Abstract
Regulation of angiotensin II type 1 receptor (AT1R) has a pathophysiological role in hypertension, atherosclerosis and heart failure. We started from an observation that the 3′-untranslated region (3′-UTR) of AT1R mRNA suppressed AT1R translation. Using affinity purification for the separation of 3′-UTR-binding proteins and mass spectrometry for their identification, we describe glyceraldehyde 3-phosphate dehydrogenase (GAPDH) as an AT1R 3′-UTR-binding protein. RNA electrophoretic mobility shift analysis with purified GAPDH further demonstrated a direct interaction with the 3′-UTR while GAPDH immunoprecipitation confirmed this interaction with endogenous AT1R mRNA. GAPDH-binding site was mapped to 1–100 of 3′-UTR. GAPDH-bound target mRNAs were identified by expression array hybridization. Analysis of secondary structures shared among GAPDH targets led to the identification of a RNA motif rich in adenines and uracils. Silencing of GAPDH increased the expression of both endogenous and transfected AT1R. Similarly, a decrease in GAPDH expression by H2O2 led to an increased level of AT1R expression. Consistent with GAPDH having a central role in H2O2-mediated AT1R regulation, both the deletion of GAPDH-binding site and GAPDH overexpression attenuated the effect of H2O2 on AT1R mRNA. Taken together, GAPDH is a translational suppressor of AT1R and mediates the effect of H2O2 on AT1R mRNA.
INTRODUCTION
Angiotensin II is a pivotal physiological regulator of blood pressure, salt and fluid homeostasis, and cardiovascular structure (1). Angiotensin II has two receptors that mediate its effects. The angiotensin II type 1 receptor (AT1R) is a G-protein-coupled receptor that confers most of the deleterious effects of angiotensin II, while the type 2 receptor confers its protective effects (2,3). Angiotensin-II-mediated triggering of AT1R has an important role in the pathogenesis of chronic renal failure, heart failure, hypertension and atherosclerosis. Moreover, numerous clinical trials have demonstrated the beneficial effects of pharmacologic therapy that reduces AT1R activity in a wide variety of cardiovascular disorders (1).
AT1R is regulated on multiple levels including transcription, protein synthesis, degradation and signal transduction. Regulation of AT1R expression is a critical mechanism that modulates the activity of renin-angiotensin system. Cell surface expression of AT1R is regulated by receptor internalization and this is modulated by multiple proteins including arrestins and ATRAP (4). Transcriptional regulators of AT1R gene include glucocorticoids and interleukin 1β (5). Posttranscriptional regulation of the AT1R mRNA transcript has been established to be an important regulatory mechanism of AT1R expression by insulin, statins and estrogen (6–11). However, the molecular mechanisms of mRNA-based regulation of AT1R expression are poorly known.
Posttranscriptional control of eukaryotic gene expression comprises several levels of regulation such as mRNA processing, export, turnover and translation. Each regulatory step involves various combinations of RNA-binding proteins that form dynamic messenger ribonucleoproteins. We found that transfer of AT1R 3′-UTR to a reporter gene leads to down-regulation of reporter gene expression by two mechanisms: inhibition of translation and decrease in mRNA stability. Therefore, 3′-UTR has elements both for the regulation of translation and mRNA turnover. In most cases of transcript selective transcriptional regulation, translational control is dictated by binding of a RNA-binding protein to a cis acting structural element in mRNA. AT1R 3′-UTR has AU-rich destabilization element in the distal part of the AT1R 3′-UTR. The binding sites for calreticulin and AUF-1 are located in this AU-rich region (6,12). Calreticulin binds to the highly conserved last 20 bases in the 3′-end of the rat AT1A receptor 3′-UTR and mediates angiotensin II-induced down-regulation of rat AT1R mRNA (6) whereas the effect of AUF1 binding remains unclear, however, in general it destabilizes mRNA (12,13). We have found that p100 regulates AT1R mRNA stability and translation (14). We focused our efforts to elucidate the molecular mechanisms of AT1R 3′-UTR mediated translational suppression.
Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) is a cellular mRNA-binding protein that suppresses the translation of viral mRNAs and influences the stability of some cellular mRNAs (15–17). GAPDH is a multifunctional protein best known for its pivotal glycolytic function (18–21) and it has been found to display a number of independent functions unrelated to its glycolytic function. These include roles in apoptosis (22), microtubule organization (23) and RNA-binding (17). It is unclear how its nonglycolytic functions are regulated but localization changes and protein–protein interactions have been suggested to play a role (19). GAPDH is involved with cellular hypoxic and oxidative response and it is down-regulated by oxidative stress (24,25).
