Abstract
The transcription regulator, tvMyb1, is the first Myb family protein identified in Trichomonas vaginalis. Using an electrophoretic mobility shift assay, we defined the amino-acid sequence from Lys35 to Ser141 (tvMyb135–141) as the minimal DNA-binding domain, encompassing two Myb-like DNA-binding motifs (designated as R2 and R3 motifs) and an extension of 10 residues at the C-terminus. NMR solution structures of tvMyb135–141 show that both the R2 and R3 motifs adopt helix-turn-helix conformations while helix 6 in the R3 motif is longer than its counterpart in vertebrate Myb proteins. The extension of helix 6 was then shown to play an important role in protein stability as well as in DNA-binding activity. The structural basis for the tvMyb135–141/DNA interaction was investigated using chemical shift perturbations, residual dipolar couplings, DNA specificity data and data-driven macromolecular docking by HADDOCK. Our data indicate that the orientation between R2 and R3 motifs dramatically changes upon binding to DNA so as to recognize the DNA major groove through a number of key contacts involving residues in helices 3 and 6. The tvMyb135–141/DNA complex model furthers our understanding of DNA recognition by Myb proteins and this approach could be applied in determining the complex structures involving proteins with multiple domains.
INTRODUCTION
Transcription factors regulate the expression of genes at the level of transcription and control many critical biological processes. These factors typically recognize DNA sequences in the promoter regions of the target genes and regulate the frequency of transcription initiation of the genes. Transcription factors contain DNA-binding domains which bind to DNA with high sequence specificity and are classified based on the sequence similarity in the DNA-binding domain. Myb is one of the largest transcription factor families in plants (1), which contains DNA-binding domains composed of one, two or three repeated motifs of approximately 50 amino acids surrounded by three conserved tryptophan residues (2). These repeated motifs adopt a helix-turn-helix conformation to recognize the major groove of target DNA sequences. Vertebrate c-Myb protein contains three tandem repeated motifs, designated as R1, R2 and R3 motifs (3). Other Myb repeated motifs are categorized according to their similarity to the R1, R2 or R3 motifs.
Myb proteins regulate myriad gene-specific transcription in a wide range of eukaryotic systems. In vertebrates, there are three cellular Myb proteins, A-Myb, B-Myb and c-Myb (4), that are composed of 630–750 amino-acid residues and contain a highly conserved N-terminal DNA-binding domain (with ∼90% identity) consisting of R1, R2 and R3 DNA-binding motifs. Vertebrate Myb proteins all recognize specific DNA stretches with a core pentanucleotide sequence, CNGTT, through three key base-contacting amino-acid residues: Lys128 in the R2 motif and Lys182 and Asn183 in the R3 motif (3). In plants such as Arabidopsis thaliana, the Myb protein family is expanded and contains more than 130 distinct members, most of which contain an R2R3 DNA-binding domain (125 genes) with at least 40% sequence identity (1). Although the key base-contacting amino-acid residues are also conserved, the sequence contexts of the Myb recognition elements in plants are not limited to those with a CNGTT core (5).
The transcription factor, tvMyb1 protein, is the first Myb family protein identified in the protozoan parasite Trichomonas vaginalis (6). Trichomonas vaginalis is responsible for the disease trichomoniasis which is one of the most common sexually transmitted diseases in humans (7). The infection of T. vaginalis is also associated with several adverse health consequences including increased human immunodeficiency virus transmission, infertility, cervical intraepithelial neoplasia development in women, and nongonoccocal urethritis and chronic prostatitis in men (8,9). With an increasing number of drug-resistant clinical T. vaginalis strains (10,11), the infection caused by T. vaginalis could become a major threat to public health. The ap65-1 gene, an iron-inducible virulence gene, encodes a 65 kDa protein that is reputed to be one of the surface adhesion proteins increasing the cytoadherence of T. vaginalis to the host cells which in turn induces the infection (12,13). Based on our previous studies, tvMyb1 protein, which contains a Myb-like R2R3 DNA-binding domain, can regulate multifarious ap65-1 gene expression by recognizing multiple Myb recognition elements, MRE-1/MRE-2r (TAACGATAT, MRE-1 in italic and MRE-2r underlined) and MRE-2f (TATCGT) with a core hexanucleotide sequence, ACGATA (6,14,15). Interestingly, the C-terminal fragment of tvMyb1 protein (residues 132–215) positively regulates binding of the R2R3 domain to MRE-1/MRE-2r but negatively regulates binding of the R2R3 domain to MRE-2f. However, the N-terminal fragment (residues 1–34) was shown to negatively regulate binding activity of the R2R3 domain to both MRE-1/MRE-2r and MRE-2f (J. H. Tai, unpublished data). Therefore, the availability of detailed structural information for the DNA-binding domain from tvMyb1 protein and its complexes with DNA target sites is essential for understanding its role and the mechanism of action in transcriptional control.
In the present study, the electrophoretic mobility shift assay (EMSA) was used to identify the DNA-binding domain of tvMyb1 spanning from residues 35 to 141 (referred as tvMyb135–141). This longer fragment showed a dramatic increase in protein stability as well as in DNA-binding ability when compared to the shorter fragment (tvMyb135–131), identified by Pfam database (16) and consisting of R2 and R3 motifs only. The NMR solution structure of tvMyb135–141 was then determined and it was found that the C-terminal 10-residue extension maintains the integrity of helix 6 at the R3 motif and thus increases both stability and DNA-binding activity of tvMyb135–141. The structural model of tvMyb135–141 in DNA-bound conformation was derived by refining the free structure with 1DNH residual dipolar couplings obtained from tvMyb135–141 in complex with DNA. The structures show that the relative orientation between R2 and R3 motifs is changed when bound to DNA. Finally, a data-driven structural model of the tvMyb135–141/DNA complex was calculated by HADDOCK (17–19) using chemical shift perturbation, residual dipolar couplings and DNA specificity data. The well-converged model reveals a number of specific contacts between the major groove of DNA and the residues at helices 3 and 6 of tvMyb135–141 as well as providing information about the DNA recognition by tvMyb1 DNA-binding domain. We also demonstrated that the inclusion of residual dipolar couplings in the data-driven macromolecular docking by HADDOCK is an efficient approach to determine the structure of complexes involving proteins with multiple domains.
MATERIALS AND METHODS
Sample preparation
The gene encoding tvMyb135–141 or tvMyb135–131 was PCR-amplified from the genomic DNA and subcloned to the pET29b (Novagen) vector. The mutants of tvMyb135–141 were constructed by using the kit QuikChange from Stratagene. Three mutants F38A, T67A and N126A were confirmed by DNA sequencing. The target protein with an N-terminal His-tag was expressed in Escherichia coli BL21(DE3) in LB broth. For labeled (15N and 15N/13C) samples, the cells were grown in M9 minimal medium containing 15NH4Cl (1 g l–1) and/or 13C-glucose (2 g l–1) at 37°C with 30 μg ml–1 kanamycin until OD600 readings reach 0.8 and then were induced with 1 mM IPTG and grown for 4 additional hours. The cells were harvested and resuspended in buffer (20 mM Tris, pH 8.0, 500 mM NaCl) and the suspension was lysed by microfluidizer and centrifuged at 12 000g for 30 min. The supernatant was applied to a nickel-nitrilotriacetic acid (Ni-NTA) affinity chromatographic column, washed with a washing buffer (20 mM Tris, pH 8.0, 500 mM NaCl and 60 mM imidazole) and eluted with an elution buffer (20 mM NaH2PO4, pH 6.0, 150 mM NaCl and 50 mM EDTA). Purity and authenticity of the recombinant proteins were verified by SDS–PAGE and mass analysis. Finally, the target protein was further dialyzed and concentrated with buffer at pH 6.0 (20 mM NaH2PO4, 50 mM NaCl, 5 mM NaN3 and 10 mM dithiothreitol) for NMR study. The single stranded DNA of MRE-1/MRE-2r (5′-AAGATAACGATATTTA-3′) and biotinylated DNA for SPR experiments were purchased from MDBio Inc., Taiwan. The double-stranded DNA was prepared by mixing equal amounts of two complementary deoxynucleotides, heating to 95°C for 10 min and cooling slowly to room temperature. The final concentrations of NMR samples are around 1.5 mM for free tvMyb135–141 and 0.7 mM for tvMyb135–141/DNA complex.
EMSA
All proein samples were incubated with γ-32P-labeled MRE-1/MRE-2r or MRE-2f. Probe labeling was performed as described previously (6). The mixture was separated in a 10% acrylamide gel by electrophoresis. The DNA–protein complexes in reaction with probes were detected by autoradiograms. A BCA protein quantification kit was used to determine protein concentration by the supplier (Pierce).
Circular dichroism (CD)
All CD spectra were measured using an Aviv CD 202 spectrometer (Lakewood, NJ) calibrated with (+)-10-camphorsulfonic acid. All spectra were acquired at 298 K with ∼20 μM protein samples in a 1 mm path-length cuvette. The signals from 195 nm to 260 nm were recorded three times with wavelength step of 0.5 nm and bandwidth of 1 mm. The average signals were converted from CD signal (millidegree) into mean residue ellipticity after subtraction of the background signals. Equilibrium thermal-denaturing experiments were obtained by measuring the change of CD signal at 222 nm from 4°C to 95°C with a 1°C interval and 3 min for equilibrium. The spectra were displayed and analyzed by SigmaPlot 8.02 (SPSS Inc.).
