (Abcam, Cambridge, UK), rabbit anti-TrkA(763) (Santa Cruz), rabbit anti-TrkA (Chemicon, Temecula, CA), mouse anti-Mint2 (BD Biosciences), rabbit anti-Myc (Sigma), mouse anti-GFP (Roche Applied Science), polyclonal GPP130 (1:400, Covance, Princeton, NJ), rabbit anti-calnexin (Sigma), rabbit anti-MAP2 (Chemicon), rabbit anti-p75NTR (Chemicon), monoclonal anti-actin (Sigma), mouse IgG (Santa Cruz), and rabbit IgG (Santa Cruz).
Immunohistochemistry and Immunocytochemistry—Adult male Sprague-Dawley rats weighing 200-250 g were anesthetized with sodium pentobarbital (50 mg/kg, intraperitoneal) and perfused transcardially with 0.1 m phosphate buffer (PB) pH 7.4, followed by perfusion with 4% paraformaldehyde with 0.2% saturated picric acid in 0.1 m phosphate-buffered saline (PBS). DRG tissues were removed, kept for 1.5 h in the same fixative at 4 °C, and then cryoprotected overnight at 4 °C in 0.01 m PBS, pH 7.4, containing 20% sucrose. Tissues were sectioned into 14-μm slices with a Leica 1900 cryostat and mounted on glass slides. Sections were washed in PBS and incubated with primary antibodies (rabbit anti-TrkA 1:200, Chemicon; mouse anti-Mint2 1:500, BD Biosciences) in PBS containing 3% bovine serum albumin and 0.3% Triton X-100 at 4 °C overnight. After three washes in PBS, sections were incubated with fluorescein isothiocyanate-conjugated donkey anti-rabbit (1:100) and rhodamine-conjugated donkey anti-mouse (1:100) antibodies (both from Jackson ImmunoResearch, West Grove, PA). Sections were washed, placed on coverslips, and examined with a Leica SP2 confocal microscope.
PC12 cells were fixed in PBS containing 4% formaldehyde for 15 min at room temperature. After fixation, cells were rinsed with PBS, blocked and permeabilized in PBS (containing 5% bovine serum albumin, 0.1% Triton), and stained with monoclonal anti-Myc (1:500, BD Biosciences) with wheat germ agglutinin (WGA)-conjugated Alexa Fluor 633 (1:1500, Invitrogen) and polyclonal anti-rabbit Myc (1:500, Sigma) in the same buffer at 4 °C overnight. A collection of fluorophore-conjugated secondary antibodies was used including donkey anti-mouse-TRITC, donkey anti-rabbit-Cy5, or donkey anti-mouse-Cy5 (Jackson ImmunoResearch).
Neurite Outgrowth Assay—PC12 cells were transfected with pEGFP-N2 alone or pEGFP-N2 and Myc-Mint2 and seeded on poly-d-lysine coated coverslips at a density of 3-5 × 105 cells/ml. NGF (100 ng/ml, Sigma) was added directly to the medium to initiate cell differentiation. Cells were then incubated for 3 days in the presence of NGF before being fixed and permeabilized for immunocytochemistry. The PC12 cell differentiation assay was performed as described before (34), and cells possessing one or more neurites of a length more than twice the diameter of the cell body were scored as positive. 100 cells per group were randomly counted for each experiment.
Primary DRG neuron cultures were generated as described before (35). Briefly, DRGs were removed from E17 Sprague-Dawley rat embryos and pooled in ice-cold Hanks' buffered salt solution. Tissue sections were dissociated by consecutive treatment with 0.25% trypsin followed by careful trituration at 37 °C for 45 min. After centrifugation, the pellet was dispersed in 10% fetal bovine serum/Dulbecco's modified Eagle's medium. Cells were then electroporated (Amaxa Biosystems, Koln, Germany) and plated directly on acid-washed coverslips precoated with 10 μg/ml mouse laminin (Invitrogen) in 10% fetal bovine serum/Dulbecco's modified Eagle's medium (35). The medium was then changed to Neurobasal media with B27 supplement (Invitrogen) with 100 ng/ml NGF (Sigma). Cells were maintained at 37 °C in 5% CO2 for 3 days, then fixed and immunostained with anti-MAP2 antibodies (Chemicon) and donkey anti-mouse-TRITC (Jackson ImmunoResearch). Neurite outgrowth was quantification as reported in our previous study (34). The longest neurite per neuron was measured by Image-pro Plus software (Olympus), and total neurite length was measured by Neurolucida software (MicroBrightField Inc.). For each experiment 150 neurons were scored.
Surface Biotinylation Assay—Transfected PC12 cells were starved overnight before being treated with 100 ng/ml NGF (Sigma) at 37 °C for 30 min. Surface biotinylation was preformed according to a previous published protocol (36). Briefly, cells were washed in PBS (Ca2+/Mg2+) containing Sulfo-NHS-LC-Biotin (0.5 mg/ml, Pierce) for 45 min at 4 °C. After extensive washing with ice-cold PBS containing 50 mm glycine, cells were lysed. Biotinylated proteins were precipitated with ImmunoPure Immobilized Streptavidin (Pierce) overnight at 4 °C. Samples were loaded on SDS-PAGE, transferred, probed with antibody against GFP (1:2000, Roche Applied Science), and visualized with enhanced chemiluminescence (ECL, Pierce).
Internalization of GFP-TrkA was also analyzed using a cell-surface biotinylation assay (37). Briefly, cells were subjected to biotinylation on ice with the reversible membrane-impermeable derivative of biotin (1.5 mg/ml sulfo-NHS-S-S-biotin in PBS, Pierce). Internalization was allowed to occur by incubation at 37 °C with media containing NGF (100 ng/ml) for 30 min. The remaining cell-surface biotin was cleaved by reducing its disulfide linkage with glutathione cleavage buffer, and cells were lysed with radioimmune precipitation assay buffer. Biotinylated proteins were precipitated using UltraLink immobilized Neutravidin beads (Pierce), eluted from the beads with boiling Laemmli buffer, resolved by SDS-PAGE, and immunoblotted with an antibody directed against GFP (1:2000, Roche Applied Science).
