Abstract
Bacteriophage T4 homologous recombination events are promoted by presynaptic filaments of UvsX recombinase bound to single-stranded DNA (ssDNA). UvsY, the phage recombination mediator protein, promotes filament assembly in a concentration-dependent manner, stimulating UvsX at stoichiometric concentrations but inhibiting at higher concentrations. Recent work demonstrated that UvsX-H195Q/A mutants exhibit decreased ssDNA-binding affinity and altered enzymatic properties. Here, we show that unlike wild-type UvsX, the ssDNA-dependent ATPase activities of UvsX-H195Q/A are strongly inhibited by both low and high concentrations of UvsY protein. This inhibition is partially relieved by UvsY mutants with decreased ssDNA-binding affinity. The UvsX-H195Q mutant retains weak DNA strand exchange activity that is inhibited by wild-type UvsY, but stimulated by ssDNA-binding compromised UvsY mutants. These and other results support a mechanism in which the formation of competent presynaptic filaments requires a hand-off of ssDNA from UvsY to UvsX, with the efficiency of the hand-off controlled by the relative ssDNA-binding affinities of the two proteins. Other results suggest that UvsY acts as a nucleotide exchange factor for UvsX, enhancing filament stability by increasing the lifetime of the high-affinity, ATP-bound form of the enzyme. Our findings reveal new details of the UvsX/UvsY relationship in T4 recombination, which may have parallels in other recombinase/mediator systems.
INTRODUCTION
Recombinases of the highly conserved RecA/Rad51 family catalyze DNA strand exchange reactions that are of central importance in pathways of homologous recombination and DNA double-strand break repair. DNA strand exchange requires the formation of a presynaptic filament consisting of recombinase cooperatively bound to single-stranded DNA (ssDNA) (1). Presynaptic filament assembly is necessary to activate enzymatic activities of RecA/Rad51 including ssDNA-stimulated ATP hydrolysis, homologous pairing and strand transfer. Defects in presynaptic filament assembly cause genome instability and sensitivity to DNA-damaging agents in all organisms, and are associated with cancer predisposition in humans (2–5).
The assembly and stability of recombinase–ssDNA presynaptic filaments are regulated by recombination mediator proteins (RMPs) and ssDNA-binding proteins (SSBs), both of which are highly conserved at the functional level (6). The assembly of recombinase onto SSB-covered ssDNA is rate limiting in many recombination processes (7,8). The RMP component stimulates DNA strand exchange by accelerating SSB displacement from ssDNA by the incoming recombinase.
Studies of the bacteriophage T4 recombination system have provided important insights on the biochemical interactions between recombinase, RMP and SSB components during presynapsis and strand exchange (6,9). UvsX protein, the RecA/Rad51 ortholog of T4 phage, catalyzes ssDNA-stimulated ATP hydrolysis and DNA strand exchange. UvsX ssDNA-stimulated ATPase activity is unusual in that both ADP and AMP are generated as products (10,11), a property unique among characterized recombinases. UvsY, the phage RMP, stimulates UvsX activities by promoting filament assembly and displacement of Gp32, the phage SSB. In vitro, UvsY is absolutely required for strand exchange along with UvsX and Gp32 under salt conditions that approximate in vivo ionic strength. In vivo, uvsY and uvsX mutants are equally deficient in recombination and repair functions, indicating that the mediator activity of UvsY is essential for UvsX biological function (12–15). Previous work demonstrated that UvsY destabilizes Gp32–ssDNA interactions while stabilizing UvsX–ssDNA interactions; data indicate that UvsY alters ssDNA structure in ways that favor UvsX binding and disfavor Gp32 binding (16–20).
Recently, our laboratory characterized missense mutants of UvsX that dramatically alter its enzymatic properties (11). UvsX-H195Q and -H195A mutants exhibit reduced DNA strand exchange activity, altered ssDNA-stimulated ATPase activity and reduced ssDNA-binding affinity compared to wild-type. We hypothesized that UvsY protein might restore wild-type-like activity to these mutants by stabilizing their filaments on ssDNA. In this report we demonstrate that, surprisingly, the opposite is true. Unlike wild-type UvsX, both mutants are strongly inhibited by UvsY protein. Their defects are partially complemented, however, by mutations in UvsY protein that reduce its ssDNA-binding affinity (19). These and other results suggest that the formation of competent presynaptic filaments requires a hand-off of ssDNA from UvsY to UvsX, with the efficiency of the hand-off controlled by the relative ssDNA-binding affinities of the two proteins. Other data suggest that UvsY promotes nucleotide exchange by UvsX protein, an activity that may be important for the observed stabilization of UvsX interactions by UvsY. Our findings provide new insights on the relationships between recombinases and recombination mediators, and on the mechanism of the presynaptic phase of homologous recombination.
MATERIALS AND METHODS
Reagents and enzymes
Chemicals, biochemicals and enzymes were purchased from Sigma unless otherwise noted. All solutions used were of analytical or enzymatic grade and made with ultrapure Barnstead water. Radiolabeled α-[32P]-ATP and γ-[32P]-ATP were purchased from Amersham Bioscience. Polyethyleneimine (PEI)–cellulose thin layer chromatography (TLC) plates were purchased from EMD Chemicals. All restriction enzymes, T4 polynucleotide kinase and calf intestinal phosphatase were purchased from New England Biolabs. Bacteriophage M13mp18 circular ssDNA and supercoiled double-stranded DNA (dsDNA) replicative form I (RFI) were purified as described (21). M13mp18 linear dsDNA (RFIII) was generated by digestion of RFI with XbaI, then 5′-end labeled with [32P] as described (10). DNA concentrations were determined by the absorbance at 260 nm using conversion factors of 36 µg/ml/A260 for ssDNA and 50 µg/ml/A260 for dsDNA. All DNA concentrations in the text and figures are expressed in units of nucleotides.
T4 recombination proteins
Wild-type UvsX (44 kDa), mutants UvsX-H195A and UvsX-H195Q, Gp32 (34 kDa), wild-type UvsY (UvsY-wt; 16 kDa), single missense mutant UvsY-K58A (UvsY-SM) and double missense mutant UvsY-K58A,R60A (UvsY-DM) proteins were purified and stored as previously described (8,11,19,22,23). All proteins were >98% pure based on SDS–PAGE analysis and all were nuclease-free according to published criteria (23). Protein stock concentrations were determined by the absorbance at 280 nm using extinction coefficients of 69 790 (M*cm)−1 for UvsX, UvsX-H195A and UvsX-H195Q; 19 180 (M*cm)−1 for UvsY, UvsY-SM and UvsY-DM; and 41 360 (M*cm)−1 for Gp32 (24) (J. Farb and S. Morrical, unpublished data).
