Abstract
Giant chromatin-modifying complexes regulate gene transcription in eukaryotes by acting on chromatin substrates and ‘setting’ the histone code. The histone deacetylase (HDAC)-associated mammalian Sin3 corepressor complex regulates a wide variety of genes involved in all aspects of cellular physiology. The recruitment of the corepressor complex by transcription factors to specific regions of the genome is mediated by Sin3 as well as 10 distinct polypeptides that comprise the corepressor complex. Here we report the solution structure of a novel CCCH zinc finger (ZnF) motif in the SAP30 polypeptide, a key component of the corepressor complex. The structure represents a novel fold comprising two β-strands and two α-helices with the zinc organizing center showing remote resemblance to the treble clef motif. In silico analysis of the structure revealed a highly conserved surface that is dominated by basic residues. NMR-based analysis of potential ligands for the SAP30 ZnF motif indicated a strong preference for nucleic acid substrates. We propose that the SAP30 ZnF functions as a double-stranded DNA-binding motif, thereby expanding the known functions of both SAP30 and the mammalian Sin3 corepressor complex. Our results also call into question the common assumption about the exclusion of DNA-binding core subunits within chromatin-modifying/remodeling complexes.
INTRODUCTION
Post-translational modifications of histones have emerged as an important epigenetic mechanism for regulating gene transcription in eukaryotes. A variety of enzymes serve to introduce or eliminate these modifications facilitating site-specific alterations of the histone code that in turn lead to distinct transcriptional outcomes. Histone deacetylases (HDACs) constitute an important class of enzymes that reverse the acetylation status of lysine residues largely to effect transcriptional repression. The specificity of these enzymes is dictated in large part through their association with transcriptional corepressors that are in turn recruited via protein–protein and protein–DNA interactions to specific sites on the genome.
The Sin3 corepressor complex is one of only a handful of HDAC-associated corepressor complexes in mammalian cells that have been biochemically characterized (1). Analogous corepressor complexes are found in a wide variety of plant and animal species, although those belonging to the Sin3 complex appear to be particularly widespread as they are also found in yeast. The mammalian Sin3 complex comprises at least 10 polypeptides besides Sin3, including HDACs HDAC1 and HDAC2, two histone-binding proteins RbAp46 and RbAp48 (Rb-associated polypeptides), and six Sin3-associated polypeptides SAP25, SAP30, SAP45/Sds3, SAP130, SAP180/BCAA and RBP1 (Rb-binding protein 1), most of whose functions, aside from presumed roles in stabilizing the Sin3 complex and providing interaction surfaces for recruitment, are largely unknown (2–9). The Sin3 protein is thought to function as a molecular scaffold for corepressor complex assembly and also as a molecular adapter bridging components of the complex with DNA-bound repressors (10,11). Underscoring its fundamental role in cellular physiology, sin3 null mutations caused embryonic lethality in Drosophila and mouse (12,13) while cell lineage-specific conditional knockouts of mouse sin3 caused a range of developmental defects (13,14). Gene expression profiling of the conditional sin3 knockouts revealed the deregulation of several genes involved in cell cycle regulation, DNA replication and repair, apoptosis and mitochondrial metabolism (14).
SAP30 was among the earliest identified components of the Sin3 corepressor complex (6,9). The protein is thought to play a role in corepressor complex function, as it is consistently detected in biochemical fractionation experiments conducted with extracts from yeast and higher organisms (3,9,15–18) while deletion of the sap30 gene in yeast produced similar effects as those resulting from the deletions of sin3 and rpd3, the yeast homologue of HDAC1 and HDAC2 (9,19–21). Alluding to a role in mammalian Sin3 corepressor complex assembly, SAP30 has been proposed to interact with RBP1 (7) and SAP180 (22)—two closely related core components of the complex—as well as the ING1 and ING2 proteins (17,23) found in a subset of Sin3 complexes in association with SWI/SNF-type chromatin-remodeling activities (16–18). Other SAP30 targets include CIR (CBF1 interacting corepressor) (24) and the yin yang 1 zinc finger (ZnF) transcription factor (25). How SAP30 performs such diverse functions is presently unknown, as no high-resolution structure–function analyses for this polypeptide have been described either in isolation or in complex with another macromolecule. Here, we describe the solution structure of an uncharacterized region of the SAP30 polypeptide that relies on zinc for proper folding. We show that this novel ZnF motif can, rather unexpectedly, bind to nucleic acids, expanding the known functions of both SAP30 and the mammalian Sin3 corepressor complex.
MATERIALS AND METHODS
SAP30 ZnF protein expression and purification
The coding sequence of SAP30 ZnF corresponding residues 64 −131 was amplified by PCR and inserted into the pMCSG7 expression vector (26). All cloned gene segments were confirmed by DNA sequencing. Escherichia coli BL21(DE3) cells (Novagen, Madison, WI) containing the vector were grown at 37°C in LB broth (EMD Chemicals Inc.). The growth temperature was shifted to 20°C when the OD600nm reached approximately 0.6. Expression of the His6-tagged protein was induced using 1 mM isopropyl-β-D-thiogalactopyranoside, and the cells were harvested 16 h thereafter. Cell pellets were suspended in 50 mM Tris–HCl buffer (pH 8) containing 0.2 M sodium chloride, 2 mM Tris–(2-carboxy-ethyl) phosphine (TCEP) hydrochloride, 1 mM phenyl-methylsulfonyl, 1 µM leupeptin, 1 mM pepstatin and 0.1% Triton X-100, lysed via sonication followed by DNAse treatment for 15 min at 4°C and centrifuged. The supernatant was incubated with the His-Select Nickel resin (Sigma, St. Louis, MO) for 30 min. The resin was washed by 50 mM Tris–HCl buffer (pH 8) containing 0.5 M sodium chloride and bound proteins were eluted using 50 mM Tris–HCl buffer (pH 8) containing 0.2 M sodium chloride, 2 mM TCEP and 300 mM Imidazole. The eluted proteins were incubated with tobacco etch virus protease for 4 h at 22°C followed by overnight incubation at 4°C. The mixture was centrifuged and the desired protein in the supernatant was purified to homogeneity via reversed phase HPLC using a C18 column (Vydac, Hesperia, CA) and a linear gradient of 0.1% trifluoroacetic acid (TFA) and 0.1% TFA in 80% acetonitrile and lyophilized. Samples uniformly labeled with 15N and/or 13C isotopes were produced using the same procedure except that cells were grown in M9 minimal medium containing 15N-ammonium sulfate and/or 13C-D-glucose (Spectra Stable Isotopes, Columbia, MD), respectively. The alanine mutants of the SAP30 ZnF were generated by QuikChange site-directed mutagenesis (Stratagene). The 15N-labeled samples for mutants C68A and H108A were produced using the same procedure as the wildtype, but the samples for C67A and C112A were solubilized from inclusion body pellets followed by purification under denaturing conditions. The identities of the proteins were confirmed by electrospray ionization-mass spectrometry.