Gradient Fractionation Assay—PC12 cells from eight 10-cm plates were collected by shake-off and centrifugation for 5 min (2000 rpm in a SA-600 rotor, Sorvall, Newtown, CT), washed once in PBS (containing 1 mm EDTA), washed once in sucrose buffer (250 mm sucrose, 10 mm triethylamine, pH 7.4, 1 mm EDTA, with protease inhibitors 1 mm phenylmethylsulfonyl fluoride, 10 mg/ml leupeptin, and 10 mg/ml pepstatin A), homogenized by 20 passages through a 25-gauge needle in 0.5 ml buffer (50 mm NaCl, 10 mm triethylamine, pH 7.4, 1 mm EDTA, with protease inhibitors), and centrifuged at 1000 × g for 2 min. The postnuclear supernatant was then fractionated on either velocity gradients or flotation gradients. In the standard velocity gradient procedure the postnuclear supernatant (0.4 ml) was layered on linear 5-25% (v/v) glycerol gradients (5 ml in 10 mm triethylamine, pH 7.4, and 1 mm EDTA on a 0.5 ml 80% sucrose cushion) and centrifuged at 150,000 × g for 30 min in a SW50.1 rotor (Beckman Instruments), and 0.4-ml fractions were collected from the top. The sedimentation coefficient was calculated using a proportionality constant value of 3.38 cm, as described previously (63). For flotation gradient analysis, the postnuclear supernatant was adjusted to 1.6 m sucrose (1.1 ml), layered on 2 m sucrose (0.5 ml), covered with three sucrose layers (1.1 ml each of 1.4, 1.2, 0.8 m), and centrifuged at 105,000 × g for 105 min in a SW50.1 rotor (Beckman), and 0.4-ml fractions were collected from the top (38). All steps were carried out at 0-4 °C. Immunoblotting was used to assay each fraction for the presence of TrkA-GFP (anti-GFP, Roche Applied Science), Myc-Mint2 (anti-Myc, Sigma) GPP130 (anti-GPP130, Covance) (39), and calnexin (anti-calnexin Sigma) (40).
RESULTS
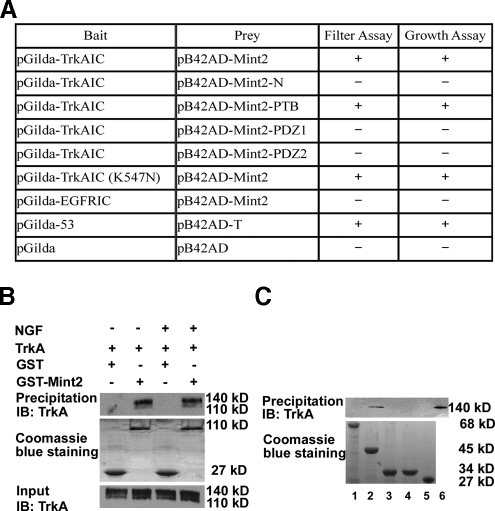
Identification of Mint2 as a Potential Binding Partner of TrkA by Yeast Two-hybrid Screening—In an attempt to identify proteins that associate with TrkA, we conducted a yeast two-hybrid screen of a human brain cDNA library using the intracellular domain of TrkA (TrkAIC) as bait (Fig. 1A). To construct the bait for the two-hybrid screen, the TrkAIC (amino acids 443-799) coding sequence was fused in-frame to the DNA binding domain of LexA using the pGilda vector. The brain cDNAs were subcloned into the B42 activation domain vector pB42AD and were cotransformed with the bait plasmid into yeast strain EGY48. About 3 × 106 cotransformants were plated on leucine-deficient medium, and activation of the LEU reporter gene was used to select for protein-protein interactions. Approximately 269 Leu-reporter gene-positive clones were isolated in the growth assay screen, and of these 78 clones scored positive for the β-galactosidase filter-lift assay; false positives were further eliminated by testing against LexA-lamin. The cDNA inserts of the remaining 11 clones were then sequenced. One of the cDNAs was identified to be a 672-bp fragment corresponding to residues 286-539 of the Mint2 gene, a region encompassing the entire PTB domain and flanking sequence, as shown in Fig. 1B.
FIGURE 1.
Schematic representation of the bait and prey used in the yeast two-hybrid screen. A, schematic representation of full-length TrkA and the pGilda-TrkAIC bait. TrkA has a single transmembrane domain and a single cytoplasmic tyrosine kinase domain. C, cysteine clusters; LRR, leucine-rich repeats; Ig, immunoglobulin-like domain; TM, transmembrane domain; TK, tyrosine kinase domain. B, the two-hybrid screen identified the region of Mint2 containing the PTB domain (amino acids 286-539) as a binding partner for TrkAIC. N, N-terminal region; PDZ, the PDZ domain. aa, amino acids.
The PTB Domain of Mint2 Mediates the Interaction with TrkA—To further confirm the presumptive interaction between Mint2 and TrkA, we tested the ability of full-length Mint2 to associate with TrkA. Both β-galactosidase activity and restricted growth assays showed that TrkA could interact with full-length Mint2 in yeast, whereas the epidermal growth factor receptor could not (Fig. 2A). Because Mint2 contains multiple domains including an N-terminal region, a central PTB domain and two C-terminal PDZ domains arranged in tandem (Fig. 1B), we subcloned fragments of rat Mint2 corresponding to these regions (N-terminal region, 1-368 amino acids (aa); PTB domain, 369-532 aa; PDZ1 domain, 566-650 aa; PDZ2 domain, 662-735 aa) into prey vectors and tested which region was critical for TrkA interaction. Our results showed that only the PTB domain was capable of binding TrkA; no interaction between TrkA and other regions of Mint2 occurred (Fig. 2A). To determine whether this interaction was dependent on the phosphorylation state of TrkA, we used a yeast two-hybrid assay to test whether Mint2 could interact with a catalytically inactive TrkAIC construct in which lysine 547, the essential ATP binding site, was substituted for glutamine (K547N (41-44)). We found that Mint2 was capable of binding to this construct as well, suggesting that the interaction is phosphorylation-independent (Fig. 2A).
FIGURE 2.
