Abstract
The helicase loader protein DnaI (the Bacillus subtilis homologue of Escherichia coli DnaC) is required to load the hexameric helicase DnaC (the B. subtilis homologue of E. coli DnaB) onto DNA at the start of replication. While the C-terminal domain of DnaI belongs to the structurally well-characterized AAA+ family of ATPases, the structure of the N-terminal domain, DnaI-N, has no homology to a known structure. Three-dimensional structure determination by nuclear magnetic resonance (NMR) spectroscopy shows that DnaI presents a novel fold containing a structurally important zinc ion. Surface plasmon resonance experiments indicate that DnaI-N is largely responsible for binding of DnaI to the hexameric helicase from B. stearothermophilus, which is a close homologue of the corresponding much less stable B. subtilis helicase.
INTRODUCTION
DNA replication in all organisms is carried out by replisomes, multiprotein machines that contain a ring-shaped DNA helicase for separation of the strands of the DNA duplex (1). In bacteria, the helicase forms a hexameric structure that encircles single-stranded (ss) DNA. Several other replication proteins bind to the helicase and its loading onto DNA is one of the earliest steps in replisome assembly (2). In Escherichia coli, loading of the DnaB helicase hexamer onto ssDNA is assisted by DnaC, a loader protein that associates with it to form a ring of six DnaC molecules on one face of the helicase ring (3). In Bacillus subtilis and some other Gram positive bacteria such as Staphylococcus aureus, the corresponding hexameric helicase is named DnaC and the helicase loader protein is DnaI. In contrast to E. coli DnaB, which is loaded onto ssDNA by ring opening (4,5), the B. subtilis helicase dissociates more readily into monomers and the role of DnaI is to assist its assembly on the ssDNA (6). This process is assisted in B. subtilis by a pair of DNA-remodelling co-loader proteins (called DnaB and DnaD), which guide the DnaC/DnaI complex to specific sites in the DNA (7–13). However, in vitro and in the presence of ATP, DnaI alone is sufficient for loading of the helicase onto ssDNA (6,14).
The B. subtilis helicase and its loader form a complex of six helicase and six DnaI molecules in analogy to the E. coli DnaB/DnaC complex (6). The amino-acid sequences of the helicase loaders from the two species indicate the presence of nucleotide-binding sites, in agreement with the requirement of ATP for helicase loading (1,15–17). In addition, experiments with DnaI (14) showed that it consists of two structured domains. The larger C-terminal AAA+ domain (1,2,16) contains the nucleotide-binding site and a cryptic site for ssDNA binding, whereas the N-terminal domain (here termed DnaI-N) seems to be primarily responsible for helicase binding and acts as a molecular switch that regulates the accessibility of the ssDNA-binding site in the C-terminal domain (14). DnaI is a larger protein than E. coli DnaC (36 versus 28 kDa) and DnaI-N is unrelated in sequence to the N-terminal portion of the E. coli homologue (15). As the B. subtilis DnaC helicase is a less stable protein that can exist as a mixture of oligomeric forms, its interactions with DnaI have mostly been inferred using the well-behaved stable hexameric helicase from B. stearothermophilus (in the following referred to as Bst DnaB), which shares 82% sequence identity and 92% similarity with the B. subtilis helicase.
The interaction between DnaI and Bst DnaB depends on the ability of DnaI to bind zinc (14). The zinc-binding site is located in DnaI-N and is lost in the mutants C67A, C70A and H84A. Remarkably, single mutations of the other two cysteine residues, C76 and C101, did not completely abolish zinc binding or helicase interaction, suggesting that side chains of these two residues might substitute for each other as zinc ligands.
To date, very little is known of the structural basis of interactions of helicase loaders with their helicase partners, and this has limited understanding of the mechanisms of helicase loading. Although no complete structure of any helicase loader has been determined to date at atomic resolution, the structure of the AAA+ domain of Aquifex aeolicus DnaC, missing the N-terminal helicase-binding domain, has recently been reported (2). Its structure shows association of domains as a helical filament that suggests a mechanism for association of the loader proteins with the DnaA replication initiator protein at chromosomal origins of replication to accomplish recruitment of the helicase (2).
The structures of several related hexameric DnaB-family helicases reported recently (18–21) all reveal unusual domain arrangements where the N-terminal primase-interacting domains form a trimer of dimers stacked on top of a hexamer of C-terminal RecA-like helicase domains. The only reported structural information on a helicase/loader protein complex comes from low-resolution single particle analysis of negative stain (3) and cryo-electron (22) microscopic images of the E. coli DnaB/DnaC complex, which revealed a ring of DnaC molecules stacked on the C-terminal face of DnaB, which in the presence of ATP results in closing of the central ssDNA-binding channel (22).
Here we present the first report of the 3D structure of the helicase-binding domain of a replicative helicase loader, the N-terminal domain of B. subtilis DnaI, and identify its zinc-coordinating residues. In addition, experiments have been conducted to delineate the boundary between the N- and C-terminal domains of DnaI to define the minimal domain required for binding to the Bst helicase.
MATERIALS AND METHODS
Expression and purification of DnaI and Bst DnaB
The production of full-length DnaI has been described earlier (14). The procedure used in the present work is provided in the Supplementary Data. The DnaI-C76A mutant protein (14) and the B. stearothermophilus DnaB helicase were prepared as described (18), using an expression strain (15) provided by Dr Panos Soultanas (University of Nottingham, UK). Concentrations of all proteins were determined spectrophotometrically at 280 nm, using calculated values of ε280 (23).
Limited proteolysis and characterization of proteolytic fragments
Limited proteolysis experiments were done for 30 min at 30°C with solutions of 126 μg of full-length DnaI in 60 μl of storage buffer (50 mM Tris–HCl, pH 7.6, 10% (w/v) glycerol, 2 mM dithiothreitol, 1 mM EDTA) using a DnaI:trypsin ratio of 125:1. Additional experiments were conducted in the presence of 3 mM ATP, 12 mM MgCl2, or both. The products were analyzed by 12% SDS–PAGE. The gels were washed in water before selected bands were excised. N-terminal sequences of fragments were determined by Edman degradation (Biomolecular Resource Facility, Australian National University).
Construction of the expression system for DnaI-N106
Plasmid pND876, which contains the dnaI gene from pET-15b-dnaI (7) under transcriptional control of strong bacteriophage λ promoters in plasmid vector pND706 (24), was used as template for the PCR amplification of the dnaI deletion mutant with primer 5′-GAGATATACATATGGAACCAATC-3′ incorporating an ATG initiation codon (italicized) as part of an NdeI site (underlined). Primer 5′-AAAAACGCGTTACTTCCGTTTGACTGGACATTCG-3′ was complementary to the 3′ end of the dnaI fragment (MluI sites underlined; complement of stop codon italicized). The PCR product was digested with NdeI and MluI and inserted into vector pND706 between the same restriction sites to yield pKL1272. The gene encoding the dnaI mutant [coding for the N-terminal 106 residues of DnaI, DnaI-N106 (Figure 1)] is under strict control of tandem heat-inducible phage λ promoters pR and pL. The plasmid pKL1272 was transformed into the E. coli strains AN1459 (25) and BL21(λDE3)recA (26) for protein expression in LB or 15N-enriched media, respectively.