ATPase assays
Rates of ssDNA-stimulated ATP hydrolysis by wild-type and mutant UvsX proteins were determined either by coupled spectrophotometric assay or by TLC assay as described (10,25). Coupled ATPase time courses were recorded on a Hitachi U-2000 spectrophotometer equipped with a water-jacketed cuvette holder to maintain a constant temperature of 37°C. Reactions contained 20 mM Tris–acetate (pH 7.4), 90 mM potassium acetate (KOAc), 10 mM magnesium acetate, 2 mM ATP, 6 U/ml pyruvate kinase (PK), 6 U/ml lactate dehydrogenase, 2.3 mM phosphoenolpyruvate, 0.46 mM NADH, 4.5 µM M13mp18 ssDNA, 0.45 µM recombinase and 0–1.5 µM UvsY depending on experiment. Reaction mixtures of 700 µl were incubated at 37°C for 5 min and then started by the addition of ATP. The reaction was allowed to continue until linear timecourses of hydrolysis were detected and velocity curves were further fit by KaleidaGraph (Synergy Software).
TLC assays were performed as follows: all reactions (100 µl final volume) contained 20 mM Tris–acetate, pH 7.4, 90 mM KOAc, 10 mM magnesium acetate, 1 mM DTT, 100 µg/ml BSA, 4.5 µM ssDNA, 0.45 µM recombinase and 0–2.7 µM of either UvsY, UvsY-SM or UvsY-DM. Reactions were started by the addition of α-[32P]-ATP to a final concentration of 4 mM and 10 µCi/ml specific activity. Aliquots (5 µl) were removed at various times and quenched with 8-mM EDTA (final concentration). Of the samples, 0.5 µl were plated onto PEI–cellulose TLC plate at 1-cm intervals, dried and developed in a 0.75 M KH2PO4 buffer. Dried, developed plates were exposed to a K-screen (Kodak) for 1 h. Images were captured using a Bio-Rad Personal Molecular Imager-FX and quantified using Quantity One v 4.5.1 (Bio-Rad) software. Timecourse data were fit to ensure linearity and rates were plotted using KaleidaGraph.
DNA strand exchange assays
DNA strand exchange assays were carried out as previously described (10). Reactions mixtures (100 µl final volume) contained 20 mM Tris–acetate, pH 7.4, 90 mM KOAc, 10 mM magnesium acetate, 1 mM DTT, 100 µg/ml BSA, 10 U/ml PK, 3.3 mM phosphoenolpyruvate, 10 µM ssDNA, 10 µM 5′-[32P]-RFIII DNA and 3 mM ATP (final concentrations). Variable components included 0.5 µM recombinase (UvsX wild-type, H195Q or H195A), 0 or 1 µM Gp32 and 0 or 0.5 µM UvsY, UvsY-SM or UvsY-DM (final concentrations) depending on reaction. Reactions at 37°C were initiated by adding ATP to a preincubated mixture containing all other components. Aliquots (20 µl) were removed at the indicated times and quenched with 1X SE stopping solution containing 1X Promega loading dye, 5% SDS and 40 mM EDTA (final volume 25 µl). Aliquots (10 µl) of the quenched samples were run on a 1% agarose gel at 130 V for 2 h in a Tris–acetate–EDTA buffer system. Gels were dried under vacuum at 45°C onto Millipore Immobilon nylon transfer membranes. Membranes were then exposed to a K-screen (Kodak) for 5 h and imaged using a Bio-Rad Personal Molecular Imager-FX. Phosphorimager data were quantified using Quantity One V4.5.1 (Bio-Rad) software.
RESULTS
Differential effects of UvsY on ATPase activities of wild-type versus mutant UvsX enzymes
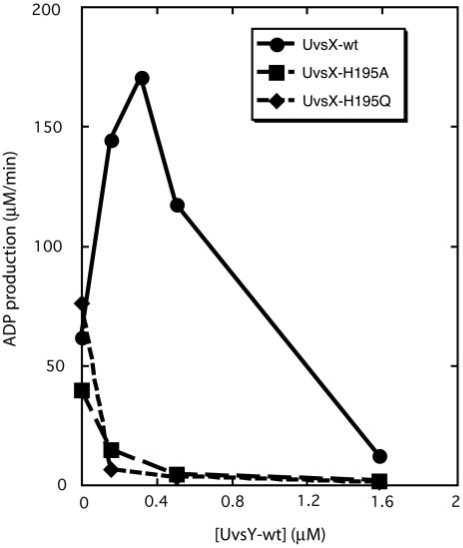
ATP hydrolysis catalyzed by UvsX protein in the presence of ssDNA is stimulated by UvsY in a concentration-dependent manner (Figure 1). Activity increases with increasing UvsY concentration until a sharp optimum activity is reached at a UvsY:UvsX molar ratio of approximately 1:1. Further increases in UvsY concentration cause activity to decrease and high concentrations are inhibitory. These results are consistent with previous reports of UvsY concentration optima in T4 recombination processes (25,26). UvsY has a dramatically different effect on the ssDNA-stimulated ATPase activities of UvsX-H195Q and -H195A mutants. Both activities decline precipitously with increasing UvsY concentration, and there are no concentration optima (Figure 1). Clearly, UvsX-H195Q/A mutants have lost the ability to be stimulated by UvsY, which now acts as their strict inhibitor.
Figure 1.
UvsY effects on ssDNA-dependent ATPase activities of UvsX, UvsX-H195A and UvsX-H195Q. Reaction velocities were measured by coupled spectrophotometric assay as described in Materials and Methods section. Reactions contained 0.4 µM UvsX (closed circles), UvsX-H195A (closed squares) or UvsX-H195Q (closed diamonds). ssDNA concentration was 4.5 µM and ATP concentration was 2 mM in all reactions, and UvsY concentration varied as indicated. All other conditions were as described in Materials and Methods section.
Changes in UvsY ssDNA-binding affinity affect UvsX ATPase activity
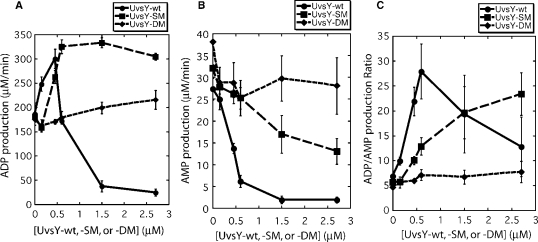
The effects of UvsY on UvsX ssDNA-stimulated ATPase activity are examined in greater detail in Figure 2. UvsY stimulates ADP production by UvsX in a highly concentration-dependent manner with an optimum at approximately 1:1 molar ratio of UvsY:UvsX (Figure 2A). Higher UvsY concentrations are inhibitory. At the same time, UvsY strongly suppresses AMP production by UvsX (Figure 2B). As a result, UvsY also affects the ADP/AMP product ratio of UvsX ssDNA-stimulated ATP hydrolysis in a concentration-dependent manner. This ratio increases from approximately 5–7 with no UvsY to approximately 25–30 at optimum UvsY before falling again at higher UvsY concentrations (Figure 2C). Therefore, the presence of stoichiometric UvsY greatly increases the specificity of UvsX for catalyzing the ATP →ADP reaction as opposed to the ATP → AMP reaction. The implications of this finding for presynaptic filament dynamics are discussed in a later section.