To delineate the molecular mechanisms of AT1R mRNA regulation, we used affinity purification and mass spectrometry to isolate and recognize mRNA-binding proteins. We identified GAPDH as an AT1R mRNA-binding protein that suppresses AT1R translation and found this suppression to be regulated by oxidative stress. In this article we describe translational suppression as a novel mechanism of AT1R mRNA regulation and identify GAPDH as a mediator of this effect.
MATERIALS AND METHODS
Cell culture, luciferase assay and protein extraction
Coronary artery vascular smooth muscle cells (VSMCs) were purchased from Lonza bioscience and cultured on smooth muscle growth medium-2 with 5% fetal bovine serum (FBS). HEK293 cells were grown on plastic plates on DMEM that was supplemented with 10% FBS, ampicillin/streptomycin and glutamine. Cells were used for 6–10 passages before replacement with fresh early passage stocks. Constructs were transiently transfected in HEK293 cells using a standard Fugene 6 protocol (Roche Diagnostics). Silencer GAPDH and negative control siRNA (Ambion) were transfected using Lipofectamine 2000 reagent (Invitrogen) according to manufacturer's instructions. VSMC were transfected as described (26). Cells were harvested 24–72 h after transfection and firefly luciferase activities were measured using the Dual Luciferase Assay System (Promega). The luciferase activity was normalized to the activity of the cotransfected renilla luciferase plasmid or total protein content. Protein lysates were prepared using NE-PER kit (Pierce) and the lysates were prepared according to manufacturer's recommendations. Protein concentrations were determined by Bradford assay (Bio-Rad). H2O2 treatment was performed as described (24).
Constructs
The constructs used have been described previously (14).
Random mutagenesis
Genemorph II EZClone Domain Mutagenesis kit (Stratagene) was used to create a library of random mutants. Mutagenesis primers were designed to subject the 1–100 3′-UTR of AT1R to random mutagenesis. Sequencing of a random sample of mutagenized constructs confirmed that the number of nucleotide substitutions per cDNA ranged from two to four. In order to search for mutants with altered steady-state mRNA levels 100 constructs were tested. Five clones showed interesting results in either increasing or decreasing steady-state luciferase activity.
Real-time PCR
In RNA half-life experiments, cells were immersed in fresh media containing 1 µg/ml (∼0.8 µM) actinomycin D (Tocris) to inhibit transcription and incubated at 37°C. At the indicated times, cells were harvested and total cellular RNA was isolated with Nucleospin RNA II kit (Macherey-Nagel). RNA was treated with DNase I (Ambion). The first-strand cDNA was synthesized with Superscript II First Strand Synthesis System for RT-PCR and oligo(dT)12–18 primer (Invitrogen). Primer sequences used for PCR amplification of luciferase, GAPDH and β-actin genes have been published earlier (27–29). AT1R oligos used were gcacaatgcttgtagccaaa (upstream) and ccctatcggaagggttgaat (downstream). After the first strand cDNA synthesis, serial dilutions were made. The PCR reactions were performed in LightCycler apparatus using an LC Fast Start DNA MasterPLUS SYBR Green Kit (Roche Diagnostics). Thermocycling for each reaction was performed according to manufacturer's instructions. The annealing temperatures were 56°C for luciferase, 59°C for GAPDH oligos and 60°C for AT1R oligos. The elongation time was calculated by dividing the size of the amplicon by 25. The fluorescence emitted by SYBR Green I was measured in every cycle at the end of the elongation step. The second derivative maximum method was used to determine the crossing-point values. The concentrations of unknown samples were then calculated by setting their crossing points to the standard curve. For normalization of the expression levels, the expressions of GAPDH or β-actin were measured. The relative expression level was obtained by dividing the values for the gene of interest with the GAPDH or β-actin value. In addition to the melting curve information obtained from the LightCycler, the PCR products were run in 1.5% agarose gel electrophoresis to ensure that only a right-sized product was amplified.
RNA probe preparation
cDNA was used as a template for PCR reactions whereby T7 RNA polymerase promoter sequence was added to the 5′-end of all fragments. RNA transcripts were synthesized from PCR-generated templates including the following constructs: 1–100, 1–300, 1–600, 1–847 as well as from 100–300, 300–600 and 600–887 (Megascript, Ambion), Table 1. In the in vitro transcription reaction, we used a ratio of biotin-UTP to standard UTP of 1:3. RNA probes with or without biotin label were generated by MEGA shortscript kit according to manufacturers instructions (Ambion). The protocol has been described in detail (14).