Surface plasmon resonance
The real-time association and dissociation of tvMyb135–131, tvMyb135–141 and three mutants with MRE-1/MRE-2r DNA duplex were measured at 25°C on a BIAcore 3000 instrument (BIAcore AB, Uppsala, Sweden). The 16-bp MRE-1/MRE-2r DNA duplex, biotinylated at the 5′ end and dissolved in PBS buffer (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4 and 2 mM KH2PO4, pH 7.4) with a concentration of 0.02 μM, was applied to the streptavidin SA sensor chip (BIAcore AB, Uppsala, Sweden) at a flow rate of 10 μl min–1, which resulted in the capture of 100–150 response units. Protein samples (25, 12.5, 6.3, 3.1, 1.7 or 0.8 nM) were injected to the SA sensor chip immobilized with DNA at a constant flow rate of 30 μl min–1 for 2 min for association and then the running buffer (10 mM HEPES, 150 mM NaCl, 3.4 mM EDTA, pH 7.4, containing 0.005% Tween 20) was applied at the same rate for 3 min for dissociation. After each binding experiment, the sensor chip was regenerated with 1 M NaCl. For fitting the binding kinetics, the sensorgrams were evaluated by BIAevaluation version 4.1 (BIAcore AB, Uppsala, Sweden) using a 1:1 (Langmuir) binding model.
NMR experiments
All NMR spectra were carried out using Bruker AVANCE 600 or 800 MHz spectrometers equipped with a triple (1H, 13C and 15N) resonance cryoprobe which included a shielded z-gradient. The triple-resonance experiments [HNCO, HN(CA)CO, CBCA(CO)NH and HNCACB] were used for backbone resonance assignment of free tvMyb135–141 and tvMyb135–141 in complex with DNA. The weighted chemical shift perturbations for backbone 15N and 1HN resonances were calculated by the equation: Δδ = [(ΔδHN)2 + (ΔδN/5)2]0.5. Side-chain resonance assignment of tvMyb135–141 was achieved following similar procedures published previously (20). To measure residual dipolar couplings (RDC) of tvMyb135–141 in complex with DNA, the filamentous bacteriophage Pf1 (Asla Biotech Ltd, Latvia) was selected as the orienting medium. Pf1 (10 mg ml–1) was added to the 15N-labeled protein/DNA complex sample at pH 7.0, to produce weak alignment of the complex. No significant perturbations in amide chemical shifts were observed in the presence of Pf1 phage, suggesting that the phage caused little structural change. 2D 1H-coupled (F1) IPAP 1H-15N HSQC spectra (21) were acquired with 256 complex t1 (15N) points and 64 scans per t1 increment for both the isotropic and anisotropic samples. All of the NMR spectra were processed using Bruker XWINNMR or NMRPipe package (22), and analyzed using NMRView 5.0 (23) or Sparky (24).
Structure determination of tvMyb135–141
The backbone dihedral angle restraints were predicted using the program TALOS (25). The hydrogen bond restraints were introduced for residues that exhibit slow amide proton exchange rates. The nuclear Overhauser effect restraints from the 1H-15N NOESY-HSQC and 1H-13C NOESY-HSQC spectra were automatically assigned by the CANDID module of CYANA (26) and checked manually. Secondary structure was identified based on chemical shift index, amide proton exchange rate and NOEs connectivities. NMR structures were calculated from all experimental restraints by dynamical simulated annealing procedure using a modified protocol of Xplor-NIH program (27). In this protocol, the final van der Waals radii in the cooling step was increased ($fin_rad = 0.80; the original value is 0.75) to reduce the numbers of close-contacts between heavy atoms. The final 20 structures with no distance restraint violation greater than 0.4 Å, and no dihedral angle restraint violations larger than 3° were chosen on the basis of total energy. The program PROCHECK-NMR (28) was applied for analyzing the generated structures.
Structure determination of tvMyb135–141 in the DNA-bound conformation
The structure of tvMyb135–141 in the DNA-bound conformation was calculated by refining the free structure against 74 1DNH RDC constraints obtained from 15N-labeled protein in complex with DNA in the presence of Pf1 (10 mg ml–1) at pH 7.0. The NOEs, dihedral angles and hydrogen bond restraints that define the helical structure of free tvMyb135–141 were used in the refinement protocol; the NOE restraints that define the distances of atoms between the R2 and R3 motifs and the restraints of loop regions were excluded. The measured 1DNH RDC values ranged from –25 to 29 Hz. The axial and rhombic components of the alignment tensor were determined by the grid search method proposed by Clore and coworkers (29) to be 15.4 Hz and 7 Hz, respectively. A set of 20 structures with no distance restraint violations greater than 0.3 Å, and no dihedral angle restraint violations larger than 5° were selected based on the energy of RDC constraints.
HADDOCK docking
The model of the tvMyb135–141/DNA complex was calculated by using the information-driven method called HADDOCK v2.0 (17–19). Chemical shift perturbations (CSP) and DNA specificity data were translated into ambiguous interaction restraints (AIRs) to drive the docking process. The AIRs can also be combined with RDC data to allow a better definition of the relative orientation of the components (18,30). The starting structures for the docking were a B form model of the 16-bp MRE-1/MRE-2r DNA duplex built by the Discovery studio 2.0 (Accelrys) and the 20 structures of tvMyb135–141 in the DNA-bound conformation. For tvMyb135–141 protein, residues having a weighted chemical shift perturbation upon complex formation greater than 0.5 ppm and displaying high solvent accessibility (>50%) were selected as active residues. Solvent accessibility for the active residues was calculated using the program NACCESS (31). The selected active residues are Val36, Phe38, Thr39, Asn69, Gln72, Glu75, Asn110, Asn122 and Asn126 and the semi-flexible regions were defined for residues 34–41, 67–77, 108–112 and 120–128. According to our previous results on MRE-1/MRE-2r DNA specificity, several base replacements disrupt the interactions between tvMyb1 protein and DNA, which include ADE6 to CYT, CYT8 to THY, GUA9 to ADE, ADE10 to CYT and THY11 to GUA (6). These nucleotides and their complementary bases were selected as active bases and the semi-flexible regions were defined for bases 6–11 and 22–27. The specific AIR restraints were defined between suitable atoms of active residues to unique base atoms of active bases. For example, the base replacement, CYT8 to THY, loses H41 of CYT8 and O6 of GUA25 for the specific hydrogen bond interaction. The AIR restraints were thus defined between H41 of CYT8 to carbonyl O of backbone, Oγ1 of Thr, or to carbonyl O of side-chains of Asn, Gln and Glu and between O6 of GUA25 to amide protons of backbone or to amide protons of side-chains of Asn and Gln. This rule was applied to define all the specific AIR restraints and a total of 31 AIRs with 2 Å distance definition were used in HADDOCK docking. Additional restraints to maintain base planarity and Watson Crick bonds were introduced for the DNA. The alignment tensor components for RDC constraints were determined as described above. Experimental RDCs were introduced as intervector projection angle restraints, VEAN energy terms (32), during stages (i) rigid body docking and (ii) semi-flexible simulated annealing. The floating alignment tensor, SANI energy term (33), was used in the stage of final water refinement. During the rigid body energy minimization, 10 000 structures were calculated and the 200 best solutions based on the intermolecular energy were used for the semi-flexible, simulated annealing followed by an explicit water refinement. Docked structures corresponding to the 200 best solutions with lowest intermolecular energies were generated. The 200 solutions were clustered using a 1.0 Å RMSD cut-off criterion. The clusters were ranked based on the averaged HADDOCK score of their top 10 structures.
Data bank accession number
The chemical shifts of tvMyb135–141 at pH 6.0 and 298K have been deposited in the BioMagResBank under accession number BMRB-15989. The best 20 structures of tvMyb135–141 and the best 10 structures of tvMyb135–141/DNA complex have been deposited in the RCSB Protein Data Bank under accession number 2k9n and 2kdz, respectively.
RESULTS
Identification of the DNA-binding domain of tvMyb1 for structural investigation
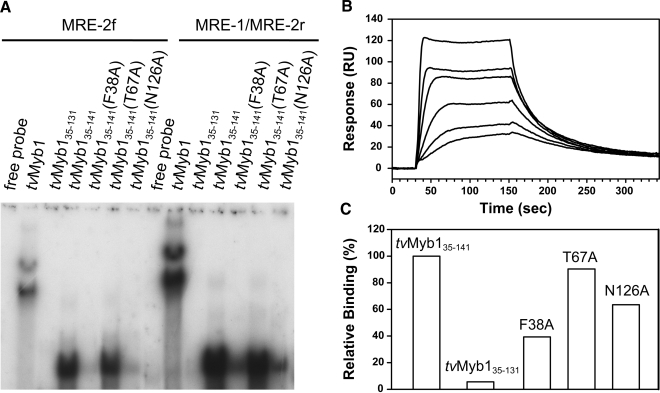
The transcription factor, tvMyb1, has been found to regulate the multifarious transcription of the ap65-1 gene by binding to two Myb recognition elements, MRE-1/MRE-2r and MRE-2f, with a core hexanucleotide sequence, ACGATA (6). Domain analysis in the Pfam database (16) indicated that the segments from Lys35 to Ile81 and Thr87 to Ile131 of tvMyb1 are classified as Myb-like DNA-binding motifs. The fragment, tvMyb135–131 was hence cloned but it was soon found to be highly susceptible to degradation and precipitation. Previous reports showed that several plant Myb proteins contain an additional C-terminal extension of the DNA-binding domain (34–36). Therefore, another fragment, tvMyb135–141 was constructed to test for the necessary DNA-binding ability and structural stability. The binding ability of these two fragments to the Myb recognition elements, MRE-1/MRE-2r and MRE-2f, were examined by EMSA. The shorter fragment, tvMyb135–131, showed little interaction with either element. In contrast, the longer fragment, that contained 10 additional C-terminal residues, exhibited the desired DNA-binding activity (Figure 1A).