Figure 2.
Effects of wild-type and mutant UvsY proteins on ADP and AMP production by UvsX ssDNA-dependent ATPase activity. Velocities of ADP and AMP production were measured by TLC assay as described in Materials and Methods section. All reactions contained 0.45 µM UvsX, 4.5 µM ssDNA and 4 mM α-[32P]-ATP. All other conditions were as described in Materials and Methods section. (A) Velocity of ADP production by wild-type UvsX protein as a function of UvsY (closed circles), UvsY-SM (closed squares) or UvsY-DM (closed diamonds) concentration. (B) Velocity of AMP production by wild-type UvsX protein as a function of UvsY (closed circles), UvsY-SM (closed squares) or UvsY-DM (closed diamonds) concentration. Note that (B) is plotted on an expanded scale compared to (A). (C) ADP/AMP product ratio for wild-type UvsX protein as a function of UvsY (closed circles), UvsY-SM (closed squares) or UvsY-DM (closed diamonds) concentration.
Previous studies described mutations in a conserved sequence motif of UvsY (residues 57–63, the so-called ‘KARL’ motif), which dramatically reduce its ssDNA-binding affinity while preserving UvsY hexameric structure and protein–protein interactions with UvsX and Gp32 (18,19). At salt concentrations relevant to UvsX/UvsY-dependent DNA strand exchange, the order of relative ssDNA-binding affinities is wild-type (UvsY-wt) > UvsY-K58A (‘single mutant’ or UvsY-SM) > UvsY-K58A,R60A (‘double mutant’ or UvsY-DM). Figure 2 shows that substitution of UvsY-SM or -DM mutants for wild-type has a major effect on UvsX ATPase profiles. UvsY-SM has a much broader concentration optima than wild-type UvsY for stimulating ADP production by UvsX (Figure 2A). Therefore, ADP production increases with increasing UvsY-SM concentration and remains high, and the inhibition of UvsX observed at high UvsY concentrations is not observed with UvsY-SM. Nor is inhibition observed with UvsY-DM, which with increasing concentration gradually increases ADP production by UvsX (Figure 2A). Thus, ‘KARL’ motif mutations relieve the inhibition of UvsX ADP production caused by high UvsY concentrations. These mutations also relieve the inhibition of UvsX AMP production caused by wild-type UvsY (Figure 2B). The relief is partial in the case of UvsY-SM and full in the case of UvsY-DM (Figure 2B). As a result, the ADP/AMP product ratio of UvsX ssDNA-stimulated ATP hydrolysis responds to UvsY-SM and UvsY-DM concentrations very differently compared to wild-type UvsY (Figure 2C). The ADP/AMP ratio increases substantially but more gradually with increasing UvsY-SM than with increasing UvsY concentration, and it does not go through an optimum within the concentration range examined. Meanwhile the ADP/AMP ratio is virtually independent of UvsY-DM concentration (Figure 2C).
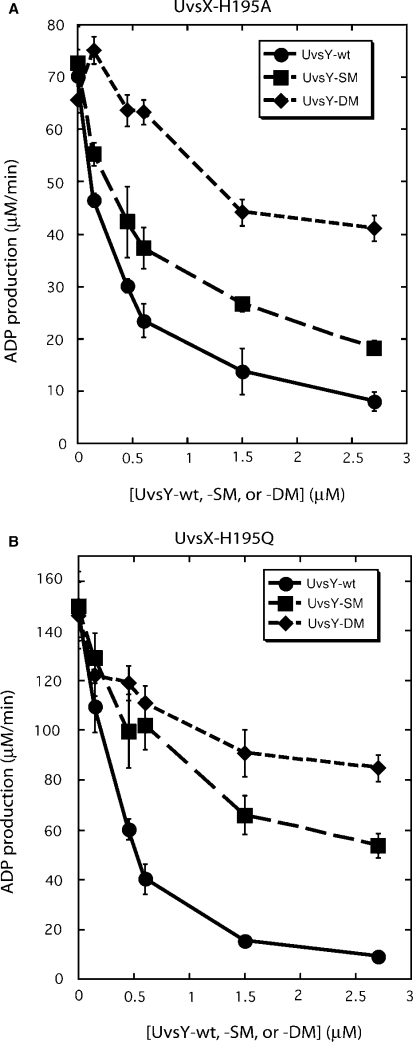
UvsY ‘KARL’ motif mutations also partially relieve UvsY's inhibition of the ATPase activities of UvsX-H195Q/A mutants (Figure 3). ADP production by UvsX-H195A decreases with increasing concentrations of UvsY, UvsY-SM or UvsY-DM, but the severity of the inhibition decreases in the order UvsY > UvsY-SM > UvsY-DM (Figure 3A). A similar pattern occurs in reactions with UvsX-H195Q (Figure 3B). Therefore, mutations that decrease UvsY ssDNA-binding affinity have the general effect of decreasing UvsY inhibitory effects on UvsX-catalyzed ATP hydrolysis, whether inhibition is caused by excessive UvsY concentration or by mutations that weaken UvsX–ssDNA interactions.
Figure 3.
Effects of wild-type and mutant UvsY proteins on ADP production by (A) UvsX-H195A mutant and (B) UvsX-H195Q mutant ssDNA-dependent ATPase activities. Velocities of ADP production were measured by TLC assay as described in Materials and Methods section. All reactions contained 0.45 µM recombinase, 4.5 µM ssDNA, 4 mM α-[32P]-ATP and variable concentrations of either UvsY (closed circles), UvsY-SM (closed squares) or UvsY-DM (closed diamonds). All other conditions were as described in Materials and Methods section.