Mint2 interacts with TrkA in yeast two-hybrid and GST pulldown assays. A, the interaction of Mint2 with TrkAIC was analyzed by filter assay for LacZ reporter gene β-galactosidase activity and leucine-deficient growth assay for LEU reporter gene expression. The indicated Mint2 domains fused in-frame into pB42AD vector were cotransformed with TrkAIC (pGilda-TrkAIC), a kinase-dead TrkAIC (pGilda-TrkAIC (K547N)), the intracellular domain of the epidermal growth factor receptor (pGilda-EGFRIC), or the vector alone (pGilda) into the yeast reporter strain EGY48. Positive and negative controls were described in methods. B, PC12 cells expressing the indicated constructs, either treated with NGF or left untreated, were lysed and subjected to GST pulldown followed by Western blotting (IB) with anti-TrkA antibody (Chemicon). The whole cell lysate is shown as an input control. C, various GST fusion proteins were incubated with PC12 cell extracts containing endogenous TrkA. After GST pulldown, GST complexes were immunoblotted with anti-TrkA (Santa Cruz) for Western blot analysis. 1, GST-Mint2-N (68 kDa); 2, GST-PTB (45 kDa); 3, GST-PDZ1 (34 kDa); 4, GST-PDZ2 (34 kDa); 5, GST (negative control; 27 kDa); 6, PC12 cell lysate (positive control).
To confirm our yeast two-hybrid assay results, we used a GST pulldown assay to examine the interaction between GST-tagged Mint2 and endogenous TrkA in PC12 cells (45). PC12 cells with or without NGF treatment were lysed and subjected to GST pulldown followed by Western blotting with anti-TrkA antibody. As shown in Fig. 2B, Mint2 could bind to TrkA in both NGF-treated and untreated PC12 cells. As shown in Fig. 2C, expression of a GST-tagged PTB domain construct was both necessary and sufficient to pull down TrkA in PC12 extract. Thus, the results from our yeast two-hybrid and GST pulldown assays indicate that TrkA can interact with Mint2, and this interaction is mediated by the PTB domain of Mint2.
Endogenous Mint2 Interacts with TrkA in Rat Tissue Homogenates—To examine whether endogenous TrkA and Mint2 can complex together in mammalian tissue in vivo, we performed immunoprecipitation assays with homogenates from different tissues of the adult rat, including DRG, cerebellum, and basal forebrain. Homogenates were immunoprecipitated with anti-TrkA or anti-Mint2 and immunoblotted with anti-Mint2 or anti-TrkA. As shown in Fig. 3, TrkA could coimmunoprecipitate Mint2 and vice versa in DRG and the basal forebrain but not the cerebellum. Neither Mint2 nor TrkA was immunoprecipitated by control IgG in DRG tissue (data not shown), further confirming the specificity of the interaction.
FIGURE 3.
Endogenous Mint2 associates with TrkA in rat tissue homogenates. Tissue homogenates from adult rat DRG, forebrain, and cerebellum were prepared as described under “Materials and Methods” and immunoprecipitated (IP) with anti-TrkA or anti-Mint2 antibodies followed by immunoblotting (IB) with anti-Mint2 or anti-TrkA antibodies. Protein expression levels in tissue homogenates were detected by immunoblotting with anti-Mint2 or anti-TrkA antibodies.
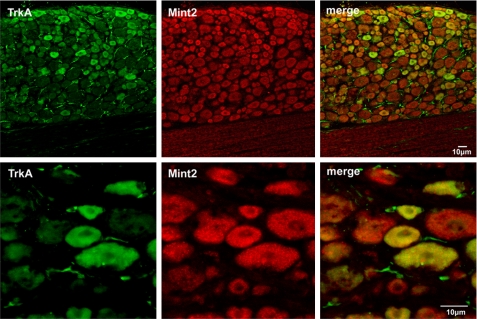
TrkA Colocalizes with Mint2 in Rat DRG Neurons—We next examined whether TrkA and Mint2 colocalize in neurons. Because TrkA is expressed in DRG neurons (46) and Mint2 is distributed ubiquitously in the nervous system (47), immunohistochemical analysis was carried out on adult rat DRG slices to determine the subcellular distribution of TrkA and Mint2 in DRG neurons. As shown in Fig. 4, fluorescent confocal imaging revealed the expression of TrkA (green) and Mint2 (red) in the DRG neurons. The merged images showed that TrkA and Mint2 could colocalize in the same neurons of DRG, and their distributions were highly overlapped. As a control, no staining was observed when the primary antibody was removed from our staining procedure, showing that the staining was specific (data not shown).
FIGURE 4.
TrkA and Mint2 colocalize in rat DRG neurons. Sections of rat DRG were coimmunostained with anti-TrkA antibodies (green) and anti-Mint2 antibodies (red). Merged images show TrkA and Mint2 colocalization in rat DRG neurons (yellow). Scale bars, 10 μm.
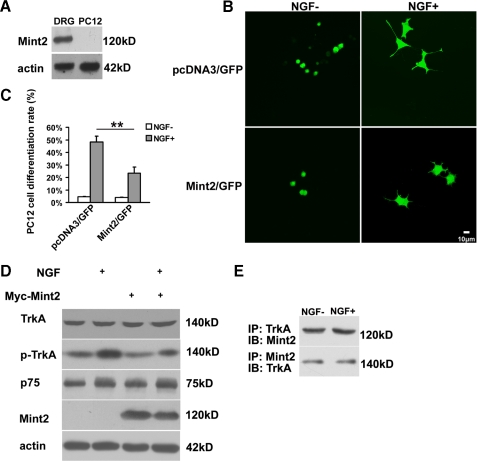
Mint2 Negatively Regulates NGF-induced Neurite Outgrowth in PC12 Cells and DRG Neurons—NGF signaling through TrkA induces PC12 cell differentiation, a process that includes the growth of neurites (45). Because Mint2 was not expressed in PC12 cells (Fig. 5A), we cotransfected PC12 cells with pEGFP and either Myc-tagged Mint2 or a control construct (pcDNA3) at a ratio of 1:10 and compared neurite outgrowth after NGF stimulation to identify a physiological role of Mint2 in TrkA signaling. Statistical analysis revealed that compared with the control group, ectopic expression of Myc-Mint2 markedly reduced the ratio of neurite-bearing cells after NGF treatment, suggesting that Mint2 negatively regulates TrkA signaling in PC12 cells (Fig. 5, B and C). To determine how Mint2 might affect NGF signaling, we first examined the expression of NGF receptors in Mint2-expressing PC12 cells both with and without NGF stimulation but did not observe any difference in the levels of TrkA or p75 in Mint2-expressing cells compared with control cells (Fig. 5D). Interestingly, the level of p-TrkA was lower in the Mint2 overexpressing group relative to the control group after exposure to NGF, suggesting that Mint2 prevents robust NGF-induced TrkA activation and thereby inhibits neurite outgrowth. We next investigated the interaction between TrkA and Mint2 after NGF treatment by coimmunoprecipitation. As shown in Fig. 5E, the interaction was not affected by NGF treatment, consistent with the results of our GST-Mint2 pulldown assays (Fig. 2B). Neither Mint2 nor TrkA was immunoprecipitated by control IgG in Mint2-transfected PC12 cells either with or without NGF stimulation (data not shown), further confirming the specificity of the interaction.
FIGURE 5.
Mint2 overexpression attenuates NGF-induced PC12 cell differentiation. A, Western blot showing Mint2 expression in DRG neurons but not in PC12 cells. B, PC12 cells were cotransfected with pEGFP-N2 together with pcDNA3 as control or with pcDNA3-Mint2. After 3 days with or without NGF, neurite outgrowth was assessed as an indicator of neuronal differentiation. Scale bar, 10 μm. C, quantification of neuronal differentiation of PC12 cells. Values represent the mean ± S.D., and the double asterisk indicates significant difference (p < 0.01, Student's t test). D, Western blotting was carried out to detect the levels of TrkA, p-TrkA, and p75 proteins in various groups. After NGF treatment, decreased levels of p-TrkA were detected in Mint2-overexpressing PC12 cells compared with those seen in wild type PC12 cells. E, coimmunoprecipitation (IP) analysis showing that the interaction between TrkA and Mint2 was not affected by NGF stimulation. IB, immunoblot.
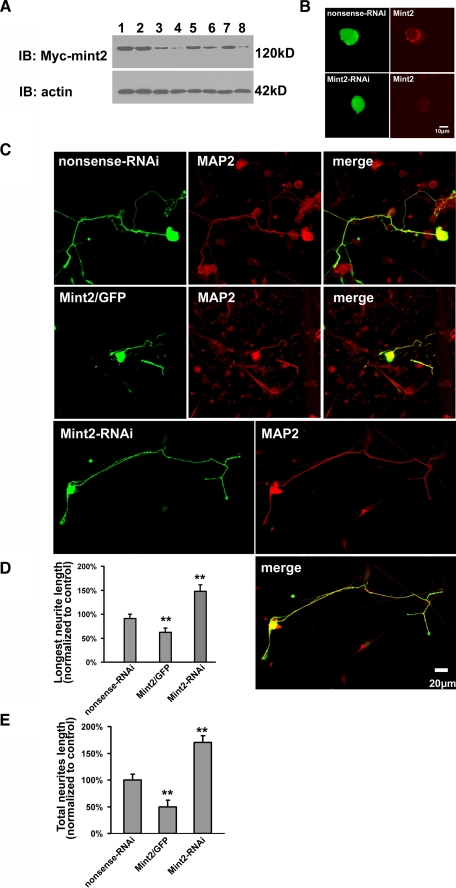
Because both Mint2 and TrkA are endogenously expressed in DRG neurons (Fig. 4), we next performed a neurite outgrowth assay on cultured DRG neurons extracted from E16 rats. We first assessed the effect of RNAi-mediated silencing of Mint2 in HEK293 cells by measuring protein levels of exogenous Mint2. Three independent Mint2-siRNA sequences were tested, among which one (#1) afforded the most robust knockdown efficacy (Fig. 6A). When the selected GFP-tagged Mint2-siRNA was transfected into DRG neuron, the endogenous Mint2, which ia mainly accumulated near the nucleus (20), was successfully knocked down (Fig. 6B), further indicating the high efficiency of the Mint2 RNAi. We then measured neurite outgrowth in cultured DRG neurons transfected with GFP-tagged Mint2-siRNA, GFP-tagged nonsense-siRNA, or Mint2/GFP after 3 days of exposure to NGF. MAP2 staining was performed to distinguish neurons from glial cells. As shown in Fig. 6C, neurite outgrowth was dramatically enhanced in DRG neurons transfected with Mint2-siRNA and attenuated in neurons transfected with Mint2. Statistical analysis revealed that NGF-induced neurite outgrowth, measured as both the length of the longest neurite and total neurite length, was significantly increased in the Mint2 RNAi group and decreased in Mint2 overexpression group compared with the control (Fig. 6, D and E). These results show that endogenous Mint2 acts as a negative regulator of NGF-induced neurite outgrowth, possibly by suppressing TrkA activation.
FIGURE 6.
Mint2 overexpression inhibits NGF-induced neurite outgrowth in cultured DRG neurons. A, Mint2 RNAi efficiency was assessed by Western blot analysis. HEK293 cells were transfected with Myc-Mint2 (lane 1), Myc-Mint2 and nonsense-RNAi (lane 2), Myc-Mint2 and the Mint2-siRNA constructs at 0.5 μg (lane 3, No.1; lane 5, No.2; lane 7, No.3) or 1 μg (lane 4, No.1; lane 6, No.2; lane 8, No.3). Mint2 was detected by blotting with a monoclonal anti-Myc antibody. Actin was used as a loading control. B, GFP-tagged Mint2-siRNA or GFP-tagged nonsense-siRNA plasmids were transfected into primary rat DRG neuron cultures (left panels). 24 h post-transfection cultures were immunostained with anti-Mint2 antibody to examine the expression of Mint2 (right panels). Scale bar, 10 μm. C, primary rat DRG neuron cultures were transfected with GFP tagged nonsense-siRNA, Mint2/GFP, or Mint2-siRNA plasmids and subsequently incubated with NGF for 3 days. The transfected neurons were visualized as both GFP (green) positive and MAP2 staining (red) positive. Scale bar, 20 μm. D and E, quantification of neurite outgrowth in cultured DRG neurons. Image-pro Plus analysis software was used to quantify the length of the longest neurite (D), and Neurolucida software analysis software was used to quantify total neurite length (E). Data are presented as the means ± S.D. The double asterisk indicates significant difference (p < 0.01, Student's t test) compared with control.
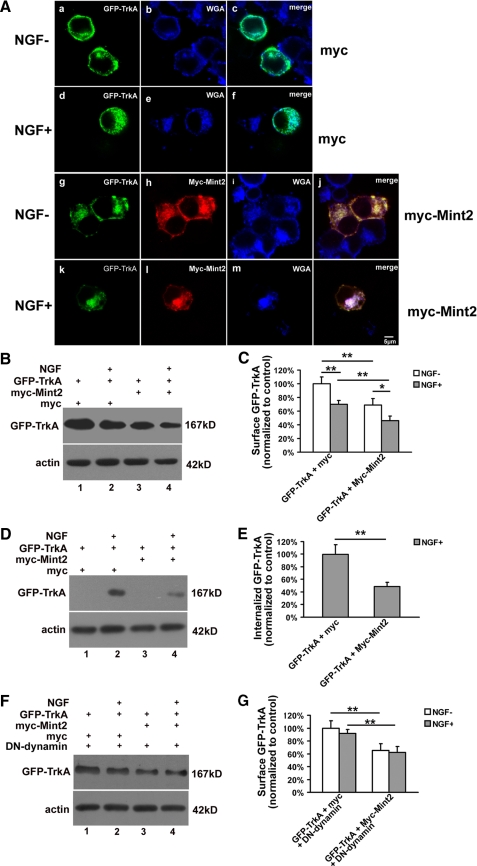
Mint2 Reduces Surface Localization of TrkA—To uncover the mechanism by which Mint2 inhibits NGF-induced neurite outgrowth, GFP-TrkA was cotransfected with either Myc-Mint2 or Myc into PC12 cells at a ratio of 1:10. 24 h after transfection, cells were treated with or without NGF for 30 min. After fixation, cells were stained with anti-Myc monoclonal antibody and the plasma membrane and Golgi apparatus marker WGA-conjugated Alexa Fluor 633 (48, 49). As shown in Fig. 7A, GFP-TrkA was mainly localized at the cell surface without NGF treatment and internalized after NGF stimulation, consistent with a previous report (50). In contrast, the majority of Myc-Mint2 colocalized with WGA, suggesting that Myc-Mint2 was mainly localized in the Golgi apparatus, as has been suggested previously (47). Moreover, GFP-TrkA was found to colocalize with Myc-Mint2 and WGA.
FIGURE 7.
Mint2 reduces surface localization of TrkA. A, PC12 cells cotransfected with GFP-TrkA (green) together with Myc or Myc-Mint2 were incubated with or without NGF for 30 min and then double-stained with anti-Myc monoclonal antibody (red) and WGA (blue). GFP-TrkA was found to colocalize with Myc-mint2 and WGA. B, PC12 cells cotransfected with GFP-TrkA together with Myc, or Myc-Mint2 were starved overnight before being treated with NGF and then subjected to cell surface biotinylation. Biotinylated proteins were recovered by incubating cell lysates with avidin-agarose. The presence of TrkA in surface fractions was analyzed by Western blotting with anti-GFP antibody and quantified as shown in C. D, before NGF treatment, cell surface biotinylation was performed on transfected PC12 cells, and cell-surface biotin was then cleaved off using glutathione cleavage buffer. The biotinylated proteins were recovered by incubating cell lysates with avidin-agarose. The internalized fraction of GFP-TrkA was analyzed by Western blotting with anti-GFP antibody and quantified as shown in E. F, DN-dynamin was cotransfected to block the internalization of TrkA. The surface fraction of GFP-TrkA was detected as described in B and quantified as shown in G. Data represent the mean ± S.D. of three independent experiments. p < 0.01 (**) and p < 0.05 (*) indicate significant difference (Student's t test) compared with the indicated group.
To quantitatively analyze the effect of Mint2 on TrkA trafficking, a surface protein biotinylation assay was performed on PC12 cells that expressed GFP-TrkA together with Myc-Mint2 or Myc. We first examined the surface GFP-TrkA. As shown in Fig. 7, B and C, NGF stimulation significantly reduced surface GFP-TrkA in PC12 cells either transfected with Myc-Mint2 or Myc, suggesting efficient internalization induced by NGF. Interestingly, in cells transfected with Myc-Mint2, the level of GFP-TrkA at the surface was significantly reduced both at rest and after NGF stimulation compared with Myc-transfected controls (Fig. 7, B and C). Further experiments by measuring internalized GFP-TrkA revealed that NGF-induced internalized GFP-TrkA was significantly decreased when Mint2 was overexpressed (Fig. 7, D and E). Previous studies have shown that the internalization of TrkA is dependent on dynamin (51, 52). We, therefore, used a dominant-negative form of dynamin (dynamin-K44A, DN-dynamin) to test whether we could suppress GFP-TrkA internalization. Indeed, when DN-dynamin was cotransfected with GFP-TrkA, NGF treatment did not lead to a reduction of surface GFP-TrkA (Fig. 7, F and G), suggesting efficient blockage of internalization. However, decreased levels of surface GFP-TrkA were still observed when Mint2 was overexpressed along with DN-dynamin and GFP-TrkA (Fig. 7, F and G). Taken together, these results suggest that Mint2 inhibited the surface sorting of GFP-TrkA.
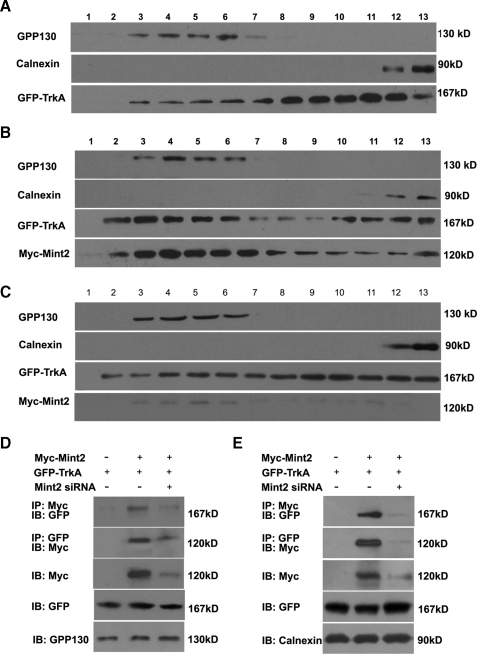
To further clarify the effect of Mint2 on TrkA trafficking, a gradient fractionation assay was carried out on transfected PC12 cells. Using the Golgi protein GPP130 and calnexin to identify Golgi and endoplasmic reticulum-enriched fractions, respectively (53), we found that the level of TrkA-GFP in the Golgi fractions was greatly increased in Myc-Mint2-expressing cells relative to control cells (Fig. 8, A and B). Golgi retention was reversed in cells coexpressing Mint2-siRNA and Myc-Mint2 (Fig. 8C). Moreover, coimmunoprecipitation revealed that Mint2 and GFP-TrkA interacted with each other in both Golgi and endoplasmic reticulum fractions (Fig. 8, D and E). Thus, these results suggest that Mint2 interacts with TrkA to promote TrkA retention in the Golgi apparatus.
FIGURE 8.
Mint2 induces retention of TrkA in the Golgi apparatus. A-C, the postnuclear supernatants of transfected PC12 cells were fractionated and immunoblotted using antibodies against GPP130 (Golgi marker) and calnexin (endoplasmic reticulum marker). Anti-GFP was used to blot for GFP-TrkA, and anti-Myc was used to blot for Myc-Mint2. PC12 cells were transfected with GFP-TrkA together with a pcDNA3 control vector (A), with Myc-Mint2 (B), or with Mint2-siRNA/Myc-Mint2 (C). D and E, the interaction between Myc-Mint2 and GFP-TrkA was detected by coimmunoprecipitation (IP) using the Golgi fractions (D) or endoplasmic reticulum fractions (E) from transfected PC12 cells as indicated. IB, immunoblot.
DISCUSSION
Despite the crucial importance of TrkA-mediated signaling in the nervous system, the mechanisms that regulate its functions and trafficking remain poorly understood. In this study we wished to identify novel TrkA-binding proteins that may represent important effectors or regulators, in the hope of obtaining a better understanding of the varied mechanisms underlying TrkA signaling. Using the intracellular domain of TrkA as bait, we performed a yeast two-hybrid screen and identified Mint2 as a candidate TrkA binding partner.
In our yeast two-hybrid screen, a fragment that included the entire PTB domain of Mint2 was isolated as a candidate binding partner of TrkA. Although structural studies have suggested that the interaction between the C-terminal tail and the PDZ domains of Mint1 has an important regulatory role in the function of Mint1 (54), it was demonstrated here by both yeast two-hybrid and GST pulldown assay that the PTB domain alone was necessary and sufficient for mediating the interaction of Mint2 with the intracellular domain of TrkA. The PTB domain is well known for phosphotyrosine recognition and preferentially binds to phosphorylated proteins containing an NPXpY motif (where pY is phosphotyrosine and X is any amino acid) (55). However, many studies have shown that PTB domain-like modules can also bind to proteins independent of tyrosine phosphorylation or even the canonical NPXY motif. For example, the PTB domains of Mint1 and Fe65 both bind with high affinity to a small peptide containing an NPXY sequence derived from a region of β-amyloid precursor protein, and this stable complex is not affected by tyrosine phosphorylation (21, 29, 56, 57). Similarly, in the present study Mint2 was capable of binding a kinase-dead form of TrkA in a yeast two-hybrid assay. Moreover, our results showed that the Mint2 PTB domain could bind directly to endogenous TrkA in PC12 cells in the absence of NGF by using GST pulldown assay, and interactions between TrkA and Mint2 were not affected by NGF treatment by immunoprecipitation analysis. Thus, our results provide another example showing that PTB domains can bind to receptor-tyrosine kinase in a ligand-independent and phosphorylation-independent manner.
Upon NGF binding to its receptor, the tyrosine kinase domain of TrkA is activated, and those phosphotyrosines become docking sites for adaptor proteins and effectors (2, 6). Thus far, to our knowledge few other studies have demonstrated phosphorylation-independent interaction of TrkA (14). Our data provided evidence that Mint2 interacted with the TrkA-receptor tyrosine kinases in an activity-independent manner, suggesting that Mint2 more likely serves as a regulator protein for TrkA as opposed to a downstream signaling molecule.
The Mint family is well characterized for its role in the formation of multiprotein complexes and its ability to regulate the signaling and trafficking of membrane proteins (20). There is evidence that Mint1 and Mint2 are enriched in the Golgi apparatus but are also distributed throughout axons and dendrites (28, 47). Mints bind to munc-18, a protein essential for synaptic vesicle exocytosis, and to CASK, which is involved in targeting and localization of synaptic membrane proteins (58). The Mint family ortholog in C. elegans, Lin-10, which is required for postsynaptic localization of the glutamate receptor GLR1 in nematodes (59), is present in dendrites and spines and also enriched in the trans-Golgi network; targeting to the Golgi is dependent on its PDZ domain (60). Mints also bind to Arf and colocalize with Arf and β-APP in regions of the Golgi/trans-Golgi network. Mints bind Arfs through a region of the PTB domain and the PDZ2 domain, and Arf-Mint interaction is necessary for increased cellular accumulation of β-APP produced by Mint overexpression (22). Recently Mint3 was reported to regulate trafficking of Furin and to play an obligate role in the traffic of APP from the trans-Golgi network to the plasma membrane (61, 62). These studies indicate that Mint2 may regulate receptor sorting and protein trafficking and that its PDZ domain may serve as a Golgi retention signal. In the present study we found that Mint2 overexpression induced the retention of TrkA in the Golgi apparatus and inhibited TrkA surface sorting.
Expression of TrkA at the cell surface is critical for proper NGF signaling; however, little is known of the mechanisms that regulate TrkA sorting to the plasma membrane. Here, we found that Mint2 inhibits TrkA sorting to the cell membrane, most likely by inducing its retention in Golgi apparatus. We also found that overexpression of Mint2 resulted in a significant decrease in NGF-induced neurite outgrowth in both PC12 cells and DRG neurons, whereas knocking down Mint2 facilitated NGF-induced neurite outgrowth. Thus, Mint2-mediated retention of TrkA in the Golgi may provide a mechanism by which functional regulation of TrkA is achieved during sensory neuron development.
Acknowledgments
We are grateful to Dr. David D. Ginty (Howard Hughes Medical Institute and the Department of Neuroscience, The Johns Hopkins University School of Medicine) for generously providing the TrkA cDNA and Dr. Thomas C. Südhof (Howard Hughes Medical Institute and the Department of Molecular Genetics, University of Texas Southwestern Medical School) for generously providing the Mint2 cDNA.
This work was supported by National Natural Science Foundation of China Grants 30400123, 30570939, 30770657, and 30530240, National Key Basic Research Program Grants 2006CB500702 and 2007CB947100, Program for Changjiang Scholars and Innovative Research Team Grant IRT0528, Shanghai Rising-Star Program Grant 05QMX1469, and Shanghai Metropolitan Fund for Research and Development Grant 07DJ14005.
Footnotes
The abbreviations used are: NGF, nerve growth factor; Arf, ADP-ribosylation factor; β-APP, β-amyloid precursor protein; GST, glutathione S-transferase; WGA, wheat germ agglutinin; TRITC, tetramethylrhodamine isothiocyanate; PTB, the phosphotyrosine binding domain; GFP, green fluorescent protein; siRNA, small interfering RNA; PBS, phosphate-buffered saline; DRG, dorsal root ganglion.
References
- 1.Huang, E. J., and Reichardt, L. F. (2001) Annu. Rev. Neurosci. 24 677-736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huang, E. J., and Reichardt, L. F. (2003) Annu. Rev. Biochem. 72 609-642 [DOI] [PubMed] [Google Scholar]
- 3.Johnson, D., Lanahan, A., Buck, C. R., Sehgal, A., Morgan, C., Mercer, E., Bothwell, M., and Chao, M. (1986) Cell 47 545-554 [DOI] [PubMed] [Google Scholar]
- 4.Kaplan, D. R., Hempstead, B. L., Martin-Zanca, D., Chao, M. V., and Parada, L. F. (1991) Science 252 554-558 [DOI] [PubMed] [Google Scholar]
- 5.Gryz, E. A., and Meakin, S. O. (2000) Oncogene 19 417-430 [DOI] [PubMed] [Google Scholar]
- 6.Kaplan, D. R., and Miller, F. D. (2000) Curr. Opin. Neurobiol. 10 381-391 [DOI] [PubMed] [Google Scholar]
- 7.Liu, H. Y., MacDonald, J. I., Hryciw, T., Li, C., and Meakin, S. O. (2005) J. Biol. Chem. 280 19461-19471 [DOI] [PubMed] [Google Scholar]
- 8.MacDonald, J. I., Gryz, E. A., Kubu, C. J., Verdi, J. M., and Meakin, S. O. (2000) J. Biol. Chem. 275 18225-18233 [DOI] [PubMed] [Google Scholar]
- 9.Meakin, S. O., MacDonald, J. I., Gryz, E. A., Kubu, C. J., and Verdi, J. M. (1999) J. Biol. Chem. 274 9861-9870 [DOI] [PubMed] [Google Scholar]
- 10.Qian, X., Riccio, A., Zhang, Y., and Ginty, D. D. (1998) Neuron 21 1017-1029 [DOI] [PubMed] [Google Scholar]
- 11.Grimes, M. L., Zhou, J., Beattie, E. C., Yuen, E. C., Hall, D. E., Valletta, J. S., Topp, K. S., LaVail, J. H., Bunnett, N. W., and Mobley, W. C. (1996) J. Neurosci. 16 7950-7964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kuruvilla, R., Ye, H., and Ginty, D. D. (2000) Neuron 27 499-512 [DOI] [PubMed] [Google Scholar]
- 13.Riccio, A., Pierchala, B. A., Ciarallo, C. L., and Ginty, D. D. (1997) Science 277 1097-1100 [DOI] [PubMed] [Google Scholar]
- 14.Yano, H., Lee, F. S., Kong, H., Chuang, J.-Z., Arevalo, J. C., Perez, P., Sung, C.-H., and Chao, M. V. (2001) J. Neurosci. 21 RC125, 1-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Geetha, T., and Wooten, M. W. (2003) J. Biol. Chem. 278 4730-4739 [DOI] [PubMed] [Google Scholar]
- 16.Valdez, G., Akmentin, W., Philippidou, P., Kuruvilla, R., Ginty, D. D., and Halegoua, S. (2005) J. Neurosci. 25 5236-5247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Okamoto, M., and Sudhof, T. C. (1997) J. Biol. Chem. 272 31459-31464 [DOI] [PubMed] [Google Scholar]
- 18.Okamoto, M., and Sudhof, T. C. (1998) Eur. J. Cell Biol. 77 161-165 [DOI] [PubMed] [Google Scholar]
- 19.Whitfield, C. W., Benard, C., Barnes, T., Hekimi, S., and Kim, S. K. (1999) Mol. Biol. Cell 10 2087-2100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rogelj, B., Mitchell, J. C., Miller, C. C., and McLoughlin, D. M. (2006) Brain Res. Rev. 52 305-315 [DOI] [PubMed] [Google Scholar]
- 21.Borg, J. P., Ooi, J., Levy, E., and Margolis, B. (1996) Mol. Cell. Biol. 16 6229-6241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hill, K., Li, Y., Bennett, M., McKay, M., Zhu, X., Shern, J., Torre, E., Lah, J. J., Levey, A. I., and Kahn, R. A. (2003) J. Biol. Chem. 278 36032-36040 [DOI] [PubMed] [Google Scholar]
- 23.Ide, N., Hata, Y., Hirao, K., Irie, M., Deguchi, M., Yao, I., Satoh, A., Wada, M., Takahashi, K., Nakanishi, H., and Takai, Y. (1998) Biochem. Biophys. Res. Commun. 244 258-262 [DOI] [PubMed] [Google Scholar]
- 24.Kitano, J., Yamazaki, Y., Kimura, K., Masukado, T., Nakajima, Y., and Nakanishi, S. (2003) J. Biol. Chem. 278 14762-14768 [DOI] [PubMed] [Google Scholar]
- 25.Penzes, P., Johnson, R. C., Sattler, R., Zhang, X., Huganir, R. L., Kambampati, V., Mains, R. E., and Eipper, B. A. (2001) Neuron 29 229-242 [DOI] [PubMed] [Google Scholar]
- 26.McLoughlin, D. M., and Miller, C. C. (1996) FEBS Lett. 397 197-200 [DOI] [PubMed] [Google Scholar]
- 27.Tanahashi, H., and Tabira, T. (1999) Neuroreport 10 2575-2578 [DOI] [PubMed] [Google Scholar]
- 28.Tomita, S., Ozaki, T., Taru, H., Oguchi, S., Takeda, S., Yagi, Y., Sakiyama, S., Kirino, Y., and Suzuki, T. (1999) J. Biol. Chem. 274 2243-2254 [DOI] [PubMed] [Google Scholar]
- 29.Zhang, Z., Lee, C. H., Mandiyan, V., Borg, J. P., Margolis, B., Schlessinger, J., and Kuriyan, J. (1997) EMBO J. 16 6141-6150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Borg, J. P., Yang, Y., De Taddeo-Borg, M., Margolis, B., and Turner, R. S. (1998) J. Biol. Chem. 273 14761-14766 [DOI] [PubMed] [Google Scholar]
- 31.McLoughlin, D. M., Irving, N. G., Brownlees, J., Brion, J. P., Leroy, K., and Miller, C. C. (1999) Eur. J. Neurosci. 11 1988-1994 [DOI] [PubMed] [Google Scholar]
- 32.Sastre, M., Turner, R. S., and Levy, E. (1998) J. Biol. Chem. 273 22351-22357 [DOI] [PubMed] [Google Scholar]
- 33.Zhang, Y., Yan, Z., Farooq, A., Liu, X., Lu, C., Zhou, M. M., and He, C. (2004) J. Mol. Biol. 343 1147-1155 [DOI] [PubMed] [Google Scholar]
