Abstract
mRNA decapping is a critical step in the control of mRNA stability and gene expression and is carried out by the Dcp2 protein. Dcp2 is an RNA-binding protein that must bind the RNA in order to recognize the cap for hydrolysis. We previously demonstrated that a 60 nucleotide (nt) element at the 5′ end of the mRNA encoding Rrp41 is preferentially bound and decapped by Dcp2. Here, we demonstrate that enhanced decapping of this element is dependent on the structural integrity of its first 33 nt and not its primary sequence. The structure consists of a stem-loop positioned <10 nt from the 5′ end of the mRNA. The generality of a stem-loop structure in enhanced Dcp2-mediated decapping was underscored by the identification of additional potential Dcp2 substrate mRNAs by a global analysis of human mRNAs containing a similar predicted stem-loop structure at their respective 5′ end. These studies suggest a general role for 5′ stem-loops in enhancing decapping activity and the utilization of this structure as a predictive tool for Dcp2 target substrates. These studies also demonstrate that Dcp2 alone in the absence of additional proteins can preferentially associate with and modulate mRNA decapping.
INTRODUCTION
mRNA decay plays an important role in the control of gene expression and response to regulatory events. In both yeast and mammalian cells, bulk mRNA decay typically initiates with the removal of 3′ poly(A) tail followed by degradation of the mRNA in a 5′–3′ direction or a 3′–5′ direction (1). Degradation from the 3′ end is carried out by the cytoplasmic RNA exosome, which is a multisubunit 3′–5′ exoribonuclease complex (2), and the resulting cap structure is hydrolyzed by the scavenger decapping enzyme DcpS (3). In the 5′–3′ decay pathway, the monomethyl guanosine (m7G) mRNA cap is cleaved first by the Dcp2 decapping enzyme (4–7) and the monophosphate RNA is degraded progressively by the 5′–3′ exoribonuclease Xrn1 (8,9).
Dcp2 is an RNA-binding protein and can only cleave cap structure that is linked to an RNA moiety (10). Uncapped RNA, but not cap analog, can inhibit Dcp2 decapping efficiently (6,7,10). These facts suggest that Dcp2 detects its cap substrate by RNA binding. The decapping activity of Dcp2 can be regulated by various protein factors. In yeast, Dcp1p forms a complex with Dcp2p and is required for optimum-decapping activity (11,12). The Edc1p, Edc2p and Edc3p proteins, as well as Dhh1p and Lsm1–7 protein complex, are all reported to stimulate Dcp2p decapping (13). In mammals, an additional protein Hedls (also known as Edc4 and Ge-1) is also a positive effector of Dcp2 decapping (14). On the other hand, negative regulators of decapping such as eIF4E cap-binding protein and the poly(A)-binding protein (PABP) can inhibit Dcp2 decapping in vitro (15–19). The primate-restricted non-canonical 5′ cap-binding protein, VCX-A, was also identified as an inhibitor of Dcp2 decapping (20).
In addition to the regulation by protein factors, decapping enzymes themselves have been shown to contain substrate specificity. The X29 protein, which is the nuclear decapping enzyme, possesses substrate-preferential decapping activity. X29 can specifically bind the U8 snoRNA (21,22) and in the presence of Mg2+, cap hydrolysis is highly specific for U8 snoRNA (21,23); in contrast, in the presence of Mn2+, all RNAs tested are decapped at high efficiency (23). Similarly, we recently reported that Dcp2 could bind RNAs differentially and preferentially decap a subset of mRNA with higher efficiency (24). Particularly, we identified the 5′ terminus of the mRNA encoding a core subunit of the exosome, Rrp41, as a specific Dcp2 substrate. A 60-nt element at the 5′ end of the Rrp41 mRNA (which we termed 2xDE) was identified and shown to confer more efficient decapping onto a heterologous RNA both in vitro and upon transfection into cells (24). Moreover, reduction of Dcp2 protein levels in cells resulted in a selective stabilization of the Rrp41 mRNA, confirming it as a downstream target of Dcp2 (24).
In this study, we further characterized the 2xDE element by mutational analysis. We found that the first 33 nt are critical for Dcp2-decapping stimulation, and this region forms a stable stem-loop structure. Mutations that disrupt the stem-loop significantly reduced decapping activity by Dcp2, which can be restored by compensatory mutations that restore the stem-loop, suggesting the secondary structure, but not the primary sequence, is critical for Dcp2 recognition. A bioinformatic search identified a subset of mRNAs that have a comparable stem-loop structure at the 5′ end as potential Dcp2 substrates in cells. These data indicate that Dcp2 has the capability to directly and specifically regulate decapping of a subset of mRNAs in the cell.
MATERIALS AND METHODS
Generation of RNA in vitro
RNAs were in vitro transcribed from PCR-generated templates that contained the T7 bacteriophage promoter sequences at the 5′ end. Cap labeling was carried out as previously described (25). The 5′ 900 nt of Stx7 mRNA used as negative control for decapping in Figures 1, 3, 4, 6 and 7 were transcribed with T7 RNA polymerase from a template generated with primers 1 and 2. 2xDE RNA mutations used in decapping assays (Figure 1A, 3, 4, 5A) were transcribed as chimeric RNAs with the coding region of the Stx7 RNA attached to the 3′ end (in order to maintain a uniform size for all the RNAs) by T7 RNA polymerase from templates generated by 3′ primer 2 and the following 5′ primers: for Figure 1A, Mut1–10 was generated with primer 3; Mut11–22 with primer 4; Mut23–33 with primer 5; Mut34–44 with primer 6; and Mut45–55 with primer 7. For Figures 3 and 4, mutant A was generated with primer 8; B with primer 9; C with primer 10; D with primer 11; E with primer 12; F with primer 13; G with primer 14; H with primer 15; and I with primer 16. The RNAs in Figure 5A, 10-2xDE and 20-2xDE were generated with 5′ primers 29 and 30, respectively, and the same 3′ primer, 2. Templates for Ndufb7 mutant RNAs in Figure 6 were transcribed with T7 RNA polymerase using the same 3′ primer (primer 17) and the following different 5′ primers: wild type with primer 18; Mut35–44 with primer 19; and Mut2–11/35–44 with primer 20. For Figure 7A, Hip1 RNA was generated with primer 21 and 22; Kcnj2 with primers 23 and 24; Zcrb1 with primers 25 and 26. Mutants of Hip1 RNA in Figure 7B were generated with 3′ primer 22 and the following 5′ primers: Mut23–31 with primer 27 and Mut4–13/23–31 with primer 28. RNA probes used in the electrophoretic mobility shift assays were in vitro transcribed in the presence of [α32P]GTP from template generated by the following primer sets: for Figure 1B, wild-type 2xDE was generated with primers 31 and 32; Mut1–10 with primers 3 and 32; Mut11–22 with primers 4 and 32; Mut23–33 with primers 5 and 33; Mut34–44 with primers 6 and 34; Mut45–55 with primers 7 and 35; Stx7 RNA with primers 1 and 36. For Figure 5B, 10-2xDE was generated with primers 29 and 36; 20-2xDE with primers 30 and 36. All primer sequences are listed in Supplementary Table 1.