NMR samples and titrations
NMR samples were prepared by dissolving the dry, lyophilized protein powder in 20 mM Tris–acetate buffer (pH 6), 2 mM DTT-d10 and 0.2% (w/v) NaN3. Protein concentrations were determined spectrophotometrically (27). The concentrations of the 15N, 13C-labeled sample and the 15N-labeled samples for titrations with various ligands were 0.56 and 0.2 mM, respectively. Titrations were performed by direct addition of the ligand to the protein at the desired molar ratio followed by adjustment of the solution to pH 6. The ligands mimicking histone modifications in Table 3 were purchased from Sigma while those mimicking phospholipids including phosphatidylinositol-5-phosphate and phosphatidic acid were purchased from Echelon Biosciences and Avanti Polar Lipids, respectively. Oligodeoxyribonucleotide sequences corresponding to the 15-mer single-stranded DNA 5′-CTGTGGCCCTGAGCC-3′, the 10-mer self-complementary duplex 5′-GCGAATTCGC-3′ and the 21-mer self-complementary duplex 5′-CCTTGGCTGACGTCAGCCAAG-3′ were purchased in lyophilized form from Integrated DNA Technologies. All ligands were used for the titrations without any additional purification.
Table 3.
SAP30 ZnF chemical shift perturbations produced by potential ligands
| Ligand | Average chemical shift deviation (ppm)a |
|---|---|
| l-Lysineb | 0.008 ± 0.002 |
| Nε-acetyl-l-lysineb | 0.006 ± 0.005 |
| Nε-trimethyl-l-lysineb | 0.011 ± 0.005 |
| Nγ-dimethyl-l-arginineb | 0.008 ± 0.003 |
| O-Phospho-l-serineb | 0.022 ± 0.009 |
| O-Phospho-l-threonineb | 0.011 ± 0.007 |
| Phosphatidylinositol-5-phosphate | 0.026 ± 0.008 |
| Phosphatidic Acid | 0.005 ± 0.002 |
| 15-mer single-stranded DNA | 0.136 ± 0.049 |
| 10-mer double-stranded DNA | 0.169 ± 0.036 |
| 21-mer double-stranded DNA | 0.203 ± 0.068 |
aCalculations based on data for the seven most strongly perturbed correlations (10% of total number of residues).
bMeasurements at 50-fold molar excess of ligand; all other measurements at equimolar protein:ligand ratios.
NMR spectroscopy and structure determination
NMR data were acquired on a Varian Inova 600 MHz spectrometer at 25°C. NMR data processing and analysis were performed using an in-house modified version of Felix 98.0 (Accelrys) and NMRView (28,29). Backbone and side chain 1H, 15N and 13C resonances for the SAP30 ZnF were assigned by analyzing 3D HNCACB, C(CO)NH-TOCSY, HNCO, H(CCO)NH-TOCSY, HCCH-TOCSY and HCCH-COSY spectra (30,31). Aromatic proton resonances were assigned from 2D 1H-13C HSQC spectra and 3D HCCH-COSY and HCCH-TOCSY spectra (32,33).
For structure determination, backbone φ and ψ torsion angle restraints were derived from an analysis of Hα, Cα, Cβ, C′ and backbone 15N chemical shifts using TALOS (34). Restraints were imposed only for those residues that exhibited TALOS reliability scores of 9. NOE-based distance restraints were derived from 3D 15N-edited NOESY (mixing time, τm = 75 ms) spectrum recorded in H2O, 3D aliphatic 13C-edited NOESY (τm = 60 ms) and 2D 1H-1H NOESY (τm = 80 ms) spectra recorded in D2O. All NOEs were calibrated and assigned iteratively and automatically by ARIA (version 1.2) (35,36) and were checked manually between successive rounds of calculations. Structures were calculated using ARIA in conjunction with CNS (37) starting from extended conformations. A total of 80 structures were computed in the final iteration and the best 20 structures with the lowest restraint energies were selected for automatic NOE assignment. The ARIA-generated restraint lists served as inputs for inferential structure determination (ISD). Structures were calculated from extended starting conformations. Calculations were terminated at 3000 samples when the total energy associated with the target distribution was distributed randomly around the median value. An ensemble of 50 ‘representative’ structures was subject to restrained energy minimization in CNS; 47 of these had good covalent geometry and were selected for final analysis. The final structures were analyzed using PROCHECK and CNS (37–39). Molecular images were generated using CHIMERA (40). Surface maps of electrostatic potentials were calculated using the APBS program while conservation scores were computed using ConSurf (41,42); both of these properties were rendered using CHIMERA.
Lipid-binding assays
Phosphatidylinositol phosphate (PIP)-strips containing membrane-immobilized lipids (100 pmol per spot; Echelon Biosciences) were incubated with blocking buffer (PBS with 3% fatty acid-free BSA (Sigma A-7030) and 0.1% (v/v) Tween-20) for 1 h at 22°C. Ni2+-affinity purified His6-SAP30 ZnF was added to the PIP-strips at 10 µg/ml final concentration and incubated for 2 h at 22°C followed by three rounds of washing by incubating with the wash buffer [PBS with 0.1% (v/v) Tween-20] for 10 min each time to remove any unbound His6-SAP30 ZnF. For sensitive detection of bound His6-SAP30 ZnF, the following series of steps were undertaken. The strip was incubated for 1 h at 22°C with an anti-His-tag antibody (1:2000, Millipore #05-949) diluted in freshly prepared blocking buffer followed by three rounds of washing with the wash buffer. The strip was incubated for another 1 h at 22°C with a goat anti-mouse horse radish peroxidase-conjugated secondary antibody directed against an IgG Fc fragment (1:5000, Millipore #AQ127P) diluted in freshly prepared blocking buffer followed by another three rounds of washing with the wash buffer. The strip was rinsed for 1 min with deionized water and incubated with a chemiluminescent substrate solution for 5 min at 22°C followed by development on film in the darkroom.
RESULTS
A novel ZnF motif in the SAP30 polypeptide
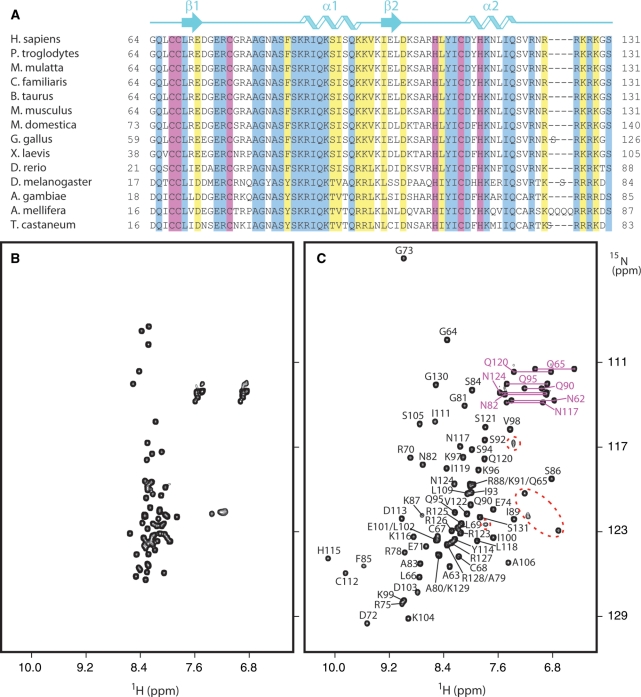
Interrogation of the Pfam and SMART databases using the SAP30 polypeptide sequence failed to yield any statistically significant matches to well-characterized domains or motifs. Previous studies had identified a ∼90 aa domain at the C-terminus of the protein that was deemed essential for Sin3 binding. The Sin3 interaction domain is conserved across a wide variety of species ranging from yeast to human. In contrast, the N-terminal segment of SAP30 spanning ∼60 aa is characterized by poor conservation and the presence of low complexity regions. The central ∼70 aa segment of the protein is surprisingly well-conserved in species ranging from flies to mammals (Figure 1A); interestingly, this region is conspicuously absent in yeast and worm. Sequence database searches conducted with a profile hidden Markov model constructed for this segment failed to yield any statistically significant matches to other proteins, implying that this novel motif might be unique to SAP30. A striking feature of this relatively short segment is the presence of six invariant cysteine and histidine residues (Figure 1A). Since these residues typically coordinate zinc thereby assisting in the proper folding and function of many proteins including transcription factors, we speculated that the SAP30 polypeptide might harbor a ZnF motif.
