Summary
Computationally predicted three-dimensional structure of protein molecules has demonstrated the usefulness in many areas of biomedicine, ranging from approximate family assignments to precise drug screening. For nearly 40 years, however, the accuracy of the predicted models has been dictated by the availability of close structural templates. Progress has recently been achieved in refining low-resolution models closer to the native ones; this has been made possible by combining knowledge-based information from multiple sources of structural templates as well as by improving the energy funnel of physics-based force fields. Unfortunately, there has been no essential progress in the development of techniques for detecting remotely homologous templates and for predicting novel protein structures.
Introduction
Determining the three-dimensional structure of protein molecules is a cornerstone for many aspects of modern biological research. Currently, over 7 million protein sequences are deposited in the UniProtKB/TrEMBL database [1] but only ~50,000 of them have experimentally solved structures [2]. These numbers can be frustrating to molecular and cell biologists who need 3D models of proteins for their research: the chance of a protein domain to have a solved structure has dropped to 0.7% by the end of 2008; this number was 2% in 2004 and 1.2% in 2007. The high demand of the community for protein structures has placed computer-based protein structure prediction, the only means to alleviate the problem, at an unprecedentedly critical position.
Here, a fundamental question arises: How useful are the computationally predicted protein models for biological research? Clearly, the answer to the question depends on how accurate the predicted models are. But is it possible to judge the accuracy of a predicted model without knowing the experimental structure or what factors determine the modeling accuracy of state-of-the-art algorithms? Can the accuracy be improved by refining low-resolution models to high-resolution ones? In this paper, I review the new progresses in the field that are relevant to answer these questions. The literature review is focused on the work published in the last two years.
Accuracy of structure prediction is essential for the biological usefulness
Algorithm and modeling accuracy
Historically, protein structure prediction methods have been divided into three general categories: comparative modeling (CM), threading, and free modeling. In CM, the protein structure is constructed by matching the target protein to an evolutionarily related protein with a solved structure (called a template), where the equivalent residues between the target and the template are found by aligning the sequences or sequence profiles. Threading is designed to match the target sequence directly to the solved 3D structures of template proteins, with the goal of recognizing similar protein folds even in the absence of an evolutionary relationship. Finally, for targets without structurally related solved proteins, models should be built from scratch by free modeling.
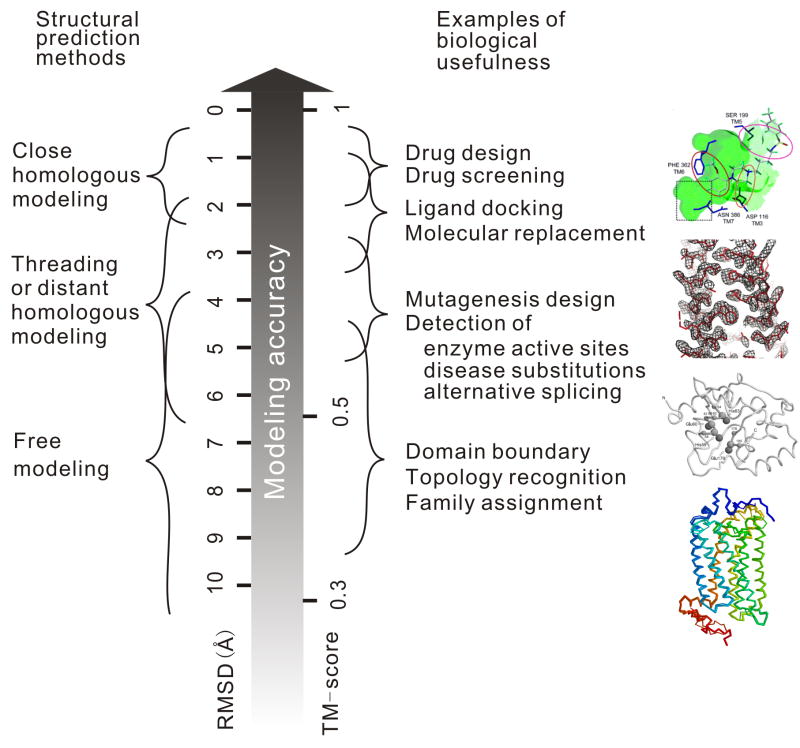
Although the boundary between these different methods is becoming increasingly blurred [3], the accuracy of a predicted model is largely determined by the availability of templates and therefore the prediction approach that can be applied (Figure 1). For proteins with close homologous templates, CM can be used, and most predicted structures have a root mean square deviation (RMSD) of 1–2 Å from the experimental structure, which in some cases achieve the accuracy of medium-resolution NMR or low-resolution X-ray structures [4]. For proteins with distant homologous or analogous templates, threading often identifies correct templates and provides models with an RMSD of 2–6 Å, with errors mainly occurring in the loop regions [5]. For target proteins without solved template structures, successful prediction by free modeling is limited to small proteins (<120 residues), with an accuracy usually in the range of 4–8 Å [6]. For low accuracy models (say, RMSD >3 Å), RMSD is no longer a meaningful measure of modeling quality because a local misorientation of tails or loops, for example, can result in a big overall RMSD even though the core region of the model may be correct. The accuracy of models in this category is usually evaluated by the GDT-score [7] or TM-score [8]. In TM-score, larger distance errors between corresponding predicted and true atom positions are scored with a smaller weight than shorter ones, thus making the score more sensitive to the correctness of the global topology than the local structural errors. By definition, TM-score lies in the [0, 1] interval, with a value >0.5 indicating a model with a roughly correct topology, and a value ≤0.17 indicating a random prediction regardless of the protein size.
Figure 1.
Approximate correspondence of the algorithms, accuracy, and the biological usefulness of protein structure predictions. The pictures in the right panel are representative examples where models of different resolutions are used for different purposes: The first picture shows the 3D model of the lead compound arylpiperazinylsulfonamide docked to the predicted structure of the serotonin receptor. The squared region highlights the interactions specified for serotonin which were exploited to design new compounds with improved selectivity over the adrenergic receptors [10]. The second picture shows the electron density map of Rv2844, a CM target in CASP7, determined from molecular replacement using ROSETTA refined models, with the sticks representing the backbone of the X-ray structure [14]. The third picture is the TASSER model for the YfcM protein from E. coli, with its active sites highlighted, which structurally match with the AFT descriptor associated with EC 3.4.24.69 (metalloendopeptidase); this functional annotation could not be obtained from homology [15]. The fourth picture is the structural superposition of the TASSER models for the orphan RDC1 receptor (thick backbone) and the chemokine CXCR1 receptor (thin backbone) [24••]; the RDC1 receptor was later deorphanized as a chemokine receptor that binds the chemokines CXCL11 and CXCL12 [26].
