Abstract
The 5-formyluracil (5-foU), a major mutagenic oxidative damage of thymine, is removed from DNA by Nth, Nei and MutM in Escherichia coli. However, DNA polymerases can also replicate past the 5-foU by incorporating C and G opposite the lesion, although the mechanism of correction of the incorporated bases is still unknown. In this study, using a borohydride-trapping assay, we identified a protein trapped by a 5-foU/C-containing oligonucleotide in an extract from E. coli mutM nth nei mutant. The protein was subsequently purified from the E. coli mutM nth nei mutant and was identified as KsgA, a 16S rRNA adenine methyltransferase. Recombinant KsgA also formed the trapped complex with 5-foU/C- and thymine glycol (Tg)/C-containing oligonucleotides. Furthermore, KsgA excised C opposite 5-foU, Tg and 5-hydroxymethyluracil (5-hmU) from duplex oligonucleotides via a β-elimination reaction, whereas it could not remove the damaged base. In contrast, KsgA did not remove C opposite normal bases, 7,8-dihydro-8-oxoguanine and 2-hydroxyadenine. Finally, the introduction of the ksgA mutation increased spontaneous mutations in E. coli mutM mutY and nth nei mutants. These results demonstrate that KsgA has a novel DNA glycosylase/AP lyase activity for C mispaired with oxidized T that prevents the formation of mutations, which is in addition to its known rRNA adenine methyltransferase activity essential for ribosome biogenesis.
INTRODUCTION
Reactive oxygen species (ROS) are generated continually in living cells during normal cellular metabolism. ROS are also generated by exogenous sources, such as ionizing radiation and various chemical oxidants. ROS can cause a number of different types of oxidative damage to purines and pyrimidines in DNA (1–3), which have been implicated in mutation, cancer induction and aging (4,5). To overcome the deleterious effects of oxidative base damage in DNA, bacteria and eukaryotes have evolved the base excision repair (BER) pathway (3,6–8). DNA glycosylases catalyze the first step of the BER pathway by removing oxidized bases from DNA. There are several DNA glycosylases that specifically recognize and remove damaged bases from DNA, generating an apurinic/apyrimidinic (AP) site (3,5,6,8,9). DNA glycosylase-associated AP lyase and AP endonuclease recognize the resultant AP site and cleave the phosphodiester backbone at the AP site, followed by DNA repair synthesis by DNA polymerases, and the nick in the DNA backbone is finally rejoined by DNA ligases (8,10).
The C8 of guanine is readily oxidized and 8-oxo-7,8-dihydroguanine (8-oxoG) is one of the most important mutagenic lesions in DNA (6,8,9) as it causes G:C to T:A transversions, due to mispairing with A during DNA replication (5,8,11). In Escherichia coli, two DNA glycosylases are involved in the repair of 8-oxoG. MutM (formamidopyrimidine–DNA glycosylase) removes 8-oxoG from 8-oxoG/C pairs generated in DNA (8,12). If unrepaired, 8-oxoG in the template directs incorporation of A to form mutagenic 8-oxoG/A mispairs (5,8,12). In this situation, MutY removes the A incorporated and thus provides a second opportunity to prevent mutations (3,5,13). Therefore, mutations in the mutM and mutY genes markedly increase G:C to T:A transversions (13).
Thymine glycol (Tg), 5-formyluracil (5-foU) and 5-hydroxymethyluracil (5-hmU) are the major oxidative damage products of thymine (1–3). These damages in DNA are primarily repaired by endonuclease III (Nth), endonuclease VIII (Nei) in E. coli and by their homologs in S. cerevisiae, S. pombe and human cells (2,3,8,14–18). 5-foU and 5-hmU are also removed by MutM and AlkA in E. coli (14,17,19).
The 5-foU has been elucidated to be a potent mutagenic lesion in vivo (14,20,21). Recently, we found that the mutation frequency of a plasmid containing a site-specific 5-foU lesion was significantly increased compared with that of a normal T-containing plasmid (14). 5-foU causes incorporation of mismatched bases C and G opposite the lesion during DNA synthesis in vitro (22,23). In this situation, bases misincorporated opposite 5-foU must be removed from DNA. Furthermore, recent studies revealed that 5-formyl-dUTP is incorporated opposite C and G during DNA replication, which also implies a significant potential for mutagenesis due to 5-foU (24,25). Indeed, the addition of 5-formyl-deoxyuridine to the culture medium promoted mutagenicity at the hprt locus in Chinese hamster fibroblast cells (26), indicating that 5-foU is incorporated into DNA and has base pairing properties different from T. Therefore, we propose that cells must contain an additional enzyme(s) that excise 5-foU incorporated opposite C and G, although such DNA glycosylases have not yet been identified. These facts led us to examine whether E. coli has DNA glycosylase activity for C and G incorporated opposite oxidatively damaged T during DNA replication and for 5-foU incorporated opposite C and G in the newly-synthesized strand from 5-formyl-dUTP generated in the nucleotide pool.
In this study, by using a borohydride-trapping assay (27), we identified and purified a protein with DNA glycosylase activity for C paired with oxidized T. The purified protein was identified as KsgA and was demonstrated to contain a novel DNA glycosylase activity to repair C/oxidized T- mispairs in DNA that prevents mutations, in addition to its rRNA adenine methyltransferase.
MATERIALS AND METHODS
Enzymes and chemicals
T4 polynucleotide kinase was obtained from New England Biolabs. Plasmid pGEX-4T-3, prepacked columns for fast protein liquid chromatography (FPLC), HiTrap SP, HiTrap Q, HiTrap Heparin, HiTrap Blue and Resource S were purchased from GE Healthcare. Hydroxyapatite column was obtained from Bio-Rad. Restriction enzymes, KOD Plus DNA polymerase and T4 DNA ligase were obtained from Toyobo. [γ-32P] ATP (>148 TBq/mmol) was obtained from ICN Biomedicals.
Synthesis of Tg-, 5-foU-, 5-hmU-, 8-oxo-G- and 2-hydroxyadenine-containing oligonucleotides
Oligonucleotides containing Tg, 5-foU, 5-hmU, 8-oxoG or 2-hydroxyadenine (2-ohA) at defined sites were synthesized as previously described (21,28–30). Other HPLC-purified oligonucleotides were purchased from Takara Shuzo. The nucleotide sequences of oligonucleotides used in this study are shown in Table 1.
Table 1.
Nucleotide sequences of oligonucleotides
| Designation | Sequence | Enzyme tested |
|---|---|---|
| 5-foU/N | 5′-GGTCGACTFAAGGTACC | KsgA, |
| 3′-CCAGCTGANTTCCATGG | MutY | |
| Tg/N | 5′-GGACGACAXAAGGAACC | KsgA |
| 3′-CCTGCTGTNTTCCTTGG | ||
| 5-hmU/C | 5′-GATCCTCTAGAGHCGACCTGCA | KsgA |
| 3′-CTAGGAGATCACCGCTGGACGT | ||
| 8-oxoG/C | 5′-GAACTAGTG8ATCCCCCGGGCTGC | KsgA |
| 3′-CTTGATCACCTAGGGGGCCCGACG | ||
| 2-ohA/C | 5′-GAACTAGTG2ATCCCCCGGGGCTGC | KsgA |
| 3′-CTTGATCACCTAGGGGGCCCCGACG | ||
| Tg/C# | 5′-CGGCGCGCXCGGGCGGC | KsgA |
| 3′-GCCGCCCGCGCGCGCCG | ||
| Tg/A# | 5′-GCCGCCCGXGCGCGCCG | Nth, Nei |
| 3′-CGGCGGGCACGCGCGGC | ||
| 8-oxoG/A | 5′-GAACTAGTG8ATCCCCCGGGCTGC | MutY |
| 3′-CTTGATCACATAGGGGGCCCGACG |
F, X, H, 8 and 2 represent 5-foU, Tg, 5-hmU, 8-oxoG and 2-ohA, respectively.
