Abstract
Selenoproteins contain the amino acid selenocysteine which is encoded by a UGA Sec codon. Recoding UGA Sec requires a complex mechanism, comprising the cis-acting SECIS RNA hairpin in the 3′UTR of selenoprotein mRNAs, and trans-acting factors. Among these, the SECIS Binding Protein 2 (SBP2) is central to the mechanism. SBP2 has been so far functionally characterized only in rats and humans. In this work, we report the characterization of the Drosophila melanogaster SBP2 (dSBP2). Despite its shorter length, it retained the same selenoprotein synthesis-promoting capabilities as the mammalian counterpart. However, a major difference resides in the SECIS recognition pattern: while human SBP2 (hSBP2) binds the distinct form 1 and 2 SECIS RNAs with similar affinities, dSBP2 exhibits high affinity toward form 2 only. In addition, we report the identification of a K (lysine)-rich domain in all SBP2s, essential for SECIS and 60S ribosomal subunit binding, differing from the well-characterized L7Ae RNA-binding domain. Swapping only five amino acids between dSBP2 and hSBP2 in the K-rich domain conferred reversed SECIS-binding properties to the proteins, thus unveiling an important sequence for form 1 binding.
INTRODUCTION
Selenoproteins are a diverse family of proteins characterized by the presence of the 21st amino acid selenocysteine (Sec). This amino acid is co-translationally incorporated into the growing peptide chain in response to a UGA Sec codon, otherwise read as a signal for termination of translation. In eukaryotes, the correct recoding event of UGA stop to UGA Sec relies on specific, conserved RNA structures and proteins. The tRNASec and the SECIS element, an RNA hairpin in the 3′UTR of selenoprotein mRNAs, and two trans-acting proteins, the specialized translation elongation factor eEFSec and the SECIS Binding Protein 2 (SBP2), are the key players of the recoding machinery (1). Specialized protein complexes that involve SECp43, the Phosphoserine tRNASec Kinase (PSTK) and the Sec synthase are recruited to the tRNASec to ensure proper selenocysteine synthesis (2–4). Ribosomal protein L30 has also been implicated in this mechanism and shown to compete with SBP2 for SECIS binding (5).
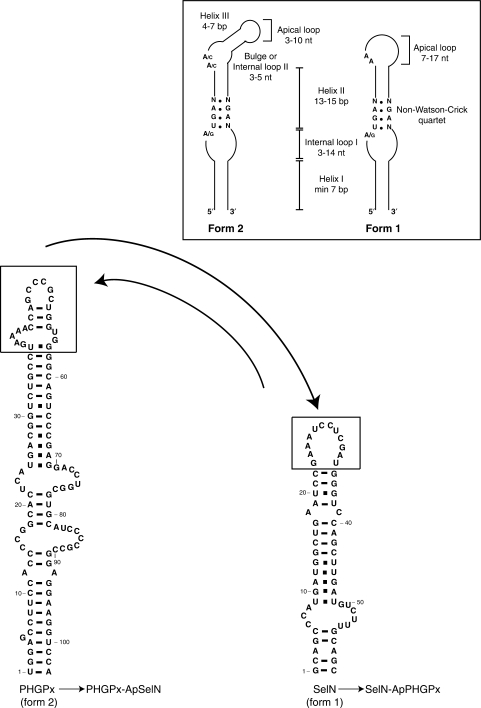
There are two types of functional SECIS RNAs, forms 1 and 2, classified according to their different apex: form 2 has an additional helix, and its apical loop is shorter than in form 1 (6,7). Structure-based alignments in the currently available eukaryotic selenoproteome identified form 2 SECIS as the most widespread element (8). Except for the apex, SECIS RNA hairpins share common structural features, in particular four consecutive non-Watson–Crick base pairs (the quartet) composed of a central tandem of sheared G.A/A.G base pairs (7–10). Such non-canonical base pairs are characteristic of K (kink)-turn motifs which are recurrent in a variety of RNAs (11–13). The SECIS RNA has been proposed to contain a K-turn like motif (14) that is essential for SBP2 interaction and selenoprotein incorporation in vivo (10). A number of proteins fulfilling different functions such as snRNPs, snoRNPs or ribosomal proteins bind K-turn RNA motifs (15,16). They all carry the L7Ae RNA-binding domain (or module) (17) that contains a restricted set of amino acids that establish base-specific contacts with the sheared G.A/A.G base pairs (11,18,19).
SBP2 also has the L7Ae module in its RNA-binding domain (20–22). In an earlier work, we predicted the human SBP2 (hSBP2) amino acids that contact the SECIS RNA at the K-turn like motif (20). However, while sharing some RNA-binding properties with other proteins of the L7Ae family, SBP2 possesses its own specificities (23). The known functions of SBP2, comprising SECIS and ribosome binding, and Sec incorporation, reside in the C-terminal two-thirds of the protein (21,22,24). However, no function has been attributed to the remaining N-terminal section which has been shown to be dispensable for Sec incorporation in rabbit reticulocyte lysate (25).
Selenoproteins exist in the three domains of life. Vertebrate genomes encode up to 25–26 selenoproteins but surprisingly larger selenoproteomes can be found in aquatic unicellular organisms (26). Only three selenoprotein genes have been discovered in Drosophila melanogaster, SPS2, SelH and SelK (27,28). SPS2 is the selenophosphate synthetase involved in selenocysteine biosynthesis. SelH and SelK are poorly characterized functionally but they seem nevertheless to play an antioxidant role (29,30). In each case however, only form 2 SECIS RNAs were found in the 3′UTR of the selenoprotein mRNAs.
Some of us have recently published the annotation and multiple sequence alignments of insect selenoprotein synthesis factors, especially in 12 Drosophila genomes (31). Among these factors, our attention was attracted by the putative Drosophila SBP2 because they lack the sequence homologous to the N-terminus of hSBP2. Although a number of SBP2 sequences from mammals, non-mammalian vertebrates or even unicellular organisms are annotated in databases, only the rat and hSBP2 have been so far isolated and functionally characterized (20–25,32,33). This prompted us to study Drosophila SBP2s and in particular Drosophila melanogaster. In this work, we report that despite its shorter length, D. melanogaster SBP2 (dSBP2) retains functional properties similar to its mammalian counterpart. However, dSBP2 exhibits selective affinities toward SECIS RNAs, being almost unable to bind form 1 SECIS. We determined that the discriminating amino acids reside in a K (lysine)-rich region that we also identified in hSBP2 as essential for SECIS RNA binding. In addition we showed that, in hSBP2, mutating the K-rich region affected form 1 and form 2 SECIS interaction differently, and that this region also plays a crucial role in 60S ribosomal subunit binding.
MATERIALS AND METHODS
Strains and growth conditions
The Escherichia coli TG2 strain was used as the host strain for plasmid construction. Growth was performed at 37°C in LB medium, complemented with 100 μg/ml ampicillin. The E. coli strain BL21(DE3)-star was used for production of Drosophila SBP2 proteins at 25°C in ZYM-5052 auto-induction medium as described by Studier et al. (34). The E. coli strain BL21(DE3)RIL (Novagen) was used for production of hSBP2 proteins at 18°C in LB medium.
Bioinformatic analyses
Alignment of human/pig/rat/insect SBP2s
Annotated SBP2 sequences from human (gb|AAK57518.1|AF380995), rat (sp|Q9QX72.1|SEBP2_RAT), pig (ef|XP_001928402.1) and chicken (ref|XP_424425.2|) were aligned against the putative SBP2 sequences found in three Drosophila species, D. melanogaster, D. pseudoobscura and D. sechelia (31) by Kalign (35).