Figure 1.
A conserved ZnF motif in the central segment of the SAP30 polypeptide. (A) CLUSTALW-based multiple sequence alignment emphasizing sequence conservation within a central ∼70 residue segment of the protein from a variety of species ranging from fly to human. Invariant and conserved residues are highlighted in blue and yellow, respectively, while invariant cysteine and histidine residues are highlighted in magenta. The cartoon on top of the alignment identifies the location of secondary structural elements as deduced from the solution structure. 1H-15N HSQC spectra of the central segment of the SAP30 polypeptide (B) before and (C) after the addition of one equivalent of zinc. Sequence-specific backbone amide assignments are indicated; the side chain amide assignments are indicated in magenta. Arginine side chain correlations are enclosed within colored oval boxes.
To test for this possibility, a recombinant protein corresponding to the central segment of SAP30 was expressed in bacteria and purified for solution NMR studies. The 1H-15N HSQC spectrum of this protein is characterized by poor dispersion of resonances (<1 ppm) in the amide proton region and uniformly intense resonances with narrow line widths (Figure 1B), indicative of an essentially unstructured and dynamically disordered polypeptide. However, the addition of zinc to the sample led to the appearance of new resonances in the spectrum whose intensities increased progressively while those of the original resonances diminished correspondingly as a function of added zinc. No further changes in the spectrum were detected once one equivalent of zinc had been added to the sample. The spectrum of the zinc-loaded form exhibited significantly enhanced dispersion of amide proton resonances (>3 ppm) which is a hallmark of folded proteins (Figure 1C). Collectively, these results indicated zinc-assisted folding of the protein and the presence of a single zinc-binding site confirming the suspicion that the SAP30 polypeptide harbored a ZnF motif.
SAP30 ZnF is a CCCH ZnF
Since the SAP30 ZnF appeared to be a novel ZnF motif (i.e. unrelated to any of the previously characterized motifs at the sequence level), we sought to determine its solution structure using NMR. Since there were six potential zinc-coordinating residues in the SAP30 ZnF and since only four of these were likely to be involved in coordinating the single zinc ion, we used NMR in conjunction with site-directed mutagenesis to identify the zinc ligands. A 1H-15N HMQC spectrum recorded using a 113Cd-loaded sample of SAP30 ZnF revealed a doublet for the His115 15Nδ resonance (unlike the singlet observed for the Zn-loaded sample) arising from 1JN-Cd spin–spin coupling, implying the involvement of this residue via the Nδ atom in zinc coordination (Supplementary Figure S1A). Conversely, no such splitting of either the 15Nδ or the 15Nε resonance was observed for His108, implying that it was not involved in zinc coordination. Also, the somewhat downfield 13Cβ shifts of Cys67, Cys76 and Cys112 (31–33 ppm) suggested the involvement of these residues in zinc binding (Supplementary Figure S1B). Further confirmation of the involvement of these residues came from NMR analyses of zinc binding by single-site alanine mutants of potential zinc-coordinating residues of SAP30 ZnF. Of the six alanine mutants, His108Ala and Cys68Ala produced enhanced spectral dispersion of amide resonances upon zinc addition, whereas Cys67Ala and Cys112Ala failed to do so (Supplementary Figure S2); attempts to express Cys76Ala and His115Ala were unsuccessful and therefore could not be tested. Collectively, these results implicated Cys67, Cys76, Cys112 and His115 in zinc coordination while Cys68 and His108 were deemed nonessential for zinc binding rendering the SAP30 ZnF as a CCCH ZnF motif. The SAP30 ZnF motif appears to be an autonomous structural unit as resonances belonging to the motif almost completely overlap with those from a construct that additionally bears the C-terminal Sin3 interaction domain (Supplementary Figure S3).
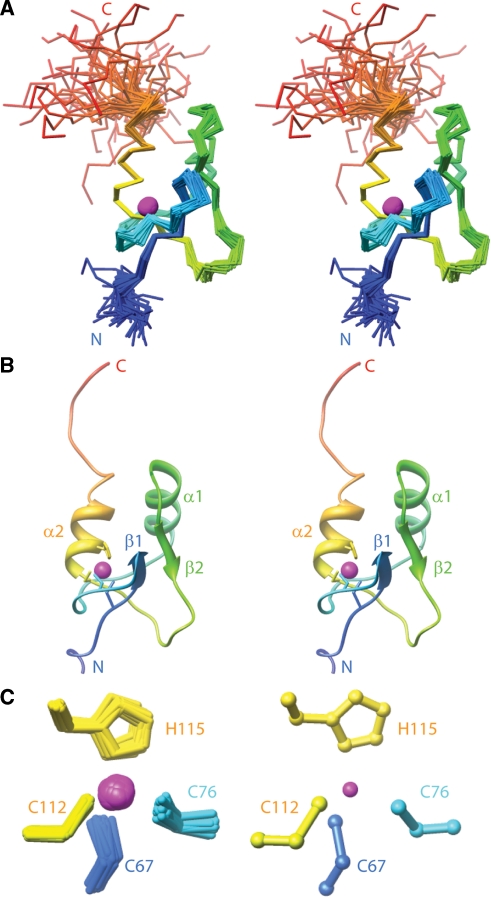
SAP30 ZnF adopts a new fold
The solution structure of the SAP30 ZnF motif was determined using 1H-1H NOEs and dihedral angle restraints that were derived from chemical shift and scalar coupling constant measurements (Table 1). Structures of good precision, reasonable geometry and in good agreement with experimental data were obtained using the ISD software package that incorporates a Bayesian inference approach (Figure 2A; Tables 1 and 2). The N- and C-termini of SAP30 ZnF are dynamically disordered as these segments exhibited greatly diminished 1H-15N heteronuclear NOE values (Figure 2A; Supplementary Figure S4). Although zinc coordination distance restraints were employed in the structure calculations, a control calculation conducted without incorporation of any of these restraints yielded essentially similar structures including similar side chain conformations for the residues involved in zinc coordination but, as expected, with a slightly reduced precision (data not shown). The distances of the histidine Nδ atom of His108 and the Sγ atom of Cys68 from the zinc are 10.6 and 7.4 Å, respectively, in the representative structure, further confirming the identities of the zinc-coordinating residues.
Table 2.
