Abstract
MurA (UDP-N-acetylglucosamine enolpyruvyl transferase) is a key enzyme involved in bacterial cell wall peptidoglycan synthesis and a target for the antimicrobial agent fosfomycin, a structural analog of the MurA substrate phosphoenol pyruvate. In this study, we identified, cloned and sequenced a novel murA gene from an environmental isolate of Vibrio fischeri that is naturally resistant to fosfomycin. The fosfomycin resistance gene was isolated from a genomic DNA library of V. fischeri. An antimicrobial agent hypersensitive strain of Escherichia coli harbouring murA from V. fischeri exhibited a high fosfomycin resistance phenotype, with an MIC of 3000 µg/ml. The cloned murA gene was 1269 bp long encoding a 422 amino acid polypeptide with an estimated pI of 5.0. The deduced amino acid sequence of the putative protein was identified as UDP-N-acetylglucosamine enolpyruvyl transferase by homology comparison. The MurA protein with an estimated molecular weight of 44.7 kDa was expressed in E. coli and purified by affinity chromatography. MurA of V. fischeri will be a useful target to identify potential inhibitors of fosfomycin resistance in pharmacological studies.
Keywords: MurA, Fosfomycin, Antibiotic resistance, Vibrio fischeri, MIC
Introduction
The cell wall of bacteria is composed of alternating units of N-acetyl glucosamine and N-acetyl muramic acid interconnected by penta-peptide cross links. The peptidoglycan cell wall determines the cell shape and provides the essential rigidity to withstand the high internal osmotic pressure. The first committed step in the synthesis of bacterial cell wall takes place in the cytoplasm, which involves the addition of enolpyruvate from phosphoenolpyruvate (PEP) to the 3'-hydroxyl group of UDP-N-acetylglucosamine (UDP-NAG) with the release of inorganic phosphate (Bugg and Walsh 1992). This reaction is catalyzed by the enzyme UDP-N-acetylglucosamine enolpyruvyl transferase (MurA) (EC 2.5.1.7). MurA is an essential enzyme in E. coli, as its inactivation is lethal to the organism due to the loss of cell integrity and susceptibility to osmotic lysis (Brown et al., 1995). This enzyme is a target for the antibiotic fosfomycin which inactivates the enzyme by irreversibly binding to the enzyme forming a covalent adduct with the cysteine 115 residue of E. coli MurA (Kahan et al. 1974; Eschenburg et al. 2005). Fosfomycin is a broad spectrum bactericidal antibiotic that is effective against a range of Gram-positive and Gram-negative bacteria. The antibiotic is used to treat uncomplicated urinary tract infections (UTIs) caused by E. coli and Enterococcus faecalis worldwide (Falagas et al. 2008). Some pathogenic bacteria such as Mycobacterium tuberculosis and Chlamydia trachomatis have been reported to be intrinsically resistant to fosfomycin, and this ability has been attributed to the cysteine-to-aspartate replacement in the active site of the MurA enzyme (De Smet et al. 1999; McCoy et al. 2003).
Bacteria develop resistance to antimicrobial agents through various mechanisms such as the enzymatic modification or degradation of antibiotics, modifications of antibiotic targets or active efflux of antibiotics (Walsh 2000). The resistance to antimicrobial drugs may be intrinsic or develop due to prolonged exposure to sub lethal concentrations of drugs over a period of time. In the majority of bacteria, fosfomycin is transported into cells via the L-α-glycerophosphate (aGP) system (Kahan et al.1974). Fosfomycin resistant mutants can appear spontaneously during the course of antibiotic treatment primarily due to mutations in chromosomal glpT, causing failure to transport fosfomycin (Tsuroka and Yamada 1975). The plasmid encoded glutathione S- transferase enzymes FosA and FosB are implicated in fosfomycin resistance in many species of bacteria (Arca et al. 1990; O'Hara 1999). The fosA gene is reported to be widely distributed among members of the Enterobacteriaceae and Pseudomonas spp. (Teran et al. 1988). The presence of fosB in Gram-positive bacteria such as Staphylococcus aureus, Bacillus subtilis and the fosX gene from Mesorhizobium loti and Listeria monocytogenes have been studied in detail with respect to their role in fosfomycin resistance (Cao et al. 2001; Etienne et al. 1991; Fillgrove et al. 2003).