Bacterial strains and media
Escherichia coli KSR7 was a mutM::Tet nth::Cm nei::Kan derivative of E. coli CSH26 (14). Escherichia coli JA200/pLC4-46 (31) was kindly gifted from Dr A. Nishimura (The National Institute of Genetics, Mishima). The ksgA::Kan mutant was kindly supplied by Dr T. Miki (Fukuoka Dental University, Fukuoka). The construction of E. coli mutants was carried out using P1 transduction. E. coli mutM::Kan mutY::Tet ksgA::Kan and nth::Kan nei::Cm ksgA::Kan mutants were isolated according to Bochner et al. (32). Escherichia coli strains used for the mutation assay are listed in Table 2. LB broth was used to culture E. coli cells. When necessary, ampicillin (Amp), chloramphenicol (Cm), tetracycline (Tet) and kanamycin (Kan) were added to the medium at a final concentration of 100, 30, 15 and 50 μg/ml, respectively.
Table 2.
Escherichia coli strains used for spontaneous mutation assay
| Strains | Relevant genotype | Construction |
|---|---|---|
| CSH101 | Wild-type | |
| CSH101m | mutM::Kan | |
| CSH101y | mutY::Tet | |
| CSH101my | mutM::Kan mutY::Tet | |
| CSH101k | ksgA::Kan | |
| CSH101c | car96::Tet | |
| CSH101kc | ksgA::Kan car96::Tet | P1(CSH101c) × CSH101k |
| CSH101yk | mutY::Tet ksgA::Kan | P1(CSH101y) × CSH101k |
| CSH101mkc | mutM::Kan ksgA::kan car96::Tet | P1(CSH101kc) × CSH101m |
| CSH101mkΔc | mutM::Kan ksgA::kan Δcar96::Tet | Deletion of car96::Tet in CSH101mkc |
| CSH101myk | mutM::Kan mutY::Tet ksgA::Kan | P1(CSH101y) × CSH101mkΔc |
| CSH101-3 | nth::Kan | |
| CSH101z | zdh925::Tet | |
| CSH101-3z | nth::Kan zdh925::Tet | P1(CSH101z) × CSH101-3 |
| CSH101-3zk | nth::Kan zdh925::Tet ksgA::Kan | P1(CSH101-3z) × CSH101k |
| CSH101-8 | nei::Cm | |
| CSH101-38k | nth::Kan nei::Cm ksgA::Kan | P1(CSH101-8) × CSH101-3zk |
The strains were constructed by P1 transduction. CC101mkΔc was isolated according to Bochner et al. (32).
Preparation of crude extract from E. coli cells
Overnight culture of E. coli KSR7 (50 l) was centrifuged, and the cell pellet (150 g) was resuspended in 1.5 l of buffer A [25 mM Tris–HCl (pH 7.5) containing 1 mM EDTA, 14 mM β-mercaptoethanol (EtSH), 5% glycerol, 1 mM PMSF, 1mg/ml of lysozyme and 100 mM NaCl] and stored at –80°C. The cells were incubated at 37°C for 15 min and then disrupted by sonication on ice after freezing and thawing. The lysate was centrifuged at 35 000g for 40 min at 4°C. The supernatant was stored at –80°C before use.
Protein purification
Proteins were assayed for the ability to form a trapped complex with Tg/C- and 5-foU/C-containing oligonucleotides. The extract prepared from E. coli KSR7 (mutM nth nei) cells (1400 ml) was loaded onto a HiTrap SP Cation Exchange column, which had been equilibrated with buffer A. Proteins were eluted with a linear gradient of NaCl (100–1000 mM). Active fractions that eluted between 340 and 580 mM NaCl were pooled and dialyzed against buffer A. The dialysate was applied to a HiTrap Heparin column and proteins were eluted with a linear gradient of NaCl (100–1000 mM). Active fractions eluted between 520 and 760 mM NaCl and were diluted with buffer A without NaCl, followed by HiTrap Blue column chromatography. Proteins were eluted with a linear gradient of NaCl (0.1–2 M). Active fractions that eluted between 0.38 and 1.24 M NaCl were pooled, applied to a desalting column, which had been equilibrated with buffer A, and then applied to a Hydroxyapatite column. Proteins were eluted with a linear gradient of NaCl (100–1000 mM). Active fractions that eluted between 520 and 580 mM NaCl were pooled, applied to a desalting column and eluted with buffer A. Finally, active fractions were applied to a Resource S column and proteins were eluted with a linear gradient of NaCl (100–1000 mM). The active fractions were eluted between 550 and 650 mM NaCl (fraction V) and stored at –80°C.
Protein sequencing
Fraction V was analyzed by SDS–PAGE on a 15% polyacrylamide gel, transferred to an Immobilon-PVDF membrane (Millipore) by electroblotting and then stained with Coomassie blue. The protein band (30 kDa) was excised from the membrane, and the sequence of the N-terminal amino acids of the protein was determined by using a gas-phase amino acid sequencer Procise 494 HT Protein Sequencing System (APRO Science).
Sodium borohydride trapping assay
Oligonucleotides were labeled at the 5′-end with [γ-32P]ATP by T4 polynucleotide kinase and then annealed to the complementary strand. DNA trapping was performed in a reaction mixture (15 μl) containing 50–100 fmol of 32P-labeled double-stranded oligonucleotide and crude extract (or purified protein) in 20 mM HEPES–KOH (pH 7.6) containing 10 mM dithiothreitol (DTT), 1.5 mM MgCl2, 5 mM KCl, 2 mM EDTA, 1% glycerol and 5 μg/ml of bovine serum albumin (BSA) in the presence of 100 mM NaBH4. The aqueous solution of NaBH4 (1 M) was prepared just before use. After incubation at 37°C for 30 min, the reaction was terminated by the addition of 2 × sample buffer. The samples were then heated at 95°C for 5 min, and subsequently subjected to 12% SDS–PAGE. The gels were exposed to imaging plates and analysed with a BAS 1800 Image Analyzer (Fuji Film).