ISD statistics for SAP30 ZnF
| Dataset | Observable | Size | Error (%)a | Violations (%)b |
|---|---|---|---|---|
| Aromatic 13C-edited NOESY | NOE | 65 | 16.6 ± 1.9 | 2 (3.1) |
| 15N-edited-NOESY | NOE | 484 | 12.8 ± 0.3 | 12 (2.5) |
| Aliphatic 13C-edited NOESY | NOE | 940 | 13.1 ± 0.5 | 32 (3.4) |
| Hydrogen bonding and zinc restraints | Distance | 24 | 2.8 ± 1.2 | 0 (0) |
aAverage percentage distance error.
bViolation statistics based on predictive distributions with threshold probability for incorrect measurement set to 95%.
Table 1.
NMR structure determination statistics for SAP30 ZnF
| Restraint statistics | |
| NOE-based distance restraints | 1279 |
| Unambiguous NOE-based restraints | 1059 |
| Intraresidue | 553 |
| Sequential (|i – j| = 1) | 170 |
| Medium range (1 < | i − j | ≤ 4) | 118 |
| Intramolecular long range (| i − j | > 4) | 218 |
| Ambiguous NOE-based restraints | 220 |
| Hydrogen bonding distance restraints | 18 |
| Zinc coordination distance restraints | 6 |
| Torsion angle restraints | 54 (24φ, 24ψ, 6χ1) |
| Structure quality of NMR ensemble | |
| Restraint satisfactiona | |
| Root-mean-square differences for distance restraints (Å) | 0.019 ± 0.007 |
| Root-mean-square differences for torsion angle restraints (°) | 0.707 ± 0.297 |
| Deviations from ideal covalent geometry | |
| Bond lengths (Å) | 0.002 ± 0.000 |
| Bond angles (°) | 0.440 ± 0.014 |
| Impropers (°) | 0.440 ± 0.025 |
| Ramachandran plot statistics (%) | |
| Residues in most favored regions | 69.3 |
| Residues in allowed regions | 29.1 |
| Residues in disallowed regions | 1.5 |
| Average atomic root-mean-square deviations from the average structure (Å) | |
| All atoms | 3.10 |
| All atoms except disordered regionsb | 1.11 |
| Backbone atoms (N, Cα, C′) | |
| All residues | 2.53 |
| All residues excluding disordered regionsb | 0.38 |
aAlso see Table 2.
bDisordered regions include residues 61–65 and 123–131.
Figure 2.
Solution NMR structure of the SAP30 ZnF motif. (A) Stereo views of the Cα traces of the ensemble of 47 representative conformers of SAP30 ZnF from ISD calculations following a best-fit superposition of the structurally ordered regions (residues 66–122) of the protein. The traces are color-ramped from blue to red from the N- to the C-terminus. Zinc ions are shown as magenta-colored spheres. (B) A stereo view of the representative structure of the ensemble shown as a ribbon diagram in the same orientation as in (A). Residues are color-ramped from blue to red also as in (A). The side chains of zinc-coordinating residues are shown in stick representation while the zinc ion is shown as a magenta-colored sphere. (C) Expanded views of the zinc coordination geometry for the NMR ensemble (left) and the representative structure (right).
SAP30 ZnF is largely devoid of secondary structural elements except for a pair of anti-parallel β-strands spanning residues Leu69 to Glu71 and Ile100 to Leu102 that pack against a pair of α-helices spanning residues Lys87 to Lys96 and Asp113 to Gln120 (Figure 2B). The side chains of Cys68, Leu69, Phe85, Ile89, Ile93, Val98, Il100, Leu102, Leu109, Ile111 and Ile119 comprise a well-defined hydrophobic core of the domain; expectedly, all of these residues are either invariant or highly conserved (Figure 1A). A DALI-based search of the PDB failed to yield any structural homologues, implying that the SAP30 ZnF defined a new fold. As expected, the geometry specified by the atoms involved in zinc coordination is approximately tetrahedral (Figure 2C). The zinc organizing center, which is located adjacent to the hydrophobic core, appears to be somewhat unique with only one of the zinc ligands His115 located in a regular secondary structural element while the others including Cys67, Cys76 and Cys112 are all drawn from loop regions. Based on visual comparisons, the zinc organizing center remotely resembles the treble clef ZnF motif (43). However, the treble clef motif is characterized by a zinc ‘knuckle’—a turn-like structure with a consensus sequence CPxCG, both of which are absent in the SAP30 ZnF. Visual comparisons with the various fold-groups in the Structural Classification of Zinc Fingers Database (44) failed to reveal significant similarities with the SAP30 ZnF motif, consistent with the inference of a novel fold.
Structure–function analysis of SAP30 ZnF
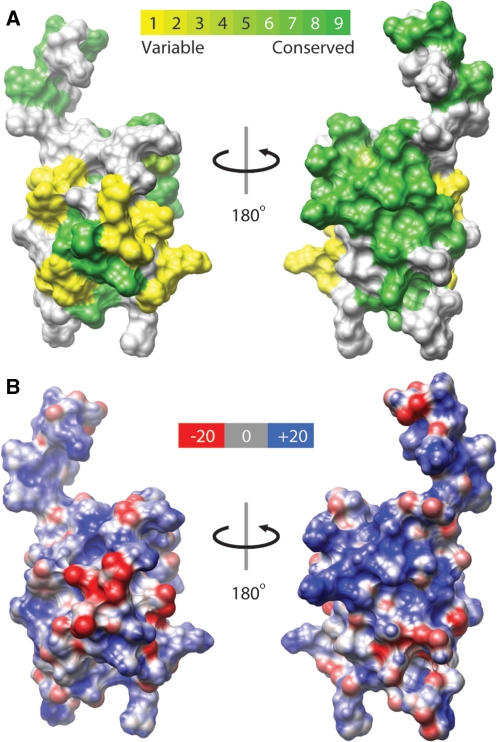
To gain insights into the molecular surface responsible for SAP30 ZnF function, evolutionary conservation scores for each residue within this segment were computed using ConSurf and mapped onto the NMR structure. Predictably, the highest degree of sequence conservation was noted for those residues that comprised the hydrophobic interior of the protein. Mapping the conservation data onto the molecular surface revealed a contiguous patch of highly conserved, yet solvent-exposed residues drawn from the two α-helices, the loop preceding helix α1 and the ‘tail’ following α2 (Figure 3A). Analysis of the electrostatic potential map revealed that the conserved surface was highly basic in character, implying a preference for ligands that are acidic in character (Figure 3B).
Figure 3.
Molecular properties of the SAP30 ZnF motif. (A) Front and back views of the molecular surface of SAP30 ZnF colored according to ConSurf-computed sequence conservation scores. The scores are distributed in nine bins and the color for each bin follows the key provided. Those residues for which scores could not be reliably computed by ConSurf are colored white. (B) Molecular surface views of the protein with the electrostatic potential mapped onto the surface. Potentials at +20, 0 and −20 kT/e are colored in blue, white and red, respectively; colors are linearly interpolated for the intermediate values.