The objective of our study was to identify and characterize the gene responsible for natural resistance of a V. fischeri strain to fosfomycin. In this report, we describe the identification, cloning and sequencing of a novel murA gene from V. fischeri that confers elevated resistance to the cell wall inhibiting antibiotic fosfomycin.
Materials and methods
Bacterial strains and plasmids
The bacterial strains and the plasmids used in this study are listed in the Table 1. V. fischeri VF107 was grown in a medium consisting of 1% peptone, 0.5% yeast extract and 2% sodium chloride at 30°C. Host E. coli strains KAM32 and SG13900 were grown in Luria Bertani (LB) broth at 37°C. LB broth was supplemented with ampicillin (100 µg/ml), kanamycin (25 µg/ml) and 1 mM IPTG (isopropyl-β-D-thiogalactopyranoside) when required. Plasmid extractions were performed using a commercial kit (Qiagen, USA). Electrotransformation was followed for introducing plasmids into E. coli host strains.
Table 1.
Bacterial strains and plasmids
| Strain/Plasmid | Description/genotype | Source/Reference |
|---|---|---|
| Vibrio fischeri VF107 | Seawater isolate | Laboratory stock |
| E. coli KAM32 | ΔacrB, ΔydhE, hsd− | Chen et al. (2002) |
| E. coli SG 13900 | [pREP4]; NaIS, StrS, RifS, F−, RecA+ | Gottesman et al. (1981) |
| KAM32/pSP72/Vf-murA | V. fischeri murA; AmpR | This study |
| SG/pQE30/Vf-murA | V. fischeri murA; AmpR KanR | This study |
Cloning and sequencing of V. fischeri murA
Genomic DNA of V. fischeri VF107 was extracted following the method of Ausubel et al. (1995). About 10 µg of the pure genomic DNA was digested with the restriction endonuclease enzyme PstI, and the fragments ranging in size from 2–10 kb were purified from an agarose gel after electrophoresis. The plasmid vector pSP72 (Promega, USA) was similarly digested, treated with calf intestinal alkaline phosphatase (Fermentas, USA), ligated with the genomic DNA fragments using a LigaFast DNA ligation kit (Promega, USA) and transformed into E. coli KAM32 (Chen et al. 2002) The transformants were screened on LB agar plates containing 100 µg/ml each of ampicillin and fosfomycin. A 1.7 kb insert was sequenced by dideoxy chain termination procedure on both strands using vector specific primers.
Determination of minimum inhibitory concentration (MIC)
The MICs of fosfomycin to V. fischeri VF107 and E. coli KAM32/pSP72/Vf-murA were determined in Mueller Hinton broth following the CLSI microbroth dilution procedure (CLSI 2006). The assay was performed in a 96-well microtitre plate with appropriate uninoculated media blanks. The test strains were grown to an O.D625 of 0.4, washed and resuspended in fresh MH broth to yield 103–104 cells/ml. A serial 10-fold dilution of the stock antibiotic was performed in MH broth in microtitre plate wells and inoculated with the test strains. The plates were incubated at 37°C for 24 h, and the growth was examined visually. The assay was repeated thrice, and the lowest concentration of fosfomycin that inhibited growth was recorded as the MIC for the test strains.
Overexpression and purification of murA in E. coli
Primers FM-F (5’-ATGGATAGCGCCGAGATTAA-3’) and FM-R (5’-TAAGTCGTCGCTATGAACT-3’) were used to amplify murA for cloning into the expression vector pQE 30 (Qiagen USA) The amplification was done in a 50 µl reaction volume consisting a 1× PCR buffer (10 mM Tris-HCl, 50 mM KCl and 2 mM MgCl2), 200 µM concentrations of each of the four dNTPs, 25 picomoles of respective primer, 3 U of Taq polymerase (MBI Fermentas) and 500 ng of the pure genomic DNA. The PCR product was purified using a PCR purification kit (Qiagen, USA), cloned into pQE-30 UA cloning vector (Qiagen, USA) and transformed into to into E. coli SG13009[pREP4] to yield SG13900/pQE30/Vf-murA.