Alignment of the K-rich domain
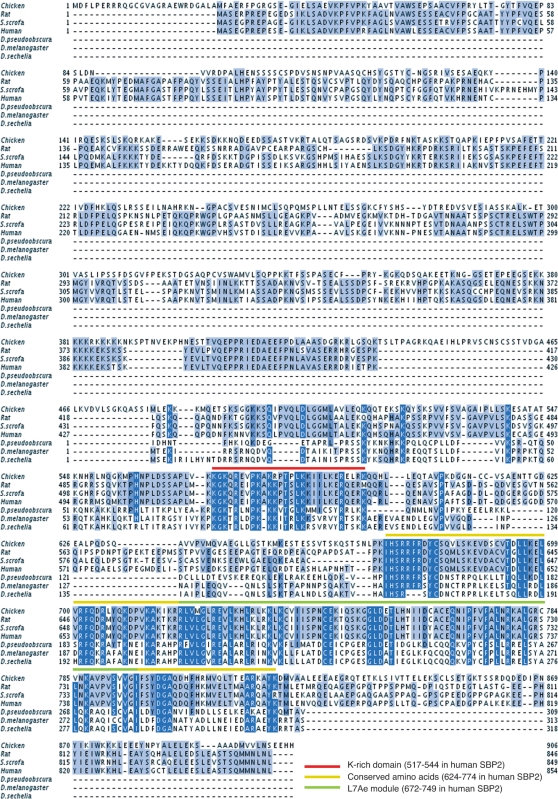
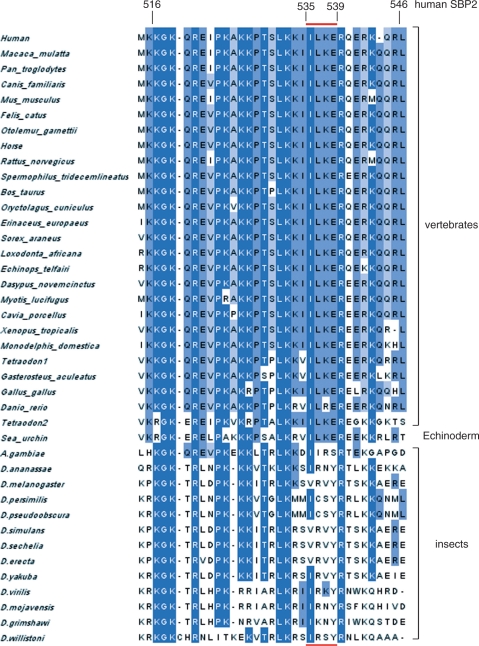
All annotated members of the SBP2 family in Ensembl (ENSF00000007674) were extracted and all those with no ‘X’ in the relevant region were kept. The search was extended by blasting the D. melanogaster SBP2 against the nr database of NCBI. Of the resulting hits, only those containing the IHSRRF motif (positions 624–629 in hSBP2) characteristic of SBP2 proteins (C.A and A.K., unpublished data) were kept. We subsequently used the L7Ae RNA-binding module of SBP2 (33) to query the nr database using Hmmer (36). Finally, we also added the insect SBP2 sequences (31). The resulting 40 sequences were aligned using mafft (37). The alignment images shown in Figures 1 and 7 were produced by Jalview (38).
Figure 1.
Alignment of SBP2 proteins. Drosophila (pseudoobscura, melanogaster and sechelia) SBP2 sequences were from (31). Accession numbers for vertebrate SBP2 are XP_424425.2| (chicken), Q9QX72.1 (rat), XP_001928402.1 (Sus scrofa), AF380995 (human). The color scheme above the sequences is explicited at the bottom of the figure.
Figure 7.
Alignment of the K-rich domain in 26 vertebrate, one echinoderm (sea urchin) and 13 insect (12 Drosophila and the mosquito Anopheles gambiae) SBP2 sequences. The numbering is that of hSBP2. The red bar above and below sequences refers to the IILKE and SVRVY pentapeptides mentioned in the text.
cDNA cloning and recombinant DNA
Drosophila melanogaster SBP2 ORF (Genebank accession # AI062219) was amplified from pOT2 cDNA clone GH01354 (Research Genetics) in a two-step PCR reaction and introduced into the pHMGWA vector (39) using the Gateway Technology (Invitrogen). The resulting pHMdSBP2 vector contains a 6× His-tag and Maltose-Binding Protein coding-frame upstream of the dSBP2 ORF and allows E. coli expression of the protein. Human SBP2 ORF was amplified by PCR from plasmid pA11 (33) and subsequently cloned into pET32b (Novagen), generating phSBP2-FL (full-length), as well as plasmids encoding the N-terminal truncated proteins phSBP2 344–854, phSBP2 399–854, phSBP2 515–854, phSBP2 525–854, phSBP2 545–854, phSBP2 625–854 and phSBP2 674–854, and C-terminal truncated proteins phSBP2 344–674, phSBP2 344–790 and phSBP2 344–820. Alanine scanning mutants in hSBP2 were generated in phSBP2 344–854 using the Kunkel mutagenesis method (40). Amino acids swapping mutants exchanging hSBP2 aa535–539 for dSBP2 aa95–99 (phSBP2-SVRVY) and dSBP2 aa95–99 for hSBP2 aa535–539 (pdSBP2-IILKE), were generated by site-directed mutagenesis of phSBP2 344–854 and pHMdSBP2, respectively, using the QuickChange XL Site-Directed Mutagenesis kit (Stratagene) according to the manufacturer's instructions.
Plasmids pT7SelN, pT7GPx1 and pT7PHGPx were used for T7 transcription of human SelN, rat GPx1 and PHGPx SECIS RNAs, respectively (7,9). To allow in vitro transcription of Drosophila SECIS RNAs, D. melanogaster SelK and SelH SECIS elements were PCR amplified from pOT2 cDNA clones GH03581 and SD09114, respectively (generously provided by M. Corominas, University of Barcelona) and introduced into the BclI-EcoRI sites of pT7-Bck vector (9) to create pT7dSelK and pT7dSelH. A point mutant in dSelH SECIS (dSelHmut), and the SECIS RNA apex-swapped mutants PHGPx-ApSelN and SelN-ApPHGPx SECIS RNAs, were generated by site-directed mutagenesis of pT7dSelH, pT7PHGPx and pT7SelN, respectively, using the QuickChange XL Site-Directed Mutagenesis kit.
To generate selenoprotein mRNA reporter constructs for in vitro translation assays, D. melanogaster SelH ORF and 3′UTR (Genebank accession #AI542675) were amplified from pOT2 cDNA clone SD09114 and cloned into the HindIII-KpnI sites of the pXJ(HA)3 eukaryotic expression vector (41) to create pHAdSelH. Rat selenoprotein reporter constructs pGPx1-GPx1SECIS and pGPx1-ΔSECIS were as described in (10). To create pGPx1-PHGPxSECIS and pGPx1-SelNSECIS, the GPx1 SECIS element of the pGPx1-GPx1SECIS reporter construct was exchanged for PHGPx and SelN SECIS elements from pT7PHGPx and pT7SelN, respectively, using BclI-EcoRI sites.
Oligonucleotides used for PCR and mutagenesis are listed in Supplementary Data.
Recombinant protein preparation
Drosophila SBP2 recombinant proteins expressed in E. coli were purified using Amylose Resin column (NEB). 6× His and MBP tags were cleaved from the dSBP2 protein by thrombin (Sigma) when used for electrophoretic mobility shift assays. hSBP2 recombinant proteins expressed in E. coli were purified using Ni-NTA agarose (Qiagen). Elution buffer was exchanged to dialysis buffer containing 20 mM Tris–HCl pH 7.5, 100 mM NaCl, 10 mM MgCl2, 20% glycerol, 1 mM DTT and Cocktail inhibitor (Sigma).
Electrophoretic mobility shift assay
Plasmids yielding PHGPx, SelN, GPx1, dSelK, dSelH, dSelHmut, PHGPx-ApSelN and SelN-ApPHGPx SECIS RNAs were linearized by EcoRI. Internally labeled SECIS RNAs were obtained by T7 transcription using 80 µCi of [α-32P]-ATP (3000 Ci/mmol). SECIS RNA-SBP2 complexes were formed as described in (20). Routinely, 30 000 cpm of 32P-labeled SECIS RNA were incubated for 30 min at 30°C with various concentrations of purified SBP2 protein (from 0 to 2000 nM), in 7.5 µl of phosphate buffer saline pH 7.4, 2 mM DTT. RNA–protein complexes were separated on 6% non-denaturing polyacrylamide gel electrophoresis in 0.5× TBE, 5% glycerol buffer. The intensities of free and bound RNAs were quantitated with the Fujifilm FLA-5100 Imaging system. Kds were determined from three independent experiments.
In vitro selenoprotein synthesis assays
In vitro translation of Drosophila (dSelH) or rat GPx1 from selenoprotein encoding plasmids carrying (or lacking) wild-type SECIS elements were performed using TNT Coupled Reticulocyte Lysate Systems (Promega). One microgram of each of the selenoprotein plasmid DNA was used as the template in 50 µl in vitro transcription/translation reactions in the presence of 25 µl rabbit reticulocyte lysate, 20 µCi of 35S-methionine and 120–240 nM of purified SBP2 protein. In vitro translated HA-tagged dSelH and hGPx1 proteins were purified using microMACS Epitope Tag Protein Isolation Kits (Miltenyi Biotec), resolved by 10% SDS–PAGE and detected with the Fujifilm FLA-5100 Imaging system. Quantification of selenoprotein synthesis was performed from two to three independent experiments.