High-resolution models
High-resolution structure models, typically generated by CM based on close homologous templates, can usually meet the highest structural requirements in the case of single-domain proteins, and are sometimes suitable for computational ligand-binding studies and virtual compound screening. There have been a number of successful examples in which computer-predicted models were used to guide the design of a new drug [9]. Of note, Becker and coworkers [10••] used the predicted structural models of the serotonin receptors to screen a compound library. The docking conformations of the lead compound with the receptor models were then used to guide the design of new compounds with a significantly improved selectivity and affinity, leading to the discovery of a novel agonist for the treatment of anxiety and depression. To benchmark the use of computer models for ligand screening, Brylinski and Skolnick [11•] recently examined the tolerance of ligand-protein docking algorithms towards the accuracy of protein structure predictions. The authors found that 62–87% of binding residues could be correctly recovered using deformed receptor structures with a RMSD of 1–3 Å by Q-DOCK, a knowledge-based reduced docking approach guided by binding restraints from threading algorithms [12].
As another application of homologous modeling, Tramontano and coworkers [13•] evaluated the usefulness of models predicted in CASP experiments for molecular replacement (MR), a procedure for recovering the phase information in X-ray diffraction studies, which is critical for determining the electron density and eventually the 3D structure of crystallized proteins. The authors found that the performance of MR depends on the overall quality of the model used, rather than on the accuracy of local structures. Consequently, models with a GDT-score >0.84 guarantee a success in the MR procedure while models with a GDT-score <0.8 never succeed; the accuracy cutoff in RMSD is blurred as both successes and failures were found with models in 1–4 Å. Moreover, the best available structural templates from threading are much less successful in MR than the complete models, demonstrating the importance of structural refinement. Qian et al. [14••] recently showed that high-resolution models refined from NMR structures, models obtained by CM from close homologous templates, and even models produced by free-modeling techniques, can be used successfully to help phase determination in the molecular replacement procedure.
Medium-resolution models
For models of medium-resolution, roughly in the RMSD range of 2.5~5 Å, typically generated by CM from distantly homologous templates or by fold recognition (Figure 1), the structural predictions can help to identify the spatial locations of functionally important residues, such as active sites and the sites of disease-associated mutations. Arakaki et al. [15] assessed the possibility of assigning the biological function of enzyme proteins by matching the structural patterns (or descriptors) of the active sites with structure decoys of various resolutions. The authors found that models with an RMSD of 3–4 Å from the experimental structure can be used to assign the first three digits of the Enzyme Commission (EC) number with an accuracy of 35%; the accuracy drops to 22% when models of 4–5 Å RMSD are used. Ye et al. [16] used models built by the threading program FFAS to study the structural characteristics of disease-related mutations in the human genome and concluded that the mutations tend to be spatially clustered on protein surfaces and interfaces. Similarly, Yue and Moult [17] performed protein stability analyses based on predicted structural models for the purpose of identifying the deleterious amino acid substitutions in human populations and found that nearly one quarter of the known non-synonymous single nucleotide polymorphisms (nsSNPs) are deleterious to the protein function in vivo. Boyd et al. [18] recently used structural models generated by the automated I-TASSER server [19••] to help interpret mutagenesis experiments with the Sec1/Munc18 (SM) proteins on the basis of the spatial clustering of the mutated residues. Wang et al. [20] used threading models produced by the PROSPECT program to identify splice sites for alternative splicing, a critical eukaryotic cellular process for producing isoform proteins through combining different portions of coding sequences in mRNA. It is found that alternative splice sites are generally in loop regions and on the surfaces of proteins.
Low-resolution models
Even models with the lowest resolution from otherwise meaningful predictions, i.e. models of approximately correct topology from free modeling approaches or based on weak hits from threading, have a number of uses including protein domain boundary identification [21,22], topology recognition, or family/superfamily assignment. Recently, Malmstrom et al. [23] performed a large-scale study of SCOP superfamily assignment based on structural models from free modeling. The authors used the ROSETTA package to predict the 3D structures of small protein domains (<150 residues), selected from the yeast (Saccharomyces cerevisiae) proteome, that are not homologous to known structures. Out of the 3,338 domains, 404 could then be assigned to SCOP superfamilies with high confidence, based on structural comparisons between the predicted models and the SCOP structures; an additional 177 were assigned after integrating the data with the Gene Ontology (GO) annotations. Zhang et al. [24••] predicted structures for all 907 putative G-protein coupled receptors (GPCRs) in the human proteome, with the majority of the targets modeled by free modeling or by assembling weakly homologous fragments. Based on an all-against-all comparison of the predicted structures, GPCRs in the same functional family were found to be more conserved in structure space than in sequence space. This finding establishes the possibility of functional annotation of orphan proteins based on topology-level comparisons of predicted structures. One such instance is the RDC1 receptor, which was considered an orphan receptor for 15 years; its closest but weak relative is the adrenomedullin receptor (AMDR) based on phylogenetic studies [25]. The TASSER structural predictions placed the RDC1 receptor in the family of chemokine receptors because the predicted RDC1 structure is closest to the predicted structure of the CXCR4 chemokine receptor [24••]. This finding was later confirmed by binding experiments [26].