Cloning of the E. coli ksgA gene
A plasmid expressing the glutathione-S-transferase (GST)-KsgA fusion protein was constructed as follows; the plasmid pLC4-46 bearing E. coli ksgA gene (31) was amplified with two PCR primers; one containing a BamHI site followed by the sequence around the putative start codon (5′-ATATGCATCCATGAATAATCGAGTCCACCAGG-3′) and the other containing an EcoRI site followed by the sequence around the stop codon (5′-TAATGAATTCTTAACTCTCCTGCAAAGGCGCGT-3′). PCR was performed in 100 μl reaction mixture containing 20 mM Tris–HCI (pH 7.5), 200 μM each dNTP, 2 mM MgCI2, 1 mM of primer and 1 unit of KOD Plus DNA polymerase. The amplified fragment was digested with BamHI and EcoRI, and then the BamHI/EcoRI fragment containing the whole coding region of the E. coli ksgA gene was inserted into BamHI/EcoRI-digested pGEX-4T-3. The resulting plasmid was named pGEX-KsgA. The sequence of the insert was verified not to contain any mutations in the region.
Expression and purification of the KsgA
Escherichia coli KSR7 (mutM nth nei) was transformed with the plasmid pGEX-KsgA. The cells were grown at 37°C in LB medium containing Amp until the optical density at 600 nm reached 0.6. After the addition of 0.4 mM of isopropyl β-D-galactopyranoside (IPTG), the culture was further incubated at 24°C for 8 h. The cells were harvested and resuspended in buffer B [50 mM sodium phosphate buffer (pH 8.0) containing 14 mM EtSH, 1 mM PMSF, 5% glycerol and 1% Triton-X-100] with 500 mM NaCl. The cells were disrupted by sonication on ice after freezing and thawing. After centrifugation of the cell lysate at 32 000g for 40 min at 4°C, the supernatant was applied to a glutathione-Sepharose 4B column and washed with 10 column volumes of 50 mM Tris–HCl (pH 8.0). The bound protein was eluted with 15 mM glutathione in 50 mM Tris–HCl (pH 8.0), followed by dialysis against buffer C [25 mM Tris–HCl (pH 7.5) containing 14 mM EtSH, 1 mM EDTA, 5% glycerol and 100 mM NaCl]. The GST–KsgA fusion protein was then cleaved by thrombin by incubation for 12 h at 4°C. The protein digest was further applied to a HiTrap SP column and eluted using a linear gradient of NaCl. KsgA protein eluted between 180–330 mM NaCl and was subsequently stored at –80°C before use. The GST–KsgA was also purified from E. coli CSH101y (MutY-deficient) and BL21 induced by IPTG as described above.
Purification of E. coli Nth, Nei and MutY
Nth, Nei and MutY of E. coli have been purified as previously described (14,18,33).
Cleavage assay for DNA glycosylase/AP lyase activity
DNA cleavage assay was carried out at 37°C in a reaction mixture (10 μl) containing 10 mM Tris–HCl (pH 7.5), 25 ∼ 100 fmol of 32P-labeled double-stranded oligonucleotide, the purified KsgA, 1 mM EDTA and 50 mM NaCl. The reaction was terminated by the addition of stop solution (95% formamide, 0.1% xylene cyanol, 20 mM EDTA). After heating at 95°C for 5 min, the samples were cooled and then loaded onto 20% polyacrylamide gels in 90 mM Tris–borate (pH 8.3) containing 7 M urea and 2 mM EDTA. After electrophoresis at 1150 V, the gels were exposed to imaging plates and analysed with a BAS 1800 Image Analyzer (Fuji Film).
Measurement of spontaneous mutant frequency
A single colony of various strains of E. coli (Table 2) were inoculated into LB medium containing the appropriate antibiotics and cultured at 37°C to stationary phase. The mutation to rifampicin resistance was assayed as follows; 0.1 ml of the culture was plated on LB plates containing 150 μg/ml of rifampicin and then incubated at 37°C for about 40 h. To determine viable cells, the overnight cultures were appropriately diluted and plated on LB plates, followed by incubation at 37°C for 18 h. The mutant frequency was estimated by the number of mutant cells/108 viable cells.
RESULTS
Identification of a DNA glycosylase activity
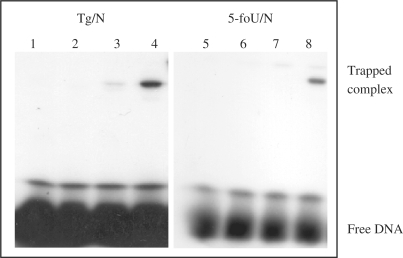
DNA glycosylase with associated AP lyase activity forms a transient Schiff base intermediate with an AP site in DNA, which can be reduced by NaBH4 or NaCNBH3 to generate a stable trapped DNA–protein complex (27). We used this trapping assay to identify and characterize protein(s) with DNA glycosylase/AP lyase activity for 5-foU/C- and Tg/C-containing oligonucleotides. A γ-32P-labeled double-stranded oligonucleotide containing a single 5-foU or Tg opposite G, A, T and C was incubated with the extract prepared from E. coli mutM nth nei mutants in the presence of 100 mM NaBH4. Trapped DNA–protein complexes were analyzed by SDS–PAGE. In Figure 1, one shifted band was detected with the oligonucleotide containing Tg/C- (lane 4) and 5-foU/C (lane 8), indicating that E. coli has a DNA glycosylase activity for Tg/C and 5-foU/C.
Figure 1.
Formation of trapped complexes of protein and Tg/C- and 5-foU/C-containing oligonucleotides in the presence of NaBH4. Oligonucleotides substrates (100 fmol) containing Tg/N and 5-foU/N mispairs were incubated at 37°C for 30 min with extract (15 μg protein) from E. coli KSR7 (mutM nth nei mutant) in the presence of 100 mM NaBH4. The samples were subjected to 12% SDS–PAGE. Lane 1, Tg/G; lane 2, Tg/A; lane 3, Tg/T, lane 4, Tg/C; lane 5, 5-foU/G; lane 6, 5-foU/A; lane 7, 5-foU/T; lane 8, 5-foU/C. The positions of the trapped complex and free DNA are indicated.
Purification of protein with 5-foU/C- and Tg/C-DNA glycosylase activity
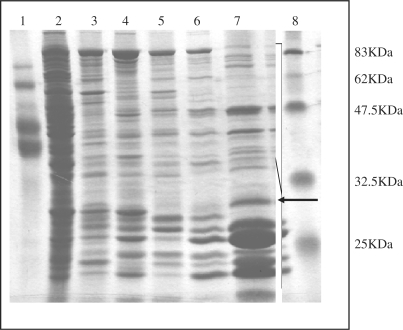
We purified the DNA glycosylase for 5-foU/C and Tg/C by standard chromatography techniques. Proteins were assayed for the ability to form trapped complexes with Tg/C-containing oligonucleotide. The purification steps for the protein are shown in Figure 2 and it was purified through HiTrap SP (fraction I), HiTrap-Heparin (fraction II), HiTrap-Blue (fraction III), Hydroxyapatite (fraction IV) and Resources S. The recovery of proteins was 5.9, 3.4 and 0.18% after HiTrap SP, the HiTrap-Heparin and the Resource S column (fraction V) chromatography, respectively.
Figure 2.
Protein purification of the sodium borohydride trapping-active protein. The borohydride-trapping active protein was purified from E. coli mutM nth nei mutant extract by standard chromatography techniques. Proteins from each purification step were separated by 12% SDS–PAGE and stained with Coomassie Blue. Lanes 1 and 8, molecular weight marker; Lane 2, crude extract; lane 3, Hi-Trap SP; lane 4, Hi-Trap Heparin; lane 5, HiTrap-Blue; lane 6, Hydroxyapatite; lane 7, Resource S. The arrow indicates the band for the candidate protein (trapping-active) used for determination of the N-terminal amino acid sequence.