The 6×His-tagged MurA fusion protein was purified by using nickel chromatography columns following the manufacturer’s protocol (Qiagen USA). Briefly, the recombinant E. coli SG13900/pQE30/Vf-murA was grown in LB broth containing 100 µg/ml ampicillin and 25 µg/ml kanamycin to an OD600 of 0.5, induced with 1 mM IPTG and further incubated for 8 h with shaking. The cells were harvested by centrifugation at 3000 × g for 15 min., and the pellet was lysed using a lysis buffer (6M GuHCl, 0.1M NaH2PO4, 0.01M Tris Cl, pH 8.0) and centrifuged at 11,000 g for 15 min. at room temperature to pellet the cellular debris. The supernatant was loaded onto Ni-NTA affinity columns, centrifuged and the column was washed twice with wash buffer (6M GuHCl, 0.1M NaH2PO4, 0.01M Tris Cl, pH 6.3). The bound recombinant protein was eluted with elution buffer (6M GuHCl, 0.1 M NaH2PO4, 0.01M Tris Cl, pH 4.5), dialyzed against Tris buffer (10 mM Tris Cl pH 8.0, 0.1% Triton X-100) overnight at 4 °C to remove guanidine hydrochloride. The recombinant protein was electrophoresed on a 12% sodium dodecyl sulphate polyacrylamide gel and visualized by staining with Coomassie brilliant blue.
Sequence analysis
The nucleotide sequence derived was subjected to a GenBank homology search using a standard nucleotide-nucleotide BLAST analysis (Altschul et al. 1997). The deduced amino acid sequence of V. fischeri MurA was analyzed for conserved domains at http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi (Marchler-Bauer et al. 2007) and was compared with other homologous sequences in GenBank using the CLUSTAL W multiple sequence alignment program (Higgins et al. 1994).
Nucleotide sequence
The nucleotide sequence derived in this study has been deposited in the GenBank with the accession number FJ172353.
Results and discussion
Vibrio fischeri is a marine bacterium that is either free living or lives in symbiotic association with some marine squids and fish (Ruby and McFall-Ngai 1999). Marine environments have diverse species of bacteria and fungi producing varieties of bioactive compounds, including antibiotics (Bernan et al. 1997). Bacteria living in such a competitive environment tend to develop natural resistance to antibiotics as a strategy to overcome the lethal effects of inhibitory compounds produced by competing bacteria in the same niche. Thus, it is likely that antibiotic resistant bacteria are encountered in the marine environment, despite the fact that bacteria are not considered clinically important and the antibiotic resistance phenotype has not evolved due to exposure to chemotherapeutic use of antibiotics. We found that V. fischeri VF107 was naturally resistant to the antibiotic fosfomycin that could not be attributed to any plasmid-borne gene. In order to identify the gene responsible for fosfomycin resistance, we created a genomic DNA library of V. fischeri in an antimicrobial agent hypersensitive strain E. coli KAM32 (Chen et al. 2002) and screened for fosfomycin resistant clones. This yielded E. coli KAM32/psp72/Vf-murA harboring a V. fischeri genomic fragment of 2400 bp. Sequence analysis of the gene revealed 2 putative open reading frames (ORFs) consisting of a 258 bp toluene tolerance gene ttg2F, followed by a gene encoding UDP-N-acetylglucosamine 1-carboxy vinyl transferase (murA). At the 3’end, a partial sequence of the gene encoding 1-acyl-sn-glycerol-3-phosphate acyltransferase was identified. The organization of the murA gene in V. fischeri was found to be identical with that of V. salmonicida, the whole genome sequences of which are currently available. In the genomes of these two species, the murA gene is preceded by predicted toluene tolerance genes ttg2A, ttg2B, ttg2C, ttg2D and ttg2F.
The minimum inhibitory concentrations (MICs) of fosfomycin was determined for the host E. coli strain KAM32, V. fischeri from which the murA gene was cloned and E. coli KAM32/pSP72/Vf-murA. The results are presented in Table 2. E. coli KAM32 was highly sensitive to fosfomycin with an MIC of 32.5 µg/ml. E. coli KAM32/pSP72/Vf-murA had an MIC of 3000 µg/ml, which was about half of the MIC obtained with V. fischeri VF107 (MIC = 6000 µg/ml). The reason for this difference in MIC levels could not be determined in this study.
Table 2.