Our attempts at generating SAP30 ZnF complexes with two putative SAP30 interactors including RBP1 and ING2 were unsuccessful (data not shown), suggesting that these interactions either involved other regions of SAP30 or were mediated by other factors. Therefore, we evaluated a range of possible ligands for the SAP30 ZnF using NMR. Two sets of ligands were analyzed, one set comprising post-translational modifications of histones associated with various forms of the ‘histone code’ and another set comprising substrates with acidic character including nucleic acids and phospholipids. The choice of ligands was guided by possible roles of SAP30 in directing the Sin3 corepressor complex to specific regions within the nucleus through interactions with post-translationally modified histones, DNA or phospholipids.
To test whether SAP30 ZnF was a chromatin-binding module, we titrated the protein samples with a vast excess of lysine, acetyllysine, trimethyllysine, dimethylarginine, phosphoserine and phosphothreonine. However, even at ligand:protein molar ratios of 50:1, these ligands produced barely detectable changes in the SAP30 ZnF NMR spectrum (Table 3; Figure 4A), indicative of extraordinarily weak or no binding. Indeed, the minute changes that were observed are likely due to changes in protein conformation caused by changes in ionic strength than to any binding event.
Figure 4.
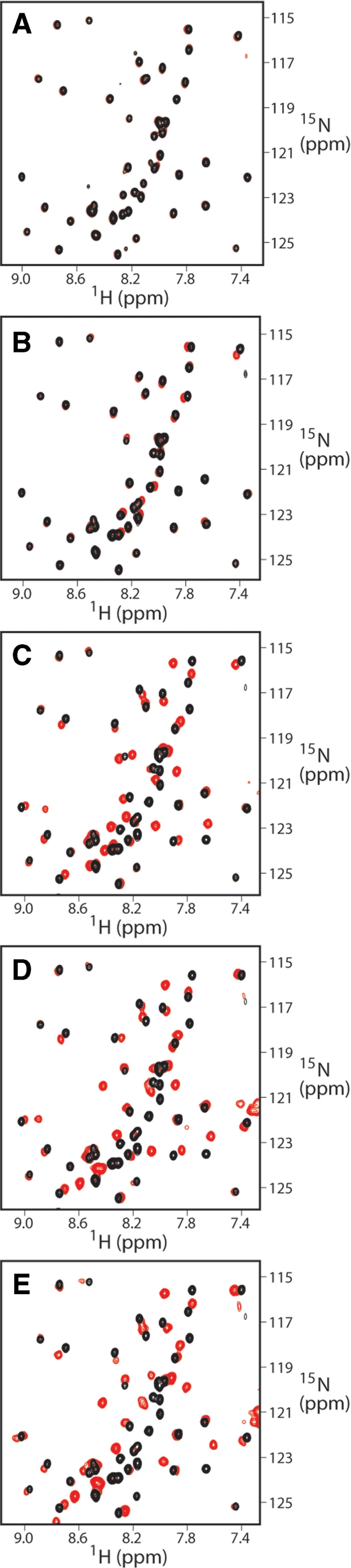
NMR titrations of the SAP30 ZnF motif with various ligands. Overlays of expanded plots of 1H-15N HSQC spectra recorded in the absence (black) and in the presence (red) of (A) trimethyllysine, (B) phosphatidylinositol-5-phosphate, (C) 15-mer single-stranded DNA, (D) 10-mer DNA duplex and (E) 21-mer DNA duplex. Protein concentrations ranged between 0.1 and 0.2 mM. The spectra in the presence of ligands were recorded at equimolar ratios except in the case of trimethyllysine, which was recorded at 50-fold excess.
To test whether the SAP30 ZnF bound to phospholipids, we initially conducted lipid binding assays in a pull-down format using the so-called PIP strips. In this assay, SAP30 ZnF exhibited preferential binding to phosphatidylinositol substrates that were phosphorylated at the 3, 4 and/or 5 positions of the inositol moiety with the strongest binding observed for PI(4)P, PI(3,5)P and PI(5)P (Supplementary Figure S5). SAP30 ZnF also appeared to bind phosphatidic acid with comparable affinity. To test whether any of these represented bona fide ligands of SAP30 ZnF, NMR titrations were conducted with PI(5)P and phosphatidic acid. Surprisingly, the addition of one equivalent of each of these ligands to SAP30 ZnF produced only small changes in the NMR spectrum (Figure 4B; Table 3) indicative of weak binding.
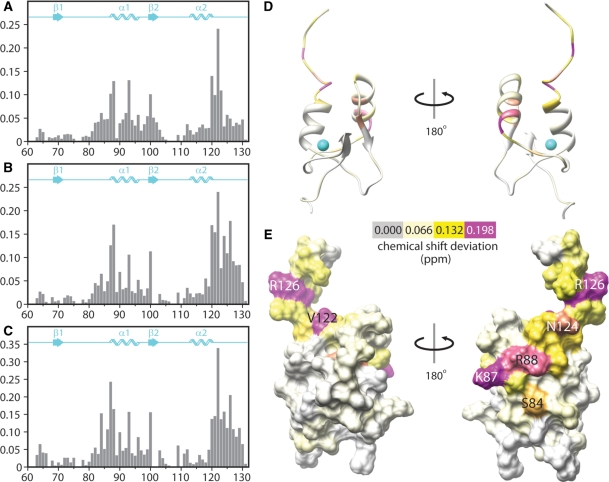
In contrast to the results obtained with the phospholipid titrations, titration of SAP30 ZnF with single-stranded 15-mer DNA and two DNA duplexes 10 and 20 bp in length produced significant changes in the NMR spectrum (Figures 4C–E and 5). In each case, the titrations produced changes in resonance positions as a function of added ligand, indicative of complexes with fast dissociation kinetics on the NMR timescale implying that these interactions were of modest affinity. Each of the oligonucleotides tested produced similar patterns of SAP30 ZnF chemical shift perturbations with a notable difference between the single- and double-stranded oligonucleotides being that the extent of the perturbations for SAP30 ZnF residues located in the C-terminal tail was more pronounced in the latter than in the former (Figure 5A–C). This is consistent with the involvement of a larger protein interface when bound to double- over single-stranded DNA. Mapping of the chemical shift perturbations induced by the 20 bp duplex onto the molecular surface of SAP30 ZnF revealed a contiguous surface that most likely constitutes the site of interaction with DNA (Figure 5D and E). This surface overlaps exceptionally well with the evolutionarily conserved patch dominated by basic residues that was presumed to be significant for SAP30 ZnF function (Figure 3A and B). The chemical shift perturbations extend well into the unstructured C-terminal tail (Figure 5D and E), implying that this region likely undergoes a significant conformational change upon binding to double-stranded DNA.
Figure 5.
Chemical shift perturbations of SAP30 ZnF induced by oligodeoxyribonucleotides. Chemical shift deviations induced by (A) a single-stranded 15-mer oligonucleotide, (B) a 10 bp duplex and (C) a 20 bp duplex graphed for each residue in the SAP30 ZnFmotif. Composite deviations of backbone amide proton and nitrogen chemical shifts for each residue were calculated using the formula:  Note that the scale for the y-axis in (C) is different from those in the other panels. Chemical shift deviation data from (C) mapped onto (D) a ribbon representation and (E) the molecular surface of the SAP30 ZnFmotif. These views share the same orientation as those shown in Figure 3. The magnitude of the deviations is color coded according to the key provided; colors are linearly interpolated for intermediate values. Some of the most perturbed residues are annotated.