Figure 1.
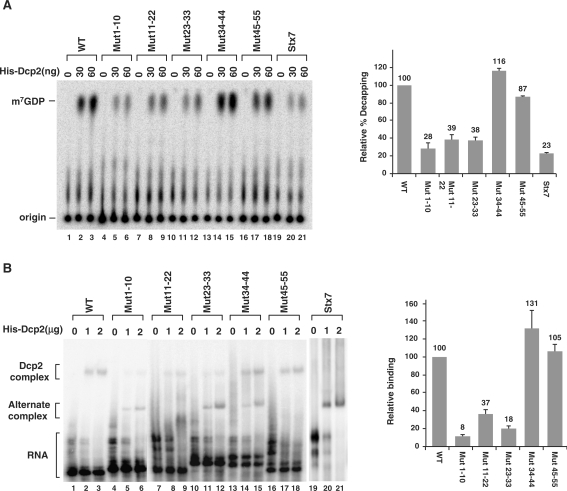
The first 33 nucleotides of the 2xDE element are critical for enhanced Dcp2 decapping and binding. (A) The first 33 nucleotides of the 2xDE are important for hDcp2 decapping. Decapping assays were carried out using increasing concentrations of bacterial expressed His-Dcp2 with cap labeled 2xDE wild-type or mutant RNAs as indicated or the Stx7 RNA negative control. Transversion mutations were generated where adenines were substituted by cytosines, thymines by guanosines, cytosines by adenines and guanosines by thymines. All RNAs were transcribed as chimeric RNAs with the coding region of Stx7 RNA linked to the 3′ end to maintain the same size as the negative control Stx7 RNA. The reaction products were resolved by PEI-TLC and the position of the capped RNA substrate and m7GDP decapping products indicated on the left. Decapping efficiency of wild-type 2xDE RNA with 60 ng of His-Dcp2 was designated as 100 and the corresponding decapping efficiencies of the 2xDE mutant RNAs obtained from three independent experiments are graphically presented on the right with standard deviation denoted by the error bars. Similar results were observed with the 30 ng protein assays. (B) The first 33 nucleotides of the 2xDE are important for binding of Dcp2. An electrophoretic mobility shift assay was carried out with increasing concentrations of His-Dcp2 and uniformly 32P-labeled uncapped RNA described in A. The slower migrating complex detected with the wild-type RNA was designated as the Dcp2-complex while the faster migrating complex detected with the Stx7 negative control was denoted as an alternate complex. Binding efficiency of wild-type 2xDE RNA with 2 µg of His-Dcp2 was designated as 100 and the corresponding binding efficiencies of the 2xDE mutant RNAs from three experiments are presented relative to the wild-type value on the right with standard deviation denoted by the error bars. A direct correlation between formation of the slower migrating complex and efficiency of decapping was observed.
Figure 3.
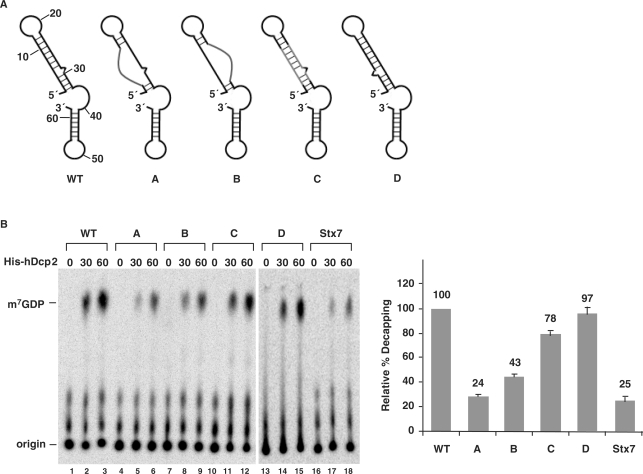
The intact stem-loop structure is critical for promoting Dcp2 decapping. (A) A schematic representation of the structures of the wild type and various mutant 2xDE RNAs are shown. WT, wild type; (A) mutations in the first strand of the stem; (B) mutations in the second strand of the stem; (C) complementary mutations in both strands of the stem; (D) the two strands of the stem are flipped. (B) Decapping assays were carried out with the indicated mutant 2xDE RNAs and the Stx7 negative control. Quantification of decapping efficiencies of each RNA obtained from three independent experiments are shown on the right with the decapping efficiency of wild-type 2xDE RNA arbitrarily set to 100 and the standard deviation (SD) denoted by the error bars. Labeling is as described in the legend to Figure 1A.
Figure 4.
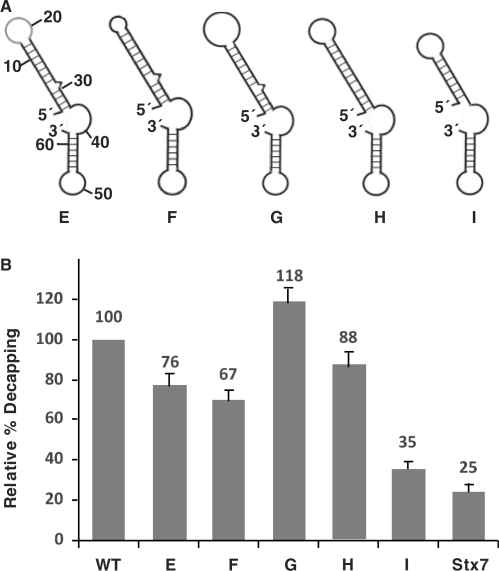
Mutational analysis of the stem-loop structure. (A) A schematic representation of the predicted structures for the various mutant 2xDE RNAs are shown. WT, wild type; E, substitution of the loop region; F, deletion of the loop to four nucleotides; G, increase the loop to 12 nucleotides; H, deletion of the bulge at nucleotide 29; I, deletion of the lower 4 bp of the stem. (B) Decapping assays were carried out with the indicated mutant 2xDE RNAs and the Stx7 negative control. Decapping efficiencies from three independent experiments were quantitated for each RNA and graphically presented. The decapping efficiency of the wild-type 2xDE RNA was arbitrarily set to 100.
Figure 6.