Minimum inhibitory concentrations (MICs) of fosfomycin to wild type and recombinant bacterial strains determined by micro broth dilution technique
| Strain/plasmid | MIC µg/ml of fosfomycin |
|---|---|
| E. coli KAM32 | 32.5 |
| V. fischeri VF107 | 6000 |
| E. coli KAM 32/pSP72 /vf-murA | 3000 |
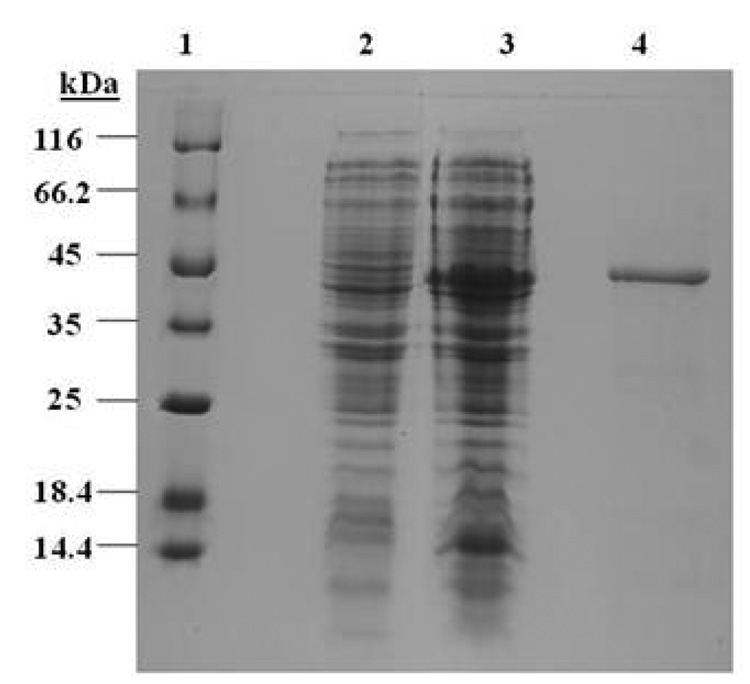
Sequence analysis of cloned the murA gene from V. fischeri revealed that the gene is 1269 bp encoding a polypeptide of 422 amino acids with a calculated pI of 5.0. The murA gene was amplified, cloned and over expressed in E. coli SG13900. The 6×Histidine tagged MurA protein was purified by metal affinity chromatography under denaturation conditions. The pure recombinant MurA exhibited a molecular mass of about 45 kDa (Fig. 1). This value corresponded well with the calculated mass of 44.7 kDa (Fig. 1). At this stage, our attempts to purify native recombinant MurA protein using imidazole in the elution buffer were not successful, possibly due to the affinity tag being hidden in the protein folds.
Fig. 1.
SDS-PAGE analysis of recombinant MurA protein purified by affinity chromatography. Lane 1, Protein molecular weight marker (Fermentas); lane 2, E. coli SG/pQE30/Vf-murA without IPTG induction; lane 3, Recombinant E. coli SG/pQE30/Vf-mur induced with 1 mM IPTG; lane 4, Pure recombinant MurA.
In E. coli fosfomycin inactivates MurA irreversibly by forming a covalent adduct with nucleophilic cysteine 115 present in the catalytic site of the enzyme (Kahan et al. 1974; Marquardt et al. 1994). Later studies have revealed that replacement of Cys-115 with aspartate renders E. coli MurA resistant to fosfomycin. Organisms with a Cys→Asp replacement at position 115 in their MurA proteins are naturally resistant to fosfomycin, such as in the case of Mycobacterium tuberculosis and Chlamydia trachomatis (De Smet et al. 1999; McCoy et al. 2003). We compared the deduced amino acid sequence of V. fischeri MurA with the homologous sequences from V. salmonicida, E. coli and M. tuberculosis. Overall, V. fischeri MurA was 95% identical with the corresponding sequence of V. salmonicida, 79% identical with the MurA of E. coli K12 and 43% with M. tuberculosis MurA (Fig. 2). The Cys-115 residue of E. coli MurA corresponds to Cys-116 of E. coli K12 and Cys-117 of M. tuberculosis. This residue is absolutely conserved in V. fischeri MurA. Based on sequence analysis, we could not determine why V. fischeri MurA was less susceptible to binding by fosfomycin. A number of highly conserved amino acids could be observed in the enzyme from diverse sources, the functions of which are not known. Speculatively, it is possible that a yet undiscovered sequence variant elsewhere along the peptide is responsible for fosfomycin resistance observed in this study. Further work will be necessary to test this hypothesis.