Note that the scale for the y-axis in (C) is different from those in the other panels. Chemical shift deviation data from (C) mapped onto (D) a ribbon representation and (E) the molecular surface of the SAP30 ZnFmotif. These views share the same orientation as those shown in Figure 3. The magnitude of the deviations is color coded according to the key provided; colors are linearly interpolated for intermediate values. Some of the most perturbed residues are annotated.
DISCUSSION
Implications for a DNA-binding activity in the mammalian Sin3 corepressor complex
Many chromatin-modifying and chromatin-remodeling enzymes derive their specificity (i.e. their ability to act on specific regions of the genome) in large part by being recruited directly or indirectly via protein–protein interactions with transcription factors that either interact with specific DNA elements or post-translationally modified forms of histones. Multi-subunit coregulator complexes with chromatin-modifying/remodeling activities are thought to be devoid of constitutively associated subunits with DNA-binding activity. However, this long-held view is being increasingly challenged as witnessed by the detection of core components with DNA-binding activity in SWI/SNF complexes (45–47) and more strikingly, in the Rpd3L complex—the yeast homologue of the mammalian Sin3 complex (15). Our findings and that of a very recent independent study implicating SAP30 in DNA binding expands the known functions of the mammalian Sin3 corepressor complex (48).
Unlike in the case of the Ash1 and Ume6 subunits of the Rpd3L complex that bind specific DNA elements, the role of SAP30 in directing the Sin3 corepressor complex is less clear. Both the present study (Figures 4 and 5; Table 3) and that of Lohi and coworkers, suggest that SAP30 can bind to unrelated DNA sequences in a non-specific manner. Whether SAP30 lacks sequence-specificity or can indeed bind to specific sequences remains to be definitively established; future studies will explore this possibility. It is conceivable that SAP30 binds specific DNA sequences in conjunction with other as yet unknown transcription factors. SAP30 has previously been shown to direct the Sin3 corepressor complex to the nucleolus (49) and its DNA-binding activity might have a similar role in directing the complex to the promoter regions of ribosomal DNA, a well-known target for Sin3/HDAC-mediated gene repression (50,51). Irrespective of the specificity of the SAP30 interaction with DNA, the polypeptide might have a role in enhancing the lifetime of the recruited complex in promoter regions even after the sequence-specific DNA-binding repressor activity involved in the initial recruitment has disappeared. Indeed, repressor-independent stable anchoring of the Sin3 corepressor complex has been reported on hypoacetylated nucleosomal substrates (52); SAP30 could conceivably have a role in this process.
Additional or alternative roles for the SAP30 ZnF motif
Although alternative roles for the SAP30 ZnF motif are plausible including a role as a phospholipid-binding module as proposed by Lohi and coworkers (49), our results suggest that nucleic acid binding activity is likely to be of greater physiological significance. This conclusion is based on the greater magnitude of chemical shift perturbations induced by both single- and double-stranded DNA when compared with phospholipid substrates (Figures 4 and 5; Table 3) and estimates of SAP30 ZnF binding affinities with dissociation constants in the low micromolar range for DNA as compared with that in the millimolar range for phospholipids (data not shown). Our results further reveal that the same subset of SAP30 ZnF resonances that are perturbed upon phospholipid binding are also affected by nucleic acid binding (compare Figures 1C and 4), suggesting that these perturbations might reflect some common feature of these substrates (i.e. phosphate and carbohydrate moieties) that is being recognized by the ZnF. Since an overlapping surface of SAP30 ZnF is likely to be mediating both phospholipid and nucleic acid interactions, it is unlikely that both sets of interactions could be supported simultaneously. The competitive binding implied by these findings is consistent with the recent results reported by Lohi and coworkers implicating phosphoinositides in regulating SAP30 ZnF DNA-binding activity and thereby affecting the function of the corepressor complex (48).
It is conceivable that the SAP30 ZnF motif has functions in addition to or in place of DNA binding such as in RNA recognition. Although the rationale for including single-stranded DNA in the titrations was to evaluate the requirement of a double-helical structure for SAP30 ZnF binding, the observation that both single- and double-stranded substrates were bound comparably keeps open the possibility that it might target either form or even higher order structures such as those found in RNA. The significance of these types of interactions is presently unclear. Alternatively, SAP30 might function in providing interaction surfaces for other factors such as RBP1 and SAP180 and/or the ING1 and ING2 proteins (7,17,22,23). Although our attempts at generating a complex with any of these proteins and SAP30 ZnF were unsuccessful, these results might be explained by the absence of additional affinity determinants such as appropriate DNA or RNA surfaces under our assay conditions or by the absence of as yet undetermined bridging proteins. Additional studies are required to clarify these issues.
Distinctive properties of the SAP30 ZnF motif
An interesting feature of the SAP30 ZnF motif is that although it is found in a broad range of organisms including fly and mammals, on the one hand, it is absent in yeast, implying that the function of this motif has been either usurped by another component of the Sin3 complex or that this particular function is dispensable in these organisms. Strikingly, the motif exhibits an extraordinarily high level of sequence conservation (Figure 1A), implying that the region is under strong selective pressure—possibly because of its small size—to preserve structure and function. Interestingly, the SAP30 ZnF motif and its relatives are virtually nonexistent in other proteins making it one of possibly a very small number of ‘singleton’ motifs. Many transcriptional co-regulator domains are found in a restricted subset of proteins possibly to limit cross-talk in nuclear signaling networks and the same may hold true for SAP30. Finally, the structure of the SAP30 ZnF motif is unique in that it appears to be a novel fold which along with the observations noted above raises questions about the antecedents of this motif. It has been over two decades since ZnF motifs were first described as DNA-binding modules (53). The structural and functional diversity exhibited by these modules has been a continued source of surprise and intrigue (54). The SAP30 ZnF motif described herein thus appears to follow this ‘tradition’.
COORDINATES
The RCSB PDB accession code for the atomic coordinates of SAP30 ZnF is 2kdp.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
This work was supported by National Institutes of Health (GM64715 to I.R.); Leukemia and Lymphoma Society (1464-05 to I.R.). Funding for open access charge: National Institutes of Health.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank Bob Eisenman for SAP30 cDNA, Don Ayer for helpful discussions regarding the functional role of the SAP30 ZnF, Heike Folsch for providing advice for the lipid-binding assays, Yongbo Zhang for assistance with NMR data collection and members of the I.R.'s laboratory for helpful discussions.