UvsY ‘KARL’ motif mutants complement partial strand exchange defect of UvsX-H195Q
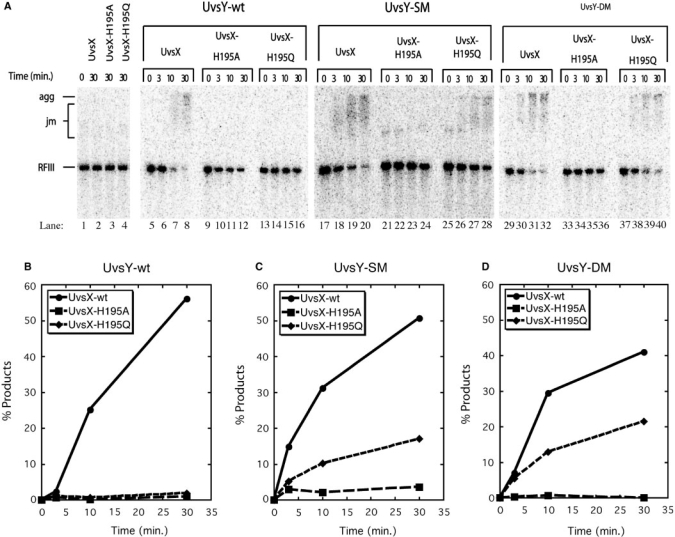
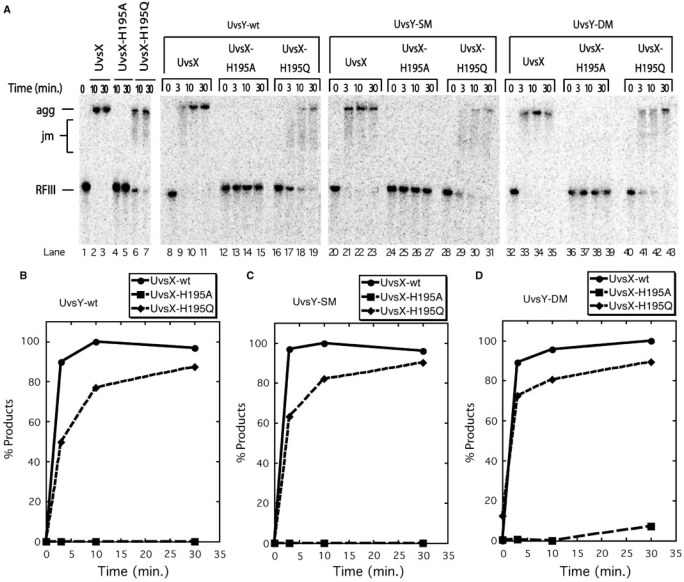
Figure 4 explores the effects of different combinations of wild-type and mutant UvsX and UvsY proteins on DNA strand exchange reactions. Typical results are shown. Note that the reaction products are DNA ‘aggregates’ that run at the top of the gel. These aggregates are a characteristic product of UvsX-catalyzed strand exchange, which generates branched networks of DNA caused by multiple synapsis events per DNA molecule (10,19,25,27). The aggregates are not resolved by Proteinase K treatment (data not shown). The reactions in Figure 4 were performed in the absence of Gp32 and under salt and protein concentration conditions in which UvsX strand exchange activity is UvsY dependent. This was verified in control reactions lacking UvsY protein (Figure 4A, lanes 1–4), in which no strand exchange products (DNA joint molecules or aggregates) and no consumption of RFIII substrate were observed after 30 min in reactions containing either UvsX, UvsX-H195A or UvsX-H195Q. The weak strand exchange activity of UvsX-H195Q (11) is not detectable under these conditions. The addition of wild-type UvsY activates the strand exchange activity of UvsX (Figure 4A, lanes 5–8), but not that of UvsX-H195A (Figure 4A, lanes 9–12) or of UvsX-H195Q (Figure 4A, lanes 13–16) (Figure 4B–D). UvsY-SM and UvsY-DM activate strand exchange by UvsX almost as well as wild-type UvsY under these reactions conditions (Figure 4A, lanes 17–20 and lanes 29–32; Figure 4B–D). This observation is consistent with previous reports that the UvsY ‘KARL’ motif mutants retain significant strand exchange-stimulatory activity toward wild-type UvsX (19). Significantly, unlike wild-type UvsY, both UvsY-SM and UvsY-DM facilitate DNA strand exchange by the UvsX-H195Q mutant (Figure 4A, lanes 25–28 and lanes 37–40, respectively; Figure 4C, D). Quantitatively, the extent of the reaction with UvsX-H195Q plus UvsY-SM or -DM is 30–50% that of the wild-type UvsX reaction in the presence of the same UvsY mutants (Figure 4C, D). In contrast, both UvsY-SM and UvsY-DM fail to rescue strand exchange by the UvsX-H195A mutant (Figure 4A, lanes 21–24 and lanes 33–36, respectively; Figure 4C, D). (The small signal plotted for the UvsX-H195A reaction in Figure 4C does not increase with time and is not due to residual enzyme activity. Instead, the signal is due to a relatively high background of slow migrating material in this reaction—see zero timepoint in Figure 4A lane 21.) These results demonstrate that UvsY ‘KARL’ motif mutants can complement the conditional DNA strand exchange defect of UvsX-H195Q, but not the more severe defect of UvsX-H195A.
Figure 4.
DNA strand exchange reactions promoted by different combinations of wild-type and mutant UvsX and UvsY proteins in the absence of Gp32. Reactions were performed at low UvsX concentration where strand exchange is codependent on UvsX and UvsY. (A) Agarose gel electrophoresis assays for DNA strand exchange were carried out as described in Materials and Methods section. All reactions contained 0.5 µM recombinase (wild-type, H195A or H195Q as indicated above the lanes), 10 µM M13mp18 ssDNA, 10 µM 5′-[32P]-labeled M13mp18 RFIII DNA and 3 mM ATP. Lanes 1–4—control reactions lacking UvsY. Lanes 5–16—reactions containing 0.5 µM UvsY. Lanes 17–28—reactions containing 0.5 µM UvsY-SM. Lanes 29–40—reactions containing 0.5 µM UvsY-DM. All other conditions were as described in Materials and Methods section. Gel mobility positions of aggregates (agg), joint molecules (jm) and linear dsDNA (RFIII) substrate are shown to the left of the gel. (B) Quantification of results for reactions containing UvsY-wt and either UvsX-wt (filled circles), UvsX-H195A (filled squares) or UvsX-H195Q (filled diamonds), as determined by phosphorimaging of gel in (A), lanes 5–16. On the y-axis, % products denote percentage of total DNA migrating as joint molecules and aggregates. (C) Quantification of results for reactions containing UvsY-SM and either UvsX-wt (filled circles), UvsX-H195A (filled squares) or UvsX-H195Q (filled diamonds), as determined by phosphorimaging of gel in (A), lanes 17–28. (D) Quantification of results for reactions containing UvsY-DM and either UvsX-wt (filled circles), UvsX-H195A (filled squares) or UvsX-H195Q (filled diamonds), as determined by phosphorimaging of gel in Panel A, lanes 29–40.