Predicting the accuracy of structural predictions – lessons from CASP experiments
Consensus of templates is a reliable indicator of final model quality
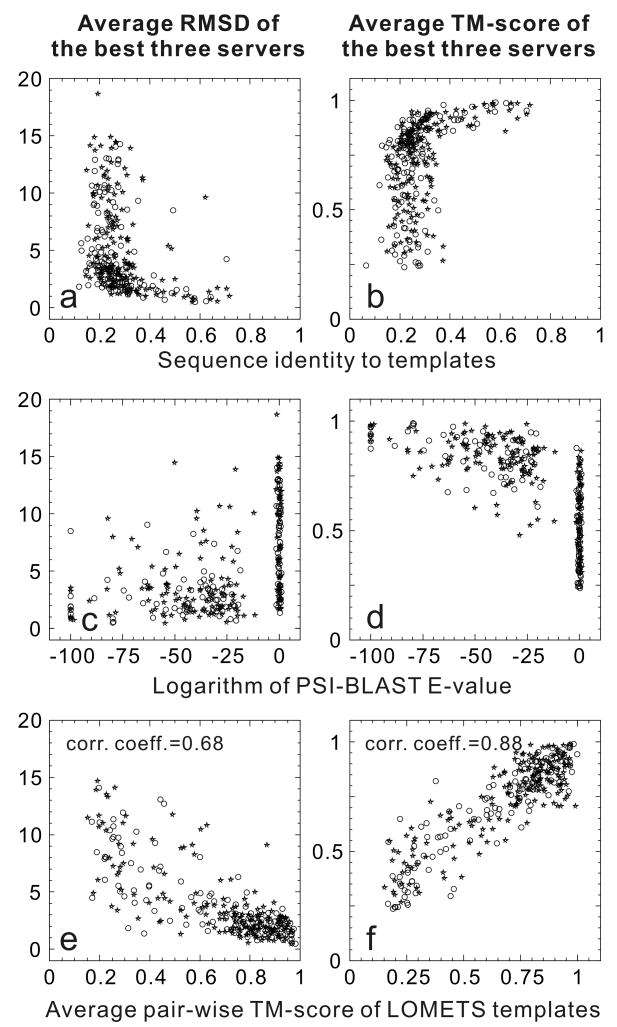
Estimating the accuracy of predicted protein structures is essential for deciding how the models will be used in biological research. Because structure predictions based on existing template structures (including comparative modeling and threading) are the most reliable approaches for producing high-resolution models, an often used and very naïve way of estimating the quality of final models is to check the percentage of identical residues between the target and the template sequences. We tested the relationship between model quality and target-template sequence identity on the 293 protein targets/domains used in the recent two CASP experiments [27] (124 from CASP7 and 169 from CASP8). Figures 2a and 2b show the average RMSD and TM-score, respectively, of the best three server predictions versus the sequence identity between each target and its closest template. To identify the closest template, the PDB structures deposited after the CASP target release date were excluded from the template library used here. As the figures show, for targets with a sequence identity higher than 35~40% with the template, the state-of-the-art algorithms can always build high-quality models with a TM-score >0.8 (or an RMSD <2 Å in the core region), and there is no significant variation in model quality for targets with a sequence identity from 40% to 70%. On the other hand, for sequence identities <35%, there is no correlation between sequence identity and the quality of final models. This is understandable because many protein families (e.g. the globin family) are diverse in sequence and the pair-wise sequence identity is low but their folds can be easily identified by sequence-profile alignment tools such as PSI-BLAST.
Figure 2.
Correlation of accuracy of state-of-the-art structure predictions with different pre-modeling parameters. (a, b) the sequence identity of targets to templates; (c, d) E-value of PSI-BLAST search; (e, f) structural consensus of the templates by LOMETS [29]. The data come from 293 targets in the CASP7 and CASP8 experiments with open circles indicating the CASP7 targets and stars the CASP8 targets. The RMSD and TM-score were calculated from the average of the best three groups for each target. Sequence identity is from the pairwise sequence alignment by BLAST. BLAST, PSI-BLAST and LOMETS were run on a target-specific template library which excludes structures published after the target was released in CASP. The left panel shows RMSD and the right panel shows TM-score as measures of model accuracy
Another often-used, simple approach to estimate model quality is based on the E-value of the templates hit by PSI-BLAST [28], which is a measure of the statistical significance of the hit and defined as the expected number of alignments to be found in the given database by chance with a score higher than the hit. As shown in Figures 2c and 2d, an E-value cut-off (say, <0.001) can pick up more targets with good predictions than a sequence identity cut-off of 40%. However, there are a number of protein targets with a high E-value that still have high-quality models, which shows that the E-value is not a good indicator of model quality when state-of-the-art prediction methods are used.
Because most CASP predictors use templates found by sophisticated threading techniques, a reliable estimate of modeling quality should come from the parameters related to the quality of the threading templates on which the modeling is based. In Figures 2e and 2d, we present the final model qualities versus the average pair-wise TM-score between the top templates identified by LOMETS, a meta-server [29] unifying the results from 8 state-of-the-art individual threading programs (FUGUE, HHsearch, MUSTER, PPA, PROSPECT2, SAM-T02, SPARKS, and SP3). Again, templates structures deposited in the PDB after the CASP target release date were removed. For each target, 8 threading alignments are collected from the first hit of each program which produces 28 pair-wise TM-scores. The average from the top half of the 14 TM-scores, 〈TM-score〉half, reflects the consens us of the threading templates and are used in Figures 2e and 2f. First, 〈TM-score〉half strongly correlates with the TM-score between the best LOMETS template and the experimental target structure, with a correlation coefficient of 0.92 (data not shown). This is understandable because each individual threading program generally has a much higher chance to produce an incorrect hit than a correct one due to the astronomically large alignment space. Thus, if the same template and alignment is identified by multiple different threading programs then it is much more likely to be correct than incorrect. Thus, a consensus measure defined as the average TM-score of multiple threading templates constitutes a more reliable estimate of final model accuracy than sequence identity or PSI-BLAST E-value. Structural consensus is also the core concept of many Model Quality Assessment Programs (MQAPs), which have been designed for ranking and selecting models from multiple predictors [30•,31]. For the CASP7 and CASP8 predictions, the correlation coefficients between 〈TM-score〉half from LOMETS and RMSD/TM-score of the final models are 0.68/0.88. If we consider models with a TM-score >0.75 or RMSD <3.5 Å as successful models and use a cutoff of 〈TM-score〉half = 0.75 to predict modeling success, then the false-positive/false-negative rates are 0.08/0.18 for the TM-score, and 0.19/0.18 for the RMSD based criterion.