Ribosome-binding assays
60S and 40S ribosomal subunits were isolated from full term human placenta according to ref. (42). Human recombinant ribosomal protein p40 was a kind gift of Dr Alexey Malygin (ICBFM, Novosibirsk, Russia). Monoclonal antibodies against human p40 were provided by Dr Valery Loktev. Binding mixtures (50 µl) containing 30 pmol of 60S or 40S subunits were reactivated at 37°C for 10 min in PBSD buffer (150 mM NaCl, 27 mM KCl, 8 mM Na2HPO4, 1.7 mM KH2PO4 and 2 mM DTT) containing 0.5 mM MgCl2. Then 3.5 µg SBP2 (or SBP2 mutants) or 2 µg ribosomal protein p40 were added and incubated at 22°C for 20 min. The mixtures were loaded onto a 15–30% linear sucrose gradient in PBSD with 0.5 mM Mg2+ and centrifuged in a SW41 rotor at 23 000 rpm for 15 h. Fractions corresponding to 60S and 40S subunits were precipitated by 10% trichloroacetic acid, and the pellet content loaded onto 10% SDS–PAGE which was blotted onto nitrocellulose membranes. SBP2 was detected with an anti-SBP2 polyclonal antibody (1/5000–1/10 000 dilution) in PBST (1× PBS containing 0.1% Tween 20, 3% dry milk), p40 with a monoclonal anti-p40 (1/3000 dilution). Membranes were treated with anti-rabbit HRP-conjugated secondary antibody (1/10 000 dilution), revealed with the ECL plus kit (GE HealthCare) and exposed to either X-ray film or ChemDoc XRS.
RESULTS
Identification and functional characterization of dSBP2
Recent comparative analysis of insect genomes have identified putative SBP2 proteins (31). In this work we set out to clone and characterize dSBP2 and compare it to hSBP2. tBlastn searches were performed at NCBI using the hSBP2 amino acid sequence (33) and one significant hit (AI062219) was obtained with a D. melanogaster sequence predicted to encode a 313 amino-acid protein. Figure 1 shows a multiple sequence alignment of this protein as well as other putative SBP2 from D. pseudoobscura and D. sechelia (31) with the characterized rat and human SBP2 (25,33) and the annotated chicken and Sus scrofa SBP2s. Strikingly, the three putative Drosophila SBP2s lack homology to the vertebrate SBP2 N-terminal region. Homologies to vertebrate SBP2s were found throughout the rest of the sequence, starting from hSBP2 residue 427. Two blocks of high sequence identity were detected, one extending from K517 to K544, the other from I624 to K774 (hSBP2 numbering), with 50% and 36% identity, respectively. The latter block contains the previously identified L7Ae RNA-binding module spanning residues R672 to I749 (21,22).
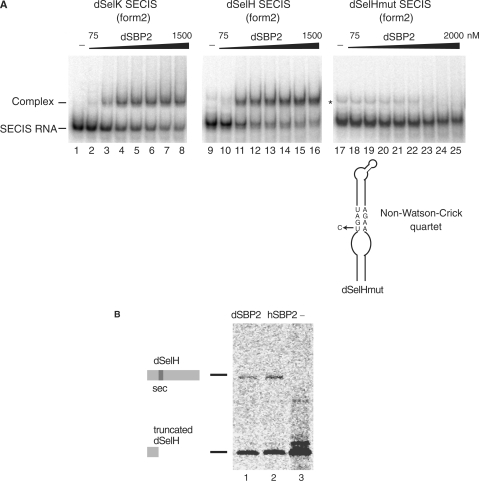
To verify that the D. melanogaster cDNA encodes a bona fide SBP2, it was cloned and expressed in E. coli and assayed for SECIS RNA binding. Gel shift assays were performed with the form 2 SECIS RNAs of the dSelK and SelH mRNAs (dSelK and dSelH SECIS RNAs), showing formation of dSBP2–dSECIS RNA complexes (Figure 2A, lanes 2–8 and 10–16). Kd values were 260 nM and 170 nM for dSelK SECIS and dSelH SECIS RNAs, respectively, indicating a slightly higher affinity for dSelH SECIS RNA. To determine whether the binding was specific, we used a mutant SECIS RNA (dSelHmut SECIS, Figure 2A) in which the conserved U in the non-Watson–Crick quartet was replaced by a C, a mutation that impedes hSBP2 binding (43). Figure 2A (lanes 18–25) showed that no retarded complex was obtained, the band marked by an asterisk containing an RNA conformer also present in the control lane 17 (see also lane 9) but not an RNA–protein complex. We therefore concluded that dSBP2 bound specifically to the cognate SECIS RNAs.
Figure 2.
Functional characterization of dSBP2. (A) Electrophoretic mobility shift assays were performed between the purified dSBP2 and the in vitro transcribed 32P-labeled form 2 SECIS RNAs (dSelK SECIS and dSelH SECIS) from Drosophila SelK and SelH selenoprotein mRNAs. Increasing concentrations (75–2000 nM) of dSBP2 were added. A dSeH mutant SECIS RNA (dSelHmut), carrying a U to C mutation in the non-Watson-Crick quartet, abolished the binding of dSBP2. dSBP2 was omitted in the control lanes 1, 9 and 17 (−). The asterisk denotes the position of a band that is also present in the control lanes 9 and 17. It often occurs and presumably contains an RNA conformer arising from T7 transcription. (B) dSBP2 can support selenoprotein H (dSelH) synthesis in rabbit reticulocyte lysate. Synthesis of full-length (35S)-Met-labeled selenoprotein dSeH was obtained by adding purified dSBP2 (lane 1) or hSBP2 (lane 2) to the lysate. Lane 3: absence of SBP2 led to premature termination of translation (Truncated). Translation products were immunopurified and resolved by SDS–PAGE.
We next investigated whether dSBP2 could support selenoprotein synthesis. To this end, rabbit reticulocyte lysate was used to synthesize D. melanogaster selenoprotein SelH (dSelH) from a reporter construct. The advantage of such an extract is that it contains all the components required for selenoprotein mRNA translation but lacks SBP2 which must be added to obtain a full-length selenoprotein (25). Figure 2B (lane 3) showed that omission of SBP2 resulted in the synthesis of a truncated dSelH selenoprotein arising from premature termination of translation at the UGA Sec codon. Full-length dSelH could be obtained only upon addition of dSBP2 (Figure 2B, lane 1), in much the same way as hSBP2 (hSBP2; Figure 2B, lane 2). The truncated form of dSelH ending at the UGA Sec codon (lanes 1 and 2) very likely originated from saturation of the selenoprotein synthesis machinery in the reticulocyte lysate.
Overall, our data established that we have identified and functionally characterized the dSBP2 protein which possesses the same selenoprotein synthesis capacities as the human counterpart.
dSBP2 manifests distinctive SECIS RNA-binding activities
The abilities of dSBP2 to bind various SECIS RNAs were further investigated and compared to hSBP2. Form 2 SECIS, the only type of SECIS found in Drosophila (31), were tested first. Figure 3 and Table 1 show that complexes were obtained between the SECIS of the rat PHGPx (phosphohydroxylipid glutathione peroxidase) mRNA and dSBP2 (Figure 3A, lanes 2–7; Kd = 220 nM) or hSBP2 (Figure 3A, lanes 23–29; Kd = 320 nM). Additionally, hSBP2 also bound dSelH and dSelK SECIS RNAs (data not shown). Major differences appeared when form 1 SECIS was tested (a schematic drawing of SECIS forms 1 and 2 is shown in the inset of Figure 4). While hSBP2, as expected, bound readily the SECISes of the rat GPx1 (glutathione peroxidase 1) and human SelN (selenoprotein N) mRNAs (Figure 3A, lanes 31–37 and 39–45), dSBP2 only weakly recognized the SelN SECIS and was unable to bind the GPx1 SECIS at all (Figure 3A, lanes 16–21 and 9–14). These findings were corroborated by selenoprotein synthesis experiments in reticulocyte lysate in which dSBP2 enabled synthesis of a selenoprotein reporter (rat GPx1) from an mRNA carrying a form 2 PHGPx but not a form 1 GPx1 SECIS in the 3′UTR (Figure 3B, compare lanes 2, 3 with lanes 7, 8). Synthesis from the form 1 SelN SECIS-containing reporter was weak (Figure 3B, lanes 12, 13), in keeping with the low affinity to SelN SECIS. In contrast, hSBP2 led to synthesis of GPx1 regardless of the type of SECIS harbored by the reporter mRNA (Figure 3B, lanes 4, 5, 9, 10, 14, 15). Control experiments with a GPx1-SECIS lacking construct (Figure 3B, lanes 17–20) indicated that selenoprotein synthesis was indeed SECIS-dependent.
Figure 3.
dSBP2 has distinct affinities for form 1 and 2 SECIS RNAs. (A) Gel shift assays were performed between in vitro transcribed 32P-labeled PHGPx (form 2), GPx1 and SelN (form 1) SECIS RNAs and either purified dSBP2 or hSBP2, with the range of protein concentration (nM) indicated above each gel. Proteins were omitted in lanes 1, 8, 15, 22, 30 and 38. The asterisk in lanes 8–14 is as in Figure 2A. (B) In vitro translation assay (reticulocyte lysate) of (35S)-Met-labeled rat glutathione peroxidase 1 (GPx1) from reporter constructs carrying the GPx1 ORF without SECIS RNA (ΔSECIS) or with either the PHGPx, GPx1 or SelN SECIS RNAs in the 3′UTR. Purified dSBP2 or hSBP2 were omitted (lanes 1, 6, 11 and 16) or added at the indicated concentrations (nM). Translation products were treated as in Figure 2B.