Figure 5 explores DNA strand exchange reactions involving different combinations of wild-type and mutant UvsX and UvsY proteins in the presence of Gp32. These reactions were performed under salt and protein concentration conditions in which UvsX strand exchange activity is typically independent of UvsY. Typical results are shown. Control reactions lacking UvsY (Figure 5A, lanes 1–7) confirm that UvsX is fully active under these conditions, whereas UvsX-H195Q is partially active and UvsX-H195A is inactive. Under these conditions, wild-type UvsX exhibits similar, high levels of strand exchange whether in the absence of UvsY (Figure 5A, lanes 1–3) or in the presence of wild-type UvsY (Figure 5, lanes 8–11), UvsY-SM (Figure 5, lanes 20–23) or UvsY-DM (Figure 5, lanes 32–35) (Figure 5B–D). Products appear very rapidly under these conditions (≥90% DNA converted to products within 3 min; Figure 5B–D). UvsX-H195A remains fully inactive whether in the absence of UvsY (Figure 5A, lanes 4–5) or in the presence of wild-type UvsY (Figure 5A, lanes 12–15), UvsY-SM (Figure 5A, lanes 24–27) or UvsY-DM (Figure 5A, lanes 36–39) (Figure 5B–D). UvsX-H195Q exhibits weaker strand exchange activity than wild-type UvsX under all of these conditions. The addition of wild-type UvsY (Figure 5A, lanes 16–19) inhibits UvsX-H195Q-catalyzed strand exchange compared to the control reaction lacking UvsY (Figure 5A, lanes 6–7), based on the slower consumption of RFIII substrate and appearance of DNA aggregates in the former reaction. Quantitatively, UvsX-H195Q generates products at about half the rate of wild-type UvsX in the presence of Gp32 and wild-type UvsY (Figure 5B). Substitution of UvsY-SM (Figure 5A, lanes 28–31) or UvsY-DM (Figure 5A, lanes 40–43) for wild-type UvsY improves the rate of UvsX-H195Q-catalyzed strand exchange (Figure 5C, D), with UvsY-DM providing the best improvement (Figure 5D). The data in Figure 5 demonstrate that even under optimal conditions for strand exchange, UvsX-H195Q still responds positively to ssDNA-binding compromising mutations in UvsY. Additionally, the data in Figures 4 and 5 demonstrate that the UvsX-H195A mutant is a knock-out for DNA strand exchange even though this mutant retains very high levels of ssDNA-stimulated ATPase activity (11).
Figure 5.
DNA strand exchange reactions promoted by different combinations of wild-type and mutant UvsX and UvsY proteins in the presence of Gp32. (A) Agarose gel electrophoresis assays for DNA strand exchange were carried out as described in Materials and Methods section. All reactions contained 0.5 µM recombinase (wild-type, H195A, or H195Q as indicated above the lanes), 1 µM Gp32, 10 µM M13mp18 ssDNA, 10 µM 5′-[32P]-labeled M13mp18 RFIII DNA and 3 mM ATP. Lanes 1–7—control reactions lacking UvsY. Lanes 8–19—reactions containing 0.5 µM UvsY. Lanes 20–31—reactions containing 0.5 µM UvsY-SM. Lanes 32–43—reactions containing 0.5 µM UvsY-DM. All other conditions were as described in Materials and Methods section. Gel mobility positions of aggregates (agg), joint molecules (jm) and linear dsDNA (RFIII) substrate are shown to the left of the gel. (B) Quantification of results for reactions containing UvsY-wt and either UvsX-wt (filled circles), UvsX-H195A (filled squares) or UvsX-H195Q (filled diamonds), as determined by phosphorimaging of gel in (A), lanes 8–19. On the y-axis,% products denote percentage of total DNA migrating as joint molecules and aggregates. (C) Quantification of results for reactions containing UvsY-SM and either UvsX-wt (filled circles), UvsX-H195A (filled squares) or UvsX-H195Q (filled diamonds), as determined by phosphorimaging of gel in (A), lanes 20–31. (D) Quantification of results for reactions containing UvsY-DM and either UvsX-wt (filled circles), UvsX-H195A (filled squares) or UvsX-H195Q (filled diamonds), as determined by phosphorimaging of gel in (A), lanes 32–43.
DISCUSSION
Studies of site-directed mutations in UvsX and UvsY proteins have provided valuable information about the mechanism of homologous recombination in the T4 system. Histidine-195 in UvsX protein is an allosteric switch residue analogous to glutamine-194 in Escherichia coli RecA protein, which coordinates ATP binding and hydrolysis activities with DNA binding and strand exchange (11). The data demonstrate that the UvsX-H195A mutation completely uncouples ATP binding/hydrolysis from strand exchange, since the mutant enzyme retains strong ssDNA-stimulated ATPase activity but is devoid of strand exchange activity under all conditions examined (Figures 1, 4 and 5). The conservative UvsX-H195Q mutant retains strong ssDNA-stimulated ATPase activity as well as partial strand exchange activity. Both mutants have reduced apparent ssDNA-binding affinity compared to wild-type UvsX (11), and unlike wild-type both proteins are inhibited by relatively low concentrations of UvsY (Figure 1).
UvsY ‘KARL’ motif mutant proteins—UvsY-SM (K58A) and UvsY-DM (K58A + R60A) have strongly reduced ssDNA-binding affinity compared to wild-type, but retain wild-type-like self- and heteroprotein-association properties (18,19). UvsY-SM and UvsY-DM mutants retain partial recombination mediator protein activity in that both partially stimulate UvsX-catalyzed DNA strand exchange reactions in the presence of Gp32 [(19) and this study] and both modestly stabilize UvsX–ssDNA complexes (18). The residual mediator activities of UvsY-SM and UvsY-DM likely result from protein–protein interactions with UvsX.
Results of this study, involving different combinations of UvsX and UvsY mutants and concentrations, demonstrate that correct assembly of presynaptic filaments requires an optimal balance between the ssDNA-binding activities of UvsX and UvsY. Conditions that strongly favor UvsY– over UvsX–ssDNA interactions, such as high UvsY/UvsX concentration ratios or UvsX-H195Q/A mutations, are generally inhibitory toward UvsX activity. These inhibitory effects are mitigated by UvsY-SM and UvsY-DM mutations, which exhibit functional complementation of UvsX-H195Q/A ssDNA-dependent ATPase and of UvsX-H195Q strand exchange activities in vitro (Figures 3 and 4). UvsY-SM/-DM mutants also relieve the UvsY concentration-dependent inhibition of wild-type UvsX ssDNA-dependent ATPase activity (Figure 2). Thus, it is possible to complement the effects of weak UvsX–ssDNA interactions by weakening UvsY–ssDNA interactions.