Figure 1.
Alignment of the amino-acid sequence of the N-terminal domain of DnaI from B. subtilis with homologous N-terminal regions of different bacterial helicase loaders. The following sequences are shown (organism, abbreviation, GenBank number): B. subtilis, Bsu, NP_390776; B. amyloliquefaciens, Bam, YP_001422191; B. licheniformis Bli, YP_092605; B. pumilus, Bpu, YP_001487763; Geobacillus sp., Gsp, ZP_02915066; Geobacillus kaustophilus, Gka, YP_148574; Geobacillus thermodenitrificans, Gth, YP_001126735; B. species SG-1, Bsp, ZP_01861306; B. cereus subsp. cytotoxis, Bcs, YP_001376478; B. thuringiensis serovar israelensis, Bts, ZP_00742835; B. halodurans, Bha, NP_244010; B. clausii, Bcl, YP_176196; B. selenitireducens Bse, ZP_02170630; Listeria welshimeri serovar, Lws, YP_849770; Lactobacillus helveticus, Lhe, YP_001577829; L. monocytogenes, Lmo, ZP_02289641; Enterococcus faecium, Efa, ZP_00604060; S. haemolyticus, Sha, YP_253156 and S. aureus, Sta, NP_646444. Residues 1–106 of B. subtilis DnaI correspond to the DnaI-N106 domain for which the 3D structure was determined by NMR spectroscopy. An extended version comprising residues 1–123 (DnaI-N123) was also investigated in the present study. The alignment includes sequences with high homology to B. subtilis DnaI-N106 as identified in a BLAST (47) search. The residue numbers and regular secondary structure elements of B. subtilis DnaI-N106 are indicated at the top. Identically conserved residues are highlighted in black. Residues with <10% side-chain solvent accessibility in the 3D structure are highlighted in blue. Four asterisks at the bottom identify the zinc-binding residues of B. subtilis DnaI.
Expression and purification of DnaI-N106
Cells from the E. coli strain AN1459/pKL1272 were grown in 4 l of LB medium containing thymine (25 mg/l) and ampicillin (50 mg/l) at 30°C to A595 = 0.61. A rapid temperature shift to 42°C induced the overproduction of DnaI-N106, and growth was continued for a further 3 h (final A595 = 1.18). The cells (5.05 g) were harvested at 11 000 × g for 6 min, frozen in liquid N2 and stored at –70°C. All purification steps following cell growth were carried out at 0–4°C. Thawed cells were resuspended in 76 ml of lysis buffer (50 mM Tris–HCl, pH 7.6, 10% w/v sucrose, 100 mM NaCl, 2 mM dithiothreitol and 10 mM spermidine) and lysed using a French press at 12 000 psi. Proteins in the soluble fraction after centrifugation (39 000 × g, 1 h) were precipitated by addition of ammonium sulfate (0.4 g/ml). After stirring for 1 h, the suspension was centrifuged (39 000 × g, 45 min); the pellet was dissolved in 30 ml of buffer A (50 mM Tris–HCl, pH 7.6, 10% w/v glycerol, 2 mM dithiothreitol, 40 mM NaCl) and dialysed against two changes (1 l each) of buffer A. The protein was applied to a Toyopearl DEAE-TSK650M column (2.5 × 14 cm) pre-equilibrated in buffer A. The column was washed with 100 ml of buffer A at a flow rate of 1 ml/min. DnaI-N106 eluted in the flow-through and solid ammonium sulfate (8.1 g) was added to the combined fractions (18 ml). The suspension was stirred for 1 h, and the pellet after centrifugation (39 000 × g, 45 min) was dissolved in 4 ml of buffer B (50 mM Tris–HCl, pH 7.6, 10% w/v glycerol, 2 mM dithiothreitol, 100 mM NaCl). The solution was applied to a column (2.5 × 62 cm) of Sephadex G50 gel filtration resin pre-equilibrated in buffer B. The protein was eluted at a flow rate of 0.5 ml/min and the pooled fractions (21 ml) yielded 15 mg of purified DnaI-N106. The purification was monitored by 15% SDS–PAGE. The identity of the protein was confirmed by ESI-MS (VG Quattro II triple quadrupole mass spectrometer) using a 0.1% formic acid solution with 1 mM 2-mercaptoethanol. The mass observed for unlabeled DnaI-N106 was 12 524 ± 1 Da (predicted 12 525 Da), indicating presence of the N-terminal methionine.
15N-labeled DnaI-N106 was produced using the E. coli strain BL21(λDE3)recA/pKL1272 in 15N-enriched medium (Silantes, Munich, Germany), yielding 2.74 g of cells from a 1 l culture. The purification process was as described above, except that the gel filtration step was replaced by chromatography over an 8 ml Mono-Q column (GE Healthcare; flow rate 1.5 ml/min) after dialysis against buffer C (50 mM Tris–HCl, pH 7.6, 2 mM dithiothreitol) containing 70 mM NaCl. 15N-labeled DnaI-N106 eluted at about 110 mM NaCl in a linear gradient (240 ml) of 70–150 mM NaCl in buffer C. The yield of purified 15N-labeled protein was 13 mg l−1 of culture.
Expression and purification of DnaI-N123 and DnaI-N132
C-terminally His6-tagged DnaI-N132 was purified as described previously (14). The DnaI-N123 construct was made using the QuikChange site-directed mutagenesis kit (Stratagene) by introduction of a TAA stop codon after that encoding Gln123 in the pET22b-dnaI plasmid described by Soultanas (14). The primers used were 5′-GCATGTATATCCAGTAAGATCTTCTTGGAG-3′ and its complement (stop codon italicized). This strategy resulted in an untagged protein that was purified in the same way as DnaI-N106.
NMR sample preparation
NMR samples were prepared by dialysis of DnaI-N proteins against NMR buffer (10 mM sodium phosphate pH 7.0, 0.1% (w/v) sodium azide, 100 mM NaCl and 1 mM dithiothreitol). Following concentration to 0.5 ml by ultrafiltration (Amicon Ultrafree-4 centrifugal filter device with a 5000 molecular weight cutoff), D2O was added to 10% (v/v). The final NMR samples contained 1.1 mM unlabeled and 0.5 mM 15N-labeled DnaI-N106.