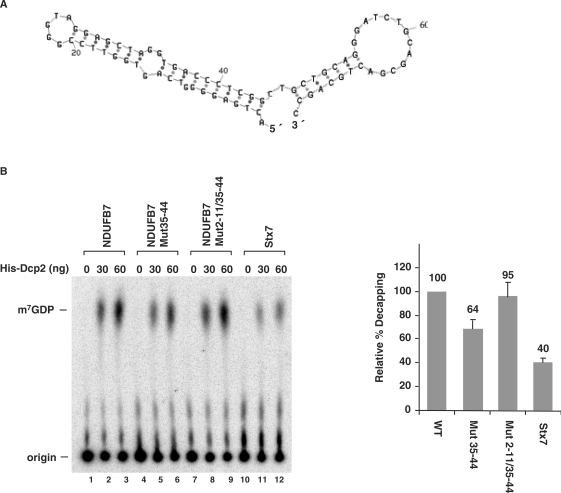
The Ndufb7 5′ UTR stem-loop structure is also capable of enhancing decapping. (A) A schematic of the Ndufb7 5′UTR predicted secondary structure is shown with the numbers denoting the nucleotides from the 5′ end. (B) Decapping assays were carried out with the indicated Ndufb7 RNAs and Stx7 negative control. WT, wild type; Mut35–44, mutations in nucleotides 35–44 to disrupt the second strand of the stem; Mut2–11/35–44, complementary mutations in both strands of the stem that maintain the same structure. Decapping efficiency quantitations for each RNA obtained from three independent experiments are shown with the decapping efficiency of wild-type Ndufb7 RNA arbitrarily set to 100. Labeling is as described in the legend to Figure 1A.
Figure 7.
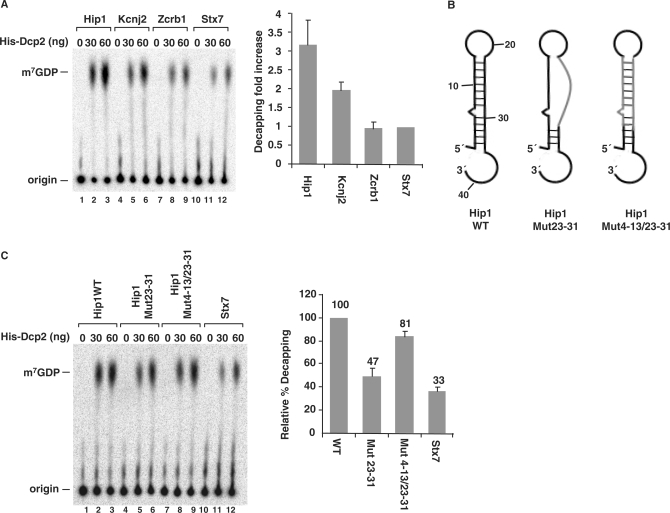
Enhancement of decapping by DBDE-like 5′ end stem-loop structure in cellular mRNAs. (A) Decapping assays were carried out with Hip1, Kcnj2 and Zcrb1 RNAs, which are all predicted to contain potential stem-loop structures at their 5′ end and the Stx7 RNA. Decapping efficiency of Stx7 RNA was arbitrarily set to one and the fold increase relative to Stx7 decapping is plotted. Quantitations were obtained from three independent experiments and the standard deviations are denoted by the error bars. (B) Schematic of the Hip1 5′ UTR wild-type and substitution mutations are shown. WT, wild type; Mut23–31, mutations in nucleotides 23–31 to disrupt the second strand of the stem; Mut4–13/23–31, complementary mutations in both strands of the stem that maintain the same structure. (C) Decapping assays were carried out with the RNAs depicted in B and resolved by PEI-TLC. Labeling is as in the legend to Figure 1A. Quantification of the decapping efficiency for each RNA obtained from three independent experiments are shown on the right with the decapping efficiency of wild-type Hip1 RNA arbitrarily set to 100.
Figure 5.
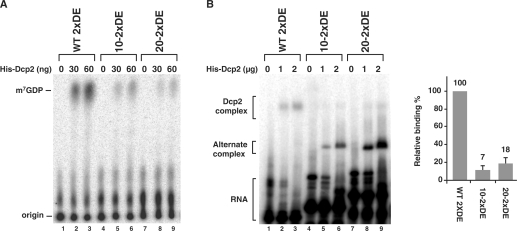
Positioning of the DBDE stem-loop structure less than 10 nucleotides from the 5′ cap is important for Dcp2-mediated enhanced decapping. (A) Decapping assays were carried out with wild type 2xDE RNA and 2xDE RNA containing 10 or 20 nucleotides of the Stx7 5′ end sequences. All RNAs were transcribed as chimeric RNAs with the coding region of Stx7 RNA attached to the 3′ end to maintain the same size as the negative control Stx7 RNA. Labeling is as described in the legend to Figure 1A. (B) Electrophoretic mobility shift assays were carried out with 32P-uniformly labeled uncapped RNAs described in A above. Binding efficiencies relative to the wild-type RNA from two experiments are presented on the right with standard deviation denoted by the error bars. Labeling is as described in the legend to Figure 1B.
In vitro RNA decapping assay
In vitro RNA decapping assays were carried out essentially as described previously (26). Reactions were carried out in 20 μl reaction volume with 25–100 ng of bacterial expressed recombinant Dcp2 protein incubated with cap-labeled RNA (2000 cpm) at 37°C for 30 min in IVDA-2 buffer (10 mM Tris–HCl at pH 7.5, 100 mM potassium acetate, 2 mM magnesium acetate, 0.5 mM MnCl2 and 2 mM dithiothreitol). An aliquot of each sample was resolved by phosphoethyleneimine-thin layer chromatography (PEI-TLC) developed in 0.45 M (NH4)2SO4 and exposed to PhosphorImager. Quantifications were carried out using a Molecular Dynamics PhosphorImager using ImageQuante-5 software. Percent decapping was determined as the level of m7GDP relative to total RNA used in the reaction.
Electrophoretic mobility shift assays
Electrophoretic mobility shift assays were carried out with in vitro transcribed [α32P]GTP uniformly labeled RNA substrate (∼4000 cpm per reaction). Binding reactions were carried out in RBB [10 mM Tris (pH 7.5), 50 mM KCl, 1.5 mM MgCl2, 0.5 mM DTT] with 1 or 2 μg of His-Dcp2 protein in a 20 µl total volume containing 1.5 μg yeast RNA and 0.25 μg heparin. Following a 25 min binding reaction at room temperature, the complexes were resolved on a 5% polyacrylamide gel (60:1 acrylamide:bis) in 0.5× Tris–borate–EDTA buffer at 8 V/cm and exposed to PhosphorImager.
RNase mapping
2xDE RNA was in vitro transcribed and incubated with Calf Intestinal Phosphatase (CIP) at 37°C for 1 h to dephosphorylate the 5′ end. A phenol/chloroform (1:1) extraction was carried out following the reaction to remove CIP. The dephosphorylated RNA was incubated with [γ-32P] ATP and T4 Polynucleotide Kinase (Ambion; Austin, TX) to label the 5′ end following the manufacturer's protocol. The 5′-end-labeled RNA (1500 cpm) was incubated with the indicated amounts of RNase A, RNase T1, RNase V1 (Ambion; Austin, TX) or alkaline hydrolysis buffer (Ambion; Austin, TX) following the manufacturer's instructions, and the digested products were resolved on a 15% polyacrylamide gel and exposed to PhosphorImager.