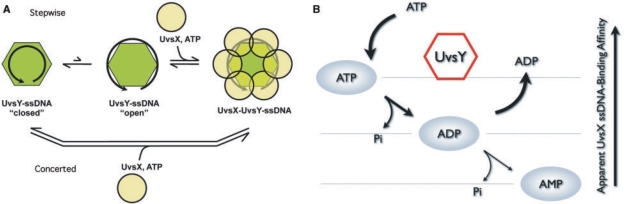
UvsY appears to modulate the ssDNA-binding activities of other proteins primarily by altering ssDNA structure, increasing UvsX–ssDNA affinity while decreasing Gp32–ssDNA affinity (16–18). Results of this study imply that UvsX itself must possess sufficiently strong ssDNA-binding activity in order to capitalize on ssDNA structural changes induced by UvsY. High-affinity UvsY–ssDNA interactions involve wrapping of ssDNA around UvsY hexamers (6,17,20). Previous studies demonstrated that UvsY co-occupies the ssDNA with either Gp32 or UvsX (16,18,28,29). Thus, UvsY forms a stoichiometric UvsY–Gp32–ssDNA complex in which Gp32–ssDNA interactions are weakened, but Gp32 is not displaced from the complex until UvsX and ATP are added (8,16). Similarly, data indicate that stoichiometric UvsY is retained in the complex after UvsX binds and Gp32 is ejected (18,29). Therefore, our complementation data cannot be explained by a simple competition for ssDNA-binding sites between UvsX and UvsY mutants with different ssDNA-binding affinities. Instead, to explain the effects of mutations and protein concentration effects described in this work, we propose that there must be a hand-off mechanism in which the ssDNA wrapped around UvsY is passed to UvsX, and that the efficiency of the hand-off is governed by the relative affinities of the two proteins for ssDNA. This model is shown in schematic form in Figure 6A. We postulate that UvsY–ssDNA is in equilibrium between a tightly wrapped or ‘closed’ complex and a loosely wrapped or ‘open’ complex, and that the closed complex is favored at higher UvsY concentrations. UvsX cannot capture ssDNA from the closed complex, but it can capture ssDNA from the open complex, effectively pulling the closed/open equilibrium to the right, as shown in the step-wise mechanism at the top of Figure 6A. UvsX mutants with weak ssDNA-binding affinity, such as H195Q or H195A, are unable to capture ssDNA from the open complex, so UvsY–ssDNA remains closed. UvsY ‘KARL’ motif mutations shift the closed/open equilibrium to the right, increasing the likelihood of ssDNA capture by UvsX mutants or at higher mediator concentrations. Alternatively, the hand-off of ssDNA from UvsY to UvsX might occur by a concerted mechanism as shown at the bottom of Figure 6A. In this case, the efficiency of the hand-off would be determined by the amount of closed versus open character in a ‘transition state’ containing both proteins and ssDNA. Changes in the relative ssDNA-binding affinities of UvsX and UvsY would determine the forward commitment of this ‘transition state’.
Figure 6.
Models for UvsY effects on UvsX–ssDNA dynamics. (A) Formation of competent presynaptic filaments requires a hand-off of ssDNA from UvsY to UvsX, with the efficiency of the hand-off controlled by the relative ssDNA-binding affinities of the two proteins. A step-wise mechanism (upper path) is postulated in which UvsY hexamers interact with ssDNA to form a tightly wrapped ‘closed’ complex in equilibrium with a loosely wrapped ‘open’ complex. High UvsY concentrations shift the equilibrium toward the closed form that is inaccessible to UvsX protein. In contrast, UvsX captures ssDNA from the open complex, effectively shifting the closed/open equilibrium to the right. Mutations that weaken UvsX–ssDNA interactions, such as H195Q/A, cannot capture ssDNA from the open complex. On the other hand, mutations that weaken UvsY–ssDNA interactions, such as K58A/R60A, shift the closed/open equilibrium to the right, complementing UvsX mutations by allowing more efficient ssDNA hand-off and neutralizing the inhibitory effects of high UvsY concentrations. Alternatively, ssDNA hand-off could occur by a concerted mechanism as shown in the lower path. See text for additional details. (B) A hypothetical model for UvsY suppression of UvsX AMP production. Kinetics data indicate that ssDNA-bound UvsX hydrolyzes ATP to AMP via a processive, step-wise mechanism (11). UvsY may short circuit this process, causing ADP to release from the active site prior to hydrolysis to AMP. UvsY may therefore act as a nucleotide exchange factor for UvsX–ssDNA presynaptic filaments, and stimulate recombination by increasing the lifetime of UvsX in a high ssDNA-binding affinity state. See text for additional details.
An alternative explanation for the inhibitory effects of high UvsY concentrations is that the excess UvsY could interfere with normal UvsX function through nonproductive protein–protein interactions occurring off of the ssDNA. The data appear to rule out this possibility, however. Under this scenario, high concentrations of UvsY-SM or -DM mutants would be expected to be as inhibitory as wild-type UvsY toward UvsX, which is not the case (Figure 2). Therefore, DNA-independent protein–protein interactions do not appear to be an important inhibitory mechanism in this system. Similarly, the strictly inhibitory effects of wild-type UvsY on UvsX-H195Q/A mutants, even at relatively low UvsY concentrations, demonstrate that the negative effects of competing protein–ssDNA interactions dominate any positive effects of protein–protein interactions on UvsX mutant activities. We conclude that the relationship between UvsX and UvsY ssDNA-binding affinities is of primary importance in determining whether the mediator stimulates or inhibits recombinase activities.
The postulated ‘closed’ and ‘open’ UvsY–ssDNA complexes have not been observed directly. Indirect evidence supports the notion that UvsY–ssDNA complexes can occupy two or more conformational states, however. Fluorescence studies of UvsY interactions with etheno-modified ssDNA (εDNA) demonstrate that there is an unusually large (4- to 6-fold) enhancement of εDNA fluorescence caused by UvsY binding at low salt (23). The addition of moderate salt reduces UvsY–εDNA fluorescence enhancement to 2- to 3-fold, a more typical value observed for many proteins that bind to ssDNA (16,22). Similarly, the observed degree of ssDNA wrapping by UvsY depends on DNA tension as well as on salt concentration (20). These observations suggest that there is a structural transition in UvsY–ssDNA complexes that is sensitive to ions, to DNA stretching or conceivably to protein–protein interactions. It is reasonable to hypothesize, as we do in our model (Figure 6A), that mutations in UvsY affect the position of equilibrium between UvsY–ssDNA conformers, and that different conformers have different accessibilities to UvsX protein.