Figure 1.
DNA-binding activity of tvMyb1, tvMyb135–131, tvMyb135–141 and the mutants probed by EMSA (A) and surface plasmon resonance (B and C). In (A), six protein samples, 100 ng tvMyb1 (lanes 2 and 9), 60 ng tvMyb135–131 (lanes 3 and 10), tvMyb135–141 (lanes 4 and 11), F38A (lanes 5 and 12), T67A (lanes 6 and 13) and N126A (lanes 7 and 14) were incubated with -32P-labeled MRE-2f probe (lanes 1–7) or MRE-1/MRE-2r probe (lanes 8–14). All protein/DNA mixtures were separated by 10% polyacrylamide gels by electrophoresis. Free probes were loaded at lanes 1 and 8 as controls. (B) The SPR sensorgrams for the binding of tvMyb135–141 to the DNA duplex. The 16-bp MRE-1/MRE-2r DNA duplex was immobilized on the streptavidin SA sensor chip. The protein concentrations are 25, 12.5, 6.3, 3.1, 1.7 and 0.8 nM. The traces were analyzed with a 1:1 Langmuir-binding model and give an equilibrium dissociation constant (KD) for the tvMyb135–141/DNA interaction to be 1.24 × 10–9 M. (C) Relative DNA-binding ability of tvMyb135–131 and three mutants as calculated from SPR responses 110 s after injection of protein (100 nM) onto the DNA immobilized SA sensor chip.
To quantitate the DNA-binding affinity of the two fragments, we performed surface plasmon resonace (SPR) experiments on a BIAcore 3000 biosensor system. The 16-bp MRE-1/MRE-2r DNA duplex (nucleotide sequence: AAGATAACGATATTTA) was immobilized onto the streptavidin SA sensor chip and probed with different concentration of proteins (from 0.78 nM to 25 nM). The sensorgrams of tvMyb135–141 showed a concentration-dependent binding (Figure 1B) and the traces were analyzed with a 1:1 Langmuir binding (with mass transfer) model by BIAevaluation software, giving an equilibrium dissociation constant (KD) of 1.24 × 10–9 M for tvMyb135–141 interacting with immobilized MRE-1/MRE-2r, indicating a high-affinity interaction that typically observed for a sequence-specific DNA-binding protein. Similar SPR experiments for the shorter fragment tvMyb135–131 did not give reliable results possibly due to its high propensity to aggregate. Hence the relative DNA-binding ability of tvMyb135–131 was calculated from SPR responses, which indicated that the DNA-binding ability of tvMyb135–131 is lower than 10% of that of tvMyb135–141 (Figure 1C), in good agreement with the results from EMSA assays. The DNA bindings of three mutants of tvMyb135–141 were also checked by EMSA and SPR experiments and are described later in this section.
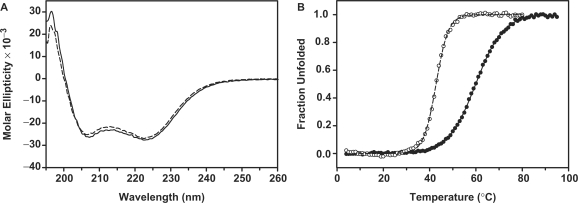
The secondary structures and structural stabilities of tvMyb135–131 and tvMyb135–141 were monitored by circular dichroism spectra. The wavelength scans of these two fragments showed similar absorptions at 222 and 208 nm, indicating that they exhibit similar helical structures (Figure 2A). However, temperature denaturation experiments indicated that tvMyb135–141 exhibits higher thermal stability than tvMyb135–131 (Figure 2B). The structures of the two fragments in solution were further checked by 2D 1H-15N heteronuclear single quantum coherence (HSQC) spectra. Both spectra showed well-dispersed cross peaks that overlap significantly with each other, indicating that each fragment is well structured in solution (Supplementary Figure S1). In addition, the cross peaks of the additional C-terminal 10 residues of tvMyb135–141 are also well dispersed, suggesting a stable conformation rather than an unstructured state. Based on all these results, the longer fragment (tvMyb135–141) was identified as the DNA-binding domain of tvMyb1 protein and was selected for further NMR structural investigation.
Figure 2.
Comparison of CD data between tvMyb135–131 (dashed line) and tvMyb135–141 (solid line). (A) CD spectra of tvMyb135–141 and tvMyb135–131 at 25°C, indicating that two proteins exhibit similar secondary structures. (B) The equilibrium thermal unfolding experiments followed at 222 nm. The Tm value of tvMyb135–141 is 59.9°C, around 17°C higher than that of tvMyb135–131.
NMR resonance assignment and structural determination of tvMyb135–141
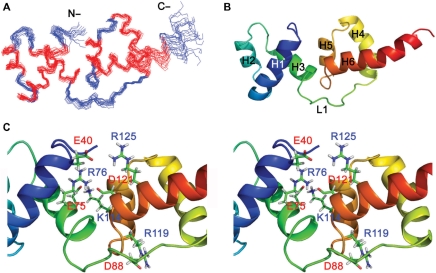
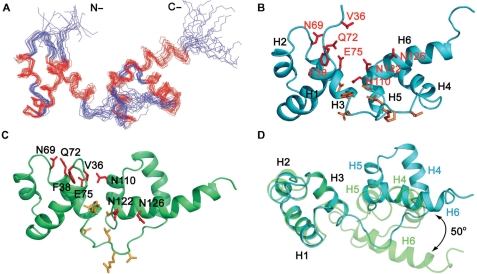
NMR resonance assignment of tvMyb135–141 was achieved following the procedures published previously (20) and is described briefly in ‘Materials and Methods’ section. All backbone resonances were clearly identified except those of Ile81 and Met129 (Supplementary Figure S2) and around 90% of the side-chain resonances were unambiguously assigned. From the consensus chemical shift indices and nuclear Overhauser effect (NOE) patterns, the secondary structure topology of tvMyb135–141 was determined. The DNA-binding domain tvMyb135–141 contains six helices, designated as: helix 1 (H1), Glu40 to Tyr53; helix 2 (H2), Trp58 to Leu64; helix 3 (H3), Pro70 to Tyr80; helix 4 (H4), Pro92 to Glu104; helix 5 (H5), Trp109 to Leu116 and helix 6 (H6), Asp121 to Arg135. The first three helices of tvMyb135–141 resemble the R2 motif of c-Myb and the last three helices correspond to the R3 motif of c-Myb. The R2 motif is connected to the R3 motif by a 11-residue loop (L1). However, it is worth noting that H6 of tvMyb135–141 is longer than the corresponding helix in c-Myb. In the preceding results, we found that the C-terminal 10-residue extension (Ala132-Ser141) increases the DNA-binding ability as well as the protein stability significantly. Accordingly, it seems that the C-terminal extension maintains the integrity of H6 and thus increases both stability and DNA-binding activity.
The tertiary structure of tvMyb135–141 was subsequently determined based on a set of 1394 NOE distance restraints, 28 hydrogen bond restraints and 144 backbone φ and ψ dihedral angle restraints. An ensemble of 20 structures with no distance restraint violations greater than 0.4 Å and no dihedral angle restraint violations greater than 3° was selected based on the total energy (Figure 3A and B). The structural statistics are listed in Table 1. The tertiary structures of the R2 and R3 motifs are in particular well defined, with root-mean-square deviations (RMSD) of 0.32 (±11) Å and 0.52 (±0.19) Å for the backbone atoms of helical residues in the R2 and R3 motifs, respectively. The helix-turn-helix structure of the R2 and R3 motifs are highly stabilized by the hydrophobic cores composed of Phe38, Leu46, Leu49, Val50, Tyr53, Trp58, Ile61 and Trp77 for R2 and Trp90, Leu98, Tyr102, Trp109, Ile112, Leu116, Ile124, Trp128, Ile131 and Ala132 for R3. It was also found that H6 of the R3 motif is amphipathic and its hydrophobic residues Ile124, Trp128, Ile131 and Ala132 constitute the major part of the hydrophobic core of the R3 motif, suggesting that H6 is important for the stability of the R3 motif.
Figure 3.