Bioinformatics search for Dcp2 substrates
The PatSearch program and UTR database provided by this link (http://www.ba.itb.cnr.it/BIG/PatSearch/) was used to search available human 5′ UTR sequences (27). The stem-loop secondary structure constraint criteria consisted of an 8–15-nt stem portion with either perfectly paired or one mismatch with no bulge or a one-base bulge with no mismatch and a 6–15 nt loop. G–U base pairing was also permitted in addition to Watson–Crick base pairing. The following pattern was submitted to the PatSearch website: r1 = {at,ta,gc,cg,gt,tg} (p1 = 8 … 15 6 … 15–p1[1,0,0] | p2 = 8 … 15 6 … 15–p2[0,1,1]). Human 5′ UTRs that adhered to the search criteria were returned with a proprietary reference ID. A Python script was developed to match and identify each reference ID to its gene description in the UTRdb from ftp://bighost.ba.itb.cnr.it/pub/Embnet/Database/UTR/.
RESULTS
The first 33 nt of 2xDE is critical for Dcp2 binding and decapping
In a previous study, we identified the Rrp41 mRNA as a substrate that is specifically bound and decapped by the human Dcp2-decapping protein. In particular, we identified the first 60 nt of the 5′ UTR of Rrp41 mRNA, which contained an ∼30 nt imperfect repeated sequence to be sufficient and necessary for enhancing Dcp2 decapping that was termed 2xDE (24). To further uncover the mechanism of how this element facilitates Dcp2 decapping, we made a series of mutations within this element. Every 10 or 11 successive nucleotides were mutated within the 2xDE element as indicated in Figure 1. Chimeric RNAs were in vitro transcribed harboring the various mutated 2xDE sequences placed upstream of the Stx7 mRNA sequence, which is normally a poor substrate for Dcp2 decapping (24). These RNAs were cap-labeled at the first phosphate following the methylguanosine (see ‘Materials and Methods’ section) and tested in an in vitro decapping assay with recombinant histidine-tagged Dcp2 (His-Dcp2) protein. Consistent with our previous report (24), the chimeric Stx7 RNA containing the wild-type (WT) 2xDE sequence at the 5′ end (lanes 1–3) displayed an enhancement of decapping of ∼4-fold relative to the level of decapping detected with the Stx7 RNA (Figure 1, compare lanes 1–3 to 19–21). RNAs containing mutations within the first 33 nt of the 2x-DE (nt 1–10, 11–22 and 23–33) were less efficiently decapped by Dcp2 to a level comparable to that observed with the Stx7 RNA (lanes 4–12); while mutations of nt 34–44 or 45–55 were decapped at approximately wild-type 2xDE decapping levels. These results suggest that the first 33 nt of the 2xDE are critical for enhancement of Dcp2-mediated decapping.
To address whether the reduced decapping of the mutated RNAs correlated with reduced Dcp2 binding, we tested the binding of Dcp2 to each 2xDE mutant by electrophoretic mobility shift assays. The same series of mutated 2xDE elements were in vitro transcribed and uniformly 32P labeled and incubated with His-Dcp2. The bound complex was resolved by native gel electrophoresis and as shown in Figure 1B, a specific Dcp2-RNA complex can be detected on the wild-type 2xDE RNA (lanes 2–3). A similarly migrating complex was inefficiently formed on the Stx7 control RNA (lanes 19–21). However, an alternate faster migrating band was observed (Figure 1B, lanes 19–21). The resistance of the slower migrating complex formed on the wild-type RNA to RNase T1 treatment (Figure S1, lane 4) and sensitivity of the alternate complex to RNase T1 treatment (Figure S1, lane 8) suggests the faster migrating complex is non-specific and is primarily detected in the absence of a high-affinity-binding site. Furthermore, formation of the slower migrating Dcp2 complex observed on the wild-type 2xDE RNA was also more prominent on mutants that retained the ability to stimulate decapping (Figure 1B, lanes 13–18). Interestingly, this complex was considerably diminished with the 2xDE RNAs containing substitutions within the first 33 nt that did not support stimulated decapping (lanes 4–12). These mutants were more likely to form the faster migrating alternate complex analogous to the one formed on the Stx7 RNA. Collectively, these data demonstrate that the first 33 nt of the 2x-DE are important for specific binding of Dcp2 that corresponded to enhanced decapping and appeared to constitute the functional unit of the 2xDE for Dcp2 activity.
The presence of a 5′-end stem-loop structure within the 2xDE
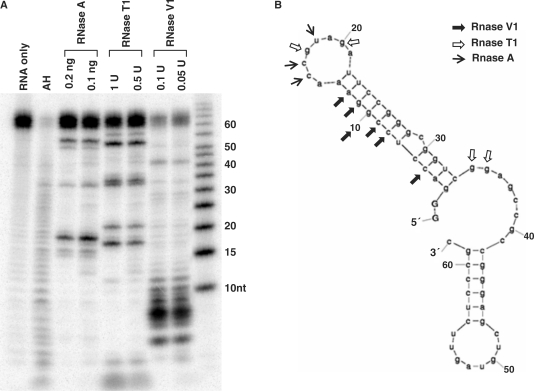
The function of an RNA element can be attributed to either its primary nucleotide sequence or its higher order structure. Examination of the 2xDE element by MFold prediction revealed that there could be a stable stem-loop structure within the critical first 33 nt region (Figure 2A). To determine whether this region can form a stable stem-loop structure, we carried out an RNase-mapping assay. The 2xDE RNA was transcribed in vitro and 32P-labeled at the 5′ terminus. The full-length RNA was gel purified and incubated with RNase A, RNase T1 or RNase V1. RNase A, which cleaves single-strand pyrimidines, showed strong cleavage of the 2xDE RNA at U18 and additional cleavages at C15 and C16 (lanes 3 and 4). RNase T1 cleaves at single-stranded guanosine residues and efficiently cleaved G17 and G20. RNase V1, which cleaves double-stranded regions of RNA, showed predominant cleavage from nt 5 to nt 12. The combined cleavage pattern of the three RNases is shown in Figure 2B and is consistent with that predicted by the Mfold program. This confirms that the 5′ end region of the 2xDE can form a stable stem-loop structure, with the first strand of the stem containing nt 3–13, second strand containing nt 22–33 and an intervening loop of eight bases in between.
Figure 2.
The first 33 nt of the 2xDE forms a stem-loop secondary structure. (A) RNase mapping of the 2xDE element. In vitro transcribed and 5′-end 32P-labeled 2xDE RNA was incubated with the indicated RNases for 15 min and resolved by a 15% polyacrylamide gel. AH designates the alkaline hydrolysis lane to generate a single nucleotide ladder. A RNA size marker is shown on the rightmost lane. (B) Deduced secondary structure of the 2xDE RNA from A. Secondary structure of the 2xDE deduced by MFold program is shown along with the indicated RNase cleavage sites denoted by arrows. The predicted secondary structure is consistent with the RNase mapping results.