An attractive corollary to our model (Figure 6A) is that the ‘closed’ UvsY–ssDNA complex interferes with Gp32–ssDNA interactions while the ‘open’ complex promotes UvsX–ssDNA interactions, allowing for a ‘double hand-off’ of ssDNA, from Gp32 to UvsY then UvsY to UvsX, during presynaptic filament assembly. There is growing evidence that the coordinated hand-off of DNA intermediates between proteins is a common feature of DNA replication and repair pathways. For example, it is proposed that ssDNA is handed off from one primosome protein to another as part of a dynamic primosome assembly process during replication fork restart (30). Evidence has also been presented for the step-wise hand-off of DNA repair intermediates between AP endonuclease, DNA polymerase beta and other elements of the base excision repair machinery (31,32). Our results indicate that DNA hand-offs also occur in homologous recombination and homology-directed repair. The biological functions of the handoff of ssDNA from an SSB protein to a mediator to a recombinase may include protecting the cell from cytotoxic effects of ssDNA and/or excluding other enzymes, such as helicases, polymerases or primases from the ssDNA until an appropriate point of the homology-directed repair pathway is reached. In fact, assembly of the T4 replicative helicase/primosome apparatus is rigidly excluded from ssDNA undergoing UvsY-mediated presynaptic filament assembly, providing evidence that ssDNA hand-off mechanisms are important for the coordination of recombination-dependent DNA replication (9). Although the coordinated transfer of ssDNA between recombination proteins appears to be driven by ssDNA structural changes, it seems likely that specific protein–protein interactions are also important for maintaining the overall integrity of the process. For instance, a seamless UvsX–UvsY–ssDNA filament contains no Gp32 and so is refractory to helicase loading by the T4 Gp59 protein; but UvsY depletion allows some Gp32 clusters to interrupt the presynaptic filament, which then become targets for untimely Gp59-mediated helicase assembly that disrupts recombination (9).
Another important finding of this study is that UvsY strongly suppresses AMP production by UvsX's ssDNA-stimulated ATPase activity (Figure 2). The ATPase activity of UvsX is unusual in that both ADP and AMP are produced as products (10,11). Differences in the steady-state kinetics of ADP and AMP production suggest that the two products are formed at two different classes of active sites within the UvsX–ssDNA presynaptic filament, one that generates ADP, and one that generates AMP (11). UvsY therefore either selectively inhibits the active sites that produce AMP, or converts them into ADP-producing active sites. Based on kinetics data, we proposed a model in which ADP and AMP production occur sequentially within one type of active site (11). According to this model (Figure 6B), UvsY could suppress AMP production at a given site by acting as an ADP/ATP nucleotide exchange factor. This would explain the large increase in ADP/AMP ratio and overall ATPase activity increase brought about by UvsY under optimal conditions. It is noteworthy that the highest ADP/AMP product ratios are observed at UvsX and UvsY concentrations that yield optimal stability of presynaptic filaments and optimal DNA strand exchange activity (Figure 6B). Under steady-state conditions for ATP hydrolysis, AMP production by the second class of sites is associated with the lowest affinity for ssDNA (11). Also, previous work demonstrated that UvsX–ssDNA presynaptic filaments are destabilized as ATP substrate is depleted and ADP and AMP products accumulate (8). Results of this study suggest that UvsY could stabilize presynaptic filaments by promoting a rapid exchange of ADP product for ATP substrate at UvsX active sites. This exchange would tend to increase the average lifetime of UvsX subunits in the ATP-occupied, high ssDNA-binding affinity state, while minimizing lifetimes of ADP- or AMP-occupied forms with lower affinity for ssDNA (Figure 6B). It is important to note that this represents a different, but ultimately congruent, method of regulating ssDNA-binding affinity than is used by many (predominantly eukaryotic) recombinases. For example, the human hRAD51 recombinase hydrolyzes ATP rapidly in the presence of ssDNA, but releases the ADP product very slowly, causing the enzyme to become trapped in an inactive hRAD51–ADP–ssDNA state (33). Ca++ ions stimulate the recombination activity of hRAD51 by slowing ATP hydrolysis, increasing its lifetime in the active hRAD51–ATP–ssDNA state (33). In the T4 system, UvsY realizes the same goal by helping UvsX to eject ADP from the active site and bind to a new ATP substrate molecule. Though carried out by different mechanisms, the net effect is the same in the human and T4 recombination systems—recombination activity is increased by increasing the average lifetime of the active recombinase–ATP–ssDNA state.
It is clear from our results that optimal assembly and activity of presynaptic filaments during homologous recombination involves a delicate interplay between the DNA-binding activities of recombinase and mediator components, which are linked in multiple ways to filament stability. The data are consistent with the notion that mediator effects on DNA structure and on recombinase nucleotidase activity are important modulators of presynaptic filament stability. The strong conservation of function between T4 UvsX/UvsY and other recombinase/mediator pairs (i.e. eukaryotic Rad51/Rad52 and bacterial RecA/RecOR) (6) suggests that similar mechanistic strategies may be used to control the assembly and activity of presynaptic filaments in many different organisms.
FUNDING
National Institutes of Health [Program Project grant number P01 CA098993]. Funding for open access charge: National Institutes of Health [Program Project grant number P01 CA098993].
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors thank Dr Hang Xu, Dr Jie Liu and Jennifer Tomczak for helpful comments, suggestions and technical assistance.
REFERENCES
- 1.Bianco PR, Tracy RB, Kowalczykowski SC. DNA strand exchange proteins: a biochemical and physical comparison. Front. Biosci. 1998;3:D570–D603. doi: 10.2741/a304. [DOI] [PubMed] [Google Scholar]
- 2.Yu VP, Koehler M, Steinlein C, Schmid M, Hanakahi LA, van Gool AJ, West SC, Venkitaraman AR. Gross chromosomal rearrangements and genetic exchange between nonhomologous chromosomes following BRCA2 inactivation. Genes Dev. 2000;14:1400–1406. [PMC free article] [PubMed] [Google Scholar]
- 3.Khanna KK, Jackson SP. DNA double-strand breaks: signaling, repair and the cancer connection. Nat. Genet. 2001;27:247–254. doi: 10.1038/85798. [DOI] [PubMed] [Google Scholar]