Table 1.
Kd values of hSBP2 and dSBP2 for form 1, form 2 and chimeric SECIS RNAs
| SECIS RNA | SBP2 protein |
|
|---|---|---|
| dSBP2 | hSBP2 | |
| PHGPx (form2) | 220 ± 50 | 320 ± 5 |
| GPx1 (form1) | No binding | 400 ± 50 |
| SeIN (form1) | nd | 340 ± 15 |
| PHGPx-APSeIN | 230 ± 30 | 530 ± 120 |
| SeIN-ApPHGPx | No binding | 470 ± 40 |
| Kd ± SD (nM) | ||
Figure 4.
Chimeric SECIS RNAs containing swapped apexes. The apical regions (boxed) of form 2 PHGPx and form 1 SelN SECIS were exchanged (curved arrows) to yield the chimeric PHGPx-ApSelN and SelN-ApPHGPx SECIS harboring the SelN and PHGPx apexes (Ap), respectively. The drawing in the inset schematizes the conserved and variable structural features occurring in forms 1 and 2 SECIS RNAs.
This unexpected result raised the possibility that the distinctive binding affinities of dSBP2 for SECIS RNAs could originate from the different apical secondary structures classifying form 1 and form 2 SECIS RNAs [Figure 4; (6,7)]. To answer the question, we decided to swap the apexes between both forms of SECIS RNAs to yield chimeric SECIS RNAs: a PHGPx-ApSelN SECIS with the apex of SelN SECIS, and a SelN-ApPHGPx SECIS with that of PHGPx SECIS (Figure 4). The binding of dSBP2 and hSBP2 was tested by gel-shift assays (data expressed as Kd values in Table 1). Surprisingly, the apex of the form 1 SECIS did not lead to abolition of dSBP2 binding to the chimeric PHGPx-ApSelN SECIS while the SelN-ApPHGPx SECIS, carrying the stem of SelN and the apex of the form 2 PHGPx, did not confer to dSBP2 the ability to bind. Indeed, the apex is not responsible for the distinctive binding of dSBP2 since replacing the genuine apexes by an ultrastable UUCG tetraloop in PHGPx and SelN SECIS did not modify the effects induced by apex swapping (data not shown). These results suggest that the difference between forms 1 and 2 SECIS may not rely solely on distinct apical structures (6) but that other determinants/discriminants could also exist elsewhere in the SECIS RNA 2D and/or 3D structures.
Point mutations in a newly mapped lysine-rich domain in hSBP2 affect binding to form 1 but not to form 2 SECIS RNAs
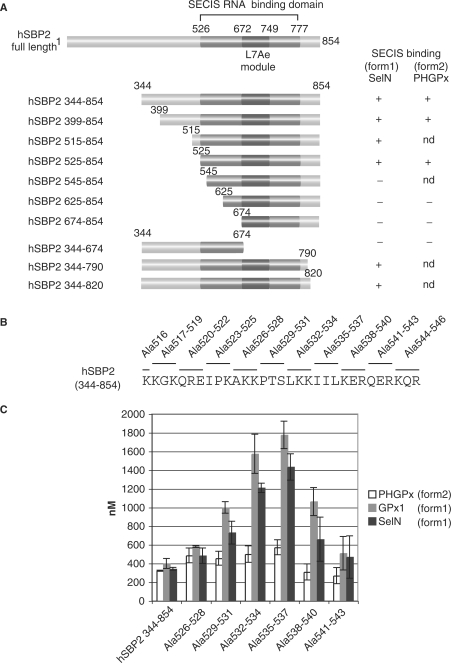
We hypothesized that a domain(s) in SBP2 might be responsible for the differential SECIS binding. First, we re-examined in-depth the boundaries of the hSBP2 RNA-binding domain. A series of N- and C-terminal deletions was performed in the construct encoding hSBP2 344–854 (Figure 5A). The resulting 6× His-tagged hSBP2 pro-teins were expressed in E. coli, purified and their binding abilities assessed by gel-shift assays with SelN (form 1) and PHGPx (form 2) SECIS RNAs (Figure 5A). Lack of binding of the 344–674 hSBP2 protein resulted from amputation of the major part of the previously mapped L7Ae RNA-binding module (20–22,25). However, the loss of binding manifested by hSBP2 proteins 545–854, 625–854 and 674–854 revealed that a region outside of the L7Ae module is also crucial. More precisely, this other domain should lie between positions 525 and 545 since hSBP2 525–854 retained SECIS binding activity. Interestingly, this region contains the conserved block of amino acids of highest sequence identity revealed by our alignments (Figure 1, residues 517–544). To characterize this domain with more accuracy, we carried out alanine scanning mutations in construct hSBP2 344–854 from positions K516 to R546, in groups of three amino acids except K516 which was singly mutated (Figure 5B). The mutant proteins were expressed in E. coli, purified and screened by gel-shift for their ability to bind the GPx1 and SelN form 1 and the PHGPx form 2 SECIS RNAs. Mutations that yielded the most significant impacts on Kd values are given in Figure 5C. While the mutations did not have a major effect on binding to the form 2 PHGPx SECIS (Kd values ranging from 480 to 580 nM, not deviating too much from the 350 nM wt value), the affinity dropped dramatically for form 1 SECIS and more particularly for GPx1 SECIS: a gradual augmentation of the Kd values was observed which peaked to a maximum for the Ala535–537 mutation, and dropped to near wt values for Ala541–543. For Ala535–537, the Kd increase for GPx1 SECIS was 4.5-fold the wt value. In conclusion, the most severely affected sequence was 526-AKKPTSLKKIILKER-540 and major effects were provoked by mutations of 532-LKK-534 and 535-IIL-537, the latter providing a culminating effect.
Figure 5.
Identification of the K-rich additional RNA binding domain in hSBP2. (A) N- and C-ter deletion mutants were engineered in the full-length hSBP2 cDNA to yield the displayed constructs. hSBP2 proteins were purified and assayed by EMSA for their binding to in vitro transcribed 32P-radiolabeled SelN (form 1) or PHGPx (form 2) SECIS RNAs. (+), binds; (−) no binding; nd, not determined. (B) Alanine scanning mutagenesis. Residues 516–546 in the hSBP2 344–854 cDNA were mutated to alanines in groups of three amino acids (with the exception of the single K516A), as shown above the sequence. Proteins were purified and assayed by gel shifts with in vitro transcribed 32P-labeled PHGPx, GPx1 or SelN SECIS RNAs. (C) Kd values derived from gel shift experiments between the indicated hSBP2 alanine scanning mutants and the form 2 PHGPx (white bars), and form 1 GPx1 (grey bars) and SelN (black bars) SECIS RNAs. Values are given in nM (mean ± SD).
Taken together, sequence comparisons, deletion and alanine scanning mutagenesis enabled us to identify a domain in hSBP2, residing between K516 and K544, that differs from the already known L7Ae module. We named it the K-rich domain because of its relatively high content in lysine residues (34%; 10 lysines out of 29 residues). This domain contains amino acids 526–540 essential for binding to form 1 SECIS RNAs but its mutation did not affect form 2 SECIS recognition.
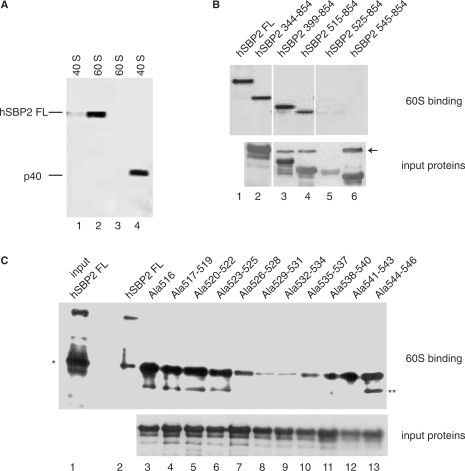
The same mutations in the K-rich domain affecting SBP2 binding to form 1 SECIS also impede interaction with the 60S ribosomal subunit
Rat SBP2 has been shown to bind purified rat 80S ribosomes (21,24,44). The down-effects that we observed upon mutating the K-rich domain prompted us to investigate whether the same mutations would affect ribosome binding. More specifically, we first determined which of the ribosomal subunits, 60S or 40S, was the target for the full-length hSBP2 (hSBP2 FL). In these and further experiments, after incubation with hSBP2, the purified ribosomal subunits were loaded onto sucrose gradients in a buffer containing 0.5 mM Mg2+, and the fractions were analyzed by western blotting with an anti-SBP2 antibody. Figure 6A shows a high intensity signal with the 60S but a very faint one with the 40S subunit (compare lane 2 with lane 1). Similar results were obtained with 80S ribosomes incubated with hSBP2 FL under these conditions (data not shown). Human recombinant ribosomal protein p40 was used as a control since it was recently shown to be capable of binding 40S subunits (45): it effectively bound 40S but not 60S subunits (Figure 6A, compare lane 4 with lane 3). Therefore, we can conclude that under the conditions used, hSBP2 FL bound to 60S subunits in a specific manner. To identify the hSBP2 domain required for 60S binding, we tested the effects of the hSBP2 deletion mutants ending at 854 and starting at 344, 399, 515, 525 and 545. Truncations until position 515 were innocuous (Figure 6B, lanes 2–4, compare with lane 1) whereas removing sequences downstream from 515 (hSBP2 525–854 and hSBP2 545–854; Figure 6B, lanes 5 and 6, respectively) led to complete inhibition of 60S binding. Next, the alanine scanning mutants were used to identify the important amino acids. None of the alanine replacements, from 516 to 525 or 541 to 546, significantly altered the binding of hSBP2 (Figure 6C, compare lanes 3–6 and 12, 13 with lane 2). In contrast, alanine scanning mutants Ala526–528 to Ala538–540 (Figure 6C, lanes 7–11) provoked a marked drop in binding, mutants Ala529–531 and Ala532–540 inducing an almost complete abolition of binding (lanes 8 and 9).