Table 1.
Primers used in the generation of PCR products that served as templates in in vitro transcription (numbering starts from the beginning of 3′-UTR)
| AT1R | 3′-UTR sense primers (number denotes the first nucleotide of the 3′-UTR included in the oligo) |
| 1 | TAATACGACTCACTATAGGG catgttc gaaacctgtc cataaagtaa ttttgtgaaa gaagg |
| 100 | TAATACGACTCACTATAGGG gaattga aggagaaaat gcattatgtgg |
| 300 | TAATACGACTCACTATAGGG agcctgc ttttgtcctg ttatttttta tttccacata aagg |
| 600 | TAATACGACTCACTATAGGG gtataat ggtgttacta aagtcacata taaaagttaa ac |
| AT1R | 3′-UTR antisense primers (number denotes the last nucleotide included in the oligo) |
| 100 | ctgaaaagta gctaatgctc atttggtagt gaag |
| 300 | cagtaaaatt tctcaaatca acacattcat cgagtttc |
| 600 | attgttttgg cagtgtaaac ctataagaca caggttg |
| 847 | tataactttg ccagatttta atcaattaac agc |
| 887 | tgcaataaaattattttattttaaagtaaatataactttgcc |
| Control | (part of the luciferase coding region) |
| S | TAATACGACTCACTATAGGG gcgccattctatccgctggaagatggaacc |
| AS | cgcccaacaccggcataaagaattgaagag |
RNA affinity purification
RNA affinity purification was performed as described previously (14).
Mass spectrometry
Silver-stained protein bands of interest were cut out of the polyacrylamide gel and ‘in-gel’ digested essentially as described by Shevchenko et al. (30). Proteins were reduced with DTT and alkylated with iodoacetamide before digestion with trypsin (Sequencing Grade Modified Trypsin, V5111, Promega). The recovered peptides were, after desalting using Millipore C18 ZipTipTM, subjected to matrix-assisted laser desorption/ionization-time of flight (MALDI-TOF) mass spectrometric analysis. MALDI-TOF mass spectra for mass fingerprinting and MALDI-TOF/TOF mass spectra for identification by fragment ion analysis were acquired using an Ultraflex TOF/TOF instrument (Bruker-Daltonik GmbH, Bremen, Germany). Protein identification with the generated data was performed using Mascot® Peptide Mass Fingerprint and MS/MS Ion Search programs (http://www.matrixscience.com).
Western blotting
Western blotting was performed as described previously (14).
RNA electromobility shift analysis (REMSA)
REMSAs were performed with biotinylated probes and purified human GAPDH protein (Sigma). In brief, 150 ng GAPDH was incubated with ∼10 fmol of 3′-UTR probe in 5 mM Hepes (pH 7.9), 7.5 mM KCl, 0.5 mM MgCl2, 0.1 mM EDTA, 0.5 mM DTT, 0.1 mg/ml yeast tRNA, 0.1 mg/ml BSA for 30 min on ice. In a control experiment, 100-fold excess of unlabeled competitor probe was used. Eight percent glycerol was added to the mixture and the samples were separated by 6% native PAGE with 0.5× Tris borate-EDTA (TBE) running buffer. REMSA was performed as described previously (14).
Protein immunoprecipitation (IP)
The assay was performed essentially as described (31). In this assay, GAPDH was immunoprecipitated from a 30 μg cytoplasmic lysate prepared from HEK293 cells with the use of 1 μg of polyclonal anti-GAPDH antibody or 1 μg of monoclonal anti-cyclooxygenase 2 (Cox-2) antibody. The samples were precleared with protein G-sepharose beads (Sepharose, Pharmacia Biotech), GAPDH was immunoprecipitated, protein G-agarose beads were added to bind with the antibody, and extensively washed with 100 mM KCl, 5 mM MgCl2, 10 mM HEPES, pH 7.0, 0.5% NP-40, 1 mM DTT and 100 U/ml RNasin RNAse inhibitor as well as 2 mM vanadyl ribonucleoside complexes solution and protease and phosphatase inhibitor cocktails. Proteins were digested with 0.1% SDS and 30 μg of proteinase K followed by RNA extraction with phenol-chloroform-isoamyl alcohol mixture. RNA was precipitated with 10 μg of yeast tRNA as a carrier. Reverse transcription of the immunoprecipitated material was performed with poly-T oligos and PCR with primers targeting coding region of AT1R mRNA for 29 cycles (as described in quantitative PCR section). The products were separated on a 2% agarose gel producing a band at 163 bp.