Comparing the size of the trapped complex with that of known DNA glycosylases such as MutM and Nth, we estimated the molecular weight of the trapping-active protein to be about 30 kDa (lane 7, indicated as arrow in Figure 2). We isolated the protein band from the gel and determined the sequence of the N-terminal amino acids, which were NNRV-QGHLA. This sequence aligned with that of a polypeptide starting at the second codon of the E. coli ksgA gene (NNRVHQGHLA) (34,35). The KsgA protein has 273 amino acid residues with a molecular weight 30.4 kDa (34–37).
Expression and purification of E. coli KsgA
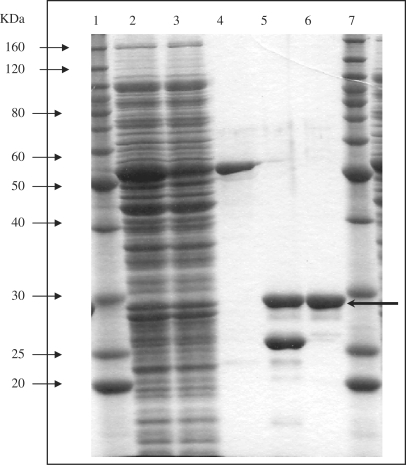
The entire open reading frame of the ksgA gene was amplified by PCR and subcloned into a plasmid vector pGEX-4T-3 to obtain the pGEX-KsgA fusion construct. The fusion protein was expressed in E. coli KSR7 mutM nth nei mutant/pGEX-KsgA induced with IPTG and then purified by a glutathione-Sepharose 4B column chromatography. The 30.4 kDa KsgA protein after cleavage of the GST–KsgA fusion protein with thrombin is shown (Figure 3). The KsgA protein was further purified by HiTrap SP chromatography.
Figure 3.
Expression and purification of E. coli KsgA. Escherichia coli KSR7 carrying pGEX-KsgA was induced by 0.4 mM of IPTG at 24°C for 8 h. Proteins were separated by 12% SDS–PAGE and stained with Coomassie Blue. Lanes 1 and 7, molecular weight marker; lane 2, soluble fraction disrupted by sonication; lane 3, flow-through fraction from glutathione-Sepharose column; lane 4, purified GST–KsgA fusion protein after elution from glutathione-Sepharose column; lane 5, fusion protein after digestion by thrombin; lane 6, purified KsgA after elution from Hi-Trap SP column.
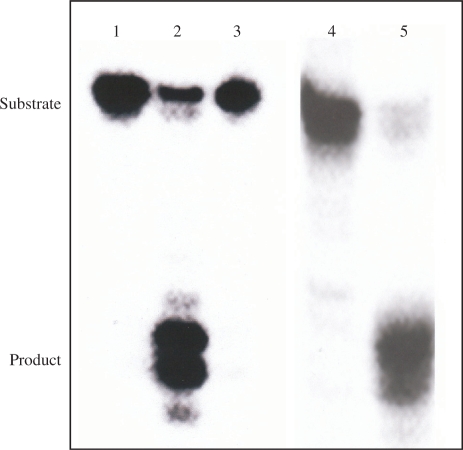
Cytosine–DNA glycosylase activity of KsgA
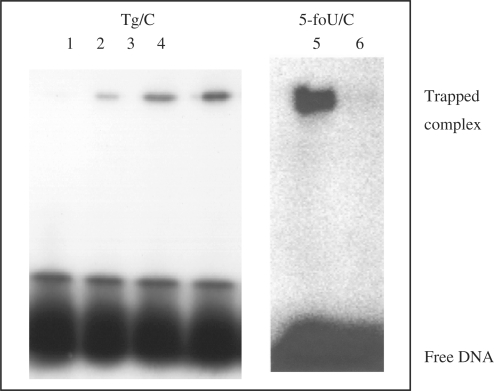
The trapping and cleavage assays were performed to demonstrate that recombinant KsgA had a DNA glycosylase activity for Tg/C and 5-foU/C. 32P-labeled Tg/C- and 5-foU/C-containing double-stranded oligonucleotides were incubated at 37°C for 60 min with purified KsgA. Figure 4 shows that KsgA could form a trapped complex with Tg/C-containing DNA (lanes 1–4) in the presence of NaBH4. The intensity of the trapped complex increased with the amount of KsgA added. The trapping activity was confirmed with 5-foU/C-containing oligonucleotide (lanes 5 and 6). These results indicated that purified KsgA has DNA glycosylase with associated AP lyase activity for C/oxidized T-containing oligonucleotides.
Figure 4.
Formation of trapped complexes of purified KsgA with Tg/C- and 5-foU/C-containing double-stranded oligonucleotides. Oligonucleotide substrates (100 fmol) containing Tg/C (lanes 1–4) and 5-foU/C (lanes 5 and 6) were incubated at 37°C for 30 min with purified KsgA in the presence of 100 mM NaBH4. The samples were separated by 12% SDS–PAGE. KsgA was added at 0 (lanes 1 and 6), 20 (lane 2), 50 (lane 3) and 100 (lanes 4 and 5) fmol, respectively.
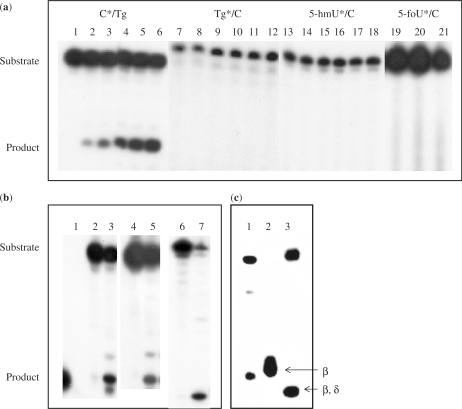
We next performed the DNA cleavage assay. The oligonucleotide containing 5-foU, Tg or 5-hmU was 32P-labeled at the 5′-end and annealed with the non-labeled complementary oligonucleotide containing C opposite the oxidized T. The double-stranded oligonucletides were incubated with purified KsgA at 37°C for 90 min and the products were analyzed on 20% polyacrylamide gels. KsgA did not excise Tg, 5-foU and 5-hmU opposite C from double-stranded oligonucleotides (Figure 5a, lanes 7–21).
Figure 5.