The 5′ stem-loop structure facilitates Dcp2 decapping
To test whether the 5′ stem-loop structure within the 2xDE contributes to the enhanced decapping activity or whether the enhanced decapping can be attributed to the primary sequence, we generated a series of mutations that disrupted or regenerated the stem-loop structure as indicated in Figure 3A and tested the in vitro decapping activity of each mutant. As shown in Figure 3B, disruption of the stem-loop structure by mutating either the first strand (mutant A) or the second strand (mutant B) of the stem significantly decreased decapping of the RNA (lanes 4–9) to ∼20–40% of the activity of that detected with the wild-type 2xDE. To distinguish whether the decrease was due to changes in the primary or secondary sequence, a compensatory set of mutants was generated such that the primary sequence was changed but the stem-loop structure was restored (mutant C). Reconstituting the stem-loop recovered decapping to 80% of the wild-type level (lanes 10–12) indicating that the stem-loop structure and not the primary sequence within the 5′ end of 2xDE is important for Dcp2 recognition. An additional confirmation of this conclusion was provided with a mutation that flipped the two strands of the stem (mutant D), such that the mutant has the same stem-loop structure as the wild-type 2xDE but with a different primary sequence. Consistently, this mutant maintained the same level of decapping activity (lanes 13–15) as the wild-type element. Collectively, these data support the conclusion that the stem-loop structure at the 5′ end of the 2xDE element enhances Dcp2 decapping and constitutes a Dcp2 binding and preferential decapping element we will refer to as the Dcp2 binding and decapping element (DBDE).
Having demonstrated that the stem-loop structure was critical for Dcp2 recognition and decapping, we next set out to further determine parameters of the loop region and the length of the stem that can be tolerated as a substrate for Dcp2. An additional set of mutations was generated that changed the sequence within the loop region of the DBDE (Figure 4A, mutant E); shortened the loop to 4 nt (mutant F) or increased it to 12 nt (mutant G); removed the single nucleotide bulge in the stem (mutant H); or shortened the stem to 7 bp (mutant I) by deleting the lower 4 bp of the stem. The decapping activity of each mutant was tested in vitro and compared to the wild-type 2xDE. As shown in Figure 4B, the mutations in the loop region (E,F,G) had a moderate effect (20–30%) on the decapping activity of Dcp2, which suggest that nucleotides in the loop are not critical for Dcp2 recognition. The bulge in the stem appears dispensable for decapping stimulation since its removal (H) did not significantly compromise Dcp2 decapping of the RNA. Importantly, when the stem was shortened to 7 bp (I), the decapping activity significantly decreased to ∼30% of the wild-type level, suggesting a stem longer than 7 bp was required for Dcp2 recognition.
Requirement for cap proximity of the DBDE
Since the DBDE exerts a function on the 5′ cap, we reasoned proximity to the cap might be an important parameter. The stem region of the Rrp41 DBDE begins 3 nt downstream of the cap. Decapping was significantly compromised when the stem-loop was positioned either 10 or 20 nt from the 5′ terminus of the RNA by the insertion of Stx7 sequences at the 5′ end (Figure 5A, compare lanes 1–3 to lanes 4–6 and 7–9). To determine whether positioning of the DBDE relative to the 5′ cap influenced Dcp2 binding to the RNA, binding of Dcp2 to the variant RNAs was tested by an electrophoretic mobility shift assay. Consistent with the decapping data, movement of the DBDE distal to the cap, an addition 7 nt resulted in reduced formation of the slower migrating Dcp2–RNA complex as determined by the gel-shift assay (Figure 5B, compare lanes 1–3 to lanes 4–6 and 7–9). These data indicate that the position of the stem-loop structure is important for facilitating enhanced Dcp2-mediated decapping and should be less than 10 nt from the 5′ end of the RNA.
Generality of decapping enhancement by a stem-loop structure at the 5′ end of an mRNA
The dependence on structure rather than sequence of the DBDE indicates that the presence of this element at the 5′ end of other mRNAs could also promote decapping by Dcp2. As an initial approach, we first examined mRNAs that we recently demonstrated are specifically copurified with Dcp2, including the mRNA encoding Rrp41 and the Ndufb7 mRNA (24). Similar to the predicted structure of the Rrp41 mRNA, MFold predictions of the Ndufb7 mRNA also revealed a stem-loop structure at the 5′ terminus (Figure 6A). We tested the decapping activity of Dcp2 on this mRNA by an in vitro decapping assay. As shown in Figure 6B, compared to the Stx7 mRNA negative control (lanes 10–12), the Nudfb7 mRNA demonstrated a 2.5-fold enhancement of decapping activity (lanes 1–3). To test whether the predicted stem-loop structure influenced the decapping, nucleotides 35–44 in the second strand of the predicted double-stranded stem region were mutated. The RNA containing this mutation was decapped with a ∼40% reduced efficiency (lanes 4–6). However, a compensatory mutation of nt 2–11 in the first strand fully restored decapping activity (lanes 7–9). This result suggests the predicted stem-loop structure contributes to decapping of the Ndufb7 mRNA and could be a general modulator of mRNA decapping.
To further test the predictive potential of the DBDE, the human 5′UTR database was searched for mRNAs that could potentially form a 5′ stem-loop structure analogous to the DBDE. The bioinformatics search parameters included a stem region ranging from 8–15 bp and the loop ranging from 6–15 nt positioned in the proximity of the 5′ cap. Two hundred and thirty-nine mRNAs with the potential of forming a stem-loop structure starting within 10 nt of the 5′ termini (Table S2) were identified. Each mRNA was subsequently subjected to MFold prediction to determine which mRNAs are likely to form the stem-loop structure within the context of the entire 5′UTR. Three representative examples were chosen for further analysis. The Hip1 and Kcnj2 mRNA 5′ ends have the potential of forming a DBDE-like stem-loop structure and are expected to be preferentially decapped by Dcp2. The third RNA, Zcrb1, also has the propensity to form a stem-loop structure, but the stem region is expected to be positioned 10 nt from the 5′ end. Based on our analysis in Figure 5, this RNA should not be recognized as a Dcp2 substrate and is not expected to be efficiently decapped. The DNA corresponding to the first 150 nt of each mRNA was transcribed in vitro with T7 polymerase, cap labeled with 32P and tested in an in vitro decapping assay with Dcp2. As expected, the Hip1 RNA was decapped at an efficiency 3-fold greater than the Stx7 control RNA (Figure 7A, compare lanes 1–3 to 10–12). Similarly, the Kcnj2 RNA was 2-fold more efficiently decapped (lanes 4–6). Interestingly, the Zcrb1 RNA, which is predicted not to be preferentially decapped, was decapped at a level comparable to that of the Stx7 negative control (lanes 7–9).