REFERENCES
- 1.Silverstein RA, Ekwall K. Sin3: a flexible regulator of global gene expression and genome stability. Curr. Genet. 2005;47:1–17. doi: 10.1007/s00294-004-0541-5. [DOI] [PubMed] [Google Scholar]
- 2.Alland L, David G, Shen-Li H, Potes J, Muhle R, Lee HC, Hou H, Jr, Chen K, DePinho RA. Identification of mammalian Sds3 as an integral component of the Sin3/histone deacetylase corepressor complex. Mol. Cell. Biol. 2002;22:2743–2750. doi: 10.1128/MCB.22.8.2743-2750.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fleischer TC, Yun UJ, Ayer DE. Identification and characterization of three new components of the mSin3A corepressor complex. Mol. Cell. Biol. 2003;23:3456–3467. doi: 10.1128/MCB.23.10.3456-3467.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hassig CA, Fleischer TC, Billin AN, Schreiber SL, Ayer DE. Histone deacetylase activity is required for full transcriptional repression by mSin3A. Cell. 1997;89:341–347. doi: 10.1016/s0092-8674(00)80214-7. [DOI] [PubMed] [Google Scholar]
- 5.Laherty CD, Yang WM, Sun JM, Davie JR, Seto E, Eisenman RN. Histone deacetylases associated with the mSin3 corepressor mediate Mad transcriptional repression. Cell. 1997;89:349–356. doi: 10.1016/s0092-8674(00)80215-9. [DOI] [PubMed] [Google Scholar]
- 6.Laherty CD, Billin AN, Lavinsky RM, Yochum GS, Bush AC, Sun JM, Mullen TM, Davie JR, Rose DW, Glass CK, et al. SAP30, a component of the mSin3 corepressor complex involved in N-CoR-mediated repression by specific transcription factors. Mol. Cell. 1998;2:33–42. doi: 10.1016/s1097-2765(00)80111-2. [DOI] [PubMed] [Google Scholar]
- 7.Lai A, Kennedy BK, Barbie DA, Bertos NR, Yang XJ, Theberge MC, Tsai SC, Seto E, Zhang Y, Kuzmichev A, et al. RBP1 recruits the mSIN3-histone deacetylase complex to the pocket of retinoblastoma tumor suppressor family proteins found in limited discrete regions of the nucleus at growth arrest. Mol. Cell. Biol. 2001;21:2918–2932. doi: 10.1128/MCB.21.8.2918-2932.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shiio Y, Rose DW, Aur R, Donohoe S, Aebersold R, Eisenman RN. Identification and characterization of SAP25, a novel component of the mSin3 corepressor complex. Mol. Cell. Biol. 2006;26:1386–1397. doi: 10.1128/MCB.26.4.1386-1397.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang Y, Sun ZW, Iratni R, Erdjument-Bromage H, Tempst P, Hampsey M, Reinberg D. SAP30, a novel protein conserved between human and yeast, is a component of a histone deacetylase complex. Mol. Cell. 1998;1:1021–1031. doi: 10.1016/s1097-2765(00)80102-1. [DOI] [PubMed] [Google Scholar]
- 10.Ayer DE, Lawrence QA, Eisenman RN. Mad-Max transcriptional repression is mediated by ternary complex formation with mammalian homologs of yeast repressor Sin3. Cell. 1995;80:767–776. doi: 10.1016/0092-8674(95)90355-0. [DOI] [PubMed] [Google Scholar]
- 11.Knoepfler PS, Eisenman RN. Sin meets NuRD and other tails of repression. Cell. 1999;99:447–450. doi: 10.1016/s0092-8674(00)81531-7. [DOI] [PubMed] [Google Scholar]
- 12.Pennetta G, Pauli D. The Drosophila Sin3 gene encodes a widely distributed transcription factor essential for embryonic viability. Dev. Genes Evol. 1998;208:531–536. doi: 10.1007/s004270050212. [DOI] [PubMed] [Google Scholar]
- 13.Cowley SM, Iritani BM, Mendrysa SM, Xu T, Cheng PF, Yada J, Liggitt HD, Eisenman RN. The mSin3A chromatin-modifying complex is essential for embryogenesis and T-cell development. Mol. Cell. Biol. 2005;25:6990–7004. doi: 10.1128/MCB.25.16.6990-7004.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dannenberg JH, David G, Zhong S, van der Torre J, Wong WH, Depinho RA. mSin3A corepressor regulates diverse transcriptional networks governing normal and neoplastic growth and survival. Genes Dev. 2005;19:1581–1595. doi: 10.1101/gad.1286905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Carrozza MJ, Florens L, Swanson SK, Shia WJ, Anderson S, Yates J, Washburn MP, Workman JL. Stable incorporation of sequence specific repressors Ash1 and Ume6 into the Rpd3L complex. Biochim. Biophys. Acta. 2005;1731:77–87. doi: 10.1016/j.bbaexp.2005.09.005. discussion 75–76. [DOI] [PubMed] [Google Scholar]
- 16.Sif S, Saurin AJ, Imbalzano AN, Kingston RE. Purification and characterization of mSin3A-containing Brg1 and hBrm chromatin remodeling complexes. Genes Dev. 2001;15:603–618. doi: 10.1101/gad.872801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kuzmichev A, Zhang Y, Erdjument-Bromage H, Tempst P, Reinberg D. Role of the Sin3-histone deacetylase complex in growth regulation by the candidate tumor suppressor p33(ING1) Mol. Cell. Biol. 2002;22:835–848. doi: 10.1128/MCB.22.3.835-848.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Doyon Y, Cayrou C, Ullah M, Landry AJ, Cote V, Selleck W, Lane WS, Tan S, Yang XJ, Cote J. ING tumor suppressor proteins are critical regulators of chromatin acetylation required for genome expression and perpetuation. Mol. Cell. 2006;21:51–64. doi: 10.1016/j.molcel.2005.12.007. [DOI] [PubMed] [Google Scholar]
- 19.Bernstein BE, Tong JK, Schreiber SL. Genomewide studies of histone deacetylase function in yeast. Proc. Natl Acad. Sci. USA. 2000;97:13708–13713. doi: 10.1073/pnas.250477697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sun ZW, Hampsey M. A general requirement for the Sin3-Rpd3 histone deacetylase complex in regulating silencing in Saccharomyces cerevisiae. Genetics. 1999;152:921–932. doi: 10.1093/genetics/152.3.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Loewith R, Smith JS, Meijer M, Williams TJ, Bachman N, Boeke JD, Young D. Pho23 is associated with the Rpd3 histone deacetylase and is required for its normal function in regulation of gene expression and silencing in Saccharomyces cerevisiae. J. Biol. Chem. 2001;276:24068–24074. doi: 10.1074/jbc.M102176200. [DOI] [PubMed] [Google Scholar]
- 22.Binda O, Roy JS, Branton PE. RBP1 family proteins exhibit SUMOylation-dependent transcriptional repression and induce cell growth inhibition reminiscent of senescence. Mol. Cell. Biol. 2006;26:1917–1931. doi: 10.1128/MCB.26.5.1917-1931.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shi X, Hong T, Walter KL, Ewalt M, Michishita E, Hung T, Carney D, Pena P, Lan F, Kaadige MR, et al. ING2 PHD domain links histone H3 lysine 4 methylation to active gene repression. Nature. 2006;442:96–99. doi: 10.1038/nature04835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hsieh JJ, Zhou S, Chen L, Young DB, Hayward SD. CIR, a corepressor linking the DNA binding factor CBF1 to the histone deacetylase complex. Proc. Natl Acad. Sci. USA. 1999;96:23–28. doi: 10.1073/pnas.96.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yoon HG, Chan DW, Huang ZQ, Li J, Fondell JD, Qin J, Wong J. Purification and functional characterization of the human N-CoR complex: the roles of HDAC3, TBL1 and TBLR1. EMBO J. 2003;22:1336–1346. doi: 10.1093/emboj/cdg120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stols L, Gu M, Dieckman L, Raffen R, Collart FR, Donnelly MI. A new vector for high-throughput, ligation-independent cloning encoding a tobacco etch virus protease cleavage site. Protein Exp. Purif. 2002;25:8–15. doi: 10.1006/prep.2001.1603. [DOI] [PubMed] [Google Scholar]