NMR solution structure of tvMyb135–141. (A) Backbone representation of the ensemble of 20 lowest energy structures. The helical residues are colored in red and others in blue. (B) Secondary structures of the lowest energy structure of tvMyb135–141 displayed in rainbow colors from N-terminus (blue) to C-terminus (red). The first three helices (H1–H3) constitute the R2 motif and the last three helices (H4–H6) form the R3 motif. Two motifs are connected by a long loop (L1). (C) Stereo view of the interface between R2 and R3 motifs. The residues involved in salt bridges between two motifs are shown and labeled.
Table 1.
Structural statistics of tvMyb135–141 and tvMyb135–141/DNA complex
| Parameter | tvMyb135–141 | DNA-bound conformation | tvMyb135–141/DNA complex |
|---|---|---|---|
| NMR restraints used | |||
| Intraresidue (i – j = 0) | 289 | 196 | |
| Sequential (i – j = 1) | 551 | 403 | |
| Medium range (1 < i – j < 5) | 4500 | 405 | |
| Long range (i – j ≥ 5) | 104 | 83 | |
| Total NOE restraints | 1394 | 1087 | |
| Total AIRs | 20 | ||
| Hydrogen bonds | 28 × 2 | 28 × 2 | |
| Dihedral angles | 144 | 120 | |
| 1DHN RDCs | 74 | 74 | |
| Energy statistics | |||
| Etotal | 335.1 ± 11.5 | 221.1 ± 9.5 | −4462.1 ± 116.5 |
| Ebond | 29.3 ± 1.6 | 10.9 ± 0.9 | 30.1 ± 3.2 |
| Eangle | 114.6 ± 3.4 | 74.6 ± 2.2 | 81.3 ± 39.1 |
| Eimproper | 16.4 ± 0.5 | 20.8 ± 1.2 | 67.4 ± 21.9 |
| Evdw | 100.9 ± 2.8 | 73.2 ± 3.1 | −629.9 ± 38.3 |
| ENOE | 70.6 ± 2.1 | 38.2 ± 1.8 | |
| Edihe | 3.4 ± 0.3 | 3.9 ± 0.4 | |
| Esani | 10.1 ± 1.4 | ||
| Eelectr | −4862.4 ± 63.2 | ||
| RDC Q-factora | 0.67 ± 0.04 | 0.029 ± 0.003 | 0.102 ± 0.007 |
| Deviation from ideal geometry (RMSD) | |||
| Bond (Å) | 0.0041 ± 0.0001 | 0.0025 ± 0.0001 | 0.0032 ± 0.0002 |
| Angle (°) | 0.48 ± 0.01 | 0.38 ± 0.007 | 0.67 ± 0.03 |
| Mean global RMSD of helical residues (Å) | |||
| Backbone atoms | 0.70 ± 0.15 | 0.96 ± 0.25 | 0.43 ± 0.09 0.68 ± 0.25b |
| Heavy atoms | 1.56 ± 0.14 | 1.89 ± 0.22 | 0.88 ± 0.16 |
| Ramachandran plot | |||
| Most favored (%) | 78.9 | 80.9 | 77.0 |
| Allowed (%) | 18.4 | 17.6 | 17.5 |
| Generously allowed (%) | 2.5 | 1.5 | 4.4 |
| Disallowed (%) | 0.2 | 0.0 | 1.1 |
aQ-factor = RMS(Dcalc – Dobs)/RMS(Dobs), where Dcalc and Dobs are calculated and observed RDC values, respectively.
bFor backbone atoms of all helical residues and all phosphate backbone atoms of DNA.
Although the R2 and R3 motifs are well defined, their relative orientation was not fixed in the beginning of structural calculation because the loop (L1) between H3 and H4 was not well defined due to fewer NOE restraints observed in this region. After the NOE assignment was finished, we found several NOE cross peaks between the R2 and R3 motifs as listed in Supplementary Table S1. The RMSD values of the 20 final NMR structures were 0.70 (±0.15) Å and 1.56 (±0.14) Å for the backbone atoms and the heavy atoms of all helical residues, respectively. Even without the addition of any direct NOE restraint between positively charged residues and negatively charged residues, four possible salt bridges of Glu40(R2)-Arg125(R3), Glu75(R2)-Lys114(R3), Arg76(R2)-Asp121(R3) and Asp88(L1)-Arg119(R3) were observed to stabilize the relative orientation between the R2 and R3 motifs as shown in Figure 3C. From our NMR solution structure, it was observed that tvMyb135–141 is highly stabilized by hydrophobic and electrostatic interactions, which contribute to its high stability toward thermal denaturation.
Mapping the tvMyb135–141/DNA interaction by NMR spectroscopy
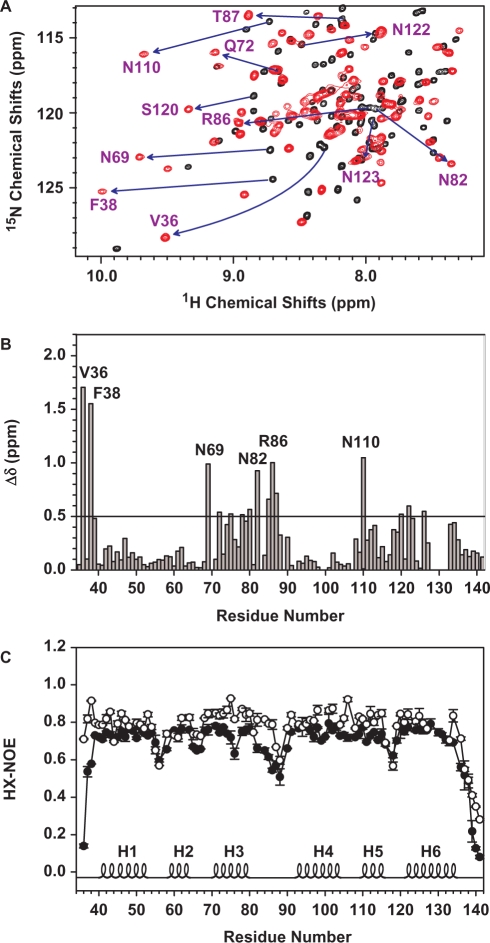
To map the DNA-binding sites of tvMyb135–141 with the 16-mer MRE-1/MRE-2r DNA duplex, the 15N-labeled protein sample was titrated with the DNA duplex in a 1:1 ratio and 2D 1H-15N HSQC spectra were acquired to monitor the chemical shift changes of backbone amide resonances. As shown in Figure 4A, the cross peaks in the 2D 1H-15N HSQC of tvMyb135–141/DNA complex in general are broader than the peaks of free tvMyb135–141 and the backbone amide resonances of Asp99, Gln100 and Trp128 to Ala132 could not be assigned due to peak broadening or peak absence, suggesting that these residues may experience intermediate exchange. A variety of experimental conditions have been tested, including different pH values, temperatures and DNA lengths, but the resulting NMR spectra did not show substantial improvements. Based on the best assignment of tvMyb135–141 in complex with DNA we can obtain, it was found that the backbone resonances undergo significant perturbations in complex with DNA (Figure 4B). The majority of residues that exhibit large changes in chemical shift (Δδ > Δδ average+SD ∼0.5 ppm) are located at the N-terminus, H3, L1 loop and H6. We mapped these residues onto the free tvMyb135–141 structure (Figure 5C) and found that these residues lie mainly in the middle part of the structure and constitute several discontinuous faces, which are unlikely to be the DNA-binding surfaces. Besides, several residues that exhibited large chemical shift changes are located in the interface between the R2 and R3 motifs (such as Asn110 and Asn122). These data suggest that the DNA-bound conformation is different from the free structure.
Figure 4.
Interaction between tvMyb135–141 and 16-bp MRE-1/MRE-2r DNA duplex analyzed by NMR. (A) Overlay of portion of the 2D 1H-15N HSQC spectra of free tvMyb135–141 (black) and tvMyb135–141 in complex with DNA (red). The residues that exhibit significant chemical shift perturbations are indicated. (B) Weighted chemical shift perturbations for backbone 15N and 1HN resonances as calculated by the equation: Δδ = [(ΔδHN)2 + (ΔδN/5)2]0.5. The solid line indicates 0.5 ppm (equal to Δδaverage + SD). (C) 1H-15N hetero-nuclear NOE of tvMyb135–141 in free form (filled circles) or in complexed with DNA (open circles). The secondary structures are annotated in the bottom.
Figure 5.
The tvMyb135–141 solution structure in free form and in DNA-bound conformation. (A) The ensemble of 20 final structures of tvMyb135–141 in DNA-bound conformation. The structures were derived from refining the free structure with 1DNH RDC constraints obtained from the tvMyb135–141/DNA complex sample. The structures with lowest RDC energies were selected. (B) Cartoon representation of tvMyb135–141 in DNA-bound conformation. The residues that exhibit significant chemical shift changes upon DNA binding are shown in stick and are colored in red for those selected as active residues in HADDOCK docking. (C) Cartoon representation of tvMyb135–141 in free form. The residues that exhibit significant chemical shift changes are also shown. (D) Structural superimposition of tvMyb135–141 R2 motifs in free form (light green) and DNA-bound form (light blue) revealed that the orientation of R3 motif in DNA-bound conformation rotated 50° relative to that in free form.