Fig. 2.
Multiple sequence alignment of the deduced amino acid sequence of V. fischeri MurA with homologous proteins from V. salmonicida (GenBank accession number CAQ78196), E. coli (GenBank accession number NP_417656), Mycobacterium tuberculosis (GenBank accession number CAA65472)
Our future work will focus on the functional characterization of the MurA protein from V. fischeri and determining amino acids responsible for fosfomycin resistance by site directed mutagenesis.
Acknowledgements
This work was made possible in part by National Institutes of Health grants 1 R15 GM070562-01 and P20 RR016480, the latter of which is from the NM-INBRE program of the National Center for Research Resources, a contribution from Calton Research Associates in honor of George and Clytie Calton, and an Internal Research Grant from ENMU. The authors are grateful to Dr. Jeffrey K. Griffith (University of New Mexico) for helpful comments and to Dr. Tomofusa Tsuchiya (Laboratory of Molecular Microbiology, University of Okayama, Japan) for kindly providing E. coli KAM32 used in this study. AP is grateful to the Director, NIO, Goa and the SIC, NIO (RC), Kochi for their kind help and support. The work at NIO was supported by grant SIP 1302.
References
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arca P, Hardisson C, Suarez JE. Purification of a glutathione S-transferase that mediates fosfomycin resistance in bacteria. Antimicrob agents Chemother. 1990;34:844–848. doi: 10.1128/aac.34.5.844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausubel FM, Brent R, Kingsten RE, Moore DD, Seidman JG, Smith JA, Struhl K. Short Protocols in Molecular Biology. 3rd edn. New York: John Wiley and Sons; 1995. [Google Scholar]
- Bernan VS, Greenstein M, Maiese WM. Marine microorganisms as a source of new natural products. Adv Appl Microbiol. 1997;43:57–90. doi: 10.1016/s0065-2164(08)70223-5. [DOI] [PubMed] [Google Scholar]
- Brown ED, Vivas EI, Walsh CT, Kolter R. MurA (MurZ), the enzyme that catalyzes the first committed step in peptidoglycan biosynthesis, is essential in Escherichia coli. J Bacteriol. 1995;177:4194–4197. doi: 10.1128/jb.177.14.4194-4197.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bugg TDH, Walsh CT. Intracellular steps of bacterial cell wall peptidoglycan biosynthesis: enzymology, antibiotics, and antibiotic resistance. Nat Prod Rep. 1992;9:199–215. doi: 10.1039/np9920900199. [DOI] [PubMed] [Google Scholar]
- Cao M, Bernat BA, Wang Z, Armstrong RN, Helmann JD. FosB, a cysteine-dependent fosfomycin resistance protein under the control of sigma(W), an extracytoplasmic-function sigma factor in Bacillus subtilis. J Bacteriol. 2001;183:2380–2383. doi: 10.1128/JB.183.7.2380-2383.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Morita Y, Huda MN, Kuroda T, Mizushima T, Tsuchiya T. VmrA, a member of a novel class of Na(+)-coupled multidrug efflux pumps from Vibrio parahaemolyticus. J Bacteriol. 2002;184:572–576. doi: 10.1128/JB.184.2.572-576.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clinical and Laboratory Standards Institute (CLSI) Approved Standard-Seventh edition. CLSI document M7-A7. Vol 26. 2006. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. No.2. [Google Scholar]
- De Smet KA, Kempsell KE, Gallagher A, Duncan K, Young DB. Alteration of a single amino acid residue reverses fosfomycin resistance of recombinant MurA from Mycobacterium tuberculosis. Microbiology. 1999;145:3177–3184. doi: 10.1099/00221287-145-11-3177. [DOI] [PubMed] [Google Scholar]