Expression profiling
Alternatively, the immunoprecipitated RNA was used for expression profiling. Biotinylated cRNA was prepared, purified and fragmented according to Affymetrix protocol (Expression Analysis Technical Manual, Affymetrix). Human Genome U133 Plus 2.0 whole human genome expression array. GeneChips were washed and stained in the Affymetrix Fluidics Station 400 and scanned using the Affymetrix GeneChip Scanner 3000. The image data were analysed with Microarray Suite version 5.0. (MAS5) using Affymetrix default analysis settings and global scaling as normalization method.
Computational analysis
Microarray data were analyzed using R (32) and Bioconductor (33). First, probes were mapped to probe sets representing Ensembl transcript IDs with custom probe set definitions (34) and then normalized with the RMA quantile normalization method (35). The transcripts were ordered according to their expression values to find the most strongly enriched putative GAPDH targets. The 3′-UTR sequences were retrieved for the transcripts when available from the Ensembl database (36). The training data set from which the motifs were derived was constructed as follows. First, we used only sequences with length between 50 and 500 bp due to computational complexity with the longer sequences. Second, if a sequence was an identical or partial subsequence of another sequence it was excluded from the analysis since almost identical sequences can severely bias the motif finding process. From these sequences we chose n transcripts having the highest expression value to the training data set. CMFinder was used to derive the motifs from the unaligned sequences (37). Sizes used for the training data set were 10, 20, 30, 50, 100, 150 and 200. The 1–100 bp sequence of ATIR was added to the training data set to guide the motif finding process. From the potential motifs those that hit the ATIR sequence with at least a bit score of 10 were selected for further analysis. The significance of the putative motifs was tested by calculating the ratio of the number of hits per kilobases between the training data set and the reference data set. The reference data set included all the available 3′-UTR sequences of the transcripts on the microarray. The program Infernal (http://infernal.janelia.org/) was used to search hits with at least a bit score of 10 against the sequence sets. The motifs with highest enrichment ratio were considered the most interesting.
Ligand-binding assay
AT1R binding was measured using subconfluent cells grown in 24-well plates. The assay was performed as described previously (38).
In vitro translation
In vitro translation reactions were performed using flexi rabbit reticulocyte lysate system (Promega) as described by manufacturer. Protein synthesized in vitro were labelled with biotinylated lysine tRNA. Purified human GAPDH was purchased from Sigma and 50 and 150 ng were added to translation reaction when appropriate. The results were evaluated by luciferase assay as well as by western blotting. In the western blot, the in vitro translation products were detected using streptavidin-HRP according to manufacturer's protocols (Promega).
RESULTS
AT1R 3′-UTR decreases protein expression
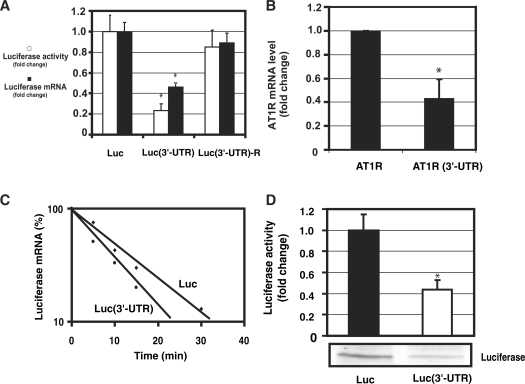
To test the hypothesis that cis-acting elements within the AT1R 3′-UTR have an important role in posttranscriptional regulation, we measured luciferase activities and mRNA levels in the presence or absence of AT1R 3′-UTR. Constructs contained luciferase coding sequence with or without AT1R 3′-UTR sequence and the SV40 late poly(A) signal. The full-length AT1R 3′-UTR reduced luciferase activity by ∼70–80% when compared with 3′-UTR-less construct. Western blotting results were similar to luciferase activities (data not shown). Quantitative PCR revealed luciferase RNA expression to be decreased ∼50% by AT1R 3′-UTR. The reverse complement of the AT1R 3′-UTR segment was cloned into the pGL3-Promoter vector downstream of the luciferase coding region. This construct reduced luciferase activity by ∼10% of promoter control or approximately one-sixth of the reduction attributable to the sense orientation of the 3′-UTR construct (Figure 1A). A pGL3 vector was constructed in which luciferase gene was replaced by AT1R coding region. Similarly to luciferase-AT1R 3′-UTR fusion construct, mRNA levels were lower with full length AT1R construct compared with AT1R coding region only (Figure 1B). These results indicate that 3′-UTR regulates both mRNA levels and protein expression.