Cleavage of oligonucleotides containing Tg/C and 5-foU/C mispairs by purified KsgA. (a) The oligonucleotides containing 50 fmol C*/Tg (lanes 1–6), Tg*/C (lanes 7–12), 5-hmU*/C (lanes 13–18) and 5-foU*/C (lanes 19–21) (asterisks indicate labeled strand) were incubated at 37°C for 90 min with increasing amounts of purified KsgA. KsgA was added at 0 (lanes 1, 7, 13 and 19), 12.5 (lanes 2, 8 and 14), 25 (lanes 3, 9 and 15), 50 (lanes 4, 10,16 and 20), 75 (lanes 5, 11, and 17) and 100 (lanes 6, 12, 18 and 21) pmol. (b) Unlabeled 5-foU, Tg or 5-hmU-containing oligonucleotides were annealed to a 32P-labeled oligonucleotide containing C opposite the oxidative modifications of thymine. The double-stranded oligonucleotides (50 fmol) were incubated at 37°C for 60 min without (lanes 2, 4 and 6) or with (lanes 3, 5 and 7) purified KsgA protein (100 pmol). Lane 1, 8-mer marker. (c) 5′-P32-labeled GCCGCCCGCGCGCGCCG/3′-CGGCGGGCTgCGCGCGGC (Tg/C#) (10 fmol) was incubated at 37°C for 60 min with purified GST-KsgA (107 pmol), and 5′-P32-labeled GCCGCCCGTgGCGCGCCG/3′-CGGCGGGCACGCGCGGC (Tg/A#) (10 fmol) with Nth (4 pmol) or Nei (6.5 pmol). The products were separated by denaturing 20% PAGE in gels containing urea. Lane 1, purified GST-KsgA; lane 2, purified GST-Nth (β-elimination), lane 3, purified Nei-his (β,δ-elimination).
Next, the oligonucleotide containing C was 32P-labeled at the 5′-end and then annealed with non-labeled complementary oligonucleotide containing Tg opposite the C. The double-stranded oligonucleotide was incubated with purified KsgA and it was evident that KsgA cleaved the oligonucleotide containing C opposite Tg (Figure 5a, lanes 1–6). The 5-foU/C* oligonucleotide was almost completely cleaved by incubating at 37°C for 180 min with GST–KsgA (107 pmol), while the enzyme could not excise Tg and 5-foU opposite C.
The oligonucleotides containing C/5-foU and C/5-hmU were also cleaved by purified KsgA (Figure 5b). KsgA cleaved the double-stranded oligonucleotides containing C opposite Tg, 5-foU and 5-hmU from the DNA at similar efficiencies. As shown in Figure 5c, KsgA cleaved the oligonucleotide at the C via a β-elimination reaction, as compared to the mobility of oligonucleotides cleaved by purified Nth (β-elimination) and Nei (β,δ-elimination). The cleavage did not require the reaction with NaOH, indicating that KsgA is a bifunctional DNA glycosylase. Therefore, we demonstrate that KsgA catalyzes both the cleavage of the glycosylic bond to release C opposite oxidized T and the incision of the phosphodiester backbone at the resulting AP site. In contrast, KsgA did not cleave C paired with normal T, G, A and C (data not shown). Furthermore, it could not excise C opposite oxidized purines, 8-oxoG and 2-ohA (data not shown).
It was also evident that the enzyme purified from E. coli mutY mutant had the activity to excise C opposite 5-foU (Figure 6). Furthermore, the purified MutY did not excise C opposite Tg and 5-foU, while it efficiently cleaved the 8-oxoG/A-containing oligonucleotide (Figure 6). The results indicated that the DNA glycosylase activity of ksgA is not due to contaminating MutY.
Figure 6.
Cleavage of oligonucleotides 5-foU/C by GST-KsgA purified from E. coli mutY mutant. The C*/5-foU-containing double-stranded oligonucleotide (10 fmol) was incubated at 37°C for 60 min with GST-KsgA purified from E. coli mutY mutant (107 pmol) or purified GST-MutY (62 pmol). Purified GST-MutY was also incubated with the 8-oxoG/A oligonucleotide under the same conditions. The products were separated by denaturing 20% PAGE in gels containing urea. Lanes 1 and 4, no enzyme; lane 2, GST-KsgA purified from E. coli mutY mutant with 5-foU/C; lane 3, GST-MutY with 5-foU/C; lane 5, GST-MutY with 8-oxoG/A.
Increase in the spontaneous mutant frequency by introduction of the ksgA mutation in E. coli mutM mutY and nth nei mutants
We examined whether spontaneous rifampicin resistance mutations are enhanced by the introduction of the ksgA::Kan mutation in E. coli CSH101 in the presence and absence of various DNA glycosylases. Table 3 shows that the spontaneous mutant frequency significantly increased in the E. coli mutM mutY ksgA mutant, compared with the mutM mutY mutant. Furthermore, the E. coli nth nei ksgA mutant also showed enhanced spontaneous mutant frequency compared with the parent strains. The increase in mutant frequency is rather modest in ksgA mutants, much less than observed in E. coli deficient in MutM, MutY or Nth, Nei.
Table 3.
Spontaneous rifampicin resistant mutations in E. coli strains with different DNA glycosylase activities
| Relevant genotype | Rifampicin resistant mutants/108 cells |
|---|---|
| wild-type | 21 ± 10 |
| mutM | 122 ± 23 |
| mutY | 255 ± 25 |
| ksgA | 72 ± 18 |
| mutM ksgA | 264 ± 30 |
| mutY ksgA | 227 ± 27 |
| mutM mutY | 985 ± 35 |
| mutM mutY ksgA | 2 691 ± 136 |
| nth nei | 122 ± 40 |
| nth nei ksgA | 402 ± 24 |
Rifampicin was added at 150 μg/ml on LB-agar plates. The values represent the mean ± SD (n = 4). The P < 0.05, indicating significant difference at the 95% confidence level.
Partial amino acid sequence alignment for E. coli KsgA, MutM and Nei
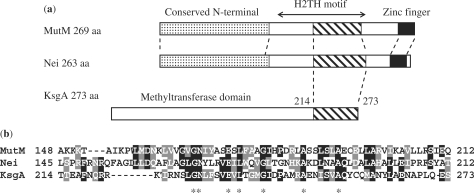
The amino acid sequence of KsgA was compared with that of E. coli MutM and Nei. MutM and Nei have the conserved N-terminal, helix–two-turns–helix (H2TH) and zinc-finger motifs (38–40). As shown in Figure 7, KsgA had a C-terminal region (amino acid 214–273) with sequence homology to the DNA binding H2TH motif of MutM (amino acid 129–229) and Nei (amino acid 126–189) (38–40).
Figure 7.
Comparison of amino acid sequences for KsgA, MutM and Nei. (a) Domain organization of E. coli MutM, Nei and KsgA. (b) Partial amino acid sequence alignment for KsgA (amino acid 214–273) and the DNA binding H2TH domains of E. coli MutM and Nei. The conserved sequences of the H2TH domains of KsgA, MutM and Nei are indicated. The identical and similar amino acids between the MutM and Nei, Nei and KsgA are boxed in black and gray, respectively. The common amino acids between these three proteins are marked as asterisks.
DISCUSSION
Thymine is vulnerable to hydroxyl radicals, which attack the 5,6-double bond of the pyrimidine ring and the exocyclic 5-methyl group, producing several kinds of oxidative damage. The primary product of the 5,6-double bond is Tg, while the oxidation of the 5-methyl group yields 5-foU and 5-hmU (1–3). 5-foU is produced by ionizing radiation in yields comparable with those of Tg and 8-oxoG (1). Several lines of evidence revealed that 5-foU is a potentially mutagenic lesion (15,20,21). DNA polymerases read through the sites of 5-foU in the template without a pause one nucleotide prior to and opposite the 5-foU sites in the template (22,23). 5-foU directs the incorporation of dCMP and sometimes dGMP opposite the lesion (15,22,23,26). These 5-foU/C and 5-foU/G mismatches must be corrected by a post-replication repair mechanism to prevent the formation of mutations. However, mismatch DNA glycosylases that excise misincorporated bases opposite 5-foU and Tg from DNA have not yet been identified.