Correlation of templates and final models
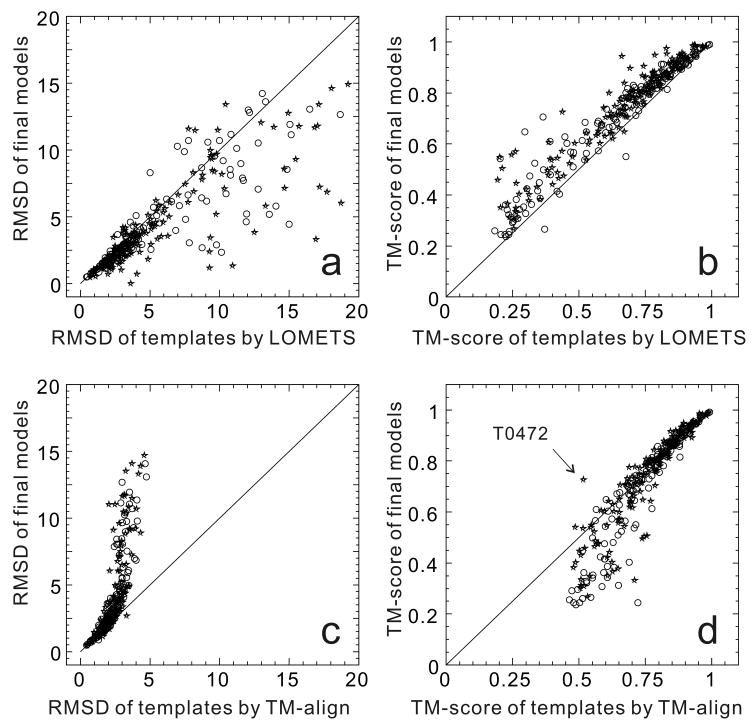
To have a clear view of how the final modeling results are affected by the quality of the initial templates, and whether the modeling procedures can yield an improvement over the templates, we make a head-to-head comparison of the quality of the final models versus the templates from threading or structural alignments. Figure 3a presents the RMSDs (from the experimental target structures) of the best three final models versus the RMSDs of the best threading templates by LOMETS [29], with both types of RMSD calculated for the residues aligned by threading. While there is a general tendency of better templates resulting in better models, the majority of the models are driven closer to the native structure than the best threading templates to the native. The same improvement can also be observed when the TM-score is used for comparison, as shown in Figure 3b. Because the TM-score of final models is calculated for the full-length chain, part of the TM-score increase in the final models is due to the lengthening of the protein chains. Nevertheless, the correlation between the qualities of the LOMETS templates and the final models is more pronounced in the TM-score comparison (correlation coefficient=0.96).
Figure 3.
Comparison of final models with templates. (a, b) templates are from meta-server threading; (c, d) templates are from structural alignment. The data is taken from 293 targets in CASP7 and CASP8 experiments with open circles indicating the CASP7 targets and stars the CASP8 targets. RMSD and TM-score of final models were calculated from the average of the best three groups for each target. RMSD was calculated based on the same aligned regions as the template alignments while TM-score was calculated along the whole chain for the final models. LOMETS and TM-align were run using template libraries excluding structures published after each target was released in CASP. The labeled point in (d) is T0472 which has a duplicated β3α two-domain structure, with the closest structural template from a domain-swapped dimer of 3bid. TM-align matches one chain of 3bid to half of the target structure while the top predictors exploit the whole dimer as a template to model the target which results in a significantly higher TM-score than that by TM-align.
The best templates identified by threading are not necessarily the best templates in the PDB library. In fact, when we use the experimental target structures to search for templates in the PDB library by the structure alignment program TM-align [32], the average TM-score of the best possible templates is 0.76 for all 293 domains; the same calculation yields only 0.65 for the templates identified by LOMETS. In the sense of structure alignments, there were actually no new fold targets in CASPs 7 and 8 because all targets have at least one template with a correct topology (TM-score>0.45). Zhang and Skolnick [33] recently showed that using the best possible templates, almost all the single-domain proteins can be folded with an overall average RMSD of 2.3 Å. It was therefore concluded that the current PDB is an almost complete template library for solving the problem of protein structure prediction at least for single-domain proteins. However, most of the structural alignments for non-homologous proteins could not be recovered by state-of-the-art threading algorithms. In Figures 3c and 3d, we present the RMSDs and TM-scores of the best CASP predictions versus the best templates obtained by structural alignment. Remarkably, for 19% of the targets, the RMSD of the final models is lower than that of the best possible templates in the aligned regions. If TM-score is considered, 54% of models are improved in comparison with the best possible templates. These data demonstrate the significant progress achieved by the community in the area of model refinement, which will be discussed in the next section.
Protein structural refinement: from low to high resolution
The approximate assignment of modeling accuracy to each category of prediction methods in Figure 1 is based on the models without refinement. Further refinement, aiming at pulling the low-resolution models closer to the experimental target structure, can increase the resolution of models, which will undoubtedly extend the scope of biological usefulness of the resulting models across all prediction categories. Protein structure refinement methods can generally be categorized into physics-based and knowledge-based approaches. While physics-based methods try to repack the backbone and side-chain atoms based on physical principles that are supposed to govern the basic atomic interactions, knowledge-based methods rely on statistical potentials and the template information obtained from solved structures in the PDB library.
Knowledge-based structure refinement
One of the most successful knowledge-based approaches to protein structure refinement is the TASSER method, developed by Zhang and Skolnick [34]. TASSER reassembles fragments excised from template structures based on threading alignments, using an energy function consisting of a variety of statistical terms derived from structures in the PDB, and energy terms representing spatial restraints from multiple threading templates. Recently, Wu et al. [35] developed a new version of I-TASSER which aims at refining the structures by an iterative implementation of the TASSER assembly procedure. In CASP7, the models generated by I-TASSER had a lower RMSD to the experimental structures in the threading-aligned regions than the initial threading templates for 86 out of 105 template-based modeling (TBM) targets, resulting in an average RMSD reduction from 4.9 to 3.8 Å [36•]. Even in comparison with the best templates identified by structural alignment search using the experimental structure as a query, the I-TASSER models have a higher GDT-score by more than half assessment units in one third of the cases, as assessed by Kopp et al. [6]. In CASP8, the first models by the automated I-TASSER server predictions [19••] is closer to the experimental structures than the best initial threading templates in 127 out of 154 TBM targets while the models are worse than (equal to) the templates in the other 24 (3) cases.
The use of composite restraint information from multiple threading templates appears to be a key factor for the success of the knowledge-based structure refinement. One advantage of using multiple templates is that the regions missing in one template can be built by borrowing information from other templates for the same regions. Second, the consensus structural information from multiple templates is in general more accurate than that from the individual templates; this information can be exploited to correct the errors in the aligned regions of the template as well [37]. There are also a number of multiple-template based MQAP approaches, which try to score and rank models from a pool of multiple structures generated from other modeling algorithms [29,30•,38–40]. MQAP is usually able to produce a set of selected models having, on average, a better quality than the models from the individual algorithms; but the individual models are not refined by MQAP.