NMR measurements
All NMR experiments were carried out at 25°C using Bruker Avance 600 and 800 MHz NMR spectrometers equipped with TCI cryoprobes. The backbone resonance assignment was obtained from the analysis of a 3D NOESY-15N-HSQC spectrum [60 ms mixing time, t1max(1H) = 20 ms, t2max(15N) = 13 ms, t3max = 106 ms] together with 3D HNHA and 2D NOESY spectra. The side-chain resonances were assigned using NOESY, TOCSY and DQF-COSY spectra. A 15N-HSQC spectrum with the INEPT delays set to 20 ms yielded correlations via 2JHN couplings of the side chain of His84. NOE restraints were collected from the 3D NOESY-15N-HSQC spectrum, 2D NOESY spectra of the 15N-labeled and unlabeled samples in 90% H2O/10% D2O solution (60 and 40 ms mixing time, respectively) and a 2D NOESY spectrum in D2O solution (40 ms mixing time).
Resonance assignments of DnaI-N123 were obtained from 3D HNCA, HN(CO)CA and CBCA(CO)NH spectra using 15N/13C doubly labeled protein, prepared as above following growth of cells in 15N/13C-enriched medium (Silantes).
Structure calculations
The cross-peaks in the NOESY spectra of DnaI were assigned and integrated using the program XEASY (27). Stereospecific resonance assignments were obtained for 21 CβH2 groups, 10 CγH2 groups, 1 CδH2 group, 5 isopropyl groups and 14 side-chain amides. The stereospecific resonance assignments were determined by using the HABAS and GLOMSA routines implemented in the program CYANA (28), including J-coupling information from HNHA, COSY and TOCSY spectra to support the stereospecific assignment of CβH2 groups. The NMR structure calculations using CYANA (28) involved eight iterations of automatic NOE assignment using the routine CANDID (29) followed by a simulated annealing procedure starting from 100 random conformers that were annealed in 20 000 steps using torsion-angle dynamics. The 20 conformers with the lowest residual restraint violations were energy minimized in a shell of water using the protocol re_h2o.inp (30) and the program CNS (31) with standard parameters.
Table 1 shows an overview of the restraints used and structural statistics. Secondary structure elements and root mean square deviation (r.m.s.d.) values were calculated using the program Molmol (32). The chemical shifts and coordinates of the structure have been deposited in the BioMagResBank (accession code 15‱926) and PDB (accession code 2K7R).
Table 1.
Structural statistics for the NMR conformers of DnaI-N106
| Parameter | Value |
|---|---|
| Number of uniquely assigned NOE cross-peaks | 2957 |
| Number of non-redundant NOE upper-distance limits | 2081 |
| Number of scalar coupling constants (HN-Hα)a | 85 |
| Number of dihedral-angle restraintsb | 524 |
| Number of distance restraints for zinc ionc | 13 |
| Intra-protein energy (kcal/mol)d | −4111.6 ± 98.2 |
| Maximum NOE-restraint violations (Å) | 0.30 ± 0.08 |
| Maximum dihedral-angle restraint violations (o) | 3.50 ± 1.01 |
| r.m.s.d. to the mean for N, Cα and C′ (Å)e | 0.78 ± 0.16 |
| r.m.s.d. to the mean for all heavy atoms (Å)e | 1.27 ± 0.15 |
| Ramachandran plot appearancef | |
| Most favoured regions (%) | 86.0 |
| Additionally allowed regions (%) | 14.0 |
| Generously allowed regions (%) | 0.0 |
| Disallowed regions (%) | 0.0 |
aFrom the 3D HNHA experiment.
bEstimated from chemical shift data using the program TALOS (43) (105 restraints), and using the grid search algorithm HABAS (44) (419 restraints).
c3 Zn – Cys Sγ (2.4 Å), 1 Zn – His Nε2 (2.05 Å), 3 Zn – Cβ (3.5 Å), 3 Cys Sγ – Cys Sγ (3.85 Å), 3 Cys Sγ – His Nε2 (3.85 Å). No angle restraints were used.
dPARALLHDG force field (45).
eFor residues 20–104.
fFrom PROCHECK-NMR (46).
Thiol titrations with DTNB
Samples of full-length DnaI, DnaI-C76A and DnaI-N123 were dialyzed extensively under N2 at 4°C against 50 mM Tris–HCl pH 8.0, 0.1 M NaCl. Reactions with 5,5′-dithiobis(2-nitrobenzoic acid) (DTNB, Sigma) were followed in matched quartz cuvettes in a UV-1700 UV-visible spectrophotometer (Shimadzu) at 23°C. The concentration of protein (∼5 μM, in 580 μl of buffer) was first determined from a UV spectrum before a 10 mM solution of DTNB (15–20 μl) was added to initiate the reaction. The release of 5-thio-2-nitrobenzoate anion was monitored spectrophotometrically at 412 nm [ε412 = 14 150 M−1 cm−1; (33)] until no further change occurred (30–40 min).
DnaI-N/Bst DnaB interaction analysis
Surface plasmon resonance (SPR) experiments were conducted in a BIAcore T100 instrument operating at 20°C at a flow rate of 5 μl/min, using streptavidin (SA) chips. Two different surfaces were used. For one, 3′-biotinylated oligonucleotide dT35, synthesized by GeneWorks (Adelaide, Australia), was immobilized on one flow cell by injection of a 20 nM solution for 19 min, which resulted in an increase of 600 response units (RU). For the other, a 100 nM solution of N-terminally biotinylated DnaI-N-peptide (DnaI residues 1–18, preceded by biotin-Ser-Gly, synthesized at the Biomolecular Resource Facility, Australian National University) was injected for 90 s, producing a 730 RU response. In each case, another flow cell remained unmodified and served as a reference. The buffer used for all experiments was 10 mM HEPES pH 7.5, 200 mM NaCl, 3 mM EDTA, 0.05% surfactant P20, 10 mM MgCl2 and 2.7 mM ATP; protein samples were prepared by direct dilution into this buffer. The interaction of Bst DnaB with the three DnaI-N domains and an unmodified peptide corresponding to residues 1–18 of DnaI (MEPIGRSLQGVTGRPDFQ) was probed by immobilization of Bst DnaB (365 RU) on dT35 and subsequent injection (120 s) of increasing concentrations of DnaI-N. The DnaI-N domains were left to dissociate in buffer for 180 s. When necessary, the flow cells were regenerated by three consecutive injections of 1 M NaCl, 50 mM NaOH (1 min) followed by one injection (1 min) of 1 M MgCl2.
The interaction of Bst DnaB (concentrations up to 200 nM, as monomer) with immobilized biotinylated DnaI-N-peptide was tested under the same conditions. The involvement of this peptide in interaction with Bst DnaB was also investigated by a competition assay where the unmodified peptide (DnaI residues 1–18) was mixed (at concentrations of 0, 20 or 50 μM) with DnaI-N106 (10 μM), DnaI-N123 (10 μM) or DnaI-N132 (5 μM) prior to injection over the dT35-Bst DnaB surface.
The equilibrium dissociation constants (KD) of all complexes, and standard errors, were derived from steady state data by least squares fits to a 1:1 interaction model. Before fitting, binding data were corrected by subtraction of the reference response due to simple refractive index changes and the small changes in equilibrium responses due to dissociation of Bst DnaB during data collection. No attempt was made to normalize data from separate injections to correct for dissociation of <5% of Bst DnaB over the course of each experiment.