To further substantiate that the increase in decapping observed with the Hip1 RNA was a consequence of the stem-loop structure, the putative stem structure was mutated. To minimize alterations of primary sequence adjacent to the 5′ cap, mutations were introduced in the cap distal strand of the stem at nt 23–31 (Figure 7B). As shown in Figure 7C, this mutation significantly reduced the decapping activity by about 50% (lanes 4–6). Compensatory changes in nt 2–11 designed to reform base pairing and formation of the stem also restored decapping activity to 80% of the wild-type level, indicating that the intact stem-loop structure is important for the enhancement of decapping in this RNA as well. Collectively, our data indicate that Dcp2 can recognize and preferentially decap mRNAs containing a stem-loop structure at their 5′ end.
DISCUSSION
In this study, we expand on our recent demonstration that Dcp2 can selectively bind to the 2xDE at the 5′ end of the Rrp41 mRNA (24). The binding region was identified as the first 33 nt of the 2xDE termed the DBDE, which forms a stable stem-loop structure. Importantly, the intact stem-loop secondary structure but not the primary sequence was responsible for enhanced Dcp2-directed decapping. Furthermore, a bioinformatics search for mRNAs that harbor a DBDE-like stem-loop in the 5′UTR identified a subset of mRNA with the propensity to form such a structure. Interestingly, the tested RNAs with a DBDE-like element positioned within 10 nt of the 5′ cap were preferentially decapped, indicating that the DBDE 5′ end stem-loop structure could be a general predictor of mRNAs that could be specifically regulated by Dcp2 decapping.
The 2xDE element, consisting of the first 60 nt of the Rrp41 mRNA 5′UTR, is an element that is specifically bound by Dcp2 and can enhance the decapping of a heterologous RNA when placed at the 5′ end (24). The element contains two stretches of similar sequences from nt 1–25 and 26–60, each of which was found to partially enhance Dcp2 decapping (24). We have now further refined these findings to show that the first 33 nt of the 2xDE termed DBDE are sufficient for the Dcp2 recognition and enhanced decapping (Figure 1A). The functional significance of the second element is currently unclear since its positioning ∼30 nt from the 5′ cap would be expected to not promote decapping. It was only when artificially positioned at the 5′ end of an RNA that its stimulatory effect was observed (24). Whether it can serve to augment the recruitment of Dcp2 in cells at its endogenous position 30 nt distal to the cap remains to be determined.
RNA-binding proteins can recognize their RNA substrates by either the primary sequence or the higher order structure of the RNA. Binding of the iron response element (IRE)-binding protein (IRP) to the IRE is one example of the importance of structural integrity to RNA binding. IRP binding to the ferritin mRNA 5′UTR is dependent on the IRE stem-loop structure (28). Similarly, the SECIS-binding proteins recognize a SECIS element, which is a structural motif that directs the cell to translate UGA codons as selenocysteines (29). In this study, we found that the intact stem-loop structure within the 5′ end of the Rrp41 mRNA 2xDE element is critical for promoting Dcp2 decapping. The Dcp2 protein appears to recognize the secondary structure of its target RNAs, but not the primary sequence, since alterations in the primary sequence of the stem-loop structure did not have a significant effect on Dcp2 decapping (Figure 3, mutants C and D).
The stem-loop structure within the first 33 nt of 2xDE consists of a double-stranded stem region of 11 bp with a single nucleotide bulge at position 29 and a single-stranded loop region of 8 nt. Mutational assays in the stem-loop region in the 2xDE indicated that the exact nucleic-acid sequences in the stem or in the loop region are not important for Dcp2 decapping. The single nucleotide bulge is also dispensable for Dcp2 decapping (Figure 4, mutant H). Increasing the size of the loop to 12 nt (Figure 4, mutant G) resulted in a slight increase in decapping activity, while decreasing the size of the loop to 4 nt (Figure 4, mutant F) modestly decreased the decapping activity, suggesting the size of the loop does not significantly contribute to the promotion of decapping. In contrast to the loop, the length of the stem was important to decapping. Decreasing the size of the stem region to less than 8 bp resulted in a significant reduction of decapping activity, indicating either the Dcp2 protein requires more than 7 bp to recognize the RNA or shortening the stem compromises the structural integrity of the overall stem-loop structure.
The position of the stem-loop structure relative to the 5′ end of the RNA was also found to be critical for Dcp2-mediated stimulation of decapping. The stem-loop structure functioned optimally in decapping stimulation when it was located at the 5′ end. We had previously reported that Dcp2 is a non-canonical cap binding protein that likely contains dual RNA-binding and cap-binding properties (10). Placing the stem-loop structure 10 nt downstream of the 5′ cap diminished decapping stimulation by the element and reduced its capacity to form the Dcp2 complex observed on the wild-type 2xDE RNA (Figure 5) indicating the distance between these two elements is important. However, Dcp2 is capable of binding to an RNA molecule lacking a 5′ cap, therefore, preferential binding when the stem-loop is proximal to the 5′ termini suggests a role for the 5′ terminus irrespective of the presence or absence of the cap (Figure 5). It is likely that the requirement is for a 5′ end, but not necessarily for a cap since the binding assays carried out in Figures 1B and 5B were carried out with uncapped RNA. Structural analysis of the nuclear X29 decapping enzyme revealed the m7G nucleobase of the cap is exposed to solvent and is surprisingly devoid of extensive contacts with the X29 protein (30). Whether a similar scenario exists for Dcp2 remains to be determined although its specificity for methylated capped RNA also implies direct contact with the cap.
An important observation from the binding studies in Figures 1 and 5 was the detection of two distinct Dcp2 complexes. RNAs that contained a DBDE within 10 nt of the 5′ end and were subjected to stimulatory decapping by Dcp2, formed a specific slower migrating complex compared to a faster migrating band on RNAs lacking a DBDE-like element within the first 10 nt (Figures 1B and 5B). The recent structural demonstration that Dcp2 can exist in a closed productive or open non-productive conformation (31,32) could be one explanation for our observations. The slower migrating Dcp2 complex could represent Dcp2 binding to the RNA in the closed productive confirmation while the faster migrating alternative complex represents binding in a non-productive open conformation. Sensitivity of the alternative complex to RNase treatment (Figure S1) further supports this premise. Future kinetic and structural analysis of Dcp2 in conjunction with RNA could address these possibilities.