- Eschenburg S, Priestman M, Schonbruns E. Evidence that the fosfomycin target Cys115 in UDP-N-acetylglucosamine enolpyruvyl transferase (MurA) is essential for product release. J Biol Chem. 2005;280:3757–3763. doi: 10.1074/jbc.M411325200. [DOI] [PubMed] [Google Scholar]
- Etienne J, Gerbaud G, Fleurette J, Courvalin P. Characterization of staphylococcal plasmids hybridizing with the fosfomycin resistance gene fosB. FEMS Microbiol Lett. 1991;68:119–122. doi: 10.1016/0378-1097(91)90406-z. [DOI] [PubMed] [Google Scholar]
- Falagas ME, Giannopoulou KP, Kokolakis GN, Rafailidis PI. Fosfomycin: use beyond urinary tract and gastrointestinal infections. Clin Infect Dis. 2008;46:1069–1077. doi: 10.1086/527442. [DOI] [PubMed] [Google Scholar]; Fillgrove KL, Pakhomova S, Newcomer ME, Armstrong RN. Mechanistic diversity of fosfomycin resistance in pathogenic microorganisms. J Am Chem Soc. 2003;125:15730–15731. doi: 10.1021/ja039307z. [DOI] [PubMed] [Google Scholar]
- Gottesman S, Halpern E, Trisler P. Role of sulA and sulB in filamentation by Ion mutants of Escherichia coli K-12. J Bacteriol. 1981;148:265–273. doi: 10.1128/jb.148.1.265-273.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins D, Thompson J, Gibson T, Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahan FM, Kahan JS, Cassidy PJ, Kropp H. The mechanism of action of fosfomycin (phosphonomycin) Ann N.Y. Acad Sci. 1974;235:364–386. doi: 10.1111/j.1749-6632.1974.tb43277.x. [DOI] [PubMed] [Google Scholar]
- Kim DH, Lees WJ, Kempsell KE, Lane WS, Duncan K, Walsh CT. Characterization of a Cys115 to Asp substitution in the Escherichia coli cell wall biosynthetic enzyme UDP-GlcNAc enolpyruvyl transferase (MurA) that confers resistance to inactivation by the antibiotic fosfomycin. Biochemistry. 1996;35:4923–4928. doi: 10.1021/bi952937w. [DOI] [PubMed] [Google Scholar]
- Marchler-Bauer A, Anderson JB, Derbyshire MK, DeWeese-Scott C, Gonzales NR, Gwadz M, Hao L, He S, Hurwitz DI, Jackson JD, Ke Z, Krylov D, Lanczycki CJ, Liebert CA, Liu C, Lu F, Lu S, Marchler GH, Mullokandov M, Song JS, Thanki N, Yamashita RA, Yin JJ, Zhang D, Bryant SH. CDD: a conserved domain database for interactive domain family analysis. Nucleic Acids Res. 2007;35:D237–D240. doi: 10.1093/nar/gkl951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marquardt JL, Siegele DA, Kolter R, Walsh CT. Cloning and sequencing of Escherichia coli murZ and purification of its product, a UDP-N-acetylglucosamine enolpyruvyl transferase. J Bacteriol. 1992;174:5748–5752. doi: 10.1128/jb.174.17.5748-5752.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy AJ, Sandlin RC, Maurelli AT. In vitro and in vivo functional activity of Chlamydia MurA, a UDP-N-acetylglucosamine enolpyruvyl transferase involved in peptidoglycan synthesis and fosfomycin resistance. J Bacteriol. 2003;185:1218–1228. doi: 10.1128/JB.185.4.1218-1228.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Hara K. Two different types of fosfomycin resistance in clinical isolates of Klebsiella pneumoniae. FEMS Microbiol Lett. 1993;114:9–16. doi: 10.1111/j.1574-6968.1993.tb06543.x. [DOI] [PubMed] [Google Scholar]
- Ruby EG, McFall-Ngai MJ. Oxygen-utilizing reactions and symbiotic colonisation of the squid light organ by Vibrio fischeri. Trends Microbiol. 1999;7:414–420. doi: 10.1016/s0966-842x(99)01588-7. [DOI] [PubMed] [Google Scholar]
- Tsuroka T, Yamada Y. Characterization of spontaneous fosfomycin (phosphonomycin)-resistant cells of Escherichia coli B in vitro. J Antibiotics. 1975;28:906–911. doi: 10.7164/antibiotics.28.906. [DOI] [PubMed] [Google Scholar]
- Walsh C. Molecular mechanisms that confer antibacterial drug resistance. Nature. 2000;406:775–781. doi: 10.1038/35021219. [DOI] [PubMed] [Google Scholar]