In this study, we identified a DNA glycosylase activity that can remove C preferentially from the oligonucleotides containing C/5-foU, C/Tg and C/5-hmU and revealed as the E. coli KsgA protein. This has been proven from the following experiments; purified KsgA was trapped by the C/5-foU- and C/Tg-containing oligonucleotides (Figure 4) and furthermore, it cleaved the double-stranded C/oxidized T-containing oligonucleotides via a β-elimination reaction (Figure 5). On the other hand, 5-foU in the template is recognized and removed from DNA by MutM, Nth and Nei in E. coli (14–18). C/5-foU-DNA glycosylase activity found in this study was not due to any contaminations from these DNA glycosylases as the KsgA protein was purified from the E. coli mutM nth nei mutant. The enzyme activity is not due to contaminating MutY (Figure 6). KsgA showed a unique substrate specificity; it removed specifically C opposite Tg, 5-foU and 5-hmU, whilst it did not excise Tg, 5-foU and 5-hmU opposite C (Figure 5a). It also did not recognize C opposite normal bases, 8-oxoG and 2-ohA. These results demonstrated that KsgA has a novel DNA glycosylase/AP lyase activity that removes C mispaired with oxidized T from DNA. KsgA thus behaves like MutY by cleaving the undamaged DNA strand. MutY excises the normal base A incorporated opposite 8-oxoG (3,5,13). This provides a second chance for incorporating C opposite 8-oxoG during repair, which could be subsequently cleaved by MutM. In the case of KsgA, cleavage of C in a duplex DNA opposite 5-foU and Tg will allow insertion of A opposite these lesions during repair, which would allow subsequent removal of the lesions themselves by MutM, Nth or Nei.
The ksgA gene has previously been shown to encode a 30.4 kDa protein with 16S rRNA adenine dimethyltransferase activity (41–43). This protein is required for ribosome biogenesis and methylates two highly conserved adjacent adenosine residues in a loop region at the 3′-end of 16S rRNA (41–43). KsgA has eight motifs (motifs I to VIII) essential for S-adenosylmethionine-dependent methyltransferase (34,37,43). KsgA has the ability to protect the E. coli Era mutant (E. coli RAS-like protein) against cold-shock stress (44). However, the methyltransferase activity of KsgA was not required for the suppression of the cold-sensitive phenotype of the era strain, suggesting that KsgA possesses a different activity other than its rRNA methyltransferase activity (35,44). Inoue et al. (35) suggested that the C-terminal domain of KsgA is essential for this additional function in E. coli. In this study, we found that KsgA has a novel activity of DNA glycosylase with associated AP lyase for C mispaired with oxidized thymine.
Structural and biochemical mechanisms underlying the DNA glycosylase activity of KsgA are still unknown. There are several well-conserved DNA binding domains in DNA glycosylases (45) and KsgA has a homologous sequence (Figure 7) comparable with the H2TH motif observed in E. coli Nei and MutM (38–40). The sequence is located in the C-terminal region of E. coli KsgA, where the motifs I to VIII essential for the activity of its rRNA methyltransferase are not contained (35). It therefore seems possible that the H2TH-homologous sequence is essential for the DNA glycosylase activity of KsgA. However, the active sites of KsgA for the DNA glycosylase activity remains uncertain. Inoue et al. (35) found using gel shift assay that purified KsgA has the ability to bind to double-stranded DNA. Interestingly, a KsgA mutant (R248A) did not show any gel shift. Structural and active sites of the DNA glycosylase activity of KsgA is currently under investigation in our laboratory.
The present experiments suggest that KsgA plays an important role in prevention of DNA against oxidative stress in vivo. This is supported by the fact that the addition of the ksgA mutation increased spontaneous mutant frequency in E. coli mutM mutY and nth nei mutants (Table 3). It is likely that KsgA acts in an MutM and MutY-independent repair pathway. Nth and Nei directly excise oxidized pyrimidines from DNA (3,8,38,39) whereas KsgA recognizes and removes C incorporated opposite oxidized T during DNA replication. Hence, the effects of mutations of the nth, nei and ksgA would be additive. The increase in mutant frequency is rather modest (about 2- to 3-fold) in ksgA mutants, much less than observed in E. coli deficient in MutM, MutY or Nth, Nei. MutM, Nth and Nei may primarily excise the 5-foU produced in DNA. To clarify the types of mutations in ksgA mutant is under investigation using a plasmid DNA containing C/oxidized T.
Escherichia coli MutT has phosphatase activity for 8-oxo-dGTP produced in the nucleotide pool under conditions of oxidative stress, preventing the incorporation of 8-oxo-dGTP into DNA during replication (5,46). 5-formyl-dUTP are efficiently incorporated into DNA and cause mutations (24,25), however, MutT cannot degrade such deleterious nucleotide triphosphates, such as 5-formyl-dUTP (data not shown). Therefore, E. coli must harbor other phosphatase activities for the 5-foU derivatives. Recently, Titz et al. (47) reported that the E. coli protein YjjG 5 is a house-cleaning nucleotidase in vivo and 5-foU was thought to be a candidate substrate for the protein (47). Therefore, it is likely that E. coli has a similar repair enzyme system for preventing mutations due to 5-foU and Tg to the ‘GO’ system for 8-oxoG (5–9).
KsgA is ubiquitous and conserved in all living cells so far examined, from bacteria to humans (29,37,48,49). The yeast KsgA ortholog (Dim1) is essential for cell growth (43). However, its essentiality is not due to its methyltransferase activity but depends on its pre-18S rRNA processing activity (35,43). A human KsgA ortholog with rRNA methyltransferase activity was originally found as human mitochondrial transcriptional factor B1 (h-mtTFB1) (48,49). The methyltransferase activity of h-mtTFB1 is not required for an essential role in its transcription regulation (35). These observations suggest that KsgA orthologs with conserved methyltransferase activity are recruited to play additional physiologically important roles within cells. We are currently examining whether human orthologs have the same DNA glycosylase activity as E. coli KsgA.
FUNDING
Global Center of Excellence Program ‘Formation of a Strategic Base for Biodiversity and Evolutionary Research (A06): from Genome to Ecosystem’ in part; the Ministry of Education, Culture, Sports and Technology, Japan; Central Research Institute of Electric Power Industry (Tokyo) and Takeda Science Foundation (Osaka) (to Q.-M., Z.-A.). Funding for open access charge: Global COE A06.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We thank Drs Akiko Nishimura and Takeyoshi Miki for kindly supplying E. coli mutants. We also thank Dr Jason Parsons for critically reading the article and helpful discussions.