RESULTS
Protein constructs
Limited proteolysis of DnaI with trypsin under native conditions showed a small number of distinct fragments corresponding to cleavage after Arg105, Lys111, Lys112 and Lys118 (Supplementary Figures S1 and S2). In the presence of ATP and MgCl2, the preferred cleavage site shifted from Lys118 to Lys111, confirming that ATP binds to DnaI and may cause conformational changes. To probe the C-terminal boundary of the N-terminal domain of DnaI, the constructs comprising residues 1–106, 1–123 and 1–132 (referred to as DnaI-N106, DnaI-N123 and DnaI-N132, respectively) were prepared, with a His6 tag at the C-terminal end of the DnaI-N132 domain. [Limited proteolysis with subtilisin indicated a cleavage site after residue 132 (14).] TOCSY spectra showed the best spectral quality for DnaI-N106, whereas the longer domains gave broader resonances indicative of aggregation. Subsequently, 15N-labeled samples were prepared of DnaI-N106 and DnaI-N123, and a 13C/15N-labeled sample of DnaI-N123.
NMR resonance assignments
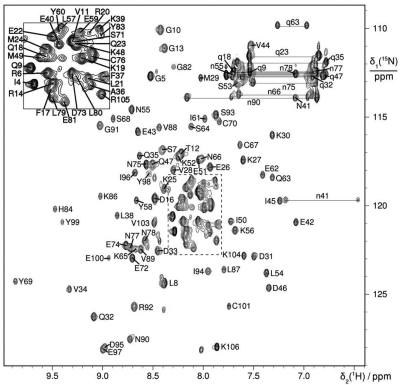
The 15N-HSQC spectrum of DnaI-N106 presented well-dispersed resonances characteristic of a monomeric folded protein (Figure 2). Virtually complete resonance assignments were obtained, except for the backbone amide of Glu2, the CεH3 group of Met1, the γ–, δ–, and ε– protons of Lys65, the CεH2 group of Lys106 and Hγ of Leu87. In the course of the 3D structure determination, stereospecific resonance assignments were obtained for 21 pairs of CβH2, five pairs of CγH2 and one pair of CδH2 protons as well as five pairs of γ-and δ-methyl groups of Leu and Val, five pairs of γ1 methylene protons of Ile, and all side-chain NH2 protons of Gln and Asn.
Figure 2.
15N-HSQC spectrum of B. subtilis DnaI-N106. The spectrum was recorded at 25°C with a 0.5 mM solution of uniformly 15N-labeled DnaI-N106 at pH 7.0. The cross-peaks are assigned using one-letter amino-acid symbols and residue numbers. Side-chain resonances are marked by small characters and cross-peaks belonging to the same NH2 group are connected by horizontal lines.
A TOCSY spectrum of DnaI-N132 indicated narrow resonances for about ten of the C-terminal residues in addition to the His6 tag, indicating increased mobility and, therefore, no participation in the structured part of the N-terminal domain (data not shown). We therefore constructed the DnaI-N123 domain, expecting it to be more fully structured.
However, resonance assignments of DnaI-N123 were much more difficult than for DnaI-N106 because the signals were broad. Only backbone resonance assignments were attempted using a 0.9 mM 13C/15N-labeled sample. Three-fold dilution did not improve the line widths of the NMR resonances, indicating that complete dissociation of the aggregates was not possible at the concentrations required for NMR. Most of the amide chemical shifts observed for this sample were the same as for DnaI-N106. For the segment with residues 107–123, tentative resonance assignments were obtained for only nine amino-acid residues.
Zinc-binding site
NMR spectroscopy cannot be used to determine directly the location of the zinc ion, so we confirmed the identity of zinc ligands by independent methods. DnaI-N106 contains one histidine, four cysteines (Figure 1) and one zinc ion (14), and all of the cysteine residues of full-length DnaI are in the DnaI-N106 domain. The 15N chemical shift of the Nε2 resonance of His84 (221 ppm; Supplementary Figure S3) was characteristic of a zinc-bound imidazole nitrogen (34), confirming that the side chain of His84 is one of the zinc ligands (14). In addition, there was no sign of structural heterogeneity, indicating that the zinc ion was present in all protein molecules and in the same coordination environment. The affinity of DnaI-N for zinc was high, as treatment of a sample with 4 mM EDTA did not alter the appearance of the 15N-HSQC spectra. Thiol titrations of full length DnaI in triplicate with DTNB (0.32 mM, at pH 8.0) gave biphasic pseudo first-order kinetics, with one exposed thiol group titrating quickly (kobs = 0.09 − 0.16 s−1), and three others about 50-fold more slowly; kobs = (1.9 – 2.7) × 10−3 s−1. Near identical stoichiometries and rate constants were observed for titration of DnaI-N123. The slowly reacting cysteines are presumed to be protected from reaction by being coordinated to zinc, and to titrate simultaneously as the metal-binding site is destroyed. In contrast, titration of the DnaI-C76A mutant protein with 0.25 mM DTNB under similar conditions showed only single-phase kinetics, with three thiols titrating slowly; kobs = (0.5–0.7) × 10−3 s−1. These data thus clearly identify His84 as a zinc ligand and Cys76 as the exposed faster-reacting cysteine residue, and suggest that the other three cysteine thiols are coordinated to the zinc ion. These conclusions were confirmed by 3D structure determination, as described below.
Structure of DnaI-N106
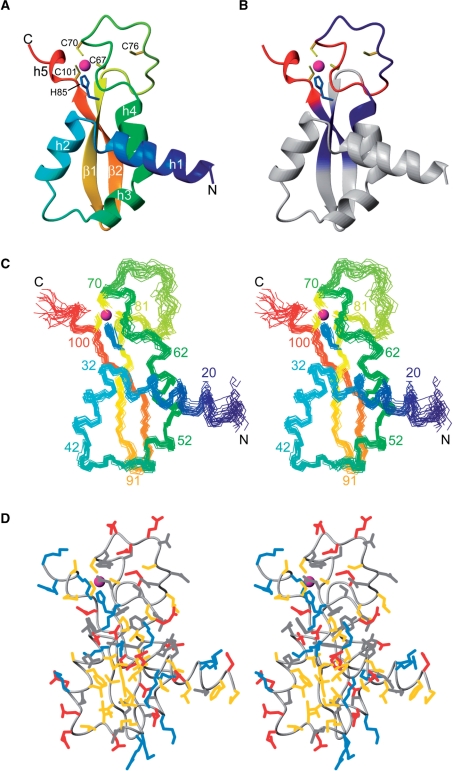
The structure of DnaI-N106 comprises four α helices and two β strands. In addition, a short 310 helix near the C-terminus was found in most of the NMR conformers (Figure 3A). The structure presents a novel fold, as a search of the protein data bank using the program Dali (35) failed to reveal a domain of similar structure. The zinc ion is coordinated by Cys67, Cys70, His84 and Cys101 (Figure 3A), whereas the side chain of Cys76 is located in the disordered loop between helix 4 and the first β strand (Figure 3 and Supplementary Figure S4). The zinc ion probably plays an important structural role, as it ties together three sequentially distant segments of the polypeptide chain in a region where the structure has few hydrophobic residues (Figure 3D). In agreement with the zinc-coordinating residues found in the present NMR study, Cys76 is not conserved in B. amyloliquefaciens and B. pumilus, while all the other zinc-coordinating residues are (Figure 1). Nonetheless, some of the DnaI homologues from other bacteria show no evidence of a zinc-binding site. In those proteins, the polypeptide segment between helix 4 and the first β strand is shorter which probably stabilizes the structure. Where this segment is longer, as in the homologue from S. aureus, it cannot be excluded that zinc binds at a similar site using a different set of cysteine and histidine residues (Figure 1).