Figure 1.
3′-UTR of AT1R is a negative regulator of AT1R expression. (A) The effect of AT1R 3′-UTR on luciferase activity (white bars). Three different constructs were studied: luciferase, luciferase with AT1R 3′-UTR and luciferase fused with AT1R 3′-UTR in reverse orientation. HEK293 cells were transfected with a luciferase and renilla luciferase plasmids. Cells were lysed and assayed for firefly and renilla luciferase activities. To evaluate mRNA levels, luciferase and GAPDH mRNA levels were measured by quantitative PCR. Corrected luciferase mRNA expression values were calculated by dividing luciferase mRNA by GAPDH mRNA expression (black bars). Values were normalized to the activity or mRNA levels of a luciferase reporter lacking the AT1R 3′-UTR to account for the effect of 3′-UTR. *P < 0.05 versus luciferase without 3′-UTR. (B) The effect of 3′-UTR on AT1R mRNA was evaluated. We compared the mRNA levels of AT1R coding region with or without 3'-UTR. AT1R and GAPDH mRNA levels were determined by quantitative PCR using AT1R coding region specific oligos. AT1R expression was corrected by GAPDH expression. Data were normalized to the activity of AT1R construct lacking the 3′-UTR. The results represent the means ± SD of an average of three independent experiments performed in triplicate for each construct. *P < 0.05 versus AT1R without 3′-UTR. (C) Measurement of the rate of degradation of luciferase mRNA. HEK293 cells were transiently transfected with luciferase construct with or without the full length AT1R 3′-UTR. Transfected cells were then treated with actinomycin D for 5 min, 10 min, 15 min and 30 min. Quantitative PCR was performed with luciferase and GAPDH specific oligos and luciferase mRNA expression was normalized by GAPDH. The mRNA measurements for each construct were normalized to the expression of the construct at time zero. Results are shown as a linear fit. Results represent the means of an average of three independent experiments performed as a triplicate for each construct. (D). Measurement of the rate of luciferase translation in rabbit reticulocyte lysates. In vitro translation of chimeric luciferase constructs with and without 3′-UTR were compared. From the in vitro translation reaction mixture luciferase activity was measured (upper panel) and western blot analysis was performed (lower panel). Translated proteins were detected with streptavidin-HRP detection system. *P < 0.05 versus luciferase without 3′-UTR.
AT1R 3′-UTR decreases mRNA half-life and suppresses mRNA translation. To understand the mechanisms of AT1R 3′-UTR-mediated decrease in protein expression, mRNA half-life and translation were evaluated. In order to study the effect of 3′-UTR on mRNA stability, transcription was halted by including actinomycin D to the medium. After various incubation periods the reaction was terminated and luciferase mRNA was extracted and analyzed by quantitative PCR. This data shows that 3′-UTR increases the rate of mRNA degradation (Figure 1C). To examine if AT1R 3′-UTR has any effect on the translation, in vitro translation was performed in the presence and absence of 3′-UTR. Luciferase activity of the translation products was measured. In addition, translation products were separated by SDS–PAGE, electroblotted and biotin-labelled translation products were detected by streptavidin-HRP. The size of the band was ∼60 kDa consistent with luciferase molecular weight. These results show a ∼60% inhibition of luciferase translation by AT1R 3′-UTR (Figure 1D). Based on these experiments, AT1R 3′-UTR decreases receptor expression by destabilizing mRNA and by suppressing mRNA translation.