We then checked the backbone rigidity of tvMyb135–141 in the presence and absence of DNA by 1H-15N hetero-nuclear NOE (HX-NOE) measurements (Figure 4C). The data showed that the N-terminal residues, Val36-Phe38, are highly flexible in the absence of DNA and adopt a much more rigid conformation upon binding to DNA. The rigidity of residues at H3, L1 loop and the C-terminus are also increased, although not as dramatically as are the N-terminal residues. In addition, almost all the residues in the DNA-bound conformation demonstrate stronger HX-NOE values and the mean values of HX-NOE were increased from 0.67 (±0.14) to 0.77 (±0.12) when the protein binds to DNA, indicating that DNA binding highly stabilized the tvMyb135–141 protein and suggesting the occurrence of certain conformational changes.
Structure of tvMyb135–141 in DNA-bound conformation
To gain more structural information of the tvMyb135–141 in complex with MRE-1/MRE-2r, we measured the 1DNH residual dipolar coupling (RDC) from the 15N-labeled tvMyb135–141/DNA complex sample partially aligned in medium containing 10 mg ml–1 pf1 phage. 74 1DNH RDC values ranging from –25 to 29 Hz were measured unambiguously. To see if the structure of tvMyb135–141 was changed when bound to DNA, the correlation between the measured 1DNH RDC values and the back-calculated values derived from the ensemble of 20 structures of tvMyb135–141 were analyzed by the program Pales and the goodness of fit was assessed with the RDC Q-factor (37). We first analyzed the residues located in the helices of the R2 motif (23 1DNH RDC values) and found that most of the back-calculated RDC values derived from the 20 structures of R2 motif fitted well to the experimental RDC values with an average Q-factor = 0.30 ± 0.03 (0.259–0.348), suggesting that the backbone conformation of the R2 motif is similar in the presence and absence of DNA. Similarly, the fitting focused on the helical residues of the R3 motif (18 1DNH RDC values) also gave a low average Q-factor (0.31 ± 0.04; 0.252–0.399), indicating that the fold of the R3 motif is also maintained upon DNA binding. However, unlike the fitting to individual R2 or R3 motifs, the fitting of the measured RDC values of all helical residues to the calculated RDC values derived from 20 structures gave a high average Q-factor (0.67 ± 0.04; 0.611–0.752). These results suggest that when tvMyb135–141 binds to DNA the backbone conformations of individual R2 and R3 motifs are mostly preserved but the relative orientation between the two motifs is dramatically changed.
The structure of DNA-bound tvMyb135–141 was generated by refining the free structure with all 1DNH RDC constraints obtained from protein sample in complex with DNA. The NOE, dihedral angle and hydrogen bond restraints of helical residues of free tvMyb135–141 were also used in the structural refinement protocol since these restraints define the individual folds of the R2 and R3 motifs, which agree with the RDC values from the DNA-bound conformation. A set of 20 structures was selected based on the energy of RDC restraints (Figure 5A) with RMSD values of 0.96 ± 0.25 Å and 1.89 ± 0.22 Å for the backbone atoms and heavy atoms of all helical residues, respectively (Table 1). The RDC values calculated from the final 20 structures correlated with observed RDC values very well with an average RMSD value = 0.37 ± 0.02 Hz. Figure 5B shows the structure with the lowest RDC energy. The angles between H1, H2 and H3 of the R2 motif in DNA-bound conformation are similar to those in the free structure as are the angles between H4, H5 and H6 in the R3 motif (Supplementary Table S2). However, the angles between the R2 helices and the R3 helices are dramatically different between the free structure and the DNA-bound conformation. Figure 5D shows that the difference between the two R3 motifs is about 50 degrees of rotation if the backbone atoms of the R2 motifs are superimposed. After the rotation, H3 and H6 form a V-shaped surface and the residues that exhibited significant chemical shift changes (residues of L1 loop are not included since their chemical shift perturbations may come from structural changes) lie mainly on this V-shaped surface. Besides, the residues located in the interface between the R2 and R3 motifs in the free structure also became accessible to DNA binding in the DNA-bound conformation. These data suggested that the V-shaped surface formed by H3 and H6 may represent the DNA-binding surface.
The structural model of tvMyb135–141/DNA complex
Although most of the backbone resonance assignment of tvMyb135–141 in complex with DNA was completed, no intermolecular NOEs could be unambiguously assigned from either a 3D [F1] 13C, 15N-filtered, [F2, F3] 15N-edited NOESY-HSQC spectrum or a 3D 15N-edited NOESY-HSQC spectrum obtained from a 95% 2H, 13C, 15N-labeled tvMyb135–141/DNA complex sample. Therefore, to gain insight into the likely DNA-bound conformation of tvMyb135–141, we used the chemical shift perturbations, 1DNH RDC values and DNA specificity data to calculate a structural model of the protein/DNA complex using the program HADDOCK (17–19). The active residues of tvMyb135–141 were defined for those that have weighted 1H and 15N chemical shift perturbations upon complex formation greater than 0.5 ppm (Δδ > Δδaverage+SD ∼0.5 ppm) and display high solvent accessibility (>50%). According to our previous results on the MRE-1/MRE-2r DNA specificity (6), the base replacements that disrupt the interactions between tvMyb1 protein and the DNA were selected as active bases, which include ADE6 and CYT8 to THY11 and their complementary bases (the strand, AAGATAACGATATTTA, is numbered 1–16 and the complementary strand, TAAATATCGTTATCTT is numbered 17–32). The specific AIR restraints (provided as Supplementary Data) were defined between suitable atoms of active residues to unique base atoms of active bases as described in ‘Materials and Methods’ section.
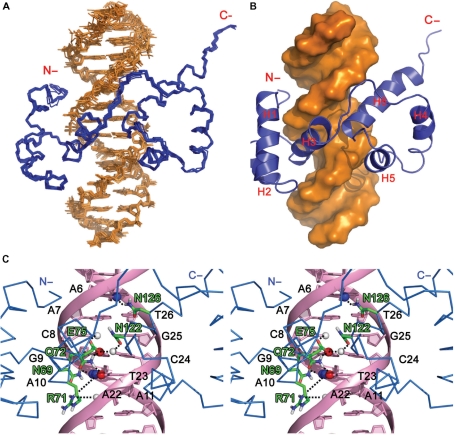
After the semi-flexible simulated annealing and explicit water refinement protocol, the final 200 structures were clustered based on the pair-wise RMSD matrix using a 1.0 Å cutoff and resulted in 11 different clusters. Table 2 shows the statistics of the top seven clusters based on the averaged HADDOCK score of their top 10 structures. Cluster 4 has the best average HADDOCK score as well as the most favorable intermolecular energies of the van der Waals’ and the electrostatic interactions. The 10 lowest energy structures from this cluster were selected to represent the model of the tvMyb135–141/DNA complex (Figure 6A). The structural ensemble has an RMSD value of 0.43 ± 0.09 Å over all backbone atoms of the secondary structural residues of the protein and an RMSD value of 0.68 ± 0.25 Å if all DNA phosphate backbone atoms are also included. The DNA-recognition surface of tvMyb135–141 comprises mainly residues of H3 and H6 that insert into the major groove of the hexanucleotide AACGAT and some residues at the N-terminus that contact the DNA minor groove (Figure 6B). A lot of hydrogen bond and hydrophobic interactions between protein and DNA molecules are observed in >40% of the final docking structures (Table 3). The side-chains of Asn69, Arg71, Gln72, Glu75, Asn122 and Asn126 play important roles in DNA specific recognition, in which the side-chains of these residues form hydrogen bonds with bases of the DNA (Figure 6C). A number of hydrogen bonds are also found mainly between the positively charged side-chains and the backbone phosphates of the DNA. In addition, two hydrogen bonds are observed between the protein backbone amide protons and the DNA backbone phosphates (Asn69-GUA9 and Asn110-THY23). The observation of plenty of hydrogen bonds and hydrophobic contacts between the protein and the DNA molecules agrees well with the high affinity found between tvMyb135–141 and the MRE-1/MRE-2r DNA duplex (KD = 1.24 × 10−9 M).
Table 2.
Statistics of the top seven clusters obtained with HADDOCK
| Clustera | Haddock scoreb | RMSD-Eminc | Nd | Evdwe | Eelece | EAIRf | Esanig | BSAh | Edesolvi |
|---|---|---|---|---|---|---|---|---|---|
| 4 | 700 ± 33 | 0.55 ± 0.2 | 19 | −64 ± 4 | −612 ± 31 | 1313 ± 21 | 23 ± 3 | 2120 ± 74 | 40 ± 3 |
| 3 | 853 ± 35 | 3.97 ± 0.1 | 23 | −50 ± 4 | −406 ± 24 | 1259 ± 22 | 27 ± 12 | 1762 ± 57 | 25 ± 7 |
| 7 | 919 ± 92 | 2.33 ± 0.1 | 8 | −67 ± 5 | −491 ± 44 | 1398 ± 39 | 39 ± 40 | 2190 ± 64 | 40 ± 4 |
| 10 | 967 ± 102 | 4.26 ± 0.1 | 4 | −52 ± 7 | −429 ± 28 | 1391 ± 71 | 29 ± 11 | 1839 ± 43 | 29 ± 4 |
| 2 | 993 ± 19 | 5.26 ± 0.1 | 36 | −73 ± 4 | −392 ± 18 | 1411 ± 17 | 24 ± 2 | 2151 ± 40 | 24 ± 4 |
| 5 | 1006 ± 31 | 17.42 ± 0.1 | 15 | −71 ± 4 | −402 ± 19 | 1430 ± 38 | 22 ± 6 | 2237 ± 72 | 26 ± 6 |
| 8 | 1015 ± 90 | 17.35 ± 0.1 | 10 | −58 ± 5 | −415 ± 40 | 1436 ± 71 | 28 ± 26 | 2195 ± 57 | 23 ± 3 |
aThe final 200 structures were clustered based on the pair-wise RMSD matrix using a 1.0 Å cutoff. The statistics are for the 10 lowest energy structures.
bThe HADDOCK score was calculated as the sum of: Evdw + Eelec + EAIR + Esani + Edesolv.
cOverall backbone RMSD from the lowest energy structure.
dNumber of structures in a given cluster.
eIntermolecular energies (kcal mol−1) were calculated with the OPLS parameters using a 8.5 Å cut-off.
fHADDOCK ambiguous interaction restraint energy (kcal mol−1).
gEnergy for the direct RDC constraints.
hBuried surface area (Å2).
iThe desolvation energy (kcal mol−1).