- 4.Moynahan ME, Pierce AJ, Jasin M. BRCA2 is required for homology-directed repair of chromosomal breaks. Mol. Cell. 2001;7:263–272. doi: 10.1016/s1097-2765(01)00174-5. [DOI] [PubMed] [Google Scholar]
- 5.Jasin M. Homologous repair of DNA damage and tumorigenesis: the BRCA connection. Oncogene. 2002;21:8981–8993. doi: 10.1038/sj.onc.1206176. [DOI] [PubMed] [Google Scholar]
- 6.Beernink HT, Morrical SW. RMPs: recombination/replication mediator proteins. Trends Biochem. Sci. 1999;24:385–389. doi: 10.1016/s0968-0004(99)01451-6. [DOI] [PubMed] [Google Scholar]
- 7.Sugiyama T, Kowalczykowski SC. Rad52 protein associates with replication protein A (RPA)-single-stranded DNA to accelerate Rad51-mediated displacement of RPA and presynaptic complex formation. J. Biol. Chem. 2002;277:31663–31672. doi: 10.1074/jbc.M203494200. [DOI] [PubMed] [Google Scholar]
- 8.Liu J, Qian N, Morrical SW. Dynamics of bacteriophage T4 presynaptic filament assembly from extrinsic fluorescence measurements of Gp32-single-stranded DNA interactions. J. Biol. Chem. 2006;281:26308–26319. doi: 10.1074/jbc.M604349200. [DOI] [PubMed] [Google Scholar]
- 9.Bleuit JS, Xu H, Ma Y, Wang T, Liu J, Morrical SW. Mediator proteins orchestrate enzyme-ssDNA assembly during T4 recombination-dependent DNA replication and repair. Proc. Natl Acad. Sci. USA. 2001;98:8298–8305. doi: 10.1073/pnas.131007498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Formosa T, Alberts BM. Purification and characterization of the T4 bacteriophage uvsX protein. J. Biol. Chem. 1986;261:6107–6118. [PubMed] [Google Scholar]
- 11.Farb JN, Morrical SW. Role of allosteric switch residue histidine-195 in maintaining active site asymmetry in presynaptic filaments of bacteriophage T4 UvsX recombinase. J. Mol. Biol. 2009;385:393–404. doi: 10.1016/j.jmb.2008.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cunningham RP, Berger H. Mutations affecting genetic recombination in bacteriophage T4D. I. Pathway analysis. Virology. 1977;80:67–82. doi: 10.1016/0042-6822(77)90381-6. [DOI] [PubMed] [Google Scholar]
- 13.Melamede RJ, Wallace SS. Properties of the nonlethal recombinational repair x and y mutants of bacteriophage T4. II. DNA synthesis. J. Virol. 1977;24:28–40. doi: 10.1128/jvi.24.1.28-40.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Melamede RJ, Wallace SS. Phenotypic differences among the alleles of the T4 recombination defective mutants. Mol. Gen. Genet. 1980;179:327–330. doi: 10.1007/BF00425460. [DOI] [PubMed] [Google Scholar]
- 15.Melamede RJ, Wallace SS. Properties of the nonlethal recombinational repair deficient mutants of bacteriophage T4. III. DNA replicative intermediates and T4w. Mol. Gen. Genet. 1980;177:501–509. doi: 10.1007/BF00271490. [DOI] [PubMed] [Google Scholar]
- 16.Sweezy MA, Morrical SW. Biochemical interactions within a ternary complex of the bacteriophage T4 recombination proteins uvsY and gp32 bound to single-stranded DNA. Biochemistry. 1999;38:936–944. doi: 10.1021/bi9817055. [DOI] [PubMed] [Google Scholar]
- 17.Beernink HT, Morrical SW. The uvsY recombination protein of bacteriophage T4 forms hexamers in the presence and absence of single-stranded DNA. Biochemistry. 1998;37:5673–5681. doi: 10.1021/bi9800956. [DOI] [PubMed] [Google Scholar]
- 18.Liu J, Bond JP, Morrical SW. Mechanism of presynaptic filament stabilization by the bacteriophage T4 UvsY recombination mediator protein. Biochemistry. 2006;45:5493–5502. doi: 10.1021/bi0525167. [DOI] [PubMed] [Google Scholar]
- 19.Bleuit JS, Ma Y, Munro J, Morrical SW. Mutations in a conserved motif inhibit single-stranded DNA binding and recombination mediator activities of bacteriophage T4 UvsY protein. J. Biol. Chem. 2004;279:6077–6086. doi: 10.1074/jbc.M311557200. [DOI] [PubMed] [Google Scholar]
- 20.Pant K, Shokri L, Karpel RL, Morrical SW, Williams MC. Modulation of T4 gene 32 protein DNA binding activity by the recombination mediator protein UvsY. J. Mol. Biol. 2008;380:799–811. doi: 10.1016/j.jmb.2008.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: a laboratory manual. 2nd. Plainview, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 22.Ando RA, Morrical SW. Single-stranded DNA binding properties of the UvsX recombinase of bacteriophage T4: binding parameters and effects of nucleotides. J. Mol. Biol. 1998;283:785–796. doi: 10.1006/jmbi.1998.2124. [DOI] [PubMed] [Google Scholar]
- 23.Sweezy MA, Morrical SW. Single-stranded DNA binding properties of the uvsY recombination protein of bacteriophage T4. J. Mol. Biol. 1997;266:927–938. doi: 10.1006/jmbi.1996.0829. [DOI] [PubMed] [Google Scholar]
- 24.Gill SC, von Hippel PH. Calculation of protein extinction coefficients from amino acid sequence data. Anal. Biochem. 1989;182:319–326. doi: 10.1016/0003-2697(89)90602-7. [DOI] [PubMed] [Google Scholar]
- 25.Yassa DS, Chou KM, Morrical SW. Characterization of an amino-terminal fragment of the bacteriophage T4 uvsY recombination protein. Biochimie. 1997;79:275–285. doi: 10.1016/s0300-9084(97)83515-8. [DOI] [PubMed] [Google Scholar]
- 26.Morrical SW, Alberts BM. The UvsY protein of bacteriophage T4 modulates recombination-dependent DNA synthesis in vitro. J. Biol. Chem. 1990;265:15096–15103. [PubMed] [Google Scholar]
- 27.Kodadek T, Wong ML, Alberts BM. The mechanism of homologous DNA strand exchange catalyzed by the bacteriophage T4 uvsX and gene 32 proteins. J. Biol. Chem. 1988;263:9427–9436. [PubMed] [Google Scholar]
- 28.Kodadek T, Gan DC, Stemke-Hale K. The phage T4 uvsY recombination protein stabilizes presynaptic filaments. J. Biol. Chem. 1989;264:16451–16457. [PubMed] [Google Scholar]
- 29.Jiang H, Salinas F, Kodadek T. The gene 32 single-stranded DNA-binding protein is not bound stably to the phage T4 presynaptic filament. Biochem. Biophys. Res. Commun. 1997;231:600–605. doi: 10.1006/bbrc.1997.6160. [DOI] [PubMed] [Google Scholar]
- 30.Lopper M, Boonsombat R, Sandler SJ, Keck JL. A hand-off mechanism for primosome assembly in replication restart. Mol. Cell. 2007;26:781–793. doi: 10.1016/j.molcel.2007.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu Y, Prasad R, Beard WA, Kedar PS, Hou EW, Shock DD, Wilson SH. Coordination of steps in single-nucleotide base excision repair mediated by apurinic/apyrimidinic endonuclease 1 and DNA polymerase beta. J. Biol. Chem. 2007;282:13532–13541. doi: 10.1074/jbc.M611295200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hitomi K, Iwai S, Tainer JA. The intricate structural chemistry of base excision repair machinery: implications for DNA damage recognition, removal, and repair. DNA Repair (Amst) 2007;6:410–428. doi: 10.1016/j.dnarep.2006.10.004. [DOI] [PubMed] [Google Scholar]
- 33.Bugreev DV, Mazin AV. Ca2+ activates human homologous recombination protein Rad51 by modulating its ATPase activity. Proc. Natl Acad. Sci. USA. 2004;101:9988–9993. doi: 10.1073/pnas.0402105101. [DOI] [PMC free article] [PubMed] [Google Scholar]