GAPDH binds to AT1R mRNA
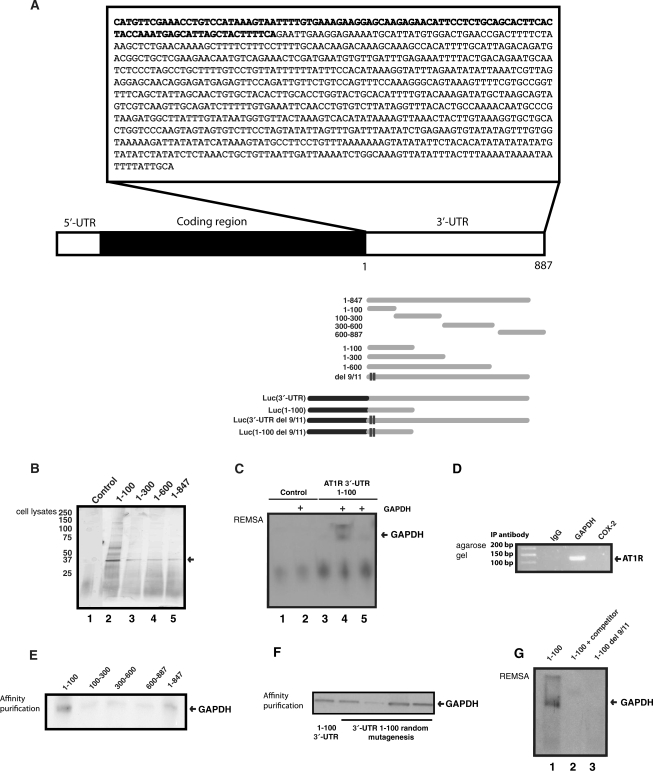
Next, we sought to identify the AT1R 3′-UTR mRNA interacting proteins that would mediate 3′-UTR-dependent effects. We assayed the existence of such RNA-binding proteins by employing affinity purification. Probes corresponding to 3′-UTR constructs 1–100, 1–300, 1–600 and 1–847 bases were transcribed and poly-A tail was included as a part of the primer used in the PCR-reaction. Due to problems in transcribing the full-length RNA construct, the longest polyadenylated RNA transcript used in the affinity purification did not include the last 40 bp in 3′-end of the 3′-UTR. Schematic representations of the probes are described in Figure 2A. Synthesized mRNAs with poly-A were bound to poly-T beads, incubated with cytoplasmic lysates followed by extensive washing. Figure 2B is a silver-stained gel showing the protein pattern of fractions eluted from AT1R 3′-UTR mRNA. The most abundant band migrated at ∼36 kDa and this band was present with all the AT1R 3′-UTR probes used. Binding was undetectable in reactions using fragments of luciferase coding region as a negative control. Protein was excised from the gel and digested ‘in gel’ by trypsin. The generated peptides were extracted and subjected to mass spectrometric analysis. Database (MSDB) search with the combined MALDI-TOF peptide mass fingerprint and MALDI-TOF/TOF peptide fragment ion data identified (Mascot score 238.0) the protein as human GAPDH. Human GAPDH is a basic protein and results in 36 tryptic peptides. Only eight have a molecular weight between 1000 and 2500 Da. Five of these peptides found in the mass fingerprinting corresponded to an intensity coverage of 59.3%. The two peptides (1 762 795 and 1 410 783 Da) subjected to MALDI-TOF/TOF analysis also individually identified the protein as human GAPDH with Mascot MS/MS Ion Search Program scores 130 and 94.70, respectively. Affinity purification results were confirmed by Western blot using polyclonal GAPDH antibodies (data not shown). As our longest probe did not have the 3′-end of the 3′-UTR bases, calreticulin was not purified by affinity purification. We did observe the binding of AUF-1 to AT1R 3′-UTR consistent with prior publications (data not shown) (12).
Figure 2.
GAPDH interacts with AT1R 3′-UTR. (A) A schematic of AT1R mRNA transcript as well as deleted and truncated transcripts used in affinity purification and gel shift assays. (B) Search for RNA-binding proteins that form a complex with AT1R 3′-UTR. The details of the purification are given under ‘Materials and Methods’ section. Lane 1: Luciferase coding region template was used as a control. Lanes 2–5: In vitro transcribed truncated fragments of AT1R 3'-UTR were used in the affinity purification. The differentially expressed 36 kDa protein was excised, digested, and the resulting peptides were recognized by mass spectrometry. (C) REMSA. After binding reaction, excess unbound RNA was digested. In lane 1A biotinylated control probe of luciferase coding region without GAPDH was used. Lane 2 has the same control probe as lane 1 but 150 ng of purified GAPDH protein was added. Lane 3 had biotinylated 3′-UTR 1–100 transcript without GAPDH, lane 4 had 150 ng of GAPDH and lane 5 had both 150 ng of GAPDH as well as 100-fold excess of unlabelled probe included in the binding mixture. (D) Binding of endogenous GAPDH with endogenous AT1R mRNA was detected by RT-qPCR assay with AT1R specific primers of material obtained by IP from cytoplasmic fractions. Changes in the level of AT1R mRNA associated with GAPDH were evaluated by measuring its abundance in the IP mixture. Immunoprecipitation was performed with preimmuno IgG, with GAPDH-specific antibody, and with anti-cox-2 antibody. PCR products were visualized after electrophoresis in 1% agarose gels stained with ethidium bromide. (E) Mapping of the GAPDH-binding site within AT1R 3′-UTR. A western blot of proteins isolated by RNA affinity purification was performed and GAPDH expression was detected by polyclonal anti-GAPDH antibodies. (F) Random mutagenesis of the 1–100 part of AT1R 3′-UTR. Mutated 3′-UTRs were generated by PCR-based mutagenesis. One hundred clones were generated and of those ten were sequenced to determine the rate of mutagenesis. Each mutant had 2 to 4 base insertions, deletions or mutations. We measured the binding of the endogenous GAPDH to the mutants by affinity purification. To reduce variability, the same HEK 293 cytoplasmic cell lysate was used for all these constructs. After protein binding, RNA treatment was performed. (G) REMSA of AT1R 3′-UTR Del 9/11. Lane 1. A biotinylated probe of 1–100 was used as a positive control for GAPDH REMSA. Cell lysate was from HEK293 cells. Lane 2. A biotinylated probe of 1–100 was used with 10-fold excess of unlabelled probe included in the binding mixture. Lane 3. A biotinylated transcript containing 1–100 del 9/11 that lacks GAPDH-binding site.