Figure 6.
Structural model of the tvMyb135–141/DNA complex. (A) Backbone illustration of the 10 lowest energy structures from HADDOCK. (B) Surface representation of the DNA and ribbon display of tvMyb135–141 clearly showed that H3 and H6 insert into the major groove of DNA and the N-terminus contacts with the DNA minor groove. (C) Stereo view of the specific hydrogen bonds in tvMyb135–141/DNA complex. The residues involved in specific hydrogen bond interactions are shown in stick and the atoms of DNA are shown as sphere (with C in green, N blue, O red and H white). The hydrogen bonds are indicated as black dash lines.
Table 3.
Intermolecular hydrogen bonds between tvMyb135–141 and DNA
| Protein | Atom | Base | Atom | Distance (Å) | No. (of 10) |
|---|---|---|---|---|---|
| Hydrogen bonds | |||||
| Lys35 | HZ* | ADE28 | O1P | 1.92 | 4 |
| Lys37 | HZ* | GUA9 | O1P | 1.88 | 7 |
| Arg68 | HE | CYT8 | O2P | 1.96 | 10 |
| Arg68 | HH* | CYT8 | O2P | 1.89 | 5 |
| Asn69 | HD2* | ADE10 | N7 | 2.13 | 10 |
| Asn69 | HN | GUA9 | O2P | 1.92 | 10 |
| Arg71 | HH* | ADE10 | N6 | 2.18 | 5 |
| Arg71 | NH* | ADE22 | H61 | 2.13 | 4 |
| Gln72 | HE2* | GUA9 | O6 | 2.21 | 10 |
| Gln72 | HE2* | THY23 | O4 | 1.90 | 5 |
| Arg74 | HH* | ADE22 | O1P | 2.08 | 10 |
| Glu75 | OE* | CYT24 | H4* | 2.26 | 8 |
| Glu75 | OE* | CYT8 | H41 | 2.01 | 8 |
| Arg76 | HH* | ADE7 | O1P | 2.02 | 10 |
| Asn110 | HD21 | ADE22 | O3’ | 1.91 | 9 |
| Asn110 | HN | THY23 | O2P | 2.21 | 4 |
| Asn122 | OD1 | CYT24 | H41 | 2.11 | 9 |
| Arg125 | HH* | CYT24 | O1P | 2.05 | 7 |
| Asn126 | HD22 | ADE6 | N7 | 2.03 | 10 |
| Arg127 | HH* | THY5 | O2P | 1.94 | 5 |
| Arg127 | HH* | ADE6 | O1P | 1.86 | 8 |
| Arg133 | HH* | ADE4 | O2P | 1.85 | 9 |
| Hydrophobic contacts | |||||
| Side chains | Bases | ||||
| Val36 | ADE28 | 3.60 | 10 | ||
| Phe38 | ADE7, CYT8 | 3.55 | 10 | ||
| Arg71 | THY21, ADE22 | 3.44 | 10 | ||
| Pro107 | CYT24 | 3.49 | 10 | ||
| Trp109 | CYT24 | 3.52 | 10 | ||
| Asp121 | THY23 | 3.62 | 10 | ||
| Asn126 | ADE6 | 3.14 | 10 | ||
| Met130 | THY5 | 3.58 | 10 |
The specific interactions are in bold.
*Indicates all possible atoms at the position.
The conformations of the MRE-1/MRE-2r DNA duplex in the final complex structures were analyzed by the program 3DNA (38). The average helical parameters for DNA bound to tvMyb135–141 are shown in Supplementary Figure S3. Basically, the overall DNA structure remains canonical double-stranded base-pairing geometry. The major groove width between ADE6-ADE7 is at maximun, the roll angles between ADE7 to ADE12 and the twist angles between CYT8 to ADE10 are deviated from standard B-form DNA, indicating the interaction between tvMyb135–141 and the DNA.
To validate our model, three point mutations of tvMyb135–141 were generated (F38A, T67A and N126A). The CD spectra showed that they are all as well-folded as tvMyb135–141 (data not shown). Their binding affinities toward the MRE-1/MRE-2r DNA duplex were checked by EMSA and SPR experiments (Figure 1A and C). The mutant F38A exhibits the lowest DNA-binding ability and N126A shows a signficant decrease in DNA binding, agreeing with our complex structure in which Phe38 and Asn126 forms contacts with the DNA. The mutant T67A binds DNA as strongly as tvMyb135–141 does and Thr67 shows no contact with DNA in the complex structure. These data highly support the accuracy of the tvMyb135–141/DNA complex structure.
DISCUSSION
In the present study, we have defined the DNA-binding domain of the first Myb family protein identified in the protozoan parasite T. vaginalis and determined its NMR solution structure with high resolution. In order to understand the rearrangement of the tertiary fold of the DNA-binding domain for DNA recognition, the 1DNH residual dipolar couplings of tvMyb135–141 in complex with DNA were acquired and used for structural refinement. Finally, based on the chemical shift perturbation, residual dipolar couplings and DNA specificity data, the molecular basis for the DNA recognition of this domain was suggested.
To define the DNA-binding domain of tvMyb1 protein, two clones were constructed and tested for DNA recognition. The shorter fragment, tvMyb135–131, which contains two classical Myb-like DNA-binding domain (one from Lys35 to Ile81 and the other Thr87 to Ile131) as suggested by the Pfam database, was found to be prone to degradation and aggregation. Its thermal stability and DNA-binding ability was also much lower than those of the longer fragment, tvMyb135–141. It then becomes important to discuss the roles of the C-terminal 10 residues A132RHRAKHQKS141 of the protein. The last helix of tvMyb135–141, H6, spanning from Asp121 to Arg135, was found to be amphipathic. Its hydrophobic residues Ile124, Trp128, Ile131 and Ala132 constitute the major part of the hydrophobic core of the R3 motif and thus the integrity of H6 is important for the hydrophobic packing of the R3 motif. It is well known that an α-helix has an overall dipole moment from C-terminus to N-terminus caused by the cumulative effect of each residue backbone unit dipole contribution. As a result, α-helices are often capped at the N-terminal end by a negatively charged amino acid or at the C-terminus with a positively charged amino acid in order to neutralize this helix dipole (39,40). In tvMyb135–141, H6 is capped and stabilized by Asp121 and Arg135 at the two termini. However, in the shorter fragment, H6 is terminated at Ile131; the C-terminal capping by Arg135 is lost, which may disrupt the integrity of H6 and destabilizes the hydrophobic core of the R3 motif. This explains why tvMyb135–131 tends to degrade and to form amorphous aggregates. For DNA-binding ability, the C-terminal 10-residues extension introduces four additional positively charged residues to the protein molecule, which may increase the electrostatic interaction between the highly positively charged protein molecule and the highly negatively charged DNA backbone. In the 1H-15N HX-NOE experiments, the C-terminal residues in the complex state showed higher HX-NOE values than those in the free form, which is likely due to electrostatic interactions between C-terminal residues and DNA. In our complex structure, Arg133 forms a hydrogen bond with the backbone phosphate of ADE4. In addition, SPR experiments also indicated that a higher salt concentration decreases the binding response between tvMyb135–141 and DNA (data not shown). Taking all these together, the C-terminal 10-residues extension can stabilize the R3 motif by hydrophobic residues and enhance the DNA binding through positively charged residues.
The structure of tvMyb135–141 in the DNA-bound form was acquired by refining the free structure with the complex 1DNH RDC constraints and the tvMyb135–141/DNA complex structural model was built based on the chemical shift perturbation, residual dipolar couplings and DNA specificity data by HADDOCK (17–19). The results showed that the relative orientation between the R2 and R3 motifs are changed upon binding to the DNA major groove. The large changes in amide chemical shifts of the residues in L1 loop suggest significant conformational rearrangements of this loop in the DNA-bound form. In the free structure, four possible salt bridges were observed to restrict the architecture of R2R3 domain. But in DNA-bound conformation, all of them were lost. Instead, four hydrogen bonds were observed between residues Arg125, Glu75 and Arg76, and DNA molecule. And after the rotation between R2 and R3 motifs, the complex conformation becomes complementary to the DNA major groove. It is then possible to calculate the complex structure by the docking program, HADDOCK.