REFERENCES
- 1.Cadet J, Douki T, Gasparutto D, Ravanat J-L. Oxidative damage to DNA: formation, measurement and biochemical features. Mutat. Res. 2003;531:5–23. doi: 10.1016/j.mrfmmm.2003.09.001. [DOI] [PubMed] [Google Scholar]
- 2.Bjelland S, Seeberg E. Mutagenicity, toxicity and repair of DNA base damage induced by oxidation. Mutat. Res. 2003;531:37–80. doi: 10.1016/j.mrfmmm.2003.07.002. [DOI] [PubMed] [Google Scholar]
- 3.Dizdaroglu M. Base-excision repair of oxidative DNA damage by DNA glycosylases. Mutat. Res. 2005;591:45–59. doi: 10.1016/j.mrfmmm.2005.01.033. [DOI] [PubMed] [Google Scholar]
- 4.Ames BN, Shigenaga MK, Hagen TM. Oxidants, antioxidants, and the degenerative diseases of aging. Proc. Natl Acad. Sci. USA. 1993;90:7915–7922. doi: 10.1073/pnas.90.17.7915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tsuzuki T, Nakatsu Y, Nakabeppu Y. Significance of error-avoiding mechanisms for oxidative DNA damage in carcinogenesis. Cancer Sci. 2007;98:465–470. doi: 10.1111/j.1349-7006.2007.00409.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.David SS, O'Shea VL, Kundu S. Base-excision repair of oxidative DNA damage. Nature. 2007;447:941–950. doi: 10.1038/nature05978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hazra TK, Das A, Das S, Choudhury S, Kow YW, Roy R. Oxidative DNA damage repair in mammalian cells: a new perspective. DNA Repair. 2007;6:470–480. doi: 10.1016/j.dnarep.2006.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zharkov DO. Base excision DNA repair. Cell. Mol. Life Sci. 2008;65:1544–1565. doi: 10.1007/s00018-008-7543-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hazra TK, Hill JW, Izumi T, Mitra S. Multiple DNA glycosylases for repair of 8-oxoguanine and their potential in vivo functions. Prog. Nucleic Acids Res. Mol. Biol. 2001;68:193–205. doi: 10.1016/s0079-6603(01)68100-5. [DOI] [PubMed] [Google Scholar]
- 10.Allinson SL, Sleeth KM, Matthewman GE, Dianov GL. Orchestration of base excision repair by controlling the rates of enzymatic activities. DNA Repair. 2004;3:23–31. doi: 10.1016/j.dnarep.2003.09.002. [DOI] [PubMed] [Google Scholar]
- 11.Hsu GW, Ober M, Carell T, Beese LS. Error-prone replication of oxidatively damaged DNA by a high-fidelity DNA polymerase. Nature. 2004;431:217–221. doi: 10.1038/nature02908. [DOI] [PubMed] [Google Scholar]
- 12.Michaels ML, Pham L, Cruz C, Miller JH. MutM, a protein that prevents G:C→T:A transversions, is formamidopyrimidine-DNA glycosylase. Nucleic Acids Res. 1991;19:3629–3632. doi: 10.1093/nar/19.13.3629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Michaels ML, Cruz C, Grollman AP, Miller JH. Evidence that MutY and MutM combine to prevent mutations by an oxidatively damaged form of guanine in DNA. Proc. Natl Acad. Sci. USA. 1992;89:7022–7025. doi: 10.1073/pnas.89.15.7022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang Q-M, Miyabe I, Matsumoto Y, Kino K, Sugiyama H, Yonei S. Identification of repair enzymes for 5-formyluracil in DNA. Nth, Nei and MutM proteins of Escherichia coli. J. Biol. Chem. 2000;275:35471–35477. doi: 10.1074/jbc.M006125200. [DOI] [PubMed] [Google Scholar]
- 15.Bjelland S, Ånensen H, Knævelsrud I, Seeberg E. Cellular effects of 5-formyluracil in DNA. Mutat. Res. 2001;486:147–154. doi: 10.1016/s0921-8777(01)00085-4. [DOI] [PubMed] [Google Scholar]
- 16.Miyabe I, Zhang Q-M, Kino K, Sugiyama H, Takao M, Yasui A, Yonei S. Identification of 5-formyluracil DNA glycosylase activity of human hNTH1 protein. Nucleic Acids Res. 2002;30:3443–3448. doi: 10.1093/nar/gkf460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hori M, Yonei S, Sugiyama H, Kino K, Yamamoto K, Zhang Q-M. Identification of high excision capacity for 5-hydroxymethyluracil mispaired with guanine in DNA of Escherichia coli MutM, Nei and Nth DNA glycosylases. Nucleic Acids Res. 2003;15:1191–1196. doi: 10.1093/nar/gkg223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang Q-M, Yonekura S, Takao M, Yasui A, Sugiyama H, Yonei S. DNA glycosylase activities for thymine residues oxidized in the methyl group are functions of the hNEIL1 and hNTH1 enzymes in human cells. DNA Repair. 2005;4:71–79. doi: 10.1016/j.dnarep.2004.08.002. [DOI] [PubMed] [Google Scholar]
- 19.Bjelland S, Birkeland NK, Benneche T, Volden G, Seeberg E. DNA glycosylase activities for thymine residues oxidized in the methyl group are functions of the AlkA enzyme in Escherichia coli. J. Biol. Chem. 1994;269:30489–30495. [PubMed] [Google Scholar]
- 20.Ånensen H, Provan F, Lian AT, Reinertsen S-HHS, Ueno Y, Matsuda A, Seeberg E, Bjelland S. Mutations induced by 5-formyl-2’-deoxyuridine in Escherichia coli include base substitutions that can arise from mispairs of 5-formyluracil with guanine, cytosine and thymine. Mutat. Res. 2001;476:99–107. doi: 10.1016/s0027-5107(01)00086-0. [DOI] [PubMed] [Google Scholar]
- 21.Miyabe I, Zhang Q-M, Sugiyama H, Kino K, Yonei S. Mutagenic effects of 5-formyluracil on a plasmid vector during replication in Escherichia coli. Int. J. Radiat. Biol. 2001;77:53–58. doi: 10.1080/095530001453113. [DOI] [PubMed] [Google Scholar]
- 22.Zhang Q-M, Sugiyama H, Miyabe I, Matsuda S, Saito I, Yonei S. Replication of DNA templates containing 5-formyluracil, a major oxidative lesion of thymine in DNA. Nucleic Acids Res. 1997;25:3969–3973. doi: 10.1093/nar/25.20.3969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang Q-M, Sugiyama H, Miyabe I, Matsuda S, Kino K, Saito I, Yonei S. Replication in vitro and cleavage by restriction endonuclease of 5-formyluracil- and 5-hydroxymethyluracil-containing oligonucleotides. Int. J. Radiat. Biol. 1999;75:59–65. doi: 10.1080/095530099140816. [DOI] [PubMed] [Google Scholar]