The C. elegens splice leader (SL) RNA that functions as a donor for the trans-splicing reaction, which incorporates the 5′ end 22 nt of this mRNA onto the 5′ end of ∼70% of C. elegans mRNAs (33), is efficiently decapped by Dcp2 (34). However, mRNAs containing the 22-nt trans spliced SL sequence at the 5′ end were inefficiently decapped (34). Consistent with our study, the initial 30 nt of the SL RNA can fold into a stem-loop structure within the context of the SL RNA (35), which is likely disrupted in the trans spliced mRNAs that only contain the first 22 nt of the SL RNA. In addition, all trypanosome mRNAs contain a 39 nt spliced leader sequence at their 5′ end, and this sequence has been shown to inhibit Dcp2-like decapping activity in trypanosome extract (36). Although this region has been proposed to adopt two forms of secondary structures containing an internal stem-loop (37), the proposed stem-loop structures are more than 10 nt downstream of the 5′ cap, which might explain their poor decapping propensity.
A second decapping enzyme, the Xenopus X29 nuclear decapping enzyme, has also been shown to possess transcript-specific decapping activity. X29 specifically binds and decaps the U8 snoRNA (21,22). Similar to the binding of Dcp2 to the 2xDE, the binding of X29 to U8 snoRNA is also independent of cap structure, since X29 binds uncapped U8 RNA with the same efficiency as the capped U8 RNA (21). The 5′ end 40 nt of U8 RNA were shown to be required, but not sufficient for X29 binding (21). Interestingly, the 5′ 40 nt of U8 RNA also form a stem-loop structure (38), which may suggest the preference for stem-loop structures at the 5′ end is not restricted to Dcp2, but also adopted by other decapping proteins. However, unlike Dcp2, which requires the stem-loop to start within 10 nt from 5′ cap, the stem-loop in U8 snoRNA starts 12 nt downstream of the cap indicating some discrepancies between Dcp2 and X29.
A search of all currently available human RNA 5′ UTRs in the 5′ UTR database (see ‘Materials and Methods’ section) revealed 239 mRNAs containing the potential to form a DBDE-like structure starting within the first 10 nt from the 5′ end of the mRNA. Although the search parameters identify local regions of the RNA capable of forming the stem-loop structure, further analysis of each mRNA within the context of the entire 5′ UTR by MFold program predicted ∼20% of these mRNAs could form a stable stem-loop structure at the 5′ end. Therefore, only a small subset of mRNAs appears to have the capacity to form into an appropriate structure that would render them as direct substrates of Dcp2 decapping in cells. Of the three mRNAs, we further characterized, Hip1 RNA was the most efficiently decapped and contains a 13-nt stem with a single nucleotide bulge and a 9-nt loop region. The Kcnj2 RNA contains a 9-nt stem with one mismatch and an 8-nt loop. It had a modest 2-fold increase in decapping activity compared to the Stx7 RNA negative control. The Zcrb1 RNA contains a 9-nt stem and a 7-nt loop but was located 10 nt downstream of the cap structure and as expected was not decapped at a level greater than that observed with the negative control. These data indicate that a general stem-loop structure formed at the 5′ terminus of an RNA, of which the primary sequence and the composition may vary, is capable of promoting Dcp2 decapping. Furthermore, this feature can be used to predict potential Dcp2 target RNAs in cells. However, considering multiple positive and negative regulators of Dcp2 decapping have been reported (13), it is likely that decapping is regulated by the combined total contributions of both the direct intrinsic properties of Dcp2 to associate with the 5′ cap as well as the decapping auxiliary proteins that will dictate the consequence of Dcp2 on an mRNA. Our studies now provide a framework for future studies addressing the direct and indirect decapping activities of Dcp2 on mRNAs in cells.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Fellowship from Rutgers IGERT program on Biointerfaces from the National Science Foundation [NSF DGE 0333196 to E.S.H] and the NIH [GM57286 to S.I.G and GM67005 to M.K.]. Funding for open access charge: GM67005.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank the members of the Kiledjian lab for helpful discussions and critical reading of the manuscript.
REFERENCES
- 1.Wilusz CJ, Wormington M, Peltz SW. The cap-to-tail guide to mRNA turnover. Nat. Rev. Mol. Cell Biol. 2001;2:237–246. doi: 10.1038/35067025. [DOI] [PubMed] [Google Scholar]
- 2.Liu Q, Greimann JC, Lima CD. Reconstitution, activities, and structure of the eukaryotic RNA exosome. Cell. 2006;127:1223–1237. doi: 10.1016/j.cell.2006.10.037. [DOI] [PubMed] [Google Scholar]
- 3.Liu H, Kiledjian M. Decapping the message: a beginning or an end. Biochem. Soc. Trans. 2006;34:35–38. doi: 10.1042/BST20060035. [DOI] [PubMed] [Google Scholar]
- 4.Dunckley T, Parker R. The DCP2 protein is required for mRNA decapping in Saccharomyces cerevisiae and contains a functional MutT motif. EMBO J. 1999;18:5411–5422. doi: 10.1093/emboj/18.19.5411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lykke-Andersen J. Identification of a human decapping complex associated with hUpf proteins in nonsense-mediated decay. Mol. Cell Biol. 2002;22:8114–8121. doi: 10.1128/MCB.22.23.8114-8121.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.van Dijk E, Cougot N, Meyer S, Babajko S, Wahle E, Seraphin B. Human Dcp2: a catalytically active mRNA decapping enzyme located in specific cytoplasmic structures. EMBO J. 2002;21:6915–6924. doi: 10.1093/emboj/cdf678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang Z, Jiao X, Carr-Schmid A, Kiledjian M. The hDcp2 protein is a mammalian mRNA decapping enzyme. Proc. Natl Acad. Sci. USA. 2002;99:12663–12668. doi: 10.1073/pnas.192445599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Decker CJ, Parker R. A turnover pathway for both stable and unstable mRNAs in yeast: evidence for a requirement for deadenylation. Genes Dev. 1993;7:1632–1643. doi: 10.1101/gad.7.8.1632. [DOI] [PubMed] [Google Scholar]