GAPDH binding to endogenous and recombinant AT1R mRNA
We first tested whether GAPDH binds directly to AT1R 3′-UTR, REMSA was performed with purified GAPDH protein. To this end, a transcript spanning 1–100 region of AT1R 3′-UTR was synthesized but no poly-A tail was added. Control transcript transcribed from luciferase coding region did not show gel shift with GAPDH. In contrast, a 1–100 fragment of AT1R 3′-UTR mRNA showed a shift with GAPDH that was competed by 100-fold excess of unlabeled probe. Gel shift with purified protein is consistent with a direct interaction (Figure 2C). REMSA performed with purified protein has two bands and it is possible that the upper band is due to GAPDH dimers or oligomers (41). This experiment also shows that polyadenylation of the transcript is not required for GAPDH's interaction with mRNA. Second, ribonucleoprotein (RNP) IP was performed to study the association of GAPDH with endogenous AT1R mRNA. The relative enrichment of AT1R mRNA in RNP IP reaction was also tested by RT, followed by conventional PCR amplification and then visualized on agarose gels. RNP IP assays revealed a strong enrichment of AT1R mRNA with GAPDH in anti-GAPDH antibody reactions relative to that of control IP reactions (IgG and cox-2) (Figure 2D). These results indicate that GAPDH forms RNP complex with endogenous AT1R mRNA. Third, we wanted to identify the AT1R 3′-UTR mRNA regions involved in associating with GAPDH. To this end, several polyadenylated transcripts spanning the different mRNA regions were synthesized. Following in vitro binding assays with cell lysates, the interaction between the transcripts and the cytosolic extract was assessed by affinity purification followed by western blotting. These observations supported a binding scheme on the AT1R 3′-UTR whereby GAPDH associated preferentially with the proximal 100 bp of the 3′-UTR (Figure 2E). To define GAPDH-binding site in more detail, we performed random mutagenesis of 1–100 region of the full length AT1R 3′-UTR. PCR-based mutagenesis was used to introduce on average two to four mutations in each clone. We isolated a clone in which residues 9 and 11 (counting from the beginning of 3′-UTR) were deleted to have a 4-fold increase in luciferase (data not shown). We hypothesized that this would be due to loss of GAPDH-binding to the 3′-UTR. In an in vitro binding assay GAPDH binding was reduced consistent with the idea that increased expression of mutant Del 9/11 is mediated by loss of GAPDH-mediated translation suppression, Figure 2F. The loss of interaction by deletion of residues 9 and 11 in the full length AT1R 3′-UTR was confirmed by gel shift, Figure 2G. Interestingly, REMSA performed with HEK293 cytoplasmic protein lysate has only one band instead of two seen with the purified GAPDH (Figure 2C). Pure GAPDH may have in the absence of other protein partners’ higher tendency to form protein dimers. Together, these findings indicated that GAPDH specifically binds the AT1R mRNA, both endogenous and in vitro transcribed, and that binding occurs in the 1–100 region of AT1R 3′-UTR.