Figure 3.
Solution structure of B. subtilis DnaI-N106. Only residues 15–106 are shown since the initial 14 residues were disordered. The zinc atom is shown as a sphere (magenta) and the side chains of coordinating residues Cys67, Cys70, His84 and Cys101 are shown in yellow (cysteine residues) and blue (histidine). (A) Ribbon representation of DnaI-N106. The secondary structure elements are labeled as in Figure 1. Helix 5 is a 310 helix found in most but not all of the NMR conformers. (B) Same as (A), but color coded to reflect the changes in backbone amide chemical shifts observed in DnaI-N106 versus those in DnaI-N123 (blue: very small chemical shift changes >0.015 ppm (1H) or >0.15 ppm (15N); red: significant chemical shift changes >0.05 ppm (1H) or >0.5 ppm (15N). (C) Stereo view of a superposition of the backbone atoms of 20 NMR conformers. (D) Stereo view of a heavy-atom representation of the conformer closest to the mean structure of DnaI-N (only residues 15–106 are shown). The side-chains are color coded in blue (Lys, Arg, His), red (Asp, Glu), yellow (Ala, Cys, Ile, Leu, Met, Phe, Pro, Trp, Val) and gray (Asn, Gln, Ser, Thr, Tyr). The figure was prepared using the program Molmol (48).
The N-terminal 14 residues were disordered. As expected for a mobile polypeptide segment, the NMR signals of the N-terminal residues were also significantly narrower than those of the structurally defined part of the protein. Similarly, the 1H NMR signals of residues 63–82 in the loop between helix 4 and the first β strand showed a reduced line width.
Conceivably, the loop residues following helix 4 could assume a single, rigid conformation in the presence of the C-terminal ATP-binding domain. Although a detailed analysis of the longer construct DnaI-N123 was hampered by broad line widths, significant chemical shift changes observed for the loop residues suggests an interaction with the segment following the C-terminus of DnaI-N106 (Figure 3B). It cannot be excluded, however, that the chemical shift change is non-specific, reflecting spatial proximity of two or several DnaI-N domains induced by self-association of the C-terminal segment of DnaI-N123. This interpretation would be consistent with the observation that the NMR signals of residues 107–123 were particularly broad yet were at chemical shifts indicative of random coil structures.
As expected for a conserved fold, the side chains of many of the conserved hydrophobic residues have low solvent accessibility because they contribute to the hydrophobic core of the protein (Figure 4). Conversely, many of the residues with low side-chain solvent accessibility are hydrophobic. For example, the completely conserved residues Leu57, Gly82 and Pro85 are buried and the side chains of Phe37 and Tyr98 have low solvent accessibilities (Figure 4).Remarkably, the positions of Ile4, Leu8 and Val11 in the unstructured N-terminus of B. subtilis DnaI-N are also occupied by hydrophobic residues in all DnaI-N domains shown in Figure 1, although those residues are highly solvent accessible (Figure 4). Phe17 is also highly conserved despite high solvent accessibility of its side chain (>75%). Their conservation suggests a functional role of these flexible residues, presumably in protein–protein interactions.
Figure 4.
Solvent accessibility of the amino-acid side chains in DnaI-N106. The solvent accessibilities are expressed as a percentage of their accessibilities calculated for the situation where the respective amino-acid residues are located in a hypothetical poly-Gly α-helix with a fully extended side chain (49). The values were averaged over the 20 NMR conformers. The locations of the helices and β-strands are indicated at the top.
Interactions with Bst DnaB
The interaction of Bst DnaB with all three DnaI-N constructs was probed by SPR, using a biotinylated oligonucleotide (dT35) immobilized on a streptavidin-coated surface. Bst DnaB was first injected over the oligonucleotide to a consistent binding level. In the presence of ATP under the conditions used, Bst DnaB binds to dT35 and dissociates very slowly; less than 9% dissociation was observed 30 min after injection. Subsequently, increasing concentrations of DnaI-N domains were injected. This experimental design allows for the real-time observation of the interaction of ssDNA-bound Bst DnaB with each of the DnaI-N constructs and direct comparison of the results.
All three DnaI-N domains (DnaI-N106, 123 and 132) interacted with Bst DnaB with association and dissociation rates that were fast, in contrast to the slower rates that full-length DnaI exhibited under similar conditions (14). Full-length DnaI is presumed to form a much more stable complex with the helicase because it oligomerizes cooperatively on the helicase surface via interactions among its C-terminal domains (2,14). This cooperativity in association and multiphasic dissociation made it impossible to obtain reliable thermodynamic data with full-length DnaI.
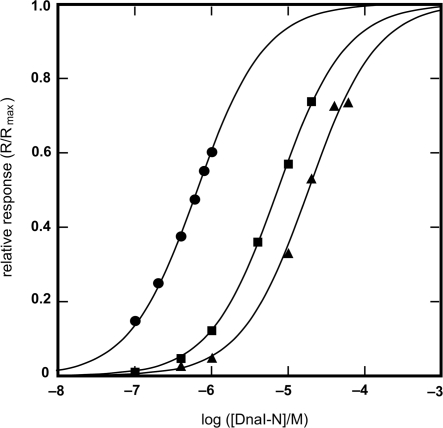
Provided that the sole contact between Bst DnaB and DnaI is in the zinc-binding region of DnaI-N106 as suggested in previous work (14), all three DnaI-N domains were anticipated to interact with similar strength. However, the experimentally determined KD values (DnaI-N106, 18.5 ± 0.9 μM; DnaI-N123, 7.3 ± 0.1 μM; DnaI-N132, 0.65 ± 0.02 μM; Figure 5) revealed significant differences, with the KD values decreasing with each longer construct, especially with DnaI-N123 in comparison with DnaI-N132. This indicates that the interaction of DnaI with Bst DnaB may extend beyond the folded core of the DnaI-N domain to the neighboring segments. Although the NMR experiments showed that this region is most likely unstructured in DnaI-N132, it appears that some residues therein may be capable of assuming a structure that contributes to interaction with the helicase.
Figure 5.