- 9.Hsu CL, Stevens A. Yeast cells lacking 5′ → 3′ exoribonuclease 1 contain mRNA species that are poly(A) deficient and partially lack the 5′ cap structure. Mol. Cell Biol. 1993;13:4826–4835. doi: 10.1128/mcb.13.8.4826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Piccirillo C, Khanna R, Kiledjian M. Functional characterization of the mammalian mRNA decapping enzyme hDcp2. RNA. 2003;9:1138–1147. doi: 10.1261/rna.5690503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.She M, Decker CJ, Sundramurthy K, Liu Y, Chen N, Parker R, Song H. Crystal structure of Dcp1p and its functional implications in mRNA decapping. Nat. Struct. Mol. Biol. 2004;11:249–256. doi: 10.1038/nsmb730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Steiger M, Carr-Schmid A, Schwartz DC, Kiledjian M, Parker R. Analysis of recombinant yeast decapping enzyme. RNA. 2003;9:231–238. doi: 10.1261/rna.2151403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Coller J, Parker R. Eukarryotic mRNA decapping. Annu. Rev. Biochem. 2004;73:861–890. doi: 10.1146/annurev.biochem.73.011303.074032. [DOI] [PubMed] [Google Scholar]
- 14.Fenger-Gron M, Fillman C, Norrild B, Lykke-Andersen J. Multiple processing body factors and the ARE binding protein TTP activate mRNA decapping. Mol. Cell. 2005;20:905–915. doi: 10.1016/j.molcel.2005.10.031. [DOI] [PubMed] [Google Scholar]
- 15.Caponigro G, Parker R. Multiple functions for the poly(A)-binding protein in mRNA decapping and deadenylation in yeast. Genes Dev. 1995;9:2421–2432. doi: 10.1101/gad.9.19.2421. [DOI] [PubMed] [Google Scholar]
- 16.Khanna R, Kiledjian M. Poly(A)-binding-protein-mediated regulation of hDcp2 decapping in vitro. EMBO J. 2004;23:1968–1976. doi: 10.1038/sj.emboj.7600213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ramirez CV, Vilela C, Berthelot K, McCarthy JE. Modulation of eukaryotic mRNA stability via the cap-binding translation complex eIF4F. J. Mol. Biol. 2002;318:951–962. doi: 10.1016/S0022-2836(02)00162-6. [DOI] [PubMed] [Google Scholar]
- 18.Schwartz DC, Parker R. Mutations in translation initiation factors lead to increased rates of deadenylation and decapping of mRNAs in Saccharomyces cerevisiae. Mol. Cell Biol. 1999;19:5247–5256. doi: 10.1128/mcb.19.8.5247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wilusz CJ, Gao M, Jones CL, Wilusz J, Peltz SW. Poly(A)-binding proteins regulate both mRNA deadenylation and decapping in yeast cytoplasmic extracts. RNA. 2001;7:1416–1424. [PMC free article] [PubMed] [Google Scholar]
- 20.Jiao X, Wang Z, Kiledjian M. Identification of an mRNA-decapping regulator implicated in X-linked mental retardation. Mol. Cell. 2006;24:713–722. doi: 10.1016/j.molcel.2006.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ghosh T, Peterson B, Tomasevic N, Peculis BA. Xenopus U8 snoRNA binding protein is a conserved nuclear decapping enzyme. Mol. Cell. 2004;13:817–828. doi: 10.1016/s1097-2765(04)00127-3. [DOI] [PubMed] [Google Scholar]
- 22.Tomasevic N, Peculis B. Identification of a U8 snoRNA-specific binding protein. J. Biol. Chem. 1999;274:35914–35920. doi: 10.1074/jbc.274.50.35914. [DOI] [PubMed] [Google Scholar]
- 23.Peculis BA, Reynolds K, Cleland M. Metal determines efficiency and substrate specificity of the nuclear nudix decapping proteins X29 and H29K(NUDT16) J. Biol. Chem. 2007;282:24792–24805. doi: 10.1074/jbc.M704179200. [DOI] [PubMed] [Google Scholar]
- 24.Li Y, Song MG, Kiledjian M. Transcript-specific decapping and regulated stability by the human Dcp2 decapping protein. Mol. Cell Biol. 2008;28:939–948. doi: 10.1128/MCB.01727-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang Z, Kiledjian M. Functional link between the mammalian exosome and mRNA decapping. Cell. 2001;107:751–762. doi: 10.1016/s0092-8674(01)00592-x. [DOI] [PubMed] [Google Scholar]
- 26.Wang Z, Day N, Trifillis P, Kiledjian M. An mRNA stability complex functions with poly(A)-binding protein to stabilize mRNA in vitro. Mol. Cell Biol. 1999;19:4552–4560. doi: 10.1128/mcb.19.7.4552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grillo G, Licciulli F, Liuni S, Sbisa E, Pesole G. PatSearch: a program for the detection of patterns and structural motifs in nucleotide sequences. Nucleic Acids Res. 2003;31:3608–3612. doi: 10.1093/nar/gkg548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hentze MW, Kuhn LC. Molecular control of vertebrate iron metabolism: mRNA-based regulatory circuits operated by iron, nitric oxide, and oxidative stress. Proc. Natl Acad. Sci. USA. 1996;93:8175–8182. doi: 10.1073/pnas.93.16.8175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Walczak R, Westhof E, Carbon P, Krol A. A novel RNA structural motif in the selenocysteine insertion element of eukaryotic selenoprotein mRNAs. RNA. 1996;2:367–379. [PMC free article] [PubMed] [Google Scholar]
- 30.Scarsdale JN, Peculis BA, Wright HT. Crystal structures of U8 snoRNA decapping nudix hydrolase, X29, and its metal and cap complexes. Structure. 2006;14:331–343. doi: 10.1016/j.str.2005.11.010. [DOI] [PubMed] [Google Scholar]
- 31.Deshmukh MV, Jones BN, Quang-Dang DU, Flinders J, Floor SN, Kim C, Jemielity J, Kalek M, Darzynkiewicz E, Gross JD. mRNA decapping is promoted by an RNA-binding channel in Dcp2. Mol. Cell. 2008;29:324–336. doi: 10.1016/j.molcel.2007.11.027. [DOI] [PubMed] [Google Scholar]
- 32.She M, Decker CJ, Svergun DI, Round A, Chen N, Muhlrad D, Parker R, Song H. Structural basis of dcp2 recognition and activation by dcp1. Mol. Cell. 2008;29:337–349. doi: 10.1016/j.molcel.2008.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Blumenthal T. Trans-splicing and polycistronic transcription in Caenorhabditis elegans. Trends Genet. 1995;11:132–136. doi: 10.1016/s0168-9525(00)89026-5. [DOI] [PubMed] [Google Scholar]
- 34.Cohen LS, Mikhli C, Jiao X, Kiledjian M, Kunkel G, Davis RE. Dcp2 Decaps m2,2,7 GpppN-capped RNAs, and its activity is sequence and context dependent. Mol. Cell Biol. 2005;25:8779–8791. doi: 10.1128/MCB.25.20.8779-8791.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Denker JA, Maroney PA, Yu YT, Kanost RA, Nilsen TW. Multiple requirements for nematode spliced leader RNP function in trans-splicing. RNA. 1996;2:746–755. [PMC free article] [PubMed] [Google Scholar]
- 36.Milone J, Wilusz J, Bellofatto V. Identification of mRNA decapping activities and an ARE-regulated 3′ to 5′ exonuclease activity in trypanosome extracts. Nucleic Acids Res. 2002;30:4040–4050. doi: 10.1093/nar/gkf521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Harris KA, Jr, Crothers DM, Ullu E. In vivo structural analysis of spliced leader RNAs in Trypanosoma brucei and Leptomonas collosoma: a flexible structure that is independent of cap4 methylations. RNA. 1995;1:351–362. [PMC free article] [PubMed] [Google Scholar]
- 38.Reddy R, Henning D, Busch H. Primary and secondary structure of U8 small nuclear RNA. J. Biol. Chem. 1985;260:10930–10935. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.