Physics-based structure refinement
Compared with knowledge-based methods, physics-based approaches have been more extensively used in the literature for refining the low-resolution models from both template-based modeling and free modeling. Early efforts in physics-based structure refinement focused on using molecular dynamics (MD)-based simulations, a computational method designed to move atoms in a protein molecule by solving Newton’s equations of motion using force fields such as AMBER and CHARMM. Except for some isolated instances, however, no systematic improvement was achieved [41]. Recently, Zhu et al. [42••] performed replica-exchange molecular dynamics (REMD) simulations using GROMACS to refine 21 models built by comparative modeling which have an initial backbone RMSD in the secondary structures (SSE-RMSD) ranging from 1.33 to 4.14 Å (with respect to the experimental structure). In the replica-exchange method, MD simulations are performed on replicas at different temperatures, with the purpose of improving the sampling at low temperatures by occasionally exchanging the conformations with those at high temperatures. The authors found that the REMD simulations could often produce structures with lower SSE-RMSD than the initial models. Selecting the structure with the lowest SSE-RMSD from 5 ns trajectories of the five lowest-temperature replicas, the average SSE-RMSD improvement relative to the initial structures was 0.82 Å. The conformations in these trajectories were then ranked by various statistical and physics-based potentials and the five best-scoring structures were selected. The SSE-RMSD improvement (relative to the initial structures) for the best of these five models was 0.24 Å. Although encouraging, the experiment highlights key problems of physics-based structure refinement. First, no atomic potential (both statistical and physics-based) could distinguish the near-native structures from the more distant non-native structures because the energy of the best near-native structure was almost always higher than some of the non-native ones. Second, however, the energy of the native structures was found to be lower than any of the structure decoys in all the tested potentials, including the RAPDF/HB and ROSETTA atomic potentials. A similar tendency was observed by Wroblewska and Skolnick [43] with the AMBER potential. These data indicate that the current energy landscape is actually similar to a golf court with the native state as the deepest hole but lacks a middle-range funnel that could guide the simulation to the target state.
To partly address this issue, Wroblewska et al. [44] recently tried to improve the funnel shape of the physics-based force fields by systematically optimizing the weight factors of individual energy terms through maximizing the correlation of the total energy with the TM-score on a large set of training structure decoys. With the optimization, the correlation of the AMBER FF03/HB energy with the TM-score increased from 0.25 to 0.59 in test decoys. When applying the optimized FF03/HB potential to refine 3,900 low-resolution models generated by TASSER for 39 small proteins (<123 residues), which had an initial C-alpha RMSD from 0 to 8 Å, the authors observed improvements in 70% of the models, and the RMSD reduction was >0.5 Å in 20% of the cases [45••]. The authors used a modified replica-exchange Monte Carlo simulation method [46] to search the conformational space, where the improvement of the energy funnel shape is of the key importance for guiding the simulations towards to the native state. The success is also partly due to the improved ability of the optimized potential to recognize near-native conformations.
Another way of resculpting the funnel shape of a physics-based energy landscape is to introduce long-range spatial restraints. Chen and Brooks [47] incorporated tertiary contact restraints and backbone phi/psi torsion angle restraints, both derived from initial near-native structures, into the CHARMM22/GB potential, and searched the conformational space by REMD. The approach was used to refine five CASP6 CM targets of 70–144 residues. In four cases, considerable improvement, with an RMSD reduction of up to 1 Å, was achieved. Misura et al. [48] also combined spatial distance restraints derived from sequence-structure alignments with all-atom ROSETTA simulations. The approach resulted in one out of the 10 lowest-energy models having an RMSD lower than the initial template in 22 out of 39 testing cases. The authors recently extended their method to the refinement of NMR structures, with the simulations focusing on the structurally variable regions. In 8 out of 10 cases, the refined models were closer to the high-resolution X-ray crystal structures than the starting NMR structures [14••]; these models were also shown to provide better molecular replacement solutions than the NMR models to the X-ray crystallographic phase problem.
Concluding remarks
The protein structure prediction problem could in principle be solved in two ways. The first is to fold all proteins by reproducing the entire folding pathway of the polypeptide chains, from their synthesis on the ribosome to their reaching their unique native states. Accomplishing this remarkable task does not appear possible in the foreseeable future unless an accurate physicochemical description of intra-protein and protein-solvent interactions is developed, not to mention the delicate interactions of proteins with associated ligands and chaperones, which dramatically complicate the problem. The second solution is more engineering-oriented rather than scientific, i.e. experimentally solving the structures of a selected set of proteins so that all proteins of unknown structures should have at least one neighbor with known structure that can be used as a template for predicting their structures by comparative modeling. This has been the goal of the various structural genomics (SG) projects [49]. It has been estimated [50] that at least 16,000 optimally selected new structures need to be determined so that CM can cover 90% of protein domain families. A total of 1,300 protein structures (65% are novel) were determined by the Protein Structure Initiative in the first five years since its launch in 2000, and 3,000 more structures (75% are novel) are planned to be solved in the next five years [51•]. Considering that traditional structural biology has contributed roughly the same number of novel structures as the SG centers in the last two years [49,51•], it will take about one more decade to determine all experimental structures needed to cover 90% of protein families.
At the preSG stage, computationally predicted protein structures, built on structural templates from a variety of threading or homology-based algorithms, have proven to be helpful for drug screening and drug design, designing mutagenesis experiments, detecting active sites, solving the phase problem by molecular replacement, and understanding the effect of disease-associated mutations. Even models based on weakly homologous templates or obtained by free modeling, with the fold correctly predicted, have been used for assigning protein families and identifying approximate domain boundaries. While experimental structures are undoubtedly the most desirable, the applications addressable with predicted models span many needs of biologists. The actual supply and demand are partly reflected by the popularity of online structure prediction servers [31], e.g. the I-TASSER server [19••] alone has generated full-length structure predictions for more than 20,000 unknown proteins submitted by about 2,000 registered scientists during the past 18 months.
One component which is often neglected by the predictors is an estimate of the modeling accuracy, which essentially determines how the predicted models are used by biologists. A number of dedicated algorithms, denoted as MQAP, have recently been developed for assessing the quality of structural models. However, the external MQAP algorithms usually do not know about the internal process by which the models were generated, and cannot utilize the information generated during the modeling, such as how the modeling simulations converged or how similar to each other the initial templates were, even though this information can provide a more precise indication about the quality of the final models [19••]. An analysis of the recent CASP predictions shows that a parameter measuring the consensus of threading template alignments has a correlation coefficient of 0.88 with the TM-score of final models from state-of-the-art predictors. On the other hand, naïve parameters such as template-target sequence identity or PSI-BLAST E-value cannot differentiate good models from bad models for most non-homologous protein targets.
Despite the fact that the PDB library has been shown to be complete at the level of structure alignments [33,52], most threading methods have difficulty in detecting the best target-template pairs when the evolutionary relationship is weak. One reason for the difficulty is that the structural similarity of non-homologous protein pairs is often only partial, spanning ~4–5 secondary structure elements [53], while threading scores based on whole-chain alignments can be confounded by the structurally irrelevant regions and therefore prevent the correct ranking and aligning of the templates. Splitting target sequences into segments and threading the spliced sequence fragments through the structure library may help pick up the correct substructure motifs (Wu and Zhang, Identifying protein substructure similarity by segmental threading, submitted).
Developing efficient algorithms for refining low-resolution models to higher resolution has become a central theme of the field. This has been motivated by several factors. First, most biological and medical applications require atomic-level, high-resolution models; thus, efficient refinement algorithms dramatically extend the scope of use of low-resolution protein structure predictions. Second, with the progress of the SG projects and structural biology, more and more structures become available as modeling templates, whereby the identification of structural templates becomes increasingly obvious and easy, and the importance of ab initio structure prediction from scratch diminishes for the purpose of structure prediction per se, although ab initio modeling on its own is an important scientific problem that is relevant to our understanding of protein folding. In fact, despite significant effort, progress in the development of new threading algorithms for detecting remotely homologous templates, as well as in ab initio structure prediction, has been slow in recent years [3]. Compared with CASP7, there is no obvious difference in the overall performance on the modeling of hard targets in the CASP8 experiment, nor are there notable novel algorithms developed to efficiently address these issues. In contrast, protein structure refinement methods demonstrate the most promising progress among many aspects of structure modeling. This is partly reflected by the fact that structure refinement based methods have dominated the blind tests of recent CASP experiments.
The success of the protein structure refinements is pursued in two aspects. For the knowledge-based approaches, the major driving force comes from the optimal use of structural information from multiple templates, and the optimization of statistical potentials. For the physics-based algorithms, improving the funnel shape of the atomic potentials from a golf-court-like energy landscape is the key where promising results have been attained by optimizing the energy weights and/or introducing external long-range spatial restraints.
However, most of the success with physics-based refinement has been limited to small protein domains (typically <150 residues). For larger proteins, the challenge is that a funnel-shaped energy landscape is more difficult to achieve because more interacting subunits and energy terms are involved; moreover, a much larger conformational space needs to be sampled where molecular dynamics and Monte Carlo simulations are prone to be trapped in local energy minima on the rugged landscape. Given the golf-court like energy landscape, there are also efforts to extend the conformational search by more aggressive sampling, which, for example, starts with a huge number of different initial conformations with the aid of worldwide-distributed computing network [54•]. However, unless the initial starting model is already near the native energy state (~1–3 Å), the atom-level refinement does not typically achieve. More efficient sampling strategies should therefore be coupled with the energy funnel optimization of the force fields. For knowledge-based approaches, despite the success in combining structure features from multiple sources of structural templates, the challenge is to generate novel structural ingredients that are not present in any of the templates or initial models. Overall, an optimal combination of knowledge-based and physics-based approaches, including the construction of a composite knowledge-based and physics-based potential of both reduced and atomic levels where the reduced knowledge-based potential is proven to be able to retain the global topology of protein structures and the atomic physics-based component serves to repack the local structural details, may help meet these challenges simultaneously.
Acknowledgments
The author wants to thank Dr. Sitao Wu for preparing the data presented in Figures 2 and 3 and Dr. Andras Szilagyi for critically reading of the manuscript. The project is supported in part by the Alfred P. Sloan Foundation, NSF Career Award (DBI 0746198), and the National Institute of General Medical Sciences (R01GM083107).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
Papers of particular interest, published within the annual period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Bairoch A, Bougueleret L, Altairac S, Amendolia V, Auchincloss A, Puy GA, Axelsen K, Baratin D, Blatter M, Boeckmann B, et al. The universal protein resource (UniProt) Nucleic Acids Res. 2008;36:D190–195. doi: 10.1093/nar/gkm895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang Y. Progress and challenges in protein structure prediction. Curr Opin Struct Biol. 2008;18:342–348. doi: 10.1016/j.sbi.2008.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Read RJ, Chavali G. Assessment of CASP7 predictions in the high accuracy template-based modeling category. Proteins. 2007;69:27–37. doi: 10.1002/prot.21662. [DOI] [PubMed] [Google Scholar]