Sequence and structure of predicted GAPDH-binding motif
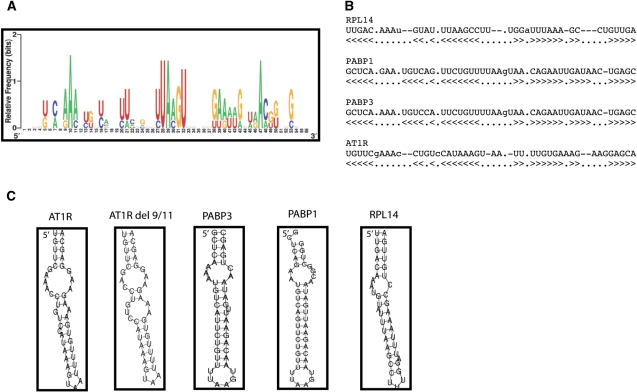
A collection of mRNAs that were GAPDH targets was identified. GAPDH was immunoprecipitated from HEK293 and the bound mRNA was exctracted and reverse transcribed, and the resulting products were hybridized to Affymetrix arrays. IgG was used as a control and the amount of mRNA immunoprecipitated under our conditions was very low. Transcripts corresponding to 6% of all genes on the array were substantially enriched in GAPDH IP compared with IgG IP. The RNA sequences were subjected to computational analysis (as described in ‘Materials and Methods’ section) to identify GAPDH motif based on secondary structures. The sequence alignment motif representation of relative frequency of nucleotides at each position and the examples of the secondary structures of this putative GAPDH motif are shown in Figure 3A and B. This analysis revealed AU-rich motif. The frequency of GAPDH motif in the database was 0.08 hits/kb which is similar to that of Hur (39). The secondary structures of representative mRNA with GAPDH-binding motif as well as del 9/11 construct are presented in Figure 3C. Our results suggest that GAPDH motif appears to be localized mostly to 3′-UTR, it is possible that in some transcripts the GAPDH motif is localized within the coding region. It should be noted that some GAPDH mRNAs identified on the arrays did not appear to contain the motif described in this study suggesting other motifs may also exist that were not identified in this analysis.
Figure 3.
Sequence and structure of the predicted GAPDH motif, as identified among GAPDH-bound transcripts. (A) Probability matrix (graphic logo) of the GAPDH motif indicating the relative frequency of finding each residue at each position within the motif, as elucidated from the array-derived experimental data set. (B) Structural alignment of four examples of GAPDH motif in specific mRNAs. (C) Secondary structure of four representative examples of the GAPDH motif in specific mRNAs. RPL14, ribosomal protein L14; PABP1, poly (A)-binding protein 1; PABP3, poly (A)-binding protein 3; AT1R, angiotensin II type 1 receptor. Furthermore, del 9/11 of AT1R 3′-UTR is shown.
GAPDH silencing increases AT1R expression
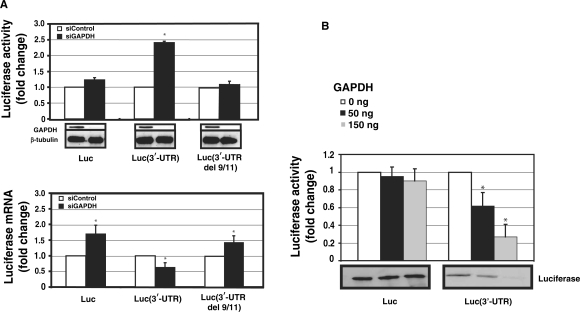
To assess the functional significance of GAPDH-AT1R 3′-UTR interactions, GAPDH levels were reduced by RNA interference. GAPDH down-regulation had very little effect (non significant) on the activity of a luciferase construct lacking GAPDH-binding site. In the luciferase construct with GAPDH-binding site, GAPDH silencing caused a 2.5-fold increase in the luciferase activity when compared to controls (Figure 4A, upper panel). When GAPDH does not bind to AT1R 3′-UTR (no 3′-UTR and del 9/11 constructs), reduction in GAPDH expression does not alter luciferase activity. These results suggest that adenines at position 9 and 11 of 3′-UTR are critical for GAPDH binding and for the functional effect of GAPDH. This increase in luciferase activity could potentially be mediated by stabilization of mRNA or by a relief of translational suppression upon GAPDH silencing. In order to evaluate the molecular mechanism of GAPDH-mediated effects, we measured the RNA-response to GAPDH silencing. To control for siRNA silencing efficiency, we measured GAPDH expression in the samples and observed an 8-fold decrease in GAPDH mRNA as compared to controls (data not shown). In contrast to protein levels, GAPDH silencing modestly decreased mRNA of luciferase fused with AT1R 3′-UTR, as measured by quantitative PCR (Figure 4A, lower panel). This suggests that the predominant effect of GAPDH on protein expression is not mