Abstract
In the mammalian hippocampus, changes in the expression of immediate early genes (IEGs) is thought to contribute to long term plastic changes in neurons brought about by learning tasks and high frequency stimulation of synapses. The phosphatase calcineurin has emerged as an important negative regulator of hippocampus-dependent learning and long term potentiation. Here we investigated the possibility that the constraining action of calcineurin on hippocampal plasticity is mediated in part by regulation of gene expression through negative control of transcription factors, such as cAMP-response element (CRE)-binding protein (CREB). We assessed the effect of calcineurin inhibitors on CREB activation by neuronal activity and show that calcineurin activity is in fact required for CREB-mediated gene expression. However, inhibition of calcineurin had disparate effects on the transcriptional induction of CREB-dependent IEGs. We find that the IEG c-fos is unaffected by suppression of calcineurin activity, the plasticity-related genes Egr1/Zif268 and Egr2/Krox-20 are up-regulated, and genes encoding the orphan nuclear hormone receptors Nor1 and Nur77 are down-regulated. We further show that the up-regulation of particular IEGs is probably due to the presence of serum response elements (SREs) in their promoters, because SRE-mediated gene expression is enhanced by calcineurin blockers. Moreover, expression of the c-fos gene, which is unaffected by calcineurin inhibitors, could be down-regulated by mutating the SRE. Conversely, SRE-mediated c-fos induction in the absence of a functional CRE was enhanced by calcineurin inhibitors. Our experiments thus implicate calcineurin as a negative regulator of SRE-dependent neuronal genes.
In the mammalian hippocampus, alterations to IEG3 expression levels induced by behavioral stimuli, such as learning tasks, or by high frequency electrical stimulations are thought to play a role in transforming electrical activity into neuronal modifications that underlie plasticity (1, 2). Synaptic activity-induced changes in IEG expression levels are triggered by intracellular Ca2+ ions that activate a network of signaling pathways involving Ca2+-sensitive protein kinases and phosphatases, which converge on neuronal transcription factors. The Ca2+/calmodulin-activated phosphatase calcineurin (PP2B), is one of the downstream effectors of Ca2+ signals (3).
Several behavioral and electrophysiological studies agree that genetic or pharmacological suppression of calcineurin activity enhances synaptic plasticity, learning, and memory (3, 4). However, the molecular mechanisms underlying this enhancement are not clear. Because calcineurin can dephosphorylate many presynaptic and postsynaptic proteins, including those involved in neurotransmission, Ca2+ homeostasis, and gene expression, it could modulate synaptic plasticity through multiple mechanisms (3, 5). A recent study showed that in the amygdala, calcineurin inhibition correlates with increased expression of the IEG Egr1/Zif268 and strengthens memory traces, making them resistant to extinction (6), suggesting that alterations in gene expression may be central to the enhancement of learning and memory. It has been speculated that calcineurin activity attenuates signaling to transcription factors by opposing the activating actions of protein kinases on transcription factors, such as CREB (6, 7). CREB activation requires its phosphorylation on serine 133, which allows CREB to associate with the coactivator CBP (8). Calcineurin has been shown to negatively modulate CREB activity in hippocampal neurons during short bursts of synaptic activity by promoting dephosphorylation of serine 133 through activation of the CREB phosphatase PP1 (9). In contrast, recent work has implicated calcineurin in positively regulating CREB-dependent gene expression in neurons by promoting nuclear translocation of the newly identified CREB coactivators called transducers of regulated CREB activity (TORCs) (10-12). Calcineurin suppression can also inhibit CREB-dependent gene expression in a TORC-independent manner (13).
These contradictory suggestions in literature pertaining to the role of calcineurin in CREB regulation prompted us to examine the effects of the calcineurin inhibitors on CREB activation. We show here that in hippocampal neurons, calcineurin activity is required for CREB-mediated gene expression induced by membrane depolarization and synaptic activity and by increases in intracellular cAMP. We further demonstrate that suppression of calcineurin activity has distinct effects on the expression of different IEGs that contain CREB binding sites. We examined the effects of calcineurin inhibitors on expression of plasticity-associated IEGs c-fos (14, 15), Nor1/Nr4a3 (16), Nur77/Nr4a1 (17), Egr1/Zif268 (18), Egr2/krox-20 (19), and Arc (15). We find that expression of the archetypal IEG c-fos is unaffected, expression of Egr1 and Egr2 is augmented, and expression of Nor1 and Nur77 is attenuated by calcineurin suppression. Furthermore, calcineurin inhibitors enhanced gene expression mediated by the serum response element (SRE) found in the promoter regions of c-fos, Egr1, and Egr2. Expression of the c-fos gene that is unaffected by calcineurin inhibitors could be down-regulated by mutating the SRE and augmented in the absence of a functional CRE. These experiments indicate that calcineurin constrains SRE-mediated gene expression.
Our findings indicate that the effect of calcineurin on expression of plasticity-associated neuronal genes is determined by combinatorial control of multiple transcription factors, some of which are activated and others of which are inhibited by calcineurin.
EXPERIMENTAL PROCEDURES
Cell Culture and Plasmids—Hippocampal neurons were cultured from newborn Wistar rats as described previously (20), except that the growth medium was Neurobasal medium (Invitrogen) containing 2% B27 (Invitrogen), 5% fetal calf serum (PAA Lab, Pasching, Austria), 1 mm l-glutamine, 35 mm glucose (Sigma), 100 units/ml penicillin, and 0.1 mg/ml streptomycin (Invitrogen). 2.4 μm cytosine arabinoside (Sigma) was added to the cultures 2-4 days after plating to inhibit proliferation of nonneuronal cells. The firefly luciferase reporter gene plasmid containing five Gal4 DNA binding sites, 5 × Gal4-E1bluc, and the Gal4CREB expression plasmid have been described previously (20). SRE2tkluc, containing two copies of the c-fos SRE upstream of the firefly luciferase gene was a gift from Prof. Alfred Nordheim (University of Tuebingen, Germany) and has been described previously (21). pRL-TK expressing Renilla luciferase was from Promega (Madison, WI). The Nur77 reporter plasmid, -1800Nur77luc (22), was provided by Prof. Talal Chatila (UCLA). The expression plasmid encoding a constitutively active form of the calcineurin catalytic subunit (pEFTAG-ΔCn) has been described previously (23) and was kindly provided by Prof. Anjana Rao (Harvard Medical School). The plasmids containing the human c-fos gene with in-context promoter mutations of the SRE (pFosΔSRFmyc) or CRE (pFosΔCREmyc) and pSVα1 encoding the human α-globin gene have been described before (24).
Transfection and Luciferase Assays—Hippocampal neurons were transfected with the indicated plasmids using Lipofectamine 2000 (Invitrogen) 8-9 days after plating, as described previously (20). 36 h after transfection, cells were either left untreated or stimulated for 6 h, as indicated, and then subjected to luciferase assays as described previously (20) using the Promega Dual Glo assay kit (Promega). Firefly luciferase activity was normalized to the Renilla luciferase signal, and all measurements were made in duplicate. To inhibit calcineurin, cells were pretreated with either 1 μm cyclosporin A (CsA; Calbiochem, Darmstadt, Germany) or 0.1 μm FK506 (Biomol, Plymouth Meeting, PA), for 10 min before stimulation with either 40 mm KCl, 10 μm forskolin (Calbiochem), 50 μm bicuculline (Sigma) with 2.5 mm 4-aminopyridine (4-AP; Calbiochem) or 50 ng/ml BDNF (Invitrogen) for 6 h.
Immunocytochemistry and Western Blotting—Neurons were fixed in 3% paraformaldehyde in phosphate-buffered saline supplemented with 4% sucrose and processed for immunocytochemistry, as described before (20). The following primary antibodies were used at the indicated dilutions: Nor1 at 1:200 (Santa Cruz Biotechnology, Inc., Santa Cruz, CA); c-Fos (Santa Cruz Biotechnology) at 1:200; HA at 1:200 (Covance, Princeton, NJ). To quantify c-Fos and Nor-1 expression levels, neurons were imaged with a Zeiss laser-scanning confocal microscope (LSM-510) or a Leica TCS SP5 confocal system (Leica Microsystems), and the fluorescence intensity was measured using Image J. A statistical significance test was performed using the SPSS statistical suite (version 13).
Western blotting was performed as described by Lam and Chawla (25). The following antibodies were used at the indicated dilutions: c-Fos (Santa Cruz) at 1:1000; phospho-CREB at 1:5000 (Upstate Biotechnology, Inc., Lake Placid, NY); β-actin at 1:10000 (Sigma); extracellular signal-regulated kinase 1/2 at 1:1000 (Cell Signaling, Beverly, MA).
Real Time Quantitative PCR—Total RNA was extracted from neurons cultured in 35-mm plates using the Qiagen RNeasy minikit according to the manufacturer's instructions (Qiagen, Hilden, Germany). First strand cDNA synthesis was performed using Superscript III RNase H- reverse transcriptase (Invitrogen) and oligo(dT)12-18 primer (Invitrogen). The cDNA prepared was then subjected to real time quantitative PCR (qPCR) analysis based on the SYBR Green I double-stranded DNA staining method using the 2× SensiMix dT kit (Quantace, London, UK). PCR was performed with gene-specific primers (all from Sigma). Primer sequences are provided in supplemental Table 1. The relative expression of genes was determined using a standard curve of threshold cycle value (CT) generated from serial dilutions of an external standard of pooled cDNAs, and the expression level was normalized with the expression of glyceraldehyde-3-phosphate dehydrogenase for comparison among samples. Where neurons were transfected with the human c-fos gene, human c-fos mRNA expression was normalized to the levels of α-globin mRNA to control for transfection efficiency. A statistical significance test was performed using the SPSS statistical suite.
RESULTS
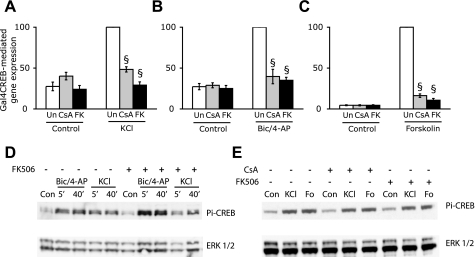
Calcineurin Activity Is Required for CREB-mediated Gene Expression—Previous studies that have tested the effects of calcineurin inhibition on CREB-dependent gene expression in cortical (13) and hippocampal neurons (10) used a luciferase reporter gene containing CREs upstream of the firefly luciferase gene. Because the related basic leucine zipper-containing transcription factors ATF1 (activating transcription factor 1) and CREMτ (cAMP-response element modulator τ) (8) can also bind the CRE, we used a Gal4CREB fusion protein, comprising the GAL4 DNA binding domain and full-length rat CREB, to directly assess the effect of calcineurin inhibitors CsA and FK506 on CREB-mediated gene expression. We found that the Gal4CREB fusion protein is able to robustly activate expression of a cotransfected luciferase reporter gene construct containing five Gal4 DNA binding sites in the promoter following membrane depolarization of cultured hippocampal neurons by 40 mm extracellular KCl (Fig. 1A). The depolarization-induced increase in reporter gene expression was strongly inhibited when neurons were pretreated with calcineurin inhibitors. CsA inhibited depolarization-induced CREB-mediated gene expression to 48.4 ± 3.2% of the KCl-induced response, whereas FK506 reduced it to 29.2 ± 4.8% of the KCl-induced response. This is in agreement with recent studies that show an attenuation of KCl-induced CRE-dependent gene expression by CsA and FK506 in hippocampal and cortical neurons (10, 13). Depolarization of neurons with KCl activates Ca2+ influx through L-type Ca2+ channels. Since CREB-mediated gene expression is also induced robustly following Ca2+ influx through NMDA receptors, we studied the effects of calcineurin inhibition on CREB-mediated gene expression by synaptic NMDA receptors by treating the cells with the GABAA antagonist bicuculline in the presence of the K+ channel blocker 4-AP. Exposure of cultured hippocampal neurons to bicuculline has previously been shown to trigger bursts of action potential firing and NMDA receptor-dependent Ca2+ transients (26). We found that Ca2+ influx through synaptic NMDA receptors resulted in a 3.7-fold increase in CREB-mediated gene expression that was inhibited by CsA and FK506 to 39.7 ± 8.8 and 35.1 ± 3.6, respectively, of the maximal response (Fig. 1B). Neither CsA nor FK506 affected basal expression of the reporter gene. Long term changes in neuronal plasticity involve signaling cascades triggered by both Ca2+ and cAMP, and either of these can stimulate CREB activity. We therefore tested the effects of CsA and FK506 on cAMP-induced CREB activation by exposing hippocampal neurons to the adenylyl cyclase activator forskolin. We found that both CsA and FK506 almost completely inhibited cAMP-induced CREB-mediated gene expression (Fig. 1C). These findings indicate that calcineurin activity is required for optimal CREB-mediated gene expression in response to both Ca2+ and cAMP signals.
FIGURE 1.
Calcineurin inhibitors attenuate CREB-mediated gene expression via a phospho-CREB independent mechanism. Inhibition of calcineurin activity by CsA or FK506 diminishes CREB-mediated gene transcription in neurons stimulated with 40 mm KCl (A), 50 μm bicuculline and 2.5 mm 4-aminopyridine (Bic/4-AP) (B), or 10 μm forskolin (C). Hippocampal neurons cultured in 12-well plates were co-transfected with 1 μg of Gal4CREB, 0.5 μg of 5×Gal4-E1bluc, and 0.1 μg of pRL-TK. 36 h after transfection, cells were pretreated for 15 min with 1 μm CsA or 0.1 μm FK506 or left untreated (Un), followed by stimulation with KCl, forskolin, or Bic/4-AP for 6 h. Firefly luciferase activity was measured and normalized to the Renilla luciferase signal. Data are from three independent transfection experiments and are shown as mean ± S.E. §, significant reduction of luciferase activity in CsA- and FK506-treated cells compared with that in the corresponding stimulation in the absence of calcineurin inhibitors; p < 0.05 (Student's t test). D, FK506 does not affect the kinetics of CREB phosphorylation on serine 133 (Pi-CREB) induced by Bic/4-AP or 50 mm KCl (KCl). Neurons were stimulated with either Bic/4-AP or KCl for 5 min or 40 min in the presence or absence of 0.1 μm FK506 and processed for immunoblotting using an antibody that recognizes CREB phosphorylated on serine 133 (top). Blots were then stripped and probed with an antibody toward extracellular signal-regulated kinase (ERK1/2)(bottom). E, CsA and FK506 do not affect CREB phosphorylation induced by 50 mm KCl or 10 μm forskolin (Fo) Neurons were pretreated for 15 min with either 1 μm CsA or 0.1 μm FK506 or left untreated followed by stimulation for 40 min with 50 mm KCl or 10 μm forskolin and processed for immunoblotting as described for D above.
The first step in CREB activation involves phosphorylation of CREB on serine 133, which is necessary for CREB-mediated gene expression. Previous work by Bito et al. (9) found that FK506 treatment increased the duration for which CREB was phosphorylated following short bursts of synaptic activity. Since sustained and not transient CREB phosphorylation correlates with CREB-mediated gene expression (27), we examined the effects of CsA and FK506 on the kinetics of CREB phosphorylation triggered by KCl and bicuculline/4-AP treatment at 5 min and 40 min. Both KCl and bicuculline/4-AP induced a sustained increase in phospho-CREB levels. FK506 did not affect the time course of CREB phosphorylation induced by either KCl or bicuculline/4-AP treatment (Fig. 1D). We also compared the effects of CsA and FK506 on phospho-CREB levels triggered by KCl and forskolin stimulation of hippocampal neurons at 40 min. Neither CsA nor FK506 had any effect on KCl-induced or forskolin-induced CREB phosphorylation (Fig. 1E). Thus, the inhibition of CREB-mediated gene expression by calcineurin inhibitors is independent of CREB phosphorylation at serine 133.
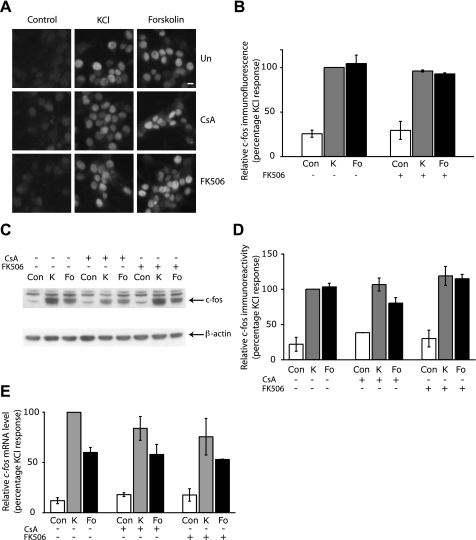
Induction of the IEG c-fos Is Unaffected by Calcineurin Inhibitors—CREB regulates the expression of many activity-induced genes that have complex promoters containing binding sites for many transcription factors. For example, expression of the IEG c-fos is regulated by neuronal activity through two Ca2+-responsive DNA-regulatory sequences, namely the CRE, which binds CREB and related basic leucine zipper transcription factors, and the SRE, which binds SRF and Elk-1 (24). Although we have shown above that CREB-mediated gene expression is attenuated by calcineurin inhibitors, it is not clear what effect the suppression of calcineurin activity would have on a gene such as c-fos that is regulated by additional transcription factors. We therefore investigated the effects of CsA and FK506 on induction of the endogenous c-fos gene by membrane depolarization and by cAMP elevation. We first assessed c-Fos protein expression in hippocampal neurons by immunofluorescence. Fig. 2A shows examples of hippocampal neurons stained with a c-Fos antibody, and quantitative analysis of c-Fos immunofluorescence data is shown in Fig. 2B. As expected, c-Fos protein expression in hippocampal neurons was induced by KCl and forskolin. However, pretreatment of neurons with CsA and FK506 had no effect on c-Fos induction by Ca2+ or cAMP (Fig. 2, A and B). Similar results were obtained when c-Fos protein expression was examined by Western blot analysis (Fig. 2, C and D). We further examined the effect of CsA and FK506 on c-fos expression at the transcriptional level by measuring c-fos mRNA levels by real time quantitative PCR. Fig. 2E shows that, similar to c-Fos protein levels, CsA and FK506 had no significant effect on the induction of c-fos transcripts by membrane depolarization. In CsA-treated neurons, KCl-induced c-fos mRNA levels were found to be 84.0 ± 11.9% of the KCl-induced levels in the absence of calcineurin inhibition, whereas in FK506-treated cells, c-fos transcripts were 75.6 ± 18.2% relative to the expression induced by KCl alone. The forskolin-induced transcription of c-fos mRNA was similarly unaffected by CsA or FK506 treatment (Fig. 2E). This is in contrast to the reported inhibition of the plasticity-related CREB-target genes TrkB and Bdnf by CsA and FK506 in cortical neurons (13). The lack of effect of CsA and FK506 on c-fos induction shows that inhibition of CREB activity by calcineurin blockers is not always sufficient to affect CREB target gene expression.
FIGURE 2.
Expression of c-fos is not affected by the suppression of calcineurin activity. A, representative example of hippocampal neurons showing nuclear c-Fos protein expression in cells pretreated with 1 μm CSA or 0.1 μm FK506 or left untreated (Un) for 15 min followed by stimulation with 50 mm KCl or 10 μm forskolin for 2 h. Scale bar, 10 μm. B, a graph showing the average level of c-Fos immunofluorescence in untreated cells (Con) or cells treated with 50 mm KCl (K) or 10 μm forskolin (Fo) for 2 h, with or without 0.1 μm FK506 pretreatment. Over 50 cells of each treatment from three independent experiments were analyzed. Values are shown as mean ± S.E. C, Western blot showing that the induction of c-Fos protein by 50 mm KCl (K) and 10 μm forskolin (Fo) is not changed by a 15-min pretreatment with either 1 μm CSA or 0.1 μm FK506. Neurons were stimulated for 2 h and processed for c-Fos immunoblotting (top). Immunoblots were stripped and probed with an antibody to β-actin (bottom). D, quantitative analyses of c-Fos protein levels from immunoblots is shown in the graph. The c-Fos and β-actin immunoreactivity was measured using Image J, and c-Fos levels were normalized to that of β-actin. Values are shown as mean ± S.E., and data are from three independent experiments. E, qPCR revealed that the induction of c-fos mRNA expression by 50 mm KCl (K) or 10 μm forskolin (Fo) in neurons stimulated for 50 min was unaffected by a 15-min pretreatment with either 1 μm CSA or 0.1 μm FK506. Values are represented as mean ± S.E., and data are from five independent experiments.
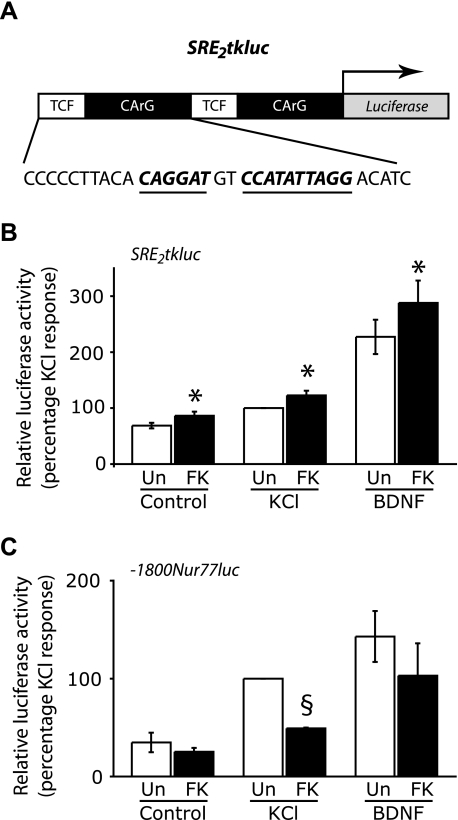
SRE-dependent Gene Expression Is Enhanced by Calcineurin Inhibitors—We hypothesized that the lack of effect of calcineurin suppression on c-fos induction might be due to the presence of an SRE. We therefore tested the effect of inhibiting calcineurin on a luciferase reporter gene driven by two copies of the c-fos SRE. The c-fos SRE, depicted in Fig. 3A, comprises a CArG box that binds SRF as a dimer (28) and a binding site for the ternary complex factor (TCF) Elk-1 (29). We found that expression of the SRE-driven luciferase reporter gene is induced by KCl and BDNF treatment, although the induction by KCl was weak (Fig. 3B). In neurons pretreated with FK506, Ca2+-induced SRE-mediated gene expression was enhanced to 122.7 ± 8.4% of the KCl response in the absence of FK506. We also observed a strong increase of BDNF-induced SRE-dependent gene expression, which was enhanced by 26.8% in FK506-treated neurons (Fig. 3B). In addition to enhanced KCl-induced and BDNF-induced SRE-mediated gene expression, we observed a significant increase (by 24.6%) in SRE-mediated gene expression in untreated hippocampal neurons (Fig. 3B). Although untreated neurons have not been stimulated, they do exhibit spontaneous synaptic activity, indicating that FK506 can enhance SRE-dependent transcription even at low levels of neuronal activity. Similar effects on SRE-dependent gene expression were seen when neurons were exposed to CsA (data not shown).
FIGURE 3.
Inhibition of calcineurin enhances SRE-mediated gene expression but reduces expression of a Nur77 reporter gene driven by CREs and MREs. A, the promoter organization of the SRE2tkluc plasmid used in this experiment. The firefly luciferase reporter gene is driven by two repeats of the c-fos SRE, each comprising a TCF binding site and an SRF binding site (CArG). The TCF binding site and the CArG box are depicted in boldface type and are also underlined. B, SRE-mediated gene expression is enhanced by FK506. Hippocampal neurons cultured in 12-well plates were co-transfected with 0.1 μg of pRL-TK and 0.5 μg of SRE2tkluc and stimulated for 6 h with either 40 mm KCl (K) or 50 ng/ml BDNF (B) in the presence or absence of a 15-min pretreatment with 0.1 μm FK506 followed by luciferase assays. The firefly luciferase activity was measured and normalized to the Renilla luciferase signal. Data are from five independent experiments, and the mean ± S.E. is shown here. *, a significant increase in luciferase activity by FK506 treatment when compared with the corresponding stimulation in the absence of FK506 p < 0.05. (Student's t test). C, FK506 inhibits KCl-induced expression of a Nur77 reporter gene. Neurons were transfected with 0.1 μg of pRL-TK and 0.5 μg of -1800Nur77luc, a firefly luciferase reporter gene driven by a Nur77 promoter that contains two MREs and four CREs. Cells were stimulated as described in B. The firefly luciferase activity was normalized to the Renilla luciferase signal. Data are from three independent experiments, and the mean ± S.E. is shown here. §, significant reduction of luciferase activity by FK506 treatment when compared with the corresponding stimulation in the absence of FK506; p < 0.05 (Student's t test).
The up-regulation of SRE-mediated gene expression may explain why c-fos expression by membrane depolarization is insensitive to CsA and FK506 despite the presence of CREB binding sites in its promoter. We next investigated the effects of CsA and FK506 on activity-induced expression of the IEG Nur77 that is also a CREB target gene. In neuronal cells, transcriptional induction of Nur77 by Ca2+ signals is mediated by CREB (30) and the MEF2 (myocyte enhancer factor-2) family of transcription factors (31). Transcriptional activity of the MEF2 family of transcription factors is activated by calcineurin through multiple mechanisms. Calcineurin enhances MEF2 DNA binding (32) and also participates in altering the sumoylation status of MEF2 transcription factors to cause derepression of MEF2-mediated gene expression (31). Since the Nur77 promoter contains four CRE sites and two MEF2 response elements (MREs) we hypothesized that its induction would be inhibited by CsA and FK506. We used a luciferase reporter construct that contains the firefly luciferase gene downstream of rat Nur77 genomic DNA from -1800 to +119 bp relative to the transcription start site that encompasses the CREs and the MREs. Expression of this Nur77 reporter gene was induced by membrane depolarization. Pretreatment with FK506 strongly inhibited KCl-induced Nur77 expression by 50.8 ± 0.8% (Fig. 3C).
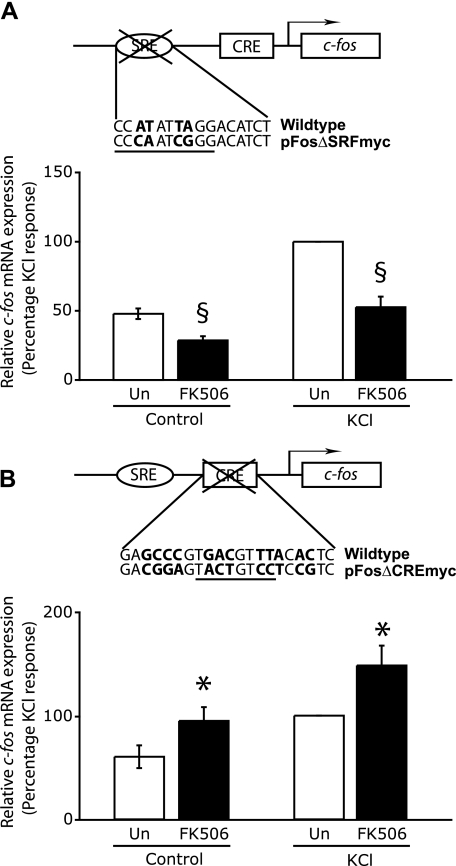
The experiments above suggest that the promoter context in which CREB binding sites are located determine what effect calcineurin inhibitors will have on the expression of a particular gene. Induction of the c-fos gene, which contains one CRE and one SRE was unaffected, whereas a Nur77 reporter gene that contains two MEF2 binding sites in addition to four CREs was down-regulated. To further support the notion that the context in which the CRE exists is an important determinant of a gene's calcineurin sensitivity, we examined the effect of calcineurin inhibition on transcriptional induction of a mutant c-fos gene that lacks either a functional SRE (Fig. 4A) or CRE (Fig. 4B). Hippocampal neurons were transfected with a plasmid containing the entire human c-fos gene, including 711 base pairs of upstream regulatory sequences, which encompasses the SRE and the CRE, with mutations in either the SRF binding site (pFosΔSRFmyc) or in the CREB binding site (pFosΔCREmyc). These plasmid constructs have been characterized previously (24), and it has been established that expression of c-fos from pFosΔSRFmyc is dependent on the CRE and that pFosΔCREmyc is an SRE-dependent c-fos gene. Because these plasmids encode the human c-fos gene, we were able to detect expression of c-fos specifically from the transfected genes by using appropriate primers. We examined the effect of calcineurin inhibitors on c-fos expressed from pFosΔSRFmyc and found that in the absence of a functional SRE, FK506 reduced CRE-mediated c-fos expression by 47.4 ± 7.7% (Fig. 4A). In contrast to this, when we analyzed the effects of FK506 on c-fos transcription when the CRE is mutated, we found that SRE-mediated c-fos expression was enhanced by 48.3 ± 19.2% in neurons treated with FK506 (Fig. 4B). These experiments demonstrate that calcineurin inhibitors have opposite effects on SRE- and CRE-mediated c-fos expression and explain why there is no net effect of calcineurin suppression on the endogenous c-fos gene.
FIGURE 4.
Differential effects of calcineurin inhibition on SRE-mediated and CRE-mediated c-fos expression. A, CRE-mediated c-fos expression is inhibited by FK506. Hippocampal neurons grown in 35-mm dishes for 8 days were transfected with 1 μg of the plasmid pFosΔSRFmyc and 0.5 μg of pSVα1. Plasmid pFosΔSRFmyc contains mutations within the c-fos CArG box (underlined) that binds SRF. The base changes in the mutant are shown in boldface type below the wild-type sequence. 24 h after transfection, cells were either left untreated or pretreated with 0.1 μm FK506 for 15 min, followed by stimulation with 50 mm KCl (KCl) for 50 min. The mRNA levels of human c-fos were determined by qPCR using primers specific for the human c-fos gene and normalized to levels of α-globin mRNA. The normalized c-fos mRNA levels from different conditions are shown in the graph as a percentage of the KCl response. Data are from five independent experiments, and the mean ± S.E. is plotted here. B, SRE-mediated c-fos expression in enhanced by FK506. Hippocampal neurons were transfected and stimulated, and RNA was quantified as in A except that the SRE-dependent reporter gene pFosΔCREmyc was used instead of pFosΔSRFmyc. Plasmid pFosΔCREmyc contains mutations within the c-fos CRE, which is underlined. The base changes in the mutant are shown in boldface type under the wild-type sequence. §, a significant reduction; *, a significant increase in relative mRNA expression in FK506-treated cells when compared with the corresponding stimulation in the absence of FK506; p < 0.05 (Student's t test).
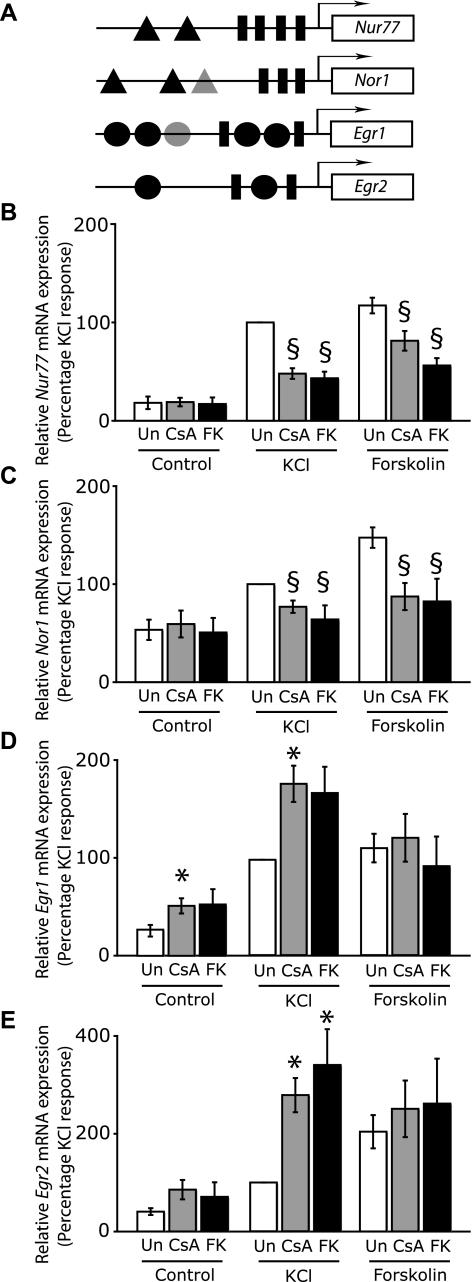
Inverse Regulation of CREB-dependent IEGs—Our results thus far show that the effect of CsA and FK506 on the expression of a particular gene is influenced by the presence of cis-regulatory elements other than the CRE. To generate some guiding rules that may help us predict a gene's sensitivity to calcineurin inhibitors, we compared six plasticity-associated CREB-dependent IEGs whose promoters contain binding sites for other neuronal transcription factors in addition to the CREB binding site(s). We first tested the effect of CsA and FK506 on the transcriptional induction of the endogenous Nur77 gene by qPCR. Similar to the Nur77 reporter gene, Nur77 mRNA levels were induced by KCl and forskolin. CsA and FK506 inhibited KCl-induced Nur77 mRNA levels by 48.0 ± 5.4 and 43.3 ± 6.6%, respectively (Fig. 5B). Forskolin-induced Nur77 induction was also attenuated by CsA and FK506 (Fig. 5B). We next examined how expression of the related orphan nuclear receptor gene Nor1, which is also a CREB target, is affected by calcineurin inhibitors. Nor1 has recently been identified as a plasticity-related gene (16), and its expression is induced by synaptic NMDA receptor activity in hippocampal neurons (33). The Nor1 promoter contains three CREs (Fig. 5A). To identify additional DNA-regulatory sequences, we performed in silico analysis of the Nor1 promoter and found three putative MREs that are conserved in the rat, human, and mouse genes (Fig. 5A and supplemental Fig. 1). Additionally, in preliminary experiments, we observed an increase in Nor1 expression by a constitutively active MEF2-VP16 fusion protein.4 We found that similar to Nur77, transcriptional induction of the Nor1 gene by membrane depolarization and by elevated cAMP was inhibited by CsA and FK506 (Fig. 5C).
FIGURE 5.
Calcineurin differentially regulates the expression of plasticity-related IEGs. A, schematic diagram showing different DNA-regulatory elements present in Nur77, Nor1, Egr1, and Egr2. A triangle represents an MRE, and a putative MRE-like sequence in the Nor1 promoter is shown here in a lighter shade. A rectangle represents a CRE, and a circle represents an SRE. Black circles indicate SREs that contain a TCF binding site within 10 bp. On the Egr1 gene, there is an SRE that is not accompanied by a TCF binding site and is shown here as a lighter circle. The MREs in the Nor1 promoter have been determined in silico, and the alignment is available in supplemental Fig. 1. B-E, hippocampal neurons at 8-9 days in culture were either left untreated or pretreated with 1 μm CsA or 0.1 μm FK506 for 15 min, followed by stimulation with 50 mm KCl (K) or 10 μm forskolin (Fo) for 50 min. The mRNA levels of Nur77 (B), Nor1 (C), Egr1 (D), and Egr2 (E) were determined by qPCR and were normalized to glyceraldehyde-3-phosphate dehydrogenase transcript levels. Data are from five independent experiments and are shown here as mean ± S.E. *, significant increase (p < 0.05) in relative mRNA expression in CsA- or FK506-treated cells when compared with the corresponding stimulation in the absence of calcineurin inhibitors. §, significant reduction in relative mRNA expression in CsA- or FK506-treated cells when compared with the corresponding stimulation in the absence of calcineurin inhibitors; p < 0.05 (Student's t test).
Whereas the c-fos gene contains one SRE, several other plasticity-associated genes, such as Egr1/Zif268, contain multiple copies of the SRE (34). Our findings that SRE-mediated gene expression is augmented by calcineurin inhibitors predict that CsA and FK506 should enhance Egr1 expression, since its promoter contains five copies of the SRE (Fig. 5A). However, the Egr1 promoter also contains two CREs (Fig. 5A) that could transmit inhibitory effects of CsA and FK506 on to Egr1 transcriptional induction. We therefore examined the effects of CsA and FK506 on Egr1 transcript levels in untreated, KCl-stimulated, and forskolin-treated neurons. We found a marked enhancement of KCl-induced Egr1 mRNA levels by calcineurin inhibitors. CsA increased KCl-induced Egr1 mRNA levels by 79.8 ± 19.1% and FK506 by 70.7 ± 28.0% (Fig. 5D). In contrast to Ca2+-activated gene expression, forskolin-induced Egr1 expression was not significantly changed by calcineurin inhibition (Fig. 5D). However, Egr1 mRNA levels were doubled by CsA in untreated hippocampal neurons that exhibit spontaneous activity (Fig. 5D). We also examined the effects of reduced calcineurin activity on transcriptional induction of the related zinc finger transcription factor Egr2/Krox-20. Similar to Egr1, Ca2+-induced Egr2 expression was strongly enhanced by calcineurin inhibitors; the mRNA levels in CsA- and FK506-treated KCl-stimulated cells were more than twice those measured in KCl-stimulated cells in the absence of calcineurin inhibitors (Fig. 5E). Forskolin-induced Egr2 expression, on the other hand, was unaffected by calcineurin inhibitors (Fig. 5E). These results indicate that endogenous calcineurin activity suppresses Egr1 and Egr2 gene expression by membrane depolarization but does not participate in cAMP-induced expression of these genes.
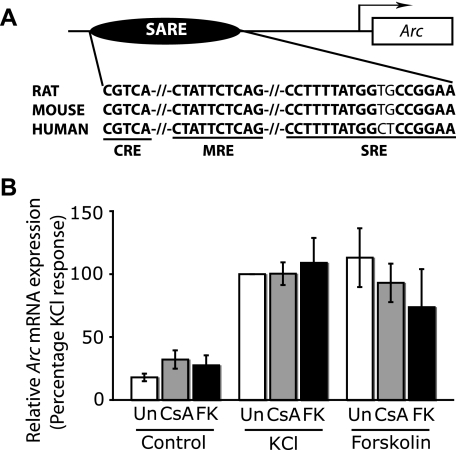
The IEGs examined above encode transcription factors that would direct secondary programs of gene expression, the products of which influence neuronal plasticity by effecting structural and functional changes in neurons. Such IEGs that control the expression of other genes are therefore classed as regulator IEGs. The products of some IEGs, however, called effector IEGs, can directly affect neuronal properties. We next tested how CsA and FK506 may influence the expression of an effector IEG, such as Arc (activity-regulated cytoskeleton-associated protein). Synaptic activity-induced Arc expression is mediated by a 100-bp distal enhancer that comprises the three cis-regulatory elements CRE, MRE, and SRE, which are conserved in rat, mouse, and human genes (Fig. 6A) (35). The Arc gene also contains two proximal SREs (36), but these make a negligible contribution to activity-induced Arc expression (35). Fig. 6 shows that Arc mRNA levels in untreated, KCl-stimulated, and forskolin-stimulated cells are unaffected by CsA and FK506. Thus, calcineurin inhibition has little effect on Arc induction by Ca2+ or cAMP signals.
FIGURE 6.
Expression of the Arc gene is not affected by the inhibition of calcineurin. A, the promoter organization of the Arc gene is depicted here. Synaptic activity-induced Arc expression is mediated by a 100-bp enhancer (SARE) composed of three DNA-regulatory elements, a CRE, an MRE, and a SRE, which are conserved in the rat, mouse, and human genes and are depicted here in boldface type and also underlined. The SRE shown here contains a 3′ CArG box and a 5′ TCF binding site. B, depolarization and cAMP-induced Arc expression are unaffected by CsA and FK506. Hippocampal neurons were either left untreated or pretreated with 1 μm CSA or 0.1 μm FK506 for 15 min, followed by stimulation with 50 mm KCl (K) or 10 μm forskolin (Fo) for 50 min. The mRNA levels of Arc were determined by qPCR and were normalized to glyceraldehyde-3-phosphate dehydrogenase.
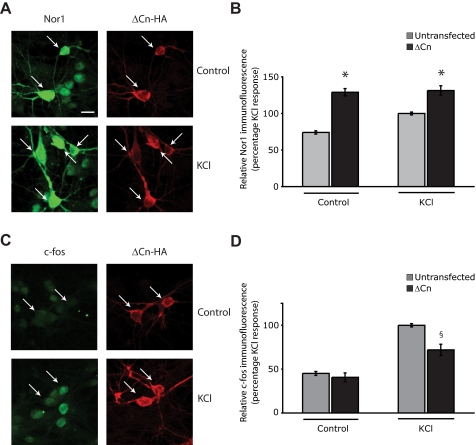
Constitutively Active Calcineurin Has Opposing Effects on Nor1 and c-Fos Expression—The experiments described above used pharmacological inhibitors of calcineurin to show that expression of CREB target genes is down-regulated by calcineurin suppression when they contain MEF2 binding sites in addition to the CRE(s) and that SRE-containing CREB targets are either up-regulated or unaffected. We next performed the converse experiment, were we examined the effect of constitutively active calcineurin on an MRE-containing gene, Nor1, and the SRE containing gene c-fos (Fig. 7, C and D) by immunofluorescence. We transfected hippocampal neurons with an HA-tagged constitutively active mutant of the catalytic subunit of calcineurin (CnAα) (23) and used immunofluorescence to compare the expression of Nor1 and c-Fos protein in cells expressing HA-tagged ΔCn with the expression of Nor1 and c-Fos in untransfected surrounding cells. We found that in untreated ΔCn-expressing hippocampal neurons, Nor1 protein expression was potentiated and was found to be 74.2 ± 6.8% higher than Nor1 expression in untransfected cells (Fig. 7, A and B). Activated calcineurin also increased Nor1 expression in KCl-stimulated neurons to 131.4 ± 6.5% of that observed in untransfected KCl-stimulated neurons (Fig. 7, A and B). In contrast to its effects on Nor1 protein expression, expression of constitutively active ΔCn inhibited c-Fos protein expression in KCl-stimulated cells by 29.1 ± 7.0% (Fig. 7, C and D). These findings further support the notion that CREB-mediated gene expression is enhanced by calcineurin activity, and SRE-mediated gene expression is inhibited by calcineurin.
FIGURE 7.
Constitutively active calcineurin up-regulates Nor1 protein expression but ameliorates c-Fos protein expression. Hippocampal neurons were transfected with 1 μg of pEFTAG-ΔCn encoding a constitutively active mutant of calcineurin. 24 h after transfection, cells were depolarized with 50 mm KCl for 1 h and stained with an antibody to Nor1 (A) or c-Fos (B) along with an HA tag antibody to detect ΔCn expression. A, representative example of neurons showing Nor1 immunoreactivity (green) in ΔCn-transfected cells (red) and surrounding untransfected neurons. The arrows indicate ΔCn-transfected neurons. Scale bar, 15 μm. B, a graph showing the average Nor1 immunofluorescence in ΔCn-transfected and surrounding untransfected cells. *, significant increase in Nor1 immunofluorescence in ΔCn-expressing cells when compared with Nor1 expression in untransfected cells p < 0.05. Over 100 ΔCn-expressing cells were analyzed. C, example of neurons showing reduced nuclear c-Fos expression (green) in ΔCn-expressing cells (red) compared with surrounding untransfected neurons. Arrows, ΔCn-transfected neurons. D, quantitative analysis of c-Fos immunofluorescence is shown in the graph. §, significant decrease in c-Fos immunofluorescence in ΔCn-expressing cells when compared with surrounding untransfected cells with p < 0.05 (Student's t test).
DISCUSSION
We have shown here that in hippocampal neurons, CREB-mediated gene expression in response to membrane depolarization is inhibited by suppression of calcineurin activity, whereas SRE-mediated gene expression is enhanced. The positive effect of calcineurin inhibitors CsA and FK506 on depolarization-induced SRE-mediated gene expression is a novel finding and has implications for the regulation of SRE-containing genes. Our experiments indicate that the effect of calcineurin inhibitors on the activity-induced expression of a particular gene is therefore determined by the overall promoter architecture, including the type of DNA-regulatory elements and their relative numbers. Here we examined six CREB-target genes and found that KCl-induced expression of genes with SREs was either up-regulated (Egr1 and Egr2) or unaffected (c-fos and Arc) by calcineurin inhibitors, depending on the relative number of SREs and CREs. Activity-induced expression of the c-fos gene, which contains one CRE and one SRE was unaffected by calcineurin inhibitors, whereas KCl-induced expression of Egr1 and Egr2, where the number of known SREs is relatively greater, was up-regulated. In contrast to SRE-regulated CREB target genes, we found that expression of CREB-dependent genes that contained MEF2 binding sites in their promoters, such as Nor1 and Nur77, was down-regulated by calcineurin blockers following membrane depolarization. The Arc gene that contains, in addition to a CRE, binding sites for both SRF and MEF2 transcription factors was unaffected by calcineurin inhibition. Thus, it appears that the relative number of binding sites for calcineurin-activated transcription factors (CREB and MEF2) versus calcineurin-inhibited transcription factors (SRE-interacting proteins) determines whether calcineurin inhibitors will inhibit or enhance expression of a gene. In the c-fos gene, the positive effect of calcineurin inhibitors on the SRE overrides the inhibitory effects on CRE-mediated gene expression. This notion is supported by experiments showing that expression of a modified c-fos gene containing an in-context deletion of the CRE is enhanced by calcineurin inhibitors and, conversely, deletion of the SRE results in a down-regulation of the c-fos gene by FK506. In addition to the relative numbers of the three cis-regulatory sequences considered here (CRE, SRE, and MRE), other factors, such as the relative positions of response elements and the presence of binding sites for other calcineurin-regulated transcription factors like NFAT3/c4 (nuclear factor of activated T-cells 3/c4), may influence whether and to what extent transcription of a gene will be affected by calcineurin suppression. It is noteworthy that SRF function is required for activity-induced expression of the genes that were either unaffected or up-regulated in our study (namely c-fos, Egr1, Egr2, and Arc) (37). The knockdown of c-fos induction in SRF mutant mice indicates that SRF activation is necessary for c-fos induction. This explains why in our experiments constitutively active calcineurin, which ameliorates SRE-dependent gene expression, inhibited c-fos induction.
We also examined the effect of calcineurin inhibitors on cAMP-induced CREB-mediated gene expression using forskolin to elevate intracellular cAMP levels. We observed that, similar to Ca2+-induced CREB activation by KCl and bicuculline/4-AP, cAMP-induced CREB activity was strongly inhibited by CsA and FK506. When we investigated the effects of calcineurin inhibitors on cAMP-induced IEG expression, we found that FK506 and CsA inhibited Nor1 and Nur77 expression but did not affect cAMP-induced expression of the other SRE-containing IEGs (c-fos, Egr1, Egr2, and Arc). The lack of effect of CsA and FK506 on forskolin-induced c-fos, Egr1, and Egr2 expression is surprising, given that their cAMP induction depends on CREB activity that is inhibited by CsA and FK506. One possible explanation is that cAMP induction of these genes involves additional transcription factors or DNA-regulatory sequences that are not affected by calcineurin inhibition. One example of a transcription factor that can contribute to cAMP induction of these genes is the CCAAT enhancer-binding protein, C/EBPβ (NF-IL6/LAP), which interacts with the SRE of the c-fos gene (38) and is also recruited to the promoters of Egr1 and Egr2 (39). The activation of C/EBPβ by cAMP is well established (40), and it is possible that in the presence of calcineurin inhibitors, the cAMP-induced transcription of c-fos, Egr1, and Egr2 is mediated by C/EBPβ.
In addition to the activity-induced IEGs examined here, inhibition of calcineurin activity has previously been reported to reduce induction of the IEGs TrkB and Bdnf in cortical neurons (13). Calcineurin also controls expression of other neuronal genes involved in Ca2+ homeostasis that could alter neuronal excitability, such as the IP3 type 1 receptor (41) and the plasma membrane Ca2+ ATPase 4CII (42). It is interesting to note that calcineurin has opposing effects on the expression of the IP3R1 receptor and the plasma membrane Ca2+ ATPase 4CII gene in cerebellar granule neurons. Although the inhibition of IP3R1 expression by calcineurin inhibitors is probably due to its regulation by NFAT3/c4 (43), the mechanism by which plasma membrane Ca2+ ATPase 4CII expression is up-regulated by calcineurin inhibitors remains to be identified. Expression of many other neuronal genes was reported to be down-regulated or up-regulated by calcineurin inhibition in cerebellar granule neurons (44). However, because Kramer et al. (44) treated neurons with calcineurin inhibitors for 2 days before assessing gene expression changes, it is likely that many of the genes identified in their study reflect secondary effects of altered regulator IEGs levels, such as those described here.
Calcineurin activity has thus far been implicated in induction of gene expression via activation of several neuronal transcription factors. Calcineurin activates CRE- and CREB-mediated gene expression by dephosphorylation of the CREB coactivators TORCs and their subsequent nuclear translocation (10, 12). Calcineurin also activates NFAT3/c4 in hippocampal neurons by promoting its nuclear translocation (43). Finally, calcineurin activates MEF2-mediated gene expression by promoting desumoylation of MEF2 transcription factors (31) and by increasing their DNA binding affinity (32). Suppression of calcineurin activity thus inhibits transcriptional activation of CREB, MEF2, and NFAT3/c4 transcription factors in neurons. The positive effect of calcineurin inhibitors on depolarization-induced SRE-mediated gene expression in hippocampal neurons is therefore novel and unusual. An interesting feature of the enhanced effects of CsA and FK506 on SRE-mediated gene expression is that it occurs in untreated spontaneously firing neurons and in cells stimulated with KCl or BDNF. Thus, the increase in SRE function is independent of the level of neuronal activity, suggesting that calcineurin constrains SRE-mediated gene expression through a constitutive mechanism. This raises questions about the molecular mechanisms underlying the increase in SRE-mediated gene expression by calcineurin blockers. The SRE binds two transcription factors, Elk-1 and SRF (45). Additionally, SRF recruits the coactivator MAL/MKL1, a myocardin-related protein (46). Of the three proteins Elk-1, SRF, and MAL, Elk-1 has previously been identified as a calcineurin substrate in nonneuronal cells (47, 48). Elk-1 is a major target of mitogen-activated protein kinases that phosphorylate it on two serine sites (serine 383/389) to positively regulate its transcriptional activity and drive expression of IEGs. Calcineurin inhibitors have been reported to prolong the duration of serine 383/389-phosphorylated Elk-1 (47, 48) and to enhance its transcriptional activity (48) in nonneuronal cell lines. In neurons, Elk-1 is phosphorylated on serines 383 and 389 by NMDA receptor activity, and this promotes its nuclear translocation and ability to induce gene expression (49). Further work will identify whether Elk-1 is dephosphorylated by calcineurin in neurons and whether this contributes to the increase in KCl and BDNF-induced SRE-mediated gene expression seen here.
Our finding that SRE-mediated gene expression is up-regulated by calcineurin inhibitors has implications for neuronal function, since SRF-mediated gene expression has been implicated in neuronal plasticity (37). We have shown here that two plasticity-associated SRE-containing target genes, Egr1/Zif268 and Egr2/Krox-20, are up-regulated by calcineurin inhibitors. Several studies support the hypothesis that endogenous calcineurin restrains long term potentiation and memory, and recent work (6) has established a link between calcineurin inhibition, increased Egr1/Zif268 expression, and establishment of emotional memory. An increase in the expression of SRE-dependent genes may thus underlie the reported enhancing effects of calcineurin inhibition on long term potentiation, learning, and memory.
Supplementary Material
Acknowledgments
We thank Profs. Anjana Rao, Alfred Nordheim, and Talal Chatila for providing plasmid constructs.
This work was supported by the Biotechnology and Biological Sciences Research Council and the Royal Society.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Table 1 and Fig. 1.
Footnotes
The abbreviations used are: IEG, immediate early gene; CREB, cAMP-response element-binding protein; CRE, cAMP-response element; MRE, MEF2 response element; SRE, serum response element; SRF, serum response factor; TORC, transducer of regulated CREB activity; 4-AP, 4-aminopyridine; HA, hemagglutinin; qPCR, quantitative PCR; TCF, ternary complex factor; CsA, cyclosporin A.
B. Y. H. Lam and S. Chawla, unpublished observations.
References
- 1.Guzowski, J. F., Lyford, G. L., Stevenson, G. D., Houston, F. P., McGaugh, J. L., Worley, P. F., and Barnes, C. A. (2000) J. Neurosci. 20 3993-4001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lanahan, A., and Worley, P. (1998) Neurobiol. Learn. Mem. 70 37-43 [DOI] [PubMed] [Google Scholar]
- 3.Mansuy, I. M. (2003) Biochem. Biophys. Res. Commun. 311 1195-1208 [DOI] [PubMed] [Google Scholar]
- 4.Zeng, H., Chattarji, S., Barbarosie, M., Rondi-Reig, L., Philpot, B. D., Miyakawa, T., Bear, M. F., and Tonegawa, S. (2001) Cell 107 617-629 [DOI] [PubMed] [Google Scholar]
- 5.Winder, D. G., and Sweatt, J. D. (2001) Nat. Rev. Neurosci. 2 461-474 [DOI] [PubMed] [Google Scholar]
- 6.Baumgartel, K., Genoux, D., Welzl, H., Tweedie-Cullen, R. Y., Koshibu, K., Livingstone-Zatchej, M., Mamie, C., and Mansuy, I. M. (2008) Nat. Neurosci. 11 572-578 [DOI] [PubMed] [Google Scholar]
- 7.Malleret, G., Haditsch, U., Genoux, D., Jones, M. W., Bliss, T. V., Vanhoose, A. M., Weitlauf, C., Kandel, E. R., Winder, D. G., and Mansuy, I. M. (2001) Cell 104 675-686 [DOI] [PubMed] [Google Scholar]
- 8.Lonze, B. E., and Ginty, D. D. (2002) Neuron 35 605-623 [DOI] [PubMed] [Google Scholar]
- 9.Bito, H., Deisseroth, K., and Tsien, R. W. (1996) Cell 87 1203-1214 [DOI] [PubMed] [Google Scholar]
- 10.Kovacs, K. A., Steullet, P., Steinmann, M., Do, K. Q., Magistretti, P. J., Halfon, O., and Cardinaux, J. R. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 4700-4705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Screaton, R. A., Conkright, M. D., Katoh, Y., Best, J. L., Canettieri, G., Jeffries, S., Guzman, E., Niessen, S., Yates, J. R., III, Takemori, H., Okamoto, M., and Montminy, M. (2004) Cell 119 61-74 [DOI] [PubMed] [Google Scholar]
- 12.Zhou, Y., Wu, H., Li, S., Chen, Q., Cheng, X. W., Zheng, J., Takemori, H., and Xiong, Z. Q. (2006) PLoS ONE 1 e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kingsbury, T. J., Bambrick, L. L., Roby, C. D., and Krueger, B. K. (2007) J. Neurochem. 103 761-770 [DOI] [PubMed] [Google Scholar]
- 14.Dragunow, M., and Robertson, H. A. (1987) Neurosci. Lett. 82 157-161 [DOI] [PubMed] [Google Scholar]
- 15.Guzowski, J. F., Setlow, B., Wagner, E. K., and McGaugh, J. L. (2001) J. Neurosci. 21 5089-5098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sun, W., Choi, S. H., Park, S. K., Kim, S. J., Noh, M. R., Kim, E. H., Kim, H. J., and Kim, H. (2007) J. Neurochem. 100 269-278 [DOI] [PubMed] [Google Scholar]
- 17.French, P. J., O'Connor, V., Voss, K., Stean, T., Hunt, S. P., and Bliss, T. V. (2001) Eur. J. Neurosci. 14 2037-2041 [DOI] [PubMed] [Google Scholar]
- 18.Cole, A. J., Saffen, D. W., Baraban, J. M., and Worley, P. F. (1989) Nature 340 474-476 [DOI] [PubMed] [Google Scholar]
- 19.Williams, J., Dragunow, M., Lawlor, P., Mason, S., Abraham, W. C., Leah, J., Bravo, R., Demmer, J., and Tate, W. (1995) Brain Res. Mol. Brain Res. 28 87-93 [DOI] [PubMed] [Google Scholar]
- 20.Belfield, J. L., Whittaker, C., Cader, M. Z., and Chawla, S. (2006) J. Biol. Chem. 281 27724-27732 [DOI] [PubMed] [Google Scholar]
- 21.Janknecht, R., Ernst, W. H., Pingoud, V., and Nordheim, A. (1993) EMBO J. 12 5097-5104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Blaeser, F., Ho, N., Prywes, R., and Chatila, T. A. (2000) J. Biol. Chem. 275 197-209 [DOI] [PubMed] [Google Scholar]
- 23.Garcia-Rodriguez, C., and Rao, A. (2000) Eur. J. Immunol. 30 2432-2436 [DOI] [PubMed] [Google Scholar]
- 24.Hardingham, G. E., Chawla, S., Johnson, C. M., and Bading, H. (1997) Nature 385 260-265 [DOI] [PubMed] [Google Scholar]
- 25.Lam, B. Y., and Chawla, S. (2007) Neurosci. Lett. 427 153-158 [DOI] [PubMed] [Google Scholar]
- 26.Hardingham, G. E., Fukunaga, Y., and Bading, H. (2002) Nat. Neurosci. 5 405-414 [DOI] [PubMed] [Google Scholar]
- 27.Hardingham, G. E., Chawla, S., Cruzalegui, F. H., and Bading, H. (1999) Neuron 22 789-798 [DOI] [PubMed] [Google Scholar]
- 28.Treisman, R. (1987) EMBO J. 6 2711-2717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shaw, P. E., Schroter, H., and Nordheim, A. (1989) Cell 56 563-572 [DOI] [PubMed] [Google Scholar]
- 30.Fass, D. M., Butler, J. E., and Goodman, R. H. (2003) J. Biol. Chem. 278 43014-43019 [DOI] [PubMed] [Google Scholar]
- 31.Shalizi, A., Gaudilliere, B., Yuan, Z., Stegmuller, J., Shirogane, T., Ge, Q., Tan, Y., Schulman, B., Harper, J. W., and Bonni, A. (2006) Science 311 1012-1017 [DOI] [PubMed] [Google Scholar]
- 32.Mao, Z., and Wiedmann, M. (1999) J. Biol. Chem. 274 31102-31107 [DOI] [PubMed] [Google Scholar]
- 33.Zhang, S. J., Steijaert, M. N., Lau, D., Schutz, G., Delucinge-Vivier, C., Descombes, P., and Bading, H. (2007) Neuron 53 549-562 [DOI] [PubMed] [Google Scholar]
- 34.Alexandre, C., Charnay, P., and Verrier, B. (1991) Oncogene 6 1851-1857 [PubMed] [Google Scholar]
- 35.Kawashima, T., Okuno, H., Nonaka, M., Adachi-Morishima, A., Kyo, N., Okamura, M., Takemoto-Kimura, S., Worley, P. F., and Bito, H. (2009) Proc. Natl. Acad. Sci. U. S. A. 106 316-321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Waltereit, R., Dammermann, B., Wulff, P., Scafidi, J., Staubli, U., Kauselmann, G., Bundman, M., and Kuhl, D. (2001) J. Neurosci. 21 5484-5493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ramanan, N., Shen, Y., Sarsfield, S., Lemberger, T., Schutz, G., Linden, D. J., and Ginty, D. D. (2005) Nat. Neurosci. 8 759-767 [DOI] [PubMed] [Google Scholar]
- 38.Metz, R., and Ziff, E. (1991) Genes Dev. 5 1754-1766 [DOI] [PubMed] [Google Scholar]
- 39.Calella, A. M., Nerlov, C., Lopez, R. G., Sciarretta, C., von Bohlen und Halbach, O., Bereshchenko, O., and Minichiello, L. (2007) Neural Dev. 2 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wilson, H. L., McFie, P. J., and Roesler, W. J. (2001) Mol. Cell. Endocrinol. 181 27-34 [DOI] [PubMed] [Google Scholar]
- 41.Genazzani, A. A., Carafoli, E., and Guerini, D. (1999) Proc. Natl. Acad. Sci. U. S. A. 96 5797-5801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Guerini, D., Wang, X., Li, L., Genazzani, A., and Carafoli, E. (2000) J. Biol. Chem. 275 3706-3712 [DOI] [PubMed] [Google Scholar]
- 43.Graef, I. A., Mermelstein, P. G., Stankunas, K., Neilson, J. R., Deisseroth, K., Tsien, R. W., and Crabtree, G. R. (1999) Nature 401 703-708 [DOI] [PubMed] [Google Scholar]
- 44.Kramer, D., Fresu, L., Ashby, D. S., Freeman, T. C., and Genazzani, A. A. (2003) Mol. Cell. Neurosci. 23 325-330 [DOI] [PubMed] [Google Scholar]
- 45.Treisman, R. (1995) EMBO J. 14 4905-4913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Miralles, F., Posern, G., Zaromytidou, A. I., and Treisman, R. (2003) Cell 113 329-342 [DOI] [PubMed] [Google Scholar]
- 47.Sugimoto, T., Stewart, S., and Guan, K. L. (1997) J. Biol. Chem. 272 29415-29418 [DOI] [PubMed] [Google Scholar]
- 48.Tian, J., and Karin, M. (1999) J. Biol. Chem. 274 15173-15180 [DOI] [PubMed] [Google Scholar]
- 49.Lavaur, J., Bernard, F., Trifilieff, P., Pascoli, V., Kappes, V., Pages, C., Vanhoutte, P., and Caboche, J. (2007) J. Neurosci. 27 14448-14458 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.