- 34.Zhang, Y., Zhu, W., Wang, Y. G., Liu, X. J., Jiao, L., Liu, X., Zhang, Z. H., Lu, C. L., and He, C. (2006) J. Cell Sci. 119 1666-1676 [DOI] [PubMed] [Google Scholar]
- 35.Brown, M., Jacobs, T., Eickholt, B., Ferrari, G., Teo, M., Monfries, C., Qi, R. Z., Leung, T., Lim, L., and Hall, C. (2004) J. Neurosci. 24 8994-9004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bao, L., Jin, S. X., Zhang, C., Wang, L. H., Xu, Z. Z., Zhang, F. X., Wang, L. C., Ning, F. S., Cai, H. J., Guan, J. S., Xiao, H. S., Xu, Z. Q., He, C., Hokfelt, T., Zhou, Z., and Zhang, X. (2003) Neuron 37 121-133 [DOI] [PubMed] [Google Scholar]
- 37.Kuruvilla, R., Zweifel, L. S., Glebova, N. O., Lonze, B. E., Valdez, G., Ye, H., and Ginty, D. D. (2004) Cell 118 243-255 [DOI] [PubMed] [Google Scholar]
- 38.Nagata-Kuno, K., Hino, Y., Nanri, H., Shibata, Y., and Minakami, S. (1990) Exp. Cell Res. 191 273-277 [DOI] [PubMed] [Google Scholar]
- 39.Linstedt, A. D., Mehta, A., Suhan, J., Reggio, H., and Hauri, H. P. (1997) Mol. Biol. Cell 8 1073-1087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Linstedt, A. D., and Hauri, H. P. (1993) Mol. Biol. Cell 4 679-693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Eggert, A., Ikegaki, N., Liu, X., Chou, T. T., Lee, V. M., Trojanowski, J. Q., and Brodeur, G. M. (2000) Oncogene 19 2043-2051 [DOI] [PubMed] [Google Scholar]
- 42.Hanks, S. K., Quinn, A. M., and Hunter, T. (1988) Science 241 42-52 [DOI] [PubMed] [Google Scholar]
- 43.Tarr, P. E., Roncarati, R., Pelicci, G., Pelicci, P. G., and D'Adamio, L. (2002) J. Biol. Chem. 277 16798-16804 [DOI] [PubMed] [Google Scholar]
- 44.Wolf, D. E., McKinnon-Thompson, C., Daou, M. C., Stephens, R. M., Kaplan, D. R., and Ross, A. H. (1998) Biochemistry 37 3178-3186 [DOI] [PubMed] [Google Scholar]
- 45.Chao, M. V., and Hempstead, B. L. (1995) Trends Neurosci. 18 321-326 [PubMed] [Google Scholar]
- 46.Carroll, S. L., Silos-Santiago, I., Frese, S. E., Ruit, K. G., Milbrandt, J., and Snider, W. D. (1992) Neuron 9 779-788 [DOI] [PubMed] [Google Scholar]
- 47.Nakajima, Y., Okamoto, M., Nishimura, H., Obata, K., Kitano, H., Sugita, M., and Matsuyama, T. (2001) Brain Res. Mol. Brain Res. 92 27-42 [DOI] [PubMed] [Google Scholar]
- 48.Gennuso, F., Fernetti, C., Tirolo, C., Testa, N., L'Episcopo, F., Caniglia, S., Morale, M. C., Ostrow, J. D., Pascolo, L., Tiribelli, C., and Marchetti, B. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 2470-2475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ramoino, P., Fronte, P., Fato, M., Beltrame, F., Robello, M., and Diaspro, A. (2001) Eur. Biophys. J. 30 305-312 [DOI] [PubMed] [Google Scholar]
- 50.Jullien, J., Guili, V., Derrington, E. A., Darlix, J. L., Reichardt, L. F., and Rudkin, B. B. (2003) J. Biol. Chem. 278 8706-8716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Deinhardt, K., Reversi, A., Berninghausen, O., Hopkins, C. R., and Schiavo, G. (2007) Traffic 8 1736-1749 [DOI] [PubMed] [Google Scholar]
- 52.Scaife, R., and Margolis, R. L. (1990) J. Cell Biol. 111 3023-3033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jesch, S. A., and Linstedt, A. D. (1998) Mol. Biol. Cell 9 623-635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Long, J. F., Feng, W., Wang, R., Chan, L. N., Ip, F. C., Xia, J., Ip, N. Y., and Zhang, M. (2005) Nat. Struct. Mol. Biol. 12 722-728 [DOI] [PubMed] [Google Scholar]
- 55.Gustafson, T. A., He, W., Craparo, A., Schaub, C. D., and O'Neill, T. J. (1995) Mol. Cell. Biol. 15 2500-2508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chien, C. T., Wang, S., Rothenberg, M., Jan, L. Y., and Jan, Y. N. (1998) Mol. Cell. Biol. 18 598-607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zambrano, N., Buxbaum, J. D., Minopoli, G., Fiore, F., De Candia, P., De Renzis, S., Faraonio, R., Sabo, S., Cheetham, J., Sudol, M., and Russo, T. (1997) J. Biol. Chem. 272 6399-6405 [DOI] [PubMed] [Google Scholar]
- 58.Butz, S., Okamoto, M., and Sudhof, T. C. (1998) Cell 94 773-782 [DOI] [PubMed] [Google Scholar]
- 59.Rongo, C., Whitfield, C. W., Rodal, A., Kim, S. K., and Kaplan, J. M. (1998) Cell 94 751-759 [DOI] [PubMed] [Google Scholar]
- 60.Stricker, N. L., and Huganir, R. L. (2003) Neuropharmacology 45 837-848 [DOI] [PubMed] [Google Scholar]
- 61.Han, J., Wang, Y., Wang, S., and Chi, C. (2008) J. Cell Sci. 121 2217-2223 [DOI] [PubMed] [Google Scholar]
- 62.Shrivastava-Ranjan, P., Faundez, V., Fang, G., Rees, H., Lah, J. J., Levey, A. I., and Kahn, R. A. (2008) Mol. Biol. Cell 19 51-64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fritsch, A. (1973) Anal. Biochem. 55 57-71 [DOI] [PubMed] [Google Scholar]