Figure 6.
Human SBP2 interacts with the 60S ribosomal subunit. (A) Detection of hSBP2 in sucrose gradient fractions of 60S and 40S ribosomal subunits. TCA-precipitated fractions were resolved by SDS–PAGE and assayed by western blot with an anti-hSBP2 antibody (lanes 1 and 2). hSBP2 FL is the full-length human SBP2, p40 (lane 4) a 40S ribosomal subunit protein used as the control. (B) hSBP2 deletion mutant proteins were assayed for ribosome binding. Fractions containing 60S bound hSBP2 were treated as in (A). Nomenclature as in (A) and Figure 5A. The lower panel (input proteins) is a western blot showing that the mutant proteins were indeed expressed and detectable by the anti-SBP2 antibody. The arrow points to a cross-reaction product arising from an E.coli protein. (C) The 60S-bound alanine scanning mutant proteins (shown in Figure 5B) were analyzed as in (A). Lane 1: hSBP2FL detected by western blot before gradient loading. The single and double asterisks denote frequently encountered hSBP2 proteolysis fragments. The lower panel (input proteins) serves the same purpose as in (B).
The two sets of mutagenesis experiments identify hSBP2 amino acids located between A526 and R543 that are crucial for both 60S ribosomal subunit interaction and to provide affinity to form 1 SECIS RNAs.
Swapping five amino acids in the lysine-rich domain between D. melanogaster and hSBP2 reversed the affinity toward form 1 or form 2 SECIS RNAs
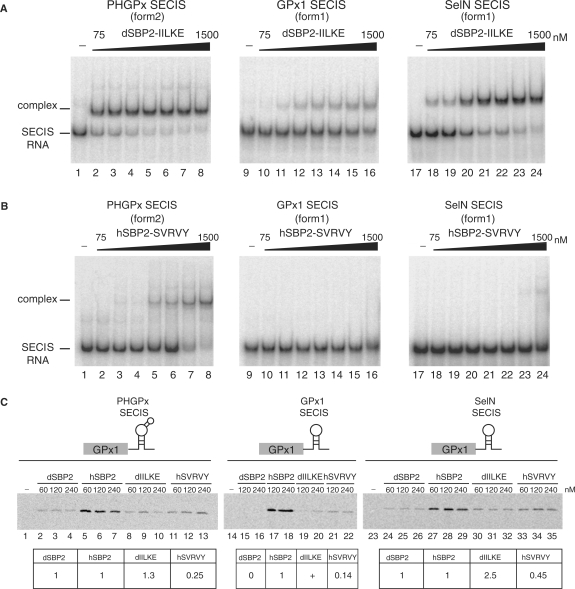
Multiple sequence alignments of 26 vertebrate, one echinoderm (sea urchin) and 13 insect SBP2 (12 Drosophila and the Anopheles gambiae mosquito) amino acid sequences were performed to obtain information about the evolutionary conservation of the K-rich domain, aligning sequences corresponding to hSBP2 positions 515–547 (Figure 7). This highlighted the sequence conservation 517-KGKXXRXXPKXKKXTXLKXXI/VXXXR-540 (numbering of hSBP2). However, inspection of non-conserved residues revealed a dichotomy. In particular, the sequence 535-I/VILKE-539 is replaced by SVRVY in dSBP2, and shows less conservation across the 12 Drosophila species and the Anopheles mosquito than between other SBP2s. The only similar amino acid is I536 in hSBP2 which is replaced by a valine residue in Drosophila. We asked whether this five amino acid change (residues 535–539) between hSBP2 and dSBP2 could be responsible for the inability of dSBP2 to bind form 1 SECIS RNAs. To test this possibility, we swapped the hSBP2 IILKE for the corresponding dSBP2 SVRVY sequence, and vice versa, to yield dSBP2-IILKE and hSBP2-SVRVY. The binding activities to form 1 and 2 SECIS RNAs of the purified chimeric SBP2 proteins were measured by gel-shift assays. Verifying our prediction, dSBP2-IILKE gained the ability to bind the GPx1 and SelN form1 SECIS RNAs (Figure 8A, lanes 10–16 and 18–24, respectively), the affinity for SelN SECIS being higher than for GPx1 (Kd = 200 nM versus Kd > 1500 nM for GPx1; Table 2). The binding affinity to form 2 PHGPx (Figure 8A, lanes 2–8) even increased by a factor of five with a Kd = 40 nM (Table 2) versus 220 nM for the wt dSBP2 (Table 1). Conversely, the five amino acid swapping led hSBP2-SVRVY to completely loose its ability to bind GPx1 and SelN SECIS (Figure 8B, lanes 10–16 and 18–24, respectively), and modified moderately (by a factor lower than 2) its binding to form 2 PHGPx SECIS (Figure 8B, lanes 2–8) with a Kd of 600 nM (Table 2) versus 320 nM for the wt hSBP2 (Table 1).
Figure 8.
Reversed SECIS RNA binding properties provided by swapping five amino acids between Drosophila and human SBP2s. (A) Increasing concentrations (75–1500 nM) of purified dSBP2 carrying the human SBP2 IILKE sequence (dSBP2-IILKE) were assayed with the indicated form 2 or form 1 32P-labeled SECIS RNAs. No protein was added in lanes 1, 9, 17. (B) The same experiments were carried out with the hSBP2 containing the D.melanogaster SVRVY sequence (hSBP2-SVRVY). (C) In vitro translation assay (reticulocyte lysate) of (35S)-Met-labeled rat glutathione peroxidase 1 (GPx1) from reporter constructs carrying the GPx1 ORF with either the PHGPx, GPx1 or SelN SECIS RNAs in the 3′UTR. Rabbit reticulocyte lysate was supplemented with purified dSBP2, dSBP2-IILKE (dIILKE), hSBP2 or hSBP2-SVRVY (hSVRVY) at the indicated concentrations (nM); SBP2 proteins were omitted in lanes 1,14,23. Translation products were treated as in Figure 2B. Values (expressed relative to dSBP2 or hSBP2 set as 1, or 0 for dSBP2 in lanes 15, 16) resulted from the mean of the values obtained with two to three protein concentrations in two to three independent experiments.
Table 2.
Kd values of chimeric Drosophila and human SBP2 for form 1 and 2 SECIS RNAs
| SECIS RNA | SBP2 protein |
|
|---|---|---|
| dSBP2-IILKE | hSBP2-SVRVY | |
| PHGPx (form2) | 40 ± 6 | 600 ± 45 |
| GPx1 (form1) | >1500 | No binding |
| SeIN (form1) | 200 ± 10 | nd |
| Kd ± SD (nM) | ||
Next, the abilities of the chimeric SBP2 proteins to support synthesis of the GPx1 selenoprotein, carrying distinct SECIS in the 3′UTR of its mRNA, were verified in rabbit reticulocyte lysate. With the form 2 PHGPx SECIS, Figure 8C shows that the amount of the synthesized reporter was not significantly altered whether wt dSBP2 or dSBP2-IILKE was employed (value = 1.3; Figure 8C, compare lanes 8–10 with lanes 2–4). However, dSBP2 acquired the ability of supporting GPx1 selenoprotein synthesis by the simple replacement of SVRVY by the homologous human IILKE sequence in the swapping mutant dSBP2-IILKE: this was the case for GPx1 (Figure 8C, lanes 19, 20; compare with the complete inability of wt dSBP2, lanes 15, 16) and SelN SECIS (Figure 8C, compare lanes 30–32 with lanes 24–26) which showed a 2.5-fold gain. In contrast, introduction of the Drosophila SVRVY sequence instead of IILKE in hSBP2 (hSBP2-SVRVY) provided an almost total (14% residual) or partial (45% residual) inhibition of selenoprotein synthesis when assayed with the form 1 SelN and GPx1 SECIS, respectively (Figure 8C; compare lanes 33–35 to lanes 27–29; lanes 21–22 with lanes 17, 18). Tested with the form 2 PHGPx SECIS, hSBP2-SVRVY led to a 0.25-fold drop of the value obtained with wt hSBP2 (Figure 8C, compare lanes 11–13 with lanes 5–7). This was unexpected but this finding nevertheless correlated with the data shown in Figure 8A and B and Table 2 establishing a loss of affinity between this chimeric SBP2 and the PHGPx SECIS (the Kd increased by a factor 2). At present, we are unable to explain why hSBP2-SVRVY did not retain close to wild-type properties in gel-shift and selenoprotein synthesis assays with form 2 SECIS RNA. Notwithstanding, this set of experiment was globally in line with the in vitro EMSA data shown in Figure 3B and Figure 8A and B and the Kd values in Tables 1 and 2.