- 27.Gill SC, von Hippel PH. Calculation of protein extinction coefficients from amino acid sequence data. Anal. Biochem. 1989;182:319–326. doi: 10.1016/0003-2697(89)90602-7. [DOI] [PubMed] [Google Scholar]
- 28.Radhakrishnan I, Perez-Alvarado GC, Parker D, Dyson HJ, Montminy MR, Wright PE. Structural analyses of CREB-CBP transcriptional activator-coactivator complexes by NMR spectroscopy: implications for mapping the boundaries of structural domains. J. Mol. Biol. 1999;287:859–865. doi: 10.1006/jmbi.1999.2658. [DOI] [PubMed] [Google Scholar]
- 29.Johnson BA. Using NMRView to visualize and analyze the NMR spectra of macromolecules. Methods Mol. Biol. 2004;278:313–352. doi: 10.1385/1-59259-809-9:313. [DOI] [PubMed] [Google Scholar]
- 30.Bax A, Grzesiek S. Methodological advances in protein NMR. Accounts Chem. Res. 1993;26:131–138. [Google Scholar]
- 31.Ferentz AE, Wagner G. NMR spectroscopy: a multifaceted approach to macromolecular structure. Q. Rev. Biophys. 2000;33:29–65. doi: 10.1017/s0033583500003589. [DOI] [PubMed] [Google Scholar]
- 32.Otting G, Wüthrich K. Heteronuclear filters in two-dimensional [1H,1H]-NMR spectroscopy: combined use with isotope labelling for studies of macromolecular conformation and intermolecular interactions. Q. Rev. Biophys. 1990;23:39–96. doi: 10.1017/s0033583500005412. [DOI] [PubMed] [Google Scholar]
- 33.Lohr F, Ruterjans H. Novel pulse sequences for the resonance assignment of aromatic side chains in 13C-labeled proteins. J. Magn. Reson. B. 1996;112:259–268. doi: 10.1006/jmrb.1996.0140. [DOI] [PubMed] [Google Scholar]
- 34.Cornilescu G, Delaglio F, Bax A. Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J. Biomol. NMR. 1999;13:289–302. doi: 10.1023/a:1008392405740. [DOI] [PubMed] [Google Scholar]
- 35.Linge JP, Habeck M, Rieping W, Nilges M. ARIA: automated NOE assignment and NMR structure calculation. Bioinformatics. 2003;19:315–316. doi: 10.1093/bioinformatics/19.2.315. [DOI] [PubMed] [Google Scholar]
- 36.Linge JP, Habeck M, Rieping W, Nilges M. Correction of spin diffusion during iterative automated NOE assignment. J. Magn. Reson. 2004;167:334–342. doi: 10.1016/j.jmr.2004.01.010. [DOI] [PubMed] [Google Scholar]
- 37.Brünger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 38.Laskowski RA, Rullmannn JA, MacArthur MW, Kaptein R, Thornton JM. AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J. Biomol. NMR. 1996;8:477–486. doi: 10.1007/BF00228148. [DOI] [PubMed] [Google Scholar]
- 39.Salerno WJ, Seaver SM, Armstrong BR, Radhakrishnan I. MONSTER: inferring non-covalent interactions in macromolecular structures from atomic coordinate data. Nucleic Acids Res. 2004;32:W566–W568. doi: 10.1093/nar/gkh434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 41.Landau M, Mayrose I, Rosenberg Y, Glaser F, Martz E, Pupko T, Ben-Tal N. ConSurf 2005: the projection of evolutionary conservation scores of residues on protein structures. Nucleic Acids Res. 2005;33:W299–W302. doi: 10.1093/nar/gki370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc. Natl Acad. Sci. USA. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Grishin NV. Treble clef finger—a functionally diverse zinc-binding structural motif. Nucleic Acids Res. 2001;29:1703–1714. doi: 10.1093/nar/29.8.1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Krishna SS, Majumdar I, Grishin NV. Structural classification of zinc fingers: survey and summary. Nucleic Acids Res. 2003;31:532–550. doi: 10.1093/nar/gkg161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tochio N, Umehara T, Koshiba S, Inoue M, Yabuki T, Aoki M, Seki E, Watanabe S, Tomo Y, Hanada M, et al. Solution structure of the SWIRM domain of human histone demethylase LSD1. Structure. 2006;14:457–468. doi: 10.1016/j.str.2005.12.004. [DOI] [PubMed] [Google Scholar]
- 46.Da G, Lenkart J, Zhao K, Shiekhattar R, Cairns BR, Marmorstein R. Structure and function of the SWIRM domain, a conserved protein module found in chromatin regulatory complexes. Proc. Natl Acad. Sci. USA. 2006;103:2057–2062. doi: 10.1073/pnas.0510949103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Qian C, Zhang Q, Li S, Zeng L, Walsh MJ, Zhou MM. Structure and chromosomal DNA binding of the SWIRM domain. Nat. Struct. Mol. Biol. 2005;12:1078–1085. doi: 10.1038/nsmb1022. [DOI] [PubMed] [Google Scholar]
- 48.Viiri KM, Janis J, Siggers T, Heinonen TY, Valjakka J, Bulyk ML, Maki M, Lohi O. DNA-binding and -bending activities of SAP30L and SAP30 are mediated by a zinc-dependent module and monophosphoinositides. Mol. Cell. Biol. 2009;29:342–356. doi: 10.1128/MCB.01213-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Viiri KM, Korkeamaki H, Kukkonen MK, Nieminen LK, Lindfors K, Peterson P, Maki M, Kainulainen H, Lohi O. SAP30L interacts with members of the Sin3A corepressor complex and targets Sin3A to the nucleolus. Nucleic Acids Res. 2006;34:3288–3298. doi: 10.1093/nar/gkl401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tsang CK, Bertram PG, Ai W, Drenan R, Zheng XF. Chromatin-mediated regulation of nucleolar structure and RNA Pol I localization by TOR. EMBO J. 2003;22:6045–6056. doi: 10.1093/emboj/cdg578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhou Y, Santoro R, Grummt I. The chromatin remodeling complex NoRC targets HDAC1 to the ribosomal gene promoter and represses RNA polymerase I transcription. EMBO J. 2002;21:4632–4640. doi: 10.1093/emboj/cdf460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vermeulen M, Walter W, Le Guezennec X, Kim J, Edayathumangalam RS, Lasonder E, Luger K, Roeder RG, Logie C, Berger SL, et al. A feed-forward repression mechanism anchors the Sin3/histone deacetylase and N-CoR/SMRT corepressors on chromatin. Mol. Cell. Biol. 2006;26:5226–5236. doi: 10.1128/MCB.00440-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Miller J, McLachlan AD, Klug A. Repetitive zinc-binding domains in the protein transcription factor IIIA from Xenopus oocytes. EMBO J. 1985;4:1609–1614. doi: 10.1002/j.1460-2075.1985.tb03825.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Laity JH, Lee BM, Wright PE. Zinc finger proteins: new insights into structural and functional diversity. Curr. Opin. Struct. Biol. 2001;11:39–46. doi: 10.1016/s0959-440x(00)00167-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.