Binding isotherms for the interaction of Bst DnaB with DnaI-N domain constructs, as measured by SPR. Experimental points obtained with: DnaI-N132 (filled circle), DnaI-N123 (filled square) and DnaI-N106 (filled triangle). The KD values for these interactions, determined by the least squares fit to a 1:1 binding model (solid lines) are: DnaI-N132, 0.65 μM; DnaI-N123, 7.3 μM; DnaI-N106, 18.5 μM. Given the difference in the KD values, different ranges of protein concentrations were used: DnaI-N132, 0.1–1 μM; DnaI-N123, 0.1–20 μM; DnaI-N106, 0.1–60 mM. The responses (R, in RU) were normalized using the computed value of the maximum response at equilibrium (Rmax), which corresponded to 0.8, 1.7 and 1.8 molecules of DnaI-N132, DnaI-N123 and DnaI-N106, respectively, bound per Bst DnaB subunit.
We also examined the possible contribution of the flexible, conserved N-terminal segment of DnaI to the interaction with Bst DnaB. An N-terminally biotinylated peptide comprising DnaI residues 1–18 was immobilized on the surface of a streptavidin chip and Bst DnaB was injected over it. We were unable to detect significant interaction at the helicase concentrations used; KD was estimated to be at least 50 μM. We also looked for evidence of direct interaction of a corresponding unmodified peptide with immobilized Bst DnaB. Under conditions where formation of a 1:1 complex should have given a response of 17.4 RU, we detected responses of 2.2 and 3.4 RU with peptide at 20 and 50 μM, respectively. These data indicate a very weak interaction, with KD > 0.14 mM. In competition experiments, the presence of peptide at 20 or 50 μM modestly reduced the response obtained when 10 μM DnaI-N123 or 5 μM DnaI-N134 were injected over immobilized Bst DnaB (responses were reduced by <10%), which again indicates a very weak interaction of the N-terminal region of DnaI with Bst DnaB. The function of the conserved, but unstructured, N-terminal segment of DnaI thus remains uncertain, though we cannot rule out that it might contribute modestly to helicase interaction.
DISCUSSION
3D structure determination of DnaI-N106, the folded core of the N-terminal domain of B. subtilis DnaI, revealed a zinc-binding module with a novel fold. High sequence conservation of DnaI-N was found only among proteins with putative primosomal functions. The zinc-binding motif of DnaI differs from conventional zinc fingers, in particular as the zinc-coordinating residues are separated in the amino-acid sequence by two relatively long polypeptide segments. The zinc-binding motif may play a purely structural role. It has been shown not to be involved in DNA binding, and study of alanine mutants of zinc-coordinating residues showed that impairment of zinc binding correlated with impaired binding to Bst DnaB (14). Although this is consistent with the zinc-binding region being involved in interaction with the helicase, it would also be readily explained if the primary role of the metal ion is to prevent unfolding of the DnaI-N domain.
By study of the binding of three different DnaI-N domain constructs to ssDNA-bound Bst DnaB helicase, we showed that the structured core domain DnaI-N106 bears most of the interacting residues, and that progressive C-terminal extension of this domain to 123 and 132 residues results in somewhat stronger binding (Figure 5). With reliable binding assays in hand, we are now in a good position to explore this interaction further by mutagenesis. Having defined the structure of DnaI-N may also facilitate structural study of the helicase/helicase loader complex by X-ray crystallography. The structural basis of this interaction in any organism is still poorly defined (2).
The stability of the DnaI-N fold clearly does not depend on conservation of hydrophobic side chains at positions 4, 8 and 11 in the mobile N-terminal segment of DnaI-N106. We have also observed these residues to be flexible in full length DnaI, indicating that they do not fold back to contact the C-terminal AAA+ domain (data not shown). Therefore we speculate that these residues, together with the highly conserved, solvent exposed hydrophobic residue at position 17, are required for binding to another protein. Specific protein–protein interactions, where a mobile terminal polypeptide segment of one protein binds to a well-structured domain of its binding partner, is a recurrent motif in bacterial replisomes, governing for example the interactions between the ψ and γ (36), τ and α (37,38), and ε and α (39) subunits of the E. coli polymerase III complex, as well as the interactions of polymerases and many other proteins with the β sliding clamp (40), and of ssDNA-binding protein with its many binding partners (41). We were unable, however, to detect a strong interaction with Bst DnaB of a peptide corresponding to the N-terminal 18 residues of DnaI, and note that a version of DnaI lacking the N-terminal seven residues still bound its natural binding partner, the B. subtilis DnaC helicase, very strongly as detected by yeast two hybrid experiments (7).
The N-terminal domain of DnaI acts as a molecular switch that regulates the accessibility of the ssDNA-binding site of the C-terminal domain (14). In agreement with this notion, the fragmentation pattern observed after limited tryptic digestion of DnaI varied depending on the presence of ATP and Mg2+, which presumably bind to the Walker A and B motifs in the C-terminal domain (Supplementary Data). The change in fragmentation pattern may reflect a conformational change affecting the accessibility of the linker segment between the N- and C-terminal domains, although it cannot be excluded that the ATP-Mg2+ complex alone is sufficient to hinder and redirect the approach of trypsin.
B. subtilis DnaI and E. coli DnaC share significant sequence homology in their C-terminal domains, but there is no similarity between DnaI-N and the N-terminal segment of E. coli DnaC (15). E. coli DnaC cannot assume the DnaI fold determined here because it does not have enough residues in the N-terminal polypeptide segment, though this segment has also been shown to comprise at least part of the helicase binding domain of DnaC (42).
In view of the homology between the DnaI-N domains of B. subtilis and S. aureus (Figure 1), breaking the interaction of DnaI-N with the helicase would be of pharmaceutical interest. The 3D structure of DnaI-N determined in this work sets the stage for directed mutagenesis experiments and further structural studies to identify the interaction surface between the two proteins.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Financial support from the Latvian Scientific Council for K.J. and E.L. is greatly acknowledged. The project and the purchase of the NMR equipment were supported by grants and Fellowships from the Australian Research Council [to G.O. and N.E.D.]. Funding for open access charge: University of Wollongong.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Drs Shigeki Moriya and Panos Soultanas for plasmids, Dr Kiyoshi Ozawa for recording preliminary NMR data and Dr Patrick Schaeffer for help with the limited proteolysis experiments. N-terminal amino-acid sequencing and peptide synthesis were carried out by the Biomolecular Research Facility at the Australian National University.
REFERENCES
- 1.Schaeffer PM, Headlam MJ, Dixon NE. Protein-protein interactions in the eubacterial replisome. IUBMB Life. 2005;57:5–12. doi: 10.1080/15216540500058956. [DOI] [PubMed] [Google Scholar]
- 2.Mott ML, Erzberger JP, Coons MM, Berger JM. Structural synergy and molecular crosstalk between bacterial helicase loaders and replication initiators. Cell. 2008;135:623–634. doi: 10.1016/j.cell.2008.09.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.San Martin C, Radermacher M, Wolpensinger B, Engel A, Miles CS, Dixon NE, Carazo J-M. Three-dimensional reconstructions from cryoelectron microscopy images reveal an intimate complex between helicase DnaB and its loading partner DnaC. Structure. 1998;6:501–509. doi: 10.1016/s0969-2126(98)00051-3. [DOI] [PubMed] [Google Scholar]
- 4.Yuzhakov A, Turner J, O’Donnell M. Replisome assembly reveals the basis for asymmetric function in leading and lagging strand replication. Cell. 1996;86:877–886. doi: 10.1016/s0092-8674(00)80163-4. [DOI] [PubMed] [Google Scholar]
- 5.Davey MJ, O’Donnell M. Replicative helicase loaders: ring breakers and ring makers. Curr. Biol. 2003;13:R594–R596. doi: 10.1016/s0960-9822(03)00523-2. [DOI] [PubMed] [Google Scholar]
- 6.Velten M, McGovern S, Marsin S, Ehrlich SD, Noirot P, Polard P. A two-protein strategy for the functional loading of a cellular replicative DNA helicase. Mol. Cell. 2003;11:1009–1020. doi: 10.1016/s1097-2765(03)00130-8. [DOI] [PubMed] [Google Scholar]
- 7.Imai Y, Ogasawara N, Ishigo-oka D, Kadoya R, Daito T, Moriya S. Subcellular localization of Dna-initiation proteins of Bacillus subtilis: evidence that chromosome replication begins at either edge of the nucleoids. Mol. Microbiol. 2000;36:1037–1048. doi: 10.1046/j.1365-2958.2000.01928.x. [DOI] [PubMed] [Google Scholar]
- 8.Bruand C, Farache M, McGovern S, Ehrlich SD, Polard P. DnaB, DnaD and DnaI proteins are components of the Bacillus subtilis replication restart primosome. Mol. Microbiol. 2001;42:245–255. doi: 10.1046/j.1365-2958.2001.02631.x. [DOI] [PubMed] [Google Scholar]
- 9.Marsin S, McGovern S, Ehrlich SD, Bruand C, Polard P. Early steps of Bacillus subtilis primosome assembly. J. Biol. Chem. 2001;276:45818–45825. doi: 10.1074/jbc.M101996200. [DOI] [PubMed] [Google Scholar]
- 10.Rokop ME, Auchtung JM, Grossman AD. Control of DNA replication initiation by recruitment of an essential initiation protein to the membrane of Bacillus subtilis. Mol. Microbiol. 2004;52:1757–1767. doi: 10.1111/j.1365-2958.2004.04091.x. [DOI] [PubMed] [Google Scholar]
- 11.Turner IJ, Scott DJ, Allen S, Roberts CJ, Soultanas P. The Bacillus subtilis DnaD protein: a putative link between DNA remodeling and initiation of DNA replication. FEBS Lett. 2004;577:460–464. doi: 10.1016/j.febslet.2004.10.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bruand C, Velten M, McGovern S, Marsin S, Sérèna C, Ehrlich D, Polard P. Functional interplay between the Bacillus subtilis DnaD and DnaB proteins essential for initiation and re-initiation of DNA replication. Mol. Microbiol. 2005;55:1138–1150. doi: 10.1111/j.1365-2958.2004.04451.x. [DOI] [PubMed] [Google Scholar]
- 13.Zhang W, Carneiro MJVM, Turner IJ, Allen S, Roberts CJ, Soultanas P. The Bacillus subtilis DnaD and DnaB proteins exhibit different DNA remodelling activities. J. Mol. Biol. 2005;351:66–75. doi: 10.1016/j.jmb.2005.05.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ioannou C, Schaeffer PM, Dixon NE, Soultanas P. Helicase binding to DnaI exposes a cryptic DNA-binding site during helicase loading in Bacillus subtilis. Nucleic Acids Res. 2006;34:5247–5258. doi: 10.1093/nar/gkl690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Soultanas P. A functional interaction between the putative primosomal protein DnaI and the main replicative DNA helicase DnaB in Bacillus. Nucleic Acids Res. 2002;30:966–974. doi: 10.1093/nar/30.4.966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Davey MJ, Jeruzalmi D, Kuriyan J, O’Donnell M. Motors and switches: AAA+ machines within the replisome. Nat. Rev. Mol. Cell Biol. 2002;3:826–835. doi: 10.1038/nrm949. [DOI] [PubMed] [Google Scholar]
- 17.Davey MJ, Fang L, McInerney P, Georgescu RE, O'Donnell M. The DnaC helicase loader is a dual ATP/ADP switch protein. EMBO J. 2002;21:3148–3159. doi: 10.1093/emboj/cdf308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bailey S, Eliason WK, Steitz TA. Structure of hexameric DnaB helicase and its complex with a domain of DnaG primase. Science. 2007;318:459–463. doi: 10.1126/science.1147353. [DOI] [PubMed] [Google Scholar]
- 19.Wang G, Klein MG, Tokonzaba E, Zhang Y, Holden LG, Chen XS. The structure of a DnaB-family replicative helicase and its interactions with primase. Nat. Struct. Mol. Biol. 2008;15:94–100. doi: 10.1038/nsmb1356. [DOI] [PubMed] [Google Scholar]
- 20.Biswas T, Tsodikov OV. Hexameric ring structure of the N-terminal domain of Mycobacterium tuberculosis DnaB helicase. FEBS J. 2008;275:3064–3071. doi: 10.1111/j.1742-4658.2008.06460.x. [DOI] [PubMed] [Google Scholar]
- 21.Lo Y-H, Tsai K-L, Sun Y-J, Chen W-T, Huang C-Y, Hsiao C-D. The crystal structure of a replicative hexameric helicase DnaC and its complex with single-stranded DNA. Nucleic Acids Res. 2009;37:804–814. doi: 10.1093/nar/gkn999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bárcena M, Ruiz T, Donate LE, Brown SE, Dixon NE, Radermacher M, Carazo JM. The DnaB·DnaC complex: a structure based on dimers assembled around an occluded channel. EMBO J. 2001;20:1462–1468. doi: 10.1093/emboj/20.6.1462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gill SC, von Hippel PH. Calculation of protein extinction coefficients from amino acid sequence data. Anal. Biochem. 1989;182:319–326. doi: 10.1016/0003-2697(89)90602-7. [DOI] [PubMed] [Google Scholar]
- 24.Love CA, Lilley PE, Dixon NE. Stable high-copy-number bacteriophage λ promoter vectors for overproduction of proteins in Escherichia coli. Gene. 1996;176:49–53. doi: 10.1016/0378-1119(96)00208-9. [DOI] [PubMed] [Google Scholar]
- 25.Vasudevan SG, Armarego WLF, Shaw DC, Lilley PE, Dixon NE, Poole RK. Isolation and nucleotide sequence of the hmp gene that encodes a haemoglobin-like protein in Escherichia coli K-12. Mol. Gen. Genet. 1991;226:49–58. doi: 10.1007/BF00273586. [DOI] [PubMed] [Google Scholar]
- 26.Williams NK, Prosselkov P, Liepinsh E, Line I, Sharipo A, Littler DR, Curmi PMG, Otting G, Dixon NE. In vivo protein cyclization promoted by a circularly permuted Synechocystis sp. PCC6803 DnaB mini-intein. J. Biol. Chem. 2002;277:7790–7798. doi: 10.1074/jbc.M110303200. [DOI] [PubMed] [Google Scholar]
- 27.Bartels C, Xia T, Billeter M, Güntert P, Wüthrich K. The program XEASY for computer-supported NMR spectral analysis of biological macromolecules. J. Biomol. NMR. 1995;6:1–10. doi: 10.1007/BF00417486. [DOI] [PubMed] [Google Scholar]
- 28.Güntert P, Mumenthaler C, Wüthrich K. Torsion angle dynamics for NMR structure calculation with the new program DYANA. J. Mol. Biol. 1997;273:283–298. doi: 10.1006/jmbi.1997.1284. [DOI] [PubMed] [Google Scholar]
- 29.Herrmann T, Güntert P, Wüthrich K. Protein NMR structure determination with automated NOE assignment using the new software CANDID and the torsion angle dynamics algorithm DYANA. J. Mol. Biol. 2002;319:209–227. doi: 10.1016/s0022-2836(02)00241-3. [DOI] [PubMed] [Google Scholar]
- 30.Linge JP, Williams MA, Spronk CAEM, Bonvin AMJJ, Nilges M. Refinement of protein structures in explicit solvent. Proteins. 2003;50:496–506. doi: 10.1002/prot.10299. [DOI] [PubMed] [Google Scholar]
- 31.Brunger AT, Adams PD, Clore GM, Delano WL, Gros P, Grosse-Kunstleve RW, Jiang J, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography and NMR System: a new software suite for macromolecular structure determination. Acta Crystallogr. D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 32.Koradi R, Billeter M, Wüthrich K. MOLMOL: a program for display and analysis of macromolecular structures. J. Mol. Graph. 1996;14:51–55. doi: 10.1016/0263-7855(96)00009-4. [DOI] [PubMed] [Google Scholar]
- 33.Riddles PW, Blakeley RL, Zerner B. Ellman's reagent: 5,5′-dithiobis(2-nitrobenzoic acid) – a reexamination. Anal. Biochem. 1979;94:75–81. doi: 10.1016/0003-2697(79)90792-9. [DOI] [PubMed] [Google Scholar]
- 34.Chen YL, Park S, Thomburg RW, Tabatabai LB, Kintanar A. Structural characterization of the active site of Brucella abortus Cu–Zn superoxide dismutase: a 15N and 1H NMR investigation. Biochemistry. 1995;34:12265–12275. doi: 10.1021/bi00038a022. [DOI] [PubMed] [Google Scholar]
- 35.Holm L, Sander C. Mapping the protein universe. Science. 1996;273:595–603. doi: 10.1126/science.273.5275.595. [DOI] [PubMed] [Google Scholar]
- 36.Ozawa K, Jergic S, Crowther JA, Thompson PR, Wijffels G, Otting G, Dixon NE. Cell-free in vitro protein synthesis in an autoinduction system for NMR studies of protein-protein interactions. J. Biomol. NMR. 2005;32:235–241. doi: 10.1007/s10858-005-7946-4. [DOI] [PubMed] [Google Scholar]
- 37.Jergic S, Ozawa K, Williams NK, Su X-C, Scott DD, Hamdan SM, Crowther JA, Otting G, Dixon NE. The unstructured C-terminus of the τ subunit of Escherichia coli DNA polymerase III holoenzyme is the site of interaction with the α subunit. Nucleic Acids Res. 2007;35:2813–2824. doi: 10.1093/nar/gkm079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Su X-C, Jergic S, Keniry MA, Dixon NE, Otting G. Solution structure of domains IVa and V of the τ subunit of Escherichia coli DNA polymerase III and interaction with the α subunit. Nucleic Acids Res. 2007;35:2825–2832. doi: 10.1093/nar/gkm080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ozawa K, Jergic S, Park AY, Dixon NE, Otting G. The proofreading exonuclease subunit ε of Escherichia coli DNA polymerase III is tethered to the polymerase subunit α via a flexible linker. Nucleic Acids Res. 2008;36:5074–5082. doi: 10.1093/nar/gkn489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dalrymple BP, Kongsuwan K, Wijffels G, Dixon NE, Jennings PA. A universal protein-protein interaction motif in the eubacterial DNA replication and repair systems. Proc. Natl Acad. Sci. USA. 2001;98:11627–11632. doi: 10.1073/pnas.191384398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shereda RD, Kozlov AG, Lohman TM, Cox MM, Keck JL. SSB as an organizer/mobilizer of genome maintenance complexes. Crit. Rev. Biochem. Mol. Biol. 2008;43:289–318. doi: 10.1080/10409230802341296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ludlam AV, McNatt MW, Carr KM, Kaguni JM. Essential amino acids of Escherichia coli DnaC protein in an N-terminal domain interact with DnaB helicase. J. Biol. Chem. 2001;276:27345–27353. doi: 10.1074/jbc.M101940200. [DOI] [PubMed] [Google Scholar]
- 43.Cornilescu G, Delaglio F, Bax A. Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J. Biomol. NMR. 1999;13:289–302. doi: 10.1023/a:1008392405740. [DOI] [PubMed] [Google Scholar]
- 44.Güntert P, Braun W, Billeter M, Wüthrich K. Automated stereospecific proton NMR assignments and their impact on the precision of protein structure determinations in solution. J. Am. Chem. Soc. 1989;111:3997–4004. [Google Scholar]
- 45.Linge JP, Nilges M. Influence of non-bonded parameters on the quality of NMR structures: a new force field for NMR structure calculation. J. Biomol. NMR. 1999;13:51–59. doi: 10.1023/a:1008365802830. [DOI] [PubMed] [Google Scholar]
- 46.Laskowski RA, Rullmann JAC, MacArthur MW, Kaptein R, Thornton JM. AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J. Biomol. NMR. 1996;8:477–486. doi: 10.1007/BF00228148. [DOI] [PubMed] [Google Scholar]
- 47.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Koradi R, Billeter M, Wüthrich K. MOLMOL: A program for display and analysis of macromolecular structures. J. Mol. Graph. 1996;14:51–55. doi: 10.1016/0263-7855(96)00009-4. [DOI] [PubMed] [Google Scholar]
- 49.Sevilla-Sierra P, Otting G, Wüthrich K. Determination of the nuclear magnetic resonance structure of the DNA-binding domain of the P22 c2 repressor (1-76) in solution and comparison with the DNA-binding domain of the 434 repressor. J. Mol. Biol. 1994;235:1003–1020. doi: 10.1006/jmbi.1994.1053. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.