- 5.Jauch R, Yeo HC, Kolatkar PR, Clarke ND. Assessment of CASP7 structure predictions for template free targets. Proteins. 2007;69:57–67. doi: 10.1002/prot.21771. [DOI] [PubMed] [Google Scholar]
- 6.Kopp J, Bordoli L, Battey JN, Kiefer F, Schwede T. Assessment of CASP7 predictions for template-based modeling targets. Proteins. 2007;69:38–56. doi: 10.1002/prot.21753. [DOI] [PubMed] [Google Scholar]
- 7.Zemla A. LGA: A method for finding 3D similarities in protein structures. Nucleic Acids Res. 2003;31:3370–3374. doi: 10.1093/nar/gkg571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang Y, Skolnick J. Scoring function for automated assessment of protein structure template quality. Proteins. 2004;57:702–710. doi: 10.1002/prot.20264. [DOI] [PubMed] [Google Scholar]
- 9.Ekins S, Mestres J, Testa B. In silico pharmacology for drug discovery: applications to targets and beyond. Br J Pharmacol. 2007;152:21–37. doi: 10.1038/sj.bjp.0707306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••10.Becker OM, Dhanoa DS, Marantz Y, Chen D, Shacham S, Cheruku S, Heifetz A, Mohanty P, Fichman M, Sharadendu A, et al. An integrated in silico 3D model-driven discovery of a novel, potent, and selective amidosulfonamide 5-HT1A agonist (PRX-00023) for the treatment of anxiety and depression. J Med Chem. 2006;49:3116–3135. doi: 10.1021/jm0508641. Computer-based models of the serotonin receptors are used to screen compound libraries and to improve the selectivity and affinity of lead compounds. This is one of the first examples for a Phase III drug candidate designed by in silico model-based methods as the primary tool. [DOI] [PubMed] [Google Scholar]
- •11.Brylinski M, Skolnick J. Q-Dock: Low-resolution flexible ligand docking with pocket-specific threading restraints. J Comput Chem. 2008;29:1574–1588. doi: 10.1002/jcc.20917. Using a knowledge-based reduced docking algorithm Q-DOCK, the authors showed that the ligand-receptor docking method can tolerate structural errors in the receptor models up to 3 Å. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brylinski M, Skolnick J. A threading-based method (FINDSITE) for ligand-binding site prediction and functional annotation. Proc Natl Acad Sci U S A. 2008;105:129–134. doi: 10.1073/pnas.0707684105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •13.Giorgetti A, Raimondo D, Miele AE, Tramontano A. Evaluating the usefulness of protein structure models for molecular replacement. Bioinformatics. 2005;21(Suppl 2):ii72–76. doi: 10.1093/bioinformatics/bti1112. The authors tested the usefulness of computer models predicted in CASP for molecular replacement (MR) and found that the MR result is more sensitive to the accuracy of global conformations than that of the local structures. [DOI] [PubMed] [Google Scholar]
- ••14.Qian B, Raman S, Das R, Bradley P, McCoy AJ, Read RJ, Baker D. High-resolution structure prediction and the crystallographic phase problem. Nature. 2007;450:259–264. doi: 10.1038/nature06249. All-atom ROSETTA is used to refine NMR structures, homologous models and free-modeling models. It is shown for the first time that a model by free modeling can be used for molecular replacement. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Arakaki AK, Zhang Y, Skolnick J. Large scale assesment of the utility of low resolution protein structures for biochemical function assignment. Bioinformatics. 2004;20:1087–1096. doi: 10.1093/bioinformatics/bth044. [DOI] [PubMed] [Google Scholar]
- 16.Ye Y, Li Z, Godzik A. Modeling and analyzing three-dimensional structures of human disease proteins. Pac Symp Biocomput. 2006:439–450. [PubMed] [Google Scholar]
- 17.Yue P, Moult J. Identification and analysis of deleterious human SNPs. J Mol Biol. 2006;356:1263–1274. doi: 10.1016/j.jmb.2005.12.025. [DOI] [PubMed] [Google Scholar]
- 18.Boyd A, Ciufo LF, Barclay JW, Graham ME, Haynes LP, Doherty MK, Riesen M, Burgoyne RD, Morgan A. A random mutagenesis approach to isolate dominant-negative yeast sec1 mutants reveals a functional role for domain 3a in yeast and mammalian Sec1/Munc18 proteins. Genetics. 2008;180:165–178. doi: 10.1534/genetics.108.090423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••19.Zhang Y. I-TASSER server for protein 3D structure prediction. BMC Bioinformatics. 2008;9:40. doi: 10.1186/1471-2105-9-40. The online I-TASSER server has been developed for automatic full-length protein structure prediction. A new scoring function is presented for predicting the accuracy of the final models. I-TASSER was ranked as the best server for tertiary structure prediction in the CASP7 and CASP8 experiments. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang P, Yan B, Guo JT, Hicks C, Xu Y. Structural genomics analysis of alternative splicing and application to isoform structure modeling. Proc Natl Acad Sci U S A. 2005;102:18920–18925. doi: 10.1073/pnas.0506770102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moult J. Comparative modeling in structural genomics. Structure. 2008;16:14–16. doi: 10.1016/j.str.2007.12.001. [DOI] [PubMed] [Google Scholar]
- 22.Tress M, Cheng J, Baldi P, Joo K, Lee J, Seo JH, Lee J, Baker D, Chivian D, Kim D, et al. Assessment of predictions submitted for the CASP7 domain prediction category. Proteins. 2007;69 (Suppl 8):137–151. doi: 10.1002/prot.21675. [DOI] [PubMed] [Google Scholar]
- 23.Malmstrom L, Riffle M, Strauss CE, Chivian D, Davis TN, Bonneau R, Baker D. Superfamily assignments for the yeast proteome through integration of structure prediction with the gene ontology. PLoS Biol. 2007;5:e76. doi: 10.1371/journal.pbio.0050076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••24.Zhang Y, Devries ME, Skolnick J. Structure modeling of all identified G protein-coupled receptors in the human genome. PLoS Comput Biol. 2006;2:e13. doi: 10.1371/journal.pcbi.0020013. 3D models are predicted by TASSER for all 907 putative GPCRs in the human genome and 820 are proven to have a correct topology in the core transmembrane regions. This is the first large-scale structure prediction for such an important membrane protein family in the human genome. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ladoux A, Frelin C. Coordinated Up-regulation by hypoxia of adrenomedullin and one of its putative receptors (RDC-1) in cells of the rat blood-brain barrier. J Biol Chem. 2000;275:39914–39919. doi: 10.1074/jbc.M006512200. [DOI] [PubMed] [Google Scholar]
- 26.Miao Z, Luker KE, Summers BC, Berahovich R, Bhojani MS, Rehemtulla A, Kleer CG, Essner JJ, Nasevicius A, Luker GD, et al. CXCR7 (RDC1) promotes breast and lung tumor growth in vivo and is expressed on tumor-associated vasculature. Proc Natl Acad Sci U S A. 2007;104:15735–15740. doi: 10.1073/pnas.0610444104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moult J, Fidelis K, Kryshtafovych A, Rost B, Hubbard T, Tramontano A. Critical assessment of methods of protein structure prediction-Round VII. Proteins. 2007;69(Suppl 8):3–9. doi: 10.1002/prot.21767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu ST, Zhang Y. LOMETS: A local meta-threading-server for protein structure prediction. Nucl Acids Res. 2007;35:3375–3382. doi: 10.1093/nar/gkm251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •30.Wallner B, Elofsson A. Prediction of global and local model quality in CASP7 using Pcons and ProQ. Proteins. 2007;69:184–193. doi: 10.1002/prot.21774. Pcons was one of the most successful MQAP methods in CASP7. This article shows that the consensus based methods are significantly better than the structure- or evolution-based methods in ranking and selecting protein models. [DOI] [PubMed] [Google Scholar]