We conclude from these experiments that the IILKE sequence confers the ability to bind form 1 SECIS RNAs.
DISCUSSION
SBP2 plays a central role in selenoprotein synthesis by binding to SECIS hairpins in the 3′UTR of selenoprotein mRNAs. Earlier domain dissection of the human and rat SBP2 established that the C-terminal 2/3 are involved in SECIS and ribosome binding as well as selenocysteine incorporation while no function has been attributed so far to its N-terminal part, which is dispensable for selenoprotein synthesis in reticulocyte lysate (20–22,24). In this work, we have isolated and functionally characterized a bona fide dSBP2 that specifically lacks the region homologous to the N-terminus of vertebrate SBP2. In addition, we report the identification in human SBP2 of a lysine-rich (K-rich) domain that is essential for SECIS binding, point mutations therein affecting form 1 but not form 2 SECIS binding. Sequence comparisons established that the K-rich domain is encountered in all the SBP2s analyzed (this work and data not shown). In the D. melanogaster K-rich domain, a five amino acid sequence difference renders Drosophila SBP2 incapable of binding form 1 SECIS RNAs with high affinity. Exchanging this sequence with that of hSBP2 enabled binding of the insect SBP2 to form 1 SECIS but impaired that of hSBP2. Moreover, we found that the K-rich sequence is also crucial for the binding of hSBP2 to the 60S ribosomal subunit.
The insect SBP2 is 313 amino acids long, a little less than a third of the mammalian counterpart. Drosophila SBP2 is shorter at the C-terminus and a small internal deletion removed positions corresponding to hSBP2 565–689 (Figure 1). However, the lack of the N-terminal region corresponding to hSBP2 1–427 accounts for most of its reduction in size. That selenoprotein synthesis can be achieved in an organism lacking this domain corroborates the finding that it is dispensable in mammalian SBP2 under the experimental conditions used (25). Although one cannot exclude the possibility that this segment of SBP2 is encoded by a separate gene in insects, this appears unlikely because a search in the Drosophila genomes failed to find significant sequence similarity to the vertebrate N-terminal region (data not shown). What might then be the function of the N-terminal extension in higher eukaryotes? Several possibilities exist that would pertain to a more complex selenoprotein synthesis mechanism in higher eukaryotes: (i) the N-terminal extension could participate in fine-tuning selenoprotein expression with a direct or indirect role in the SBP2 nuclear/cytoplasmic shuttling, as it contains a nuclear localization signal (46); (ii) we established by structure prediction and experimental data that SBP2 is an Intrinsically Disordered Protein and that the N-terminal extension is widely unstructured (Olieric et al., manuscript in preparation). It is very possible that this region acquires its proper folding in the presence of yet to be discovered protein partners, consistent with the role of the Hsp90 chaperone and co-chaperones in the folding and assembly of proteins bearing an L7Ae RNA-binding module (47) and (iii) finally, the N-terminal domain could be involved in an SBP2 function different from selenocysteine incorporation, as inferred very recently from the finding that several SBP2 isoforms arise from splice variants in the N-terminal region (48).
We found a region in hSBP2, the K-rich (lysine-rich) domain 516-KKGKQREIPKAKKPTSLKKIILKERQER-543, that is essential for SECIS binding. This sequence is highly conserved across vertebrates and is distinct from the L7Ae RNA-binding module which is therefore insufficient on its own to provide SECIS recognition. Surprisingly, however, alanine scanning mutagenesis of sequence 516–543 manifested adverse effects on SECIS binding. While it did not significantly prohibit binding to form 2, dramatic effects were observed on form 1, especially when altering the sequence 526-AKKPTSLKKIILKER-540 and most prominently 532-LKKIIL-537 which profoundly disabled SBP2.
A multiple sequence alignment identified KGKTRLDPKKKITRLKKSVRVYR (D. melanogaster sequence) as the homolog of the human K-rich domain. This sequence analysis highlighted a characteristic feature distinguishing the insect and other K-rich domains, i.e. the five amino-acid substitution I/VILKE in vertebrates to SVRVY in D. melanogaster. dSBP2 can bind Drosophila and mammalian form 2 SECIS RNAs but was unable—or with very low affinity—to bind form 1 SECIS RNAs, consistent with Drosophila having selenoprotein mRNAs with form 2 SECIS exclusively (8,31). The IILKE/SVRVY swapping experiment enabled dSBP2 to bind form 1 SECIS but led hSBP2 to loose this ability. It is very unlikely that the SVRVY sequence per se is inhibitory to form 1 binding since replacement of IIL and KER by alanines in hSBP2 yielded the same down effect as introduction of SVRVY in hSBP2. It seems more plausible that the ability to bind form 1 SECIS requires the occurrence of IILKE because of its different amino-acid composition versus SVRVY, giving rise to a different charge and hydrophobicity. The SBP2 functional RNA-binding domain (526–777 in humans) consists therefore of two subdomains, the K-rich (this work) and the previously characterized conserved L7Ae module extending from Arginine 672 to Isoleucine 749 (20,49). The intervening sequence exhibits less conservation with the exception of the 624-IHSRRFR-630 block (positions relative to human SBP2). Bubenik and Driscoll (22) reported in rat SBP2 the existence of a bipartite RNA binding domain in which R531 (R540 in hSBP2) appeared important for discriminating forms 1 and 2 SECIS hairpins. Indeed, this arginine is universal in SBP2 (Figure 7) and a mutation to glutamine, described in SBP2 patients with thyroid dysfunctions, impaired binding to the form 1 SECIS of the iodothyronine deiodinase mRNA (50). However, we did not observe such a dramatic effect of the R540 mutation in human SBP2 since the Ala 538–540 mutation only mildly affected form 1 binding.
Additionally, and very interestingly, we showed the importance of that same 526-AKKPTSLKKIILKER-540 sequence for binding to the 60S ribosomal subunit. These findings establish the ability of SBP2 to stably bind the 60S subunit and the identification of the PTSLKK motif that contributes in a very important manner to the binding. While this work was in progress, Donovan et al. (51) reported in rat SBP2 the identification of the additional domain SID, required for SECIS and 80S ribosome binding. The boundaries of the SID are similar to those we delineated for the K-rich domain but the authors did not report a distinctive effect of the SID on form 1 or form 2. Moreover, the mutation of the IILKE sequence by these authors significantly affected binding to form 2 SECIS, in contrast to our alanine scanning and swapping experiments. Also, we determined by sucrose gradient centrifugation that SBP2 interacts with the 60S but not the 40S ribosomal subunit. This is consistent with a proposal that SBP2 could bind the 28S ribosomal RNA (24). Lastly, our finding that the same amino acids in the K-rich domain are involved in SECIS RNA and 60S ribosomal subunit recognition strengthens the model establishing that SBP2 cannot bind both simultaneously (44).
Worthy of note is that SBP2, with two domains crucial for SECIS binding, is set apart from the other proteins of the L7Ae family which are shorter and for which the L7Ae domain is itself sufficient to ensure specific interactions with the cognate RNA (11,18,19,52,53). In a previous work, we provided a very precise definition of the RNA-binding specificity of hSBP2 (23). In particular, we identified nucleotide determinants in the SECIS RNA that are unique to SBP2 among the L7Ae family members: while the 15.5 kD and L7Ae ribosomal proteins have a rather broad specificity and can recognize the SECIS RNA, the converse does not hold true since SBP2 is unable to bind the U3 snoRNA or the archaeal sRNA (23,54). As a matter of fact, footprinting experiments and interference of binding established the requirement of helix I and internal loop 1 in SECIS RNAs for SBP2 recognition (23,55). These structural features are idiosyncratic to SECIS RNAs and therefore not found in other Kink-turn containing RNAs, leading to our proposal that SECIS hairpins contain a Kink-turn like rather than a canonical Kink-turn motif (14). We therefore propose a model rationalizing the necessity of two domains in SBP2 for complex formation with the SECIS RNA: (i) a restricted number of amino acids in the L7Ae module establish contacts with the guanine bases of the G.A/A.G base pairs as in other L7Ae proteins, and the conserved U in the SECIS non-Watson-Crick quartet (11,20); (ii) the lysines in the K-rich domain contribute positive charges for electrostatic interactions with the phosphates of the SECIS-specific structural features and thus increase the affinity of SBP2 for SECIS RNAs. In this domain, the amino acid composition of the IILKE sequence could have a direct impact on SECIS binding or indirectly lead to an L7Ae conformational change allowing form 1 SECIS recognition.