- 24.Yoshida M, Makino K, Morita H, Terato H, Ohyama Y, Ide H. Substrate and mispairing properties of 5-formyl-2′-deoxyuridine 5′-triphosphate assessed by in vitro DNA polymerase reactions. Nucleic Acids Res. 1997;25:1570–1577. doi: 10.1093/nar/25.8.1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fujikawa K, Kamiya H, Kasai H. The mutations induced by oxidatively damaged nucleotides, 5-formyl-dUTP and 5-hydroxy-dCTP, in Escherichia coli. Nucleic Acids Res. 1998;26:4582–4587. doi: 10.1093/nar/26.20.4582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Klungland A, Paulsen R, Rolseth V, Yamada Y, Ueno Y, Wiik P, Matsuda A, Seeberg E, Bjelland S. 5-Formyluracil and its nucleoside derivatives confer toxicity and mutagenicity to mammalian cells by interfering with normal RNA and DNA metabolism. Toxicol. Lett. 2001;119:71–78. doi: 10.1016/s0378-4274(00)00308-8. [DOI] [PubMed] [Google Scholar]
- 27.Rabow LE, Kow YW. Mechanism of action of base release by Escherichia coli Fpg protein: role of lysine 155 in catalysis. Biochemistry. 1997;36:5084–5096. doi: 10.1021/bi963005a. [DOI] [PubMed] [Google Scholar]
- 28.Sugiyama H, Matsuda S, Zhang Q-M, Yonei S, Saito I. New synthetic method of 5-formyluracil-containing oligonucleotides and their melting behavior. Tetrahedron Lett. 1996;37:9067–9070. [Google Scholar]
- 29.Sugiyama H, Ikeda S, Saito I. Remarkably stable parallel-stranded oligonucleotides containing 5-methylisocytosine and isoguanine. J. Am. Chem. Sci. 1996;118:9994–9995. [Google Scholar]
- 30.Dianov GL, Thybo T, Dianova II, Lipinski LJ, Bohr VA. Single strand patch base excision repair is the major pathway for removal of thymine glycol from DNA in human cell extracts. J. Biol. Chem. 2000;275:11809–11813. doi: 10.1074/jbc.275.16.11809. [DOI] [PubMed] [Google Scholar]
- 31.Nishimura A, Akiyama K, Kohara Y, Horiuchi K. Correlation of a subset of the pLC plasmids to the physical map of Escherichia coli. Microbiol. Rev. 1992;56:137–151. doi: 10.1128/mr.56.1.137-151.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bochner BR, Huang H-C, Schieven GL, Ames BN. Positiveselection for loss of tetracycline resistance. J. Bacteriol. 1980;143:926–933. doi: 10.1128/jb.143.2.926-933.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang Q-M, Ishikawa N, Nakahara T, Yonei S. Escherichia coli MutY protein has a guanine-DNA glycosylase that acts on 7,8-dihydro-8-oxoguanine:guanine mispair to prevent spontaneous G:C→C:G transversions. Nucleic Acids Res. 1998;26:4669–4675. doi: 10.1093/nar/26.20.4669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.O’Farrell HC, Scarsdale JN, Rife JP. Crystal structure of KsgA, a universally conserved rRNA adenine dimethyltransferase in Escherichia coli. J. Mol. Biol. 2004;28:337–353. doi: 10.1016/j.jmb.2004.02.068. [DOI] [PubMed] [Google Scholar]
- 35.Inoue K, Basu S, Inouye M. Dissection of 16S rRNA methyltransferase (KsgA) function in Escherichia coli. J. Bacteriol. 2007;189:8510–8518. doi: 10.1128/JB.01259-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Helser TL, Davies JE, Dahlberg JE. Change in methylation of 16S ribosomal RNA associated with mutation to kasugamycin resistance in Escherichia coli. Nature. 1971;233:12–14. doi: 10.1038/newbio233012a0. [DOI] [PubMed] [Google Scholar]
- 37.Xu Z, O’Farrell HC, Rife JP, Culver GM. A conserved rRNA methyltransferase regulates ribosome biogenesis. Nat. Struct. Mol. Biol. 2008;15:534–536. doi: 10.1038/nsmb.1408. [DOI] [PubMed] [Google Scholar]
- 38.Jiang D, Hatahet Z, Blaisdell JO, Melamede RJ, Wallace SS. Escherichia coli endonuclease VIII: cloning, sequencing, and overexpression of the nei structural gene and characterization of nei and nei nth mutants. J. Bacteriol. 1997;179:3773–3782. doi: 10.1128/jb.179.11.3773-3782.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wallace SS, Bandaru V, Kathe SD, Bond JP. The epigma of endonuclease VIII. DNA Repair. 2003;2:441–453. doi: 10.1016/s1568-7864(02)00182-9. [DOI] [PubMed] [Google Scholar]
- 40.Sugahara M, Mikawa T, Kumasaka T, Yamamoto M, Kato R, Fukuyama K, Inoue Y, Kuramitsu S. Crystal structure of a repair enzyme of oxidatively damaged DNA, MutM (Fpg), from an extreme thermophile, Thermus thermophilus HB8. EMBO J. 2000;19:3857–3869. doi: 10.1093/emboj/19.15.3857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Buul CP, van Knippenberg PH. Nucleotide sequence of the ksgA gene of Escherichia coli: comparison of methyltransferases effecting dimethylation in ribosomal RNA. Gene. 1985;38:65–72. doi: 10.1016/0378-1119(85)90204-5. [DOI] [PubMed] [Google Scholar]
- 42.van Gemen B, Koets HJ, Bodlaender J, van Knippenberg PH. Characterization of the ksgA gene of Escherichia coli determining kasugamycin sensitivity. Biochimie. 1987;69:841–848. doi: 10.1016/0300-9084(87)90210-0. [DOI] [PubMed] [Google Scholar]
- 43.O’Farrell HC, Pullicherla N, Desai PM, Rife JP. Recognition of a complex substrate by the KsgA/Dim1 family of enzymes has been conserved throughout evolution. RNA. 2006;12:725–733. doi: 10.1261/rna.2310406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lu Q, Inouye M. The gene for 16S rRNA methyltransferase (ksgA) functions as a multicopy suppressor for a cold-sensitive mutant of Era, an essential RAS-like GTP-binding protein in Escherichia coli. J. Bacterial. 1998;180:5243–5246. doi: 10.1128/jb.180.19.5243-5246.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Aravind L, Walker DR, Koonin EV. Conserved domains in DNA repair proteins and evolution of repair systems. Nucleic Acids Res. 1999;27:1223–1242. doi: 10.1093/nar/27.5.1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Maki H, Sekiguchi M. MutT protein specifically hydrolyses a potent mutagenic substrate for DNA synthesis. Nature. 1992;355:273–275. doi: 10.1038/355273a0. [DOI] [PubMed] [Google Scholar]
- 47.Titz B, Hauser R, Engelbrecher A, Uetz P. The Escherichia coli protein YjjG is a house-cleaning nucleotidase in vivo. FEMS Microbiol. Lett. 2007;270:49–57. doi: 10.1111/j.1574-6968.2007.00646.x. [DOI] [PubMed] [Google Scholar]
- 48.McCulloch V, Seidel-Rogol BL, Shadel GS. A human mitochondrial transcription factor is related to RNA adenine methyltransferases and binds S-adenosylmethionine. Mol. Cell. Biol. 2002;22:1116–1125. doi: 10.1128/MCB.22.4.1116-1125.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Seidel-Rogol BL, McCulloch V, Shadel GS. Human mitochondrial transcription factor B1 methylates ribosomal RNA at a conserved stem-loop. Nature Genet. 2003;33:23–24. doi: 10.1038/ng1064. [DOI] [PubMed] [Google Scholar]