- 31.Fischer D. Servers for protein structure prediction. Curr Opin Struct Biol. 2006;16:178–182. doi: 10.1016/j.sbi.2006.03.004. [DOI] [PubMed] [Google Scholar]
- 32.Zhang Y, Skolnick J. TM-align: a protein structure alignment algorithm based on the TM-score. Nucleic Acids Res. 2005;33:2302–2309. doi: 10.1093/nar/gki524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang Y, Skolnick J. The protein structure prediction problem could be solved using the current PDB library. Proc Natl Acad Sci U S A. 2005;102:1029–1034. doi: 10.1073/pnas.0407152101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang Y, Skolnick J. Automated structure prediction of weakly homologous proteins on a genomic scale. Proc Natl Acad Sci U S A. 2004;101:7594–7599. doi: 10.1073/pnas.0305695101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu S, Skolnick J, Zhang Y. Ab initio modeling of small proteins by iterative TASSER simulations. BMC Biol. 2007;5:17. doi: 10.1186/1741-7007-5-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •36.Zhang Y. Template-based modeling and free modeling by I-TASSER in CASP7. Proteins. 2007;69:108–117. doi: 10.1002/prot.21702. The I-TASSER method is tested in CASP7 where 86 out of 105 template-based modeling targets have the final models closer to the experimental structures than the initial templates. [DOI] [PubMed] [Google Scholar]
- 37.Cheng J. A multi-template combination algorithm for protein comparative modeling. BMC Struct Biol. 2008;8:18. doi: 10.1186/1472-6807-8-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McGuffin LJ. Benchmarking consensus model quality assessment for protein fold recognition. BMC Bioinformatics. 2007;8:345. doi: 10.1186/1471-2105-8-345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhou H, Skolnick J. Protein model quality assessment prediction by combining fragment comparisons and a consensus C(alpha) contact potential. Proteins. 2008;71:1211–1218. doi: 10.1002/prot.21813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Archie J, Karplus K. Applying undertaker cost functions to model quality assessment. Proteins: Structure, Function, and Bioinformatics. 2008 doi: 10.1002/prot.22288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee MR, Tsai J, Baker D, Kollman PA. Molecular dynamics in the endgame of protein structure prediction. J Mol Biol. 2001;313:417–430. doi: 10.1006/jmbi.2001.5032. [DOI] [PubMed] [Google Scholar]
- ••42.Zhu J, Fan H, Periole X, Honig B, Mark AE. Refining homology models by combining replica-exchange molecular dynamics and statistical potentials. Proteins. 2008;72:1171–1188. doi: 10.1002/prot.22005. This article presents a systematic study using the replica-exchange MD simulation to refine 21 low-resolution models which are then ranked by statistical potentials. The study highlights the problem of current atomic potentials which have no middle-range funnel towards the native state. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wroblewska L, Skolnick J. Can a physics-based, all-atom potential find a protein’s native structure among misfolded structures? I. Large scale AMBER benchmarking. J Comput Chem. 2007;28:2059–2066. doi: 10.1002/jcc.20720. [DOI] [PubMed] [Google Scholar]
- 44.Wroblewska L, Jagielska A, Skolnick J. Development of a physics-based force field for the scoring and refinement of protein models. Biophysical Journal. 2008;94:3227–3240. doi: 10.1529/biophysj.107.121947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••45.Jagielska A, Wroblewska L, Skolnick J. Protein model refinement using an optimized physics-based all-atom force field. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:8268–8273. doi: 10.1073/pnas.0800054105. The authors performed a large-scale test of structure refinement applied to 39 × 100 models using an optimized atomic potential. 70% of the models are drawn closer to the native conformation. The research shows that the funnel shape of physics-based potentials can be improved by adjusting the weight factors. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang Y, Kihara D, Skolnick J. Local energy landscape flattening: Parallel hyperbolic Monte Carlo sampling of protein folding. Proteins. 2002;48:192–201. doi: 10.1002/prot.10141. [DOI] [PubMed] [Google Scholar]
- 47.Chen J, Brooks CL., 3 Can molecular dynamics simulations provide high-resolution refinement of protein structure? Proteins. 2007;67:922–930. doi: 10.1002/prot.21345. [DOI] [PubMed] [Google Scholar]
- 48.Misura KM, Chivian D, Rohl CA, Kim DE, Baker D. Physically realistic homology models built with ROSETTA can be more accurate than their templates. Proc Natl Acad Sci U S A. 2006;103:5361–5366. doi: 10.1073/pnas.0509355103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chandonia JM, Brenner SE. The impact of structural genomics: expectations and outcomes. Science. 2006;311:347–351. doi: 10.1126/science.1121018. [DOI] [PubMed] [Google Scholar]
- 50.Vitkup D, Melamud E, Moult J, Sander C. Completeness in structural genomics. Nat Struct Biol. 2001;8:559–566. doi: 10.1038/88640. [DOI] [PubMed] [Google Scholar]
- •51.Norvell JC, Berg JM. Update on the protein structure initiative. Structure. 2007;15:1519–1522. doi: 10.1016/j.str.2007.11.004. The authors summarize the accomplishments of the Protein Structure Initiative in solving novel protein structures during the first five years, and the plans for the next five years. [DOI] [PubMed] [Google Scholar]
- 52.Zhang Y, Hubner I, Arakaki A, Shakhnovich E, Skolnick J. On the origin and completeness of highly likely single domain protein structures. Proc Natl Acad Sci U S A. 2006;103:2605–2610. doi: 10.1073/pnas.0509379103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Harrison A, Pearl F, Mott R, Thornton J, Orengo C. Quantifying the similarities within fold space. J Mol Biol. 2002;323:909–926. doi: 10.1016/s0022-2836(02)00992-0. [DOI] [PubMed] [Google Scholar]
- •54.Das R, Qian B, Raman S, Vernon R, Thompson J, Bradley P, Khare S, Tyka MD, Bhat D, Chivian D, et al. Structure prediction for CASP7 targets using extensive all-atom refinement with Rosetta@home. Proteins. 2007;69:118–128. doi: 10.1002/prot.21636. Aggressive sampling using the atomic ROSETTA potential was performed in CASP7 with the aid of a worldwide-distributed computing network. Despite the success in refining some small proteins, conformational sampling appears to be one of major issues in atomic-level structure refinement. [DOI] [PubMed] [Google Scholar]