In our docking structure, many hydrogen bonds and hydrophobic interactions are observed between tvMyb135–141 and the DNA duplex which correlate well with our experimental results. From the analysis of the 1H-15N HSQC titration and 1H-15N HX-NOE experiments, we found that the residues located in the N-terminus, H3, H5 and H6 exhibit large chemical shift perturbations and increased HX-NOE values in the presence of DNA. These residues make hydrogen bonds and hydrophobic contacts with the DNA in our structure. Especially for the residues in the N-terminus, they are flexible in the free structure but showed the highest chemical shift perturbation and largest increase in HX-NOE value in complex with DNA, implying that this region adopts a much more rigid conformation compared to the free structure. In the complex model, the N-terminal residues do display several hydrogen bonds and hydrophobic interactions with DNA. It is also notable that the chemical shifts of the amide resonances of Asn69 and Asn110 are greatly perturbed with downfield shifting, which agrees well with the observation that the amide protons of Asn69 and Asn110 form direct hydrogen bonds with the phosphate backbone of DNA. In addition, based on our previous studies on MRE-1/MRE-2f specificity (6), the tvMyb1 protein binds specifically to ADE6 and CYT8 to THY11. These nucleotides display nine hydrogen bonds between the bases and the protein in the complex structure. Although we failed to assign the inter-molecular NOEs between the protein and the DNA duplex, the complex model agrees well with all our experimental results and may reflect the specific DNA recognition by tvMyb135–141.
From the searching of RCSB protein data bank, there is only one R2R3 domain/DNA complex structure determined by NMR, the c-Myb R2R3 domain/DNA complex structure (3). The main difference in the DNA recognition domain between tvMyb1 and c-Myb is the length of H6. The H6 in tvMyb135–141 is much longer than that in c-Myb (Supplementary Figure S4) and we have demonstrated that this longer helix is important in protein stability and DNA-binding activity for tvMyb135–141. In our complex structure, some of the intermolecular contacts between tvMyb135–141 and DNA are very similar to those observed in the homologous c-Myb R2R3 domain/DNA complex structure, but some are unique which correlates with DNA specificity of tvMyb135–141. For clarity, our DNA sequence (ATAACGAT) is numbered from 4 to 11 (the complementary strand, ATCGTTAT, is numbered from 22 to 29), and the DNA sequence in c-Myb/DNA complex (CTAACTGA) is numbered from 2 to 9 (the complementary strand, TCAGTTAG, is numbered from 14 to 21). The underlined sequence is the binding sequence of the two proteins. In c-Myb/DNA complex structure, Glu132 forms hydrogen bonds with CYT6 and CYT15; Asn183 forms a hydrogen bond with ADE4; Arg133 forms a hydrogen bond with phosphate of ADE5; Arg131 forms a hydrogen bond with phosphate of THY14; and Ala167 forms a hydrogen bond with phosphate of ADE16. Similar hydrogen bond contacts are also observed in our complex model; the aforementioned interaction between Glu75, Asn126, Arg76, Arg74 and Asn110 of tvMyb135–141 with DNA. However, three hydrogen bonds in c-Myb complex, Asn186 with THY19 and THY18 and Ser187 with THY3, which are important for specific binding of c-Myb, are not observed in our model because the corresponding residues in tvMyb135–141 are Met129 and Met130. In our complex structure, three additional hydrogen bonds are observed; Asn69 with ADE10 and Gln72 with GUA9 and THY23. The similarities and differences of hydrogen bond contacts observed in c-Myb and tvMyb135–141 complexes explain the difference in DNA-binding specificity.
The structure determination of multi-domain proteins in complex with DNA is biologically relevant. However, this kind of complex normally does not provide NMR spectra of enough quality for complex structure determination. The residues at binding interfaces may experience intermediate exchange and give broadened peaks or no peaks for assignment. Different buffer conditions, protein lengths or even mutations must be tested in order to improve the quality of the NMR spectra. In addition, the identification of inter-molecular NOEs is tedious and time consuming. In this study, we obtained several intermolecular NOEs especially in the 3D 15N-edited NOESY-HSQC spectrum obtained from a 95% 2H, 13C, 15N-labeled tvMyb135–141/DNA complex sample. However, the assignment of proton resonances for the protein/DNA complex could not be finished due to the limited stability of the complex sample, the lack of stable isotope labeling on the DNA duplex, peak overlapping, and the short T2 relaxation time. The average transverse relaxation time, T2, of the amide protons of tvMyb135–141/DNA complex sample at 298K, which was measured by the 1-1 echo sequence (41), is around 12.9 ms, similar to what is observed for a protein with molecular weight of 30 kDa. The sensitivity and resolution of NMR spectra of the complex sample were therefore largely reduced. Hence, the assignments for the proton resonances of the complex sample and for the intermolecular NOEs could not be well established. Here, we used an alternative approach, combining the free form structure, 1DNH RDC values from the complex sample, chemical shift perturbations upon binding, DNA specificity data from EMSA and data-driven macro-molecular docking, to understand the molecular basis for DNA recognition by tvMyb135–141. This method could be especially useful for revealing the structural information of a multi-domain protein in complex with macro-molecules. First, it is much easier to assign the backbone resonances in HSQC or TROSY spectra of the complex than to identify the inter-molecular NOEs. Second, residual dipolar couplings have been shown to be powerful sources of long-range structural information, especially domain orientations (42,43). The RDCs have been successfully applied in determining the first solution structure of Lys48-linked di-ubiquitin by HADDOCK (18). In our case, although we did not provide any direct inter-molecular NOE restraints, the AIRs from chemical shift perturbations and the DNA specificity data are sufficient to define many reasonable interactions between tvMyb135–141 and the DNA that correlate well with all the experimental results. These data further our understanding of DNA recognition by Myb protein. This approach should be very useful when applied to determine the complex structures involving proteins with multiple domains.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Academia Sinica; National Science Council, Taiwan, ROC (NSC 95-2320-B-001-040-MY2). Funding for open access charge: Academia Sinica, Taiwan.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We would like to thank the High-field Biomacromolecular NMR Core Facility at the Academia Sinica supported by the National Research Program for Genomic Medicine for obtaining all the NMR spectra. We also thank D. Victoria Williams from Department of Chemistry, University of Washington, Seattle, USA for proofreading the manuscript.
REFERENCES
- 1.Stracke R, Werber M, Weisshaar B. The R2R3-MYB gene family in Arabidopsis thaliana. Curr. Opin. Plant Biol. 2001;4:447–456. doi: 10.1016/s1369-5266(00)00199-0. [DOI] [PubMed] [Google Scholar]
- 2.Lipsick JS. One billion years of Myb. Oncogene. 1996;13:223–235. [PubMed] [Google Scholar]
- 3.Ogata K, Morikawa S, Nakamura H, Sekikawa A, Inoue T, Kanai H, Sarai A, Ishii S, Nishimura Y. Solution structure of a specific DNA complex of the Myb DNA-binding domain with cooperative recognition helices. Cell. 1994;79:639–648. doi: 10.1016/0092-8674(94)90549-5. [DOI] [PubMed] [Google Scholar]
- 4.Nomura N, Takahashi M, Matsui M, Ishii S, Date T, Sasamoto S, Ishizaki R. Isolation of human cDNA clones of myb-related genes, A-myb and B-myb. Nucleic Acids Res. 1988;16:11075–11089. doi: 10.1093/nar/16.23.11075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Solano R, Fuertes A, Sanchez-Pulido L, Valencia A, Paz-Ares J. A single residue substitution causes a switch from the dual DNA binding specificity of plant transcription factor MYB.Ph3 to the animal c-MYB specificity. J. Biol. Chem. 1997;272:2889–2895. doi: 10.1074/jbc.272.5.2889. [DOI] [PubMed] [Google Scholar]
- 6.Ong SJ, Hsu HM, Liu HW, Chu CH, Tai JH. Multifarious transcriptional regulation of adhesion protein gene ap65-1 by a novel Myb1 protein in the protozoan parasite Trichomonas vaginalis. Eukaryot. Cell. 2006;5:391–399. doi: 10.1128/EC.5.2.391-399.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Honigberg BM. In Parasitic Protozoa. Inc., New York: Academic Press; 1978. [Google Scholar]