Validation of these models definitely requires that the crystal structure of the SECIS RNA-SBP2 complex be solved.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Action Concertée Incitative (BCMS 226) and Programme InterOrganismes (Tox.Nuc-E) [to A.K.]; the Spanish Ministry of Education (BIO2006-03380) and Biosapiens LSHG-CT-2003-503265 (FP6 Programme of the European Commission) [to R.G.]; the Russian Foundation for Basic Research (grant #08-04-00508) [to G.K.]; Pre-doctoral fellowship from the Japanese Ministry of Education, Culture, Sports, Science and Technology [to A.T.]; Pre-doctoral fellowship of the Spanish Ministry of Education and Science [to C.C.]; EMBO short-term fellowship (ASTF 91-2007) [to E.B.]. Funding for open access charge: CNRS.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to A. Beniaminov for providing plasmid GPx1-SelN SECIS, to S. Baudrey and A. Schweigert for skillful technical assistance. We thank A. Beniaminov, P. Carbon, M. Leichter, E. Myslinski and L. Wurth for reading the manuscript and helpful discussions.
REFERENCES
- 1.Allmang C, Krol A. Selenoprotein synthesis: UGA does not end the story. Biochimie. 2006;88:1561–1571. doi: 10.1016/j.biochi.2006.04.015. [DOI] [PubMed] [Google Scholar]
- 2.Carlson BA, Xu X-M, Kryukov GV, Rao M, Berry MJ, Gladyshev VN, Hatfield DL. Identification and characterization of phosphoseryl-tRNA[Ser]Sec kinase. Proc. Natl Acad. Sci. USA. 2004;101:12848–12853. doi: 10.1073/pnas.0402636101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xu X-M, Mix H, Carlson BA, Grabowski PJ, Gladyshev VN, Berry MJ, Hatfield DL. Evidence for direct roles of two additional factors, SECp43 and soluble liver antigen, in the selenoprotein synthesis machinery. J. Biol. Chem. 2005;280:41568–41575. doi: 10.1074/jbc.M506696200. [DOI] [PubMed] [Google Scholar]
- 4.Small-Howard A, Morozova N, Stoytcheva Z, Forry EP, Mansell JB, Harney JW, Carlson BA, Xu XM, Hatfield DL, Berry MJ. Supramolecular complexes mediate selenocysteine incorporation in vivo. Mol. Cell Biol. 2006;26:2337–2346. doi: 10.1128/MCB.26.6.2337-2346.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chavatte L, Brown BA, Driscoll DM. Ribosomal protein L30 is a component of the UGA-selenocysteine recoding machinery in eukaryotes. Nat. Struct. Mol. Biol. 2005;12:408–416. doi: 10.1038/nsmb922. [DOI] [PubMed] [Google Scholar]
- 6.Grundner-Culemann E, Martin GW, 3rd, Harney JW, Berry MJ. Two distinct SECIS structures capable of directing selenocysteine incorporation in eukaryotes. RNA. 1999;5:625–635. doi: 10.1017/s1355838299981542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fagegaltier D, Lescure A, Walczak R, Carbon P, Krol A. Structural analysis of new local features in SECIS RNA hairpins. Nucleic Acids Res. 2000;28:2679–2689. doi: 10.1093/nar/28.14.2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chapple CE, Guigo R, Krol A. SECISaln, a web-based tool for the creation of structure-based alignments of eukaryotic SECIS elements. Bioinformatics. 2009 doi: 10.1093/bioinformatics/btp020. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Walczak R, Westhof E, Carbon P, Krol A. A novel RNA structural motif in the selenocysteine insertion element of eukaryotic selenoprotein mRNAs. RNA. 1996;2:367–379. [PMC free article] [PubMed] [Google Scholar]
- 10.Walczak R, Carbon P, Krol A. An essential non-Watson-Crick base pair motif in 3'UTR to mediate selenoprotein translation. RNA. 1998;4:74–84. [PMC free article] [PubMed] [Google Scholar]
- 11.Vidovic I, Nottrott S, Hartmuth K, Lührmann R, Ficner R. Crystal structure of the spliceosomal 15.5kD protein bound to a U4 snRNA fragment. Mol. Cell. 2000;6:1331–1342. doi: 10.1016/s1097-2765(00)00131-3. [DOI] [PubMed] [Google Scholar]
- 12.Klein DJ, Schmeing TM, Moore PB, Steitz TA. The kink-turn: a new RNA secondary structure motif. EMBO J. 2001;20:4214–4221. doi: 10.1093/emboj/20.15.4214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lescoute A, Leontis NB, Massire C, Westhof E. Recurrent structural RNA motifs, Isostericity Matrices and sequence alignments. Nucleic Acids Res. 2005;33:2395–2409. doi: 10.1093/nar/gki535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Allmang C, Krol A. SECIS RNAs and K-turn binding proteins. A survey of evolutionary conserved RNA and protein motifs. In. In: Hatfield DL, editor. Selenium, its Molecular Biology and Role in Human Health 2nd edition. Vol. 5. New York, USA: Kluwer Academic Publishers; 2006. pp. 51–61. [Google Scholar]
- 15.Watkins NJ, Segault V, Charpentier B, Nottrott S, Fabrizio P, Bachi A, Wilm M, Rosbash M, Branlant C, Luhrmann R. A common core RNP structure shared between the small nucleoar box C/D RNPs and the spliceosomal U4 snRNP. Cell. 2000;103:457–466. doi: 10.1016/s0092-8674(00)00137-9. [DOI] [PubMed] [Google Scholar]
- 16.Rozhdestvensky TS, Tang TH, Tchirkova IV, Brosius J, Bachellerie JP, Hüttenhofer A. Binding of L7Ae protein to the K-turn of archaeal snoRNAs: a shared RNA binding motif for C/D and H/ACA box snoRNAs in Archaea. Nucleic Acids Res. 2003;31:869–877. doi: 10.1093/nar/gkg175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Koonin EV, Bork P, Sander C. A novel RNA-binding motif in omnipotent suppressors of translation termination, ribosomal proteins and a ribosome modification enzyme? Nucleic Acids Res. 1994;22:2166–2167. doi: 10.1093/nar/22.11.2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chao JA, Williamson JR. Joint X-ray and NMR refinement of the yeast L30e-mRNA complex. Structure. 2004;12:1165–1176. doi: 10.1016/j.str.2004.04.023. [DOI] [PubMed] [Google Scholar]
- 19.Moore T, Zhang Y, Fenley MO, Li H. Molecular basis of box C/D RNA-protein interactions; cocrystal structure of archaeal L7Ae and a box C/D RNA. Structure. 2004;12:807–818. doi: 10.1016/j.str.2004.02.033. [DOI] [PubMed] [Google Scholar]
- 20.Allmang C, Carbon P, Krol A. The SBP2 and 15.5 kD/Snu13p proteins share the same RNA binding domain: identification of SBP2 amino acids important to SECIS RNA binding. RNA. 2002;8:1308–1318. doi: 10.1017/s1355838202020034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Caban K, Kinzy SA, Copeland PR. The L7Ae RNA binding motif is a multifunctional domain required for the ribosome-dependent Sec incorporation activity of Sec insertion sequence binding protein 2. Mol. Cell Biol. 2007;27:6350–6360. doi: 10.1128/MCB.00632-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bubenik JL, Driscoll DM. Altered RNA binding activity underlies abnormal thyroid hormone metabolism linked to a mutation in selenocysteine insertion sequence-binding protein 2. J. Biol. Chem. 2007;282:34653–34662. doi: 10.1074/jbc.M707059200. [DOI] [PubMed] [Google Scholar]