- 8.Sorvillo F, Smith L, Kerndt P, Ash L. Trichomonas vaginalis, HIV, and African-Americans. Emerg. Infect. Dis. 2001;7:927–932. doi: 10.3201/eid0706.010603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Soper D. Trichomoniasis: under control or undercontrolled? Am. J. Obstet. Gynecol. 2004;190:281–290. doi: 10.1016/j.ajog.2003.08.023. [DOI] [PubMed] [Google Scholar]
- 10.Cudmore SL, Delgaty KL, Hayward-McClelland SF, Petrin DP, Garber GE. Treatment of infections caused by metronidazole-resistant Trichomonas vaginalis. Clin. Microbiol. Rev. 2004;17:783–793. doi: 10.1128/CMR.17.4.783-793.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dunne RL, Dunn LA, Upcroft P, O’Donoghue PJ, Upcroft JA. Drug resistance in the sexually transmitted protozoan Trichomonas vaginalis. Cell Res. 2003;13:239–249. doi: 10.1038/sj.cr.7290169. [DOI] [PubMed] [Google Scholar]
- 12.Alderete JF, O’Brien JL, Arroyo R, Engbring JA, Musatovova O, Lopez O, Lauriano C, Nguyen J. Cloning and molecular characterization of two genes encoding adhesion proteins involved in Trichomonas vaginalis cytoadherence. Mol. Microbiol. 1995;17:69–83. doi: 10.1111/j.1365-2958.1995.mmi_17010069.x. [DOI] [PubMed] [Google Scholar]
- 13.O’Brien JL, Lauriano CM, Alderete JF. Molecular characterization of a third malic enzyme-like AP65 adhesin gene of Trichomonas vaginalis. Microb. Pathog. 1996;20:335–349. doi: 10.1006/mpat.1996.0032. [DOI] [PubMed] [Google Scholar]
- 14.Ong SJ, Huang SC, Liu HW, Tai JH. Involvement of multiple DNA elements in iron-inducible transcription of the ap65-1 gene in the protozoan parasite Trichomonas vaginalis. Mol. Microbiol. 2004;52:1721–1730. doi: 10.1111/j.1365-2958.2004.04088.x. [DOI] [PubMed] [Google Scholar]
- 15.Tsai CD, Liu HW, Tai JH. Characterization of an iron-responsive promoter in the protozoan pathogen Trichomonas vaginalis. J. Biol. Chem. 2002;277:5153–5162. doi: 10.1074/jbc.M110234200. [DOI] [PubMed] [Google Scholar]
- 16.Finn RD, Tate J, Mistry J, Coggill PC, Sammut SJ, Hotz HR, Ceric G, Forslund K, Eddy SR, Sonnhammer EL, et al. The Pfam protein families database. Nucleic Acids Res. 2008;36:D281–D288. doi: 10.1093/nar/gkm960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dominguez C, Boelens R, Bonvin AM. HADDOCK: a protein-protein docking approach based on biochemical or biophysical information. J. Am. Chem. Soc. 2003;125:1731–1737. doi: 10.1021/ja026939x. [DOI] [PubMed] [Google Scholar]
- 18.van Dijk AD, Fushman D, Bonvin AM. Various strategies of using residual dipolar couplings in NMR-driven protein docking: application to Lys48-linked di-ubiquitin and validation against 15N-relaxation data. Proteins. 2005;60:367–381. doi: 10.1002/prot.20476. [DOI] [PubMed] [Google Scholar]
- 19.van Dijk M, van Dijk AD, Hsu V, Boelens R, Bonvin AM. Information-driven protein-DNA docking using HADDOCK: it is a matter of flexibility. Nucleic Acids Res. 2006;34:3317–3325. doi: 10.1093/nar/gkl412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lin TH, Chen C, Huang RF, Lee YL, Shaw JF, Huang TH. Multinuclear NMR resonance assignments and the secondary structure of Escherichia coli thioesterase/protease I: a member of a new subclass of lipolytic enzymes. J. Biomol. NMR. 1998;11:363–380. doi: 10.1023/a:1008226515482. [DOI] [PubMed] [Google Scholar]
- 21.Ottiger M, Delaglio F, Bax A. Measurement of J and dipolar couplings from simplified two-dimensional NMR spectra. J. Magn. Reson. 1998;131:373–378. doi: 10.1006/jmre.1998.1361. [DOI] [PubMed] [Google Scholar]
- 22.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 23.Johnson BA. Using NMRView to visualize and analyze the NMR spectra of macromolecules. Methods. Mol. Biol. 2004;278:313–352. doi: 10.1385/1-59259-809-9:313. [DOI] [PubMed] [Google Scholar]
- 24.Goddard TD, Kneller DG. SPARKY 3. University of California, San Francisco: 2004. [Google Scholar]
- 25.Cornilescu G, Delaglio F, Bax A. Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J. Biomol. NMR. 1999;13:289–302. doi: 10.1023/a:1008392405740. [DOI] [PubMed] [Google Scholar]
- 26.Herrmann T, Guntert P, Wuthrich K. Protein NMR structure determination with automated NOE assignment using the new software CANDID and the torsion angle dynamics algorithm DYANA. J. Mol. Biol. 2002;319:209–227. doi: 10.1016/s0022-2836(02)00241-3. [DOI] [PubMed] [Google Scholar]
- 27.Schwieters CD, Kuszewski JJ, Tjandra N, Clore GM. The Xplor-NIH NMR molecular structure determination package. J. Magn. Reson. 2003;160:65–73. doi: 10.1016/s1090-7807(02)00014-9. [DOI] [PubMed] [Google Scholar]
- 28.Laskowski RA, Rullmannn JA, MacArthur MW, Kaptein R, Thornton JM. AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J. Biomol. NMR. 1996;8:477–486. doi: 10.1007/BF00228148. [DOI] [PubMed] [Google Scholar]
- 29.Clore GM, Gronenborn AM, Tjandra N. Direct structure refinement against residual dipolar couplings in the presence of rhombicity of unknown magnitude. J. Magn. Reson. 1998;131:159–162. doi: 10.1006/jmre.1997.1345. [DOI] [PubMed] [Google Scholar]
- 30.Clore GM, Schwieters CD. Docking of protein-protein complexes on the basis of highly ambiguous intermolecular distance restraints derived from 1HN/15N chemical shift mapping and backbone 15N-1H residual dipolar couplings using conjoined rigid body/torsion angle dynamics. J. Am. Chem. Soc. 2003;125:2902–2912. doi: 10.1021/ja028893d. [DOI] [PubMed] [Google Scholar]
- 31.Hubbard SJ, Thornton JM. University College London: Department of Biochemistry and Molecular Biology; 1993. ‘NACCESS’, Computer Program. [Google Scholar]
- 32.Meiler J, Blomberg N, Nilges M, Griesinger C. A new approach for applying residual dipolar couplings as restraints in structure elucidation. J. Biomol. NMR. 2000;16:245–252. doi: 10.1023/a:1008378624590. [DOI] [PubMed] [Google Scholar]
- 33.Tjandra N, Omichinski JG, Gronenborn AM, Clore GM, Bax A. Use of dipolar 1H-15N and 1H-13C couplings in the structure determination of magnetically oriented macromolecules in solution. Nat. Struct. Biol. 1997;4:732–738. doi: 10.1038/nsb0997-732. [DOI] [PubMed] [Google Scholar]
- 34.Kissinger CR, Liu BS, Martin-Blanco E, Kornberg TB, Pabo CO. Crystal structure of an engrailed homeodomain-DNA complex at 2.8 Å resolution: a framework for understanding homeodomain-DNA interactions. Cell. 1990;63:579–590. doi: 10.1016/0092-8674(90)90453-l. [DOI] [PubMed] [Google Scholar]
- 35.Hosoda K, Imamura A, Katoh E, Hatta T, Tachiki M, Yamada H, Mizuno T, Yamazaki T. Molecular structure of the GARP family of plant Myb-related DNA binding motifs of the Arabidopsis response regulators. Plant Cell. 2002;14:2015–2029. doi: 10.1105/tpc.002733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stevenson CE, Burton N, Costa MM, Nath U, Dixon RA, Coen ES, Lawson DM. Crystal structure of the MYB domain of the RAD transcription factor from Antirrhinum majus. Proteins. 2006;65:1041–1045. doi: 10.1002/prot.21136. [DOI] [PubMed] [Google Scholar]
- 37.Zweckstetter M, Bax A. Prediction of sterically induced alignment in a dilute liquid crystalline phase: aid to protein structure determination by NMR. J. Am. Chem. Soc. 2000;122:3791–3792. [Google Scholar]
- 38.Lu XJ, Olson WK. 3DNA: a software package for the analysis, rebuilding and visualization of three-dimensional nucleic acid structures. Nucleic Acids Res. 2003;31:5108–5121. doi: 10.1093/nar/gkg680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bodkin MJ, Goodfellow JM. Competing interactions contributing to alpha-helical stability in aqueous solution. Protein Sci. 1995;4:603–612. doi: 10.1002/pro.5560040402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Serrano L, Fersht AR. Capping and alpha-helix stability. Nature. 1989;342:296–299. doi: 10.1038/342296a0. [DOI] [PubMed] [Google Scholar]
- 41.Anglister J, Grzesiek S, Ren H, Klee CB, Bax A. Isotope-edited multidimensional NMR of calcineurin B in the presence of the non-deuterated detergent CHAPS. J. Biomol. NMR. 1993;3:121–126. doi: 10.1007/BF00242480. [DOI] [PubMed] [Google Scholar]
- 42.Bax A, Kontaxis G, Tjandra N. Dipolar couplings in macromolecular structure determination. Methods Enzymol. 2001;339:127–174. doi: 10.1016/s0076-6879(01)39313-8. [DOI] [PubMed] [Google Scholar]
- 43.Bax A. Weak alignment offers new NMR opportunities to study protein structure and dynamics. Protein Sci. 2003;12:1–16. doi: 10.1110/ps.0233303. [DOI] [PMC free article] [PubMed] [Google Scholar]