- 23.Cléry A, Bourguignon-Igel V, Allmang C, Krol A, Branlant C. An improved definition of the RNA-binding specificity of SECIS-binding protein 2, an essential component of the selenocysteine incorporation machinery. Nucleic Acids Res. 2007;35:1868–1884. doi: 10.1093/nar/gkm066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Copeland PR, Stepanik VA, Driscoll DM. Insight into mammalian selenocysteine insertion: domain structure and ribosome binding properties of Sec insertion sequence binding protein 2. Mol. Cell Biol. 2001;21:1491–1498. doi: 10.1128/MCB.21.5.1491-1498.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Copeland PR, Fletcher JE, Carlson BA, Hatfield DL, Driscoll DM. A novel RNA binding protein, SBP2, is required for the translation of mammalian selenoprotein mRNAs. EMBO J. 2000;19:306–314. doi: 10.1093/emboj/19.2.306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lobanov AV, Fomenko DE, Zhang Y, Sengupta A, Hatfield DL, Gladyshev VN. Evolutionary dynamics of eukaryotic selenoproteomes: large selenoproteomes may associate with aquatic life and small with terrestrial life. Genome Biol. 2007;8:R198. doi: 10.1186/gb-2007-8-9-r198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hirosawa-Takamori M, Jackle H, Vorbruggen G. The class 2 selenophosphate synthetase gene of Drosophila contains a functional mammalian-type SECIS. EMBO Rep. 2000;1:441–446. doi: 10.1093/embo-reports/kvd087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Castellano S, Morozova N, Morey M, Berry MJ, Serras F, Corominas M, Guigó R. In silico identification of novel selenoproteins in the Drosophila melanogaster genome. EMBO Rep. 2001;2:697–702. doi: 10.1093/embo-reports/kve151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lu C, Qiu F, Zhou H, Peng Y, Hao W, Xu J, Yuan J, Wang S, Qiang B, Xu C, et al. Identification and characterization of selenoprotein K: an antioxidant in cardiomyocytes. FEBS Lett. 2006;580:5189–5197. doi: 10.1016/j.febslet.2006.08.065. [DOI] [PubMed] [Google Scholar]
- 30.Dikiy A, Novoselov SV, Fomenko DE, Sengupta A, Carlson BA, Cerny RL, Ginalski K, Grishin NV, Hatfield DL, Gladyshev VN. SelT, SelW, SelH, and Rdx12: genomics and molecular insights into the functions of selenoproteins of a novel thioredoxin-like family. Biochemistry. 2007;46:6871–6882. doi: 10.1021/bi602462q. [DOI] [PubMed] [Google Scholar]
- 31.Chapple CE, Guigo R. Relaxation of selective constraints causes independent selenoprotein extinction in insect genomes. PLoS ONE. 2008;3:e2968. doi: 10.1371/journal.pone.0002968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Copeland PR, Driscoll DM. Purification, redox sensitivity, and RNA binding properties of SECIS- binding protein 2, a protein involved in selenoprotein biosynthesis. J. Biol. Chem. 1999;274:25447–25454. doi: 10.1074/jbc.274.36.25447. [DOI] [PubMed] [Google Scholar]
- 33.Lescure A, Allmang C, Yamada K, Carbon P, Krol A. cDNA cloning, expression pattern and RNA binding analysis of human selenocysteine insertion sequence (SECIS) binding protein 2. Gene. 2002;291:279–285. doi: 10.1016/s0378-1119(02)00629-7. [DOI] [PubMed] [Google Scholar]
- 34.Studier FW. Protein production by auto-induction in high density shaking cultures. Protein Expr. Purif. 2005;41:207–234. doi: 10.1016/j.pep.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 35.Lassmann T, Sonnhammer EL. Kalign – an accurate and fast multiple sequence alignment algorithm. BMC Bioinformatics. 2005;6:298. doi: 10.1186/1471-2105-6-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Eddy SR. Profile hidden Markov models. Bioinformatics. 1998;14:755–763. doi: 10.1093/bioinformatics/14.9.755. [DOI] [PubMed] [Google Scholar]
- 37.Katoh K, Misawa K, Kuma K, Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002;30:3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Clamp M, Cuff J, Searle SM, Barton GJ. The Jalview Java alignment editor. Bioinformatics. 2004;20:426–427. doi: 10.1093/bioinformatics/btg430. [DOI] [PubMed] [Google Scholar]
- 39.Busso D, Delagoutte-Busso B, Moras D. Construction of a set Gateway-based destination vectors for high-throughput cloning and expression screening in Escherichia coli. Anal. Biochem. 2005;343:313–321. doi: 10.1016/j.ab.2005.05.015. [DOI] [PubMed] [Google Scholar]
- 40.Kunkel TA, Roberts JD, Zakour RA. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- 41.Lescure A, Gautheret D, Carbon P, Krol A. Novel selenoproteins identified in silico and in vivo by using a conserved RNA structural motif. J. Biol. Chem. 1999;274:38147–38154. doi: 10.1074/jbc.274.53.38147. [DOI] [PubMed] [Google Scholar]
- 42.Matasova NB, Myltseva SV, Zenkova MA, Graifer DM, Vladimirov SN, Karpova GG. Isolation of ribosomal subunits containing intact rRNA from human placenta: estimation of functional activity of 80S ribosomes. Anal. Biochem. 1991;198:219–223. doi: 10.1016/0003-2697(91)90416-q. [DOI] [PubMed] [Google Scholar]
- 43.Allamand V, Richard P, Lescure A, Ledeuil C, Desjardin D, Petit N, Gartioux C, Ferreiro A, Krol A, Pellegrini N, et al. A single homozygous point mutation in a 3'untranslated region motif of selenoprotein N mRNA causes SEPN1-related myopathy. EMBO Rep. 2006;7:450–454. doi: 10.1038/sj.embor.7400648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kinzy SA, Caban K, Copeland PR. Characterization of the SECIS binding protein 2 complex required for the co-translational insertion of selenocysteine in mammals. Nucleic Acids Res. 2005;33:5172–5180. doi: 10.1093/nar/gki826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kossinova OA, Malygin AA, Babailova ES, Karpova GG. Binding of human ribosomal protein p40 and its truncated mutants to the small ribosomal subunit. Molecular Biology. 2008;42:911–916. [PubMed] [Google Scholar]
- 46.Papp LV, Lu J, Striebel F, Kennedy D, Holmgren A, Khanna KK. The redox state of SECIS binding protein 2 controls its localization and selenocysteine incorporation function. Mol. Cell Biol. 2006;26:4895–4910. doi: 10.1128/MCB.02284-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Boulon S, Marmier-Gourrier N, Pradet-Balade B, Wurth L, Verheggen C, Jady BE, Rothe B, Pescia C, Robert MC, Kiss T, et al. The Hsp90 chaperone controls the biogenesis of L7Ae RNPs through conserved machinery. J. Cell Biol. 2008;180:579–595. doi: 10.1083/jcb.200708110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Papp LV, Wang J, Kennedy D, Boucher D, Zhang Y, Gladyshev VN, Singh RN, Khanna KK. Functional characterization of alternatively spliced human SECISBP2 transcript variants. Nucleic Acids Res. 2008;36:7192–7206. doi: 10.1093/nar/gkn829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lescure A, Fagegaltier D, Carbon P, Krol A. Protein factors mediating selenoprotein synthesis. Curr. Protein Pept. Sci. 2002;3:143–151. doi: 10.2174/1389203023380783. [DOI] [PubMed] [Google Scholar]
- 50.Dumitrescu AM, Liao XH, Abdullah MS, Lado-Abeal J, Majed FA, Moeller LC, Boran G, Schomburg L, Weiss RE, Refetoff S. Mutations in SECISBP2 result in abnormal thyroid hormone metabolism. Nat. Genet. 2005;37:1247–1252. doi: 10.1038/ng1654. [DOI] [PubMed] [Google Scholar]
- 51.Donovan J, Caban K, Ranaweera R, Gonzalez-Flores JN, Copeland PR. A novel protein domain induces high affinity selenocysteine insertion sequence binding and elongation factor recruitment. J. Biol. Chem. 2008;283:35129–35139. doi: 10.1074/jbc.M806008200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nottrott S, Urlaub H, Lührmann R. Hierarchical, clustered protein interactions with U4/U6 snRNA: a biochemical role for U4/U6 proteins. EMBO J. 2002;21:5527–5538. doi: 10.1093/emboj/cdf544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Marmier-Gourrier N, Clery A, Senty-Segault V, Charpentier B, Schlotter F, Leclerc F, Fournier R, Branlant C. A structural, phylogenetic, and functional study of 15.5-kD/Snu13 protein binding on U3 small nucleolar RNA. RNA. 2003;9:821–838. doi: 10.1261/rna.2130503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Charron C, Manival X, Clery A, Senty-Segault V, Charpentier B, Marmier-Gourrier N, Branlant C, Aubry A. The archaeal sRNA binding protein L7Ae has a 3D structure very similar to that of its eukaryal counterpart while having a broader RNA-binding specificity. J. Mol. Biol. 2004;342:757–773. doi: 10.1016/j.jmb.2004.07.046. [DOI] [PubMed] [Google Scholar]
- 55.Fletcher JE, Copeland PR, Driscoll DM, Krol A. The selenocysteine incorporation machinery: interactions between the SECIS RNA and the SECIS-binding protein SBP2. RNA. 2001;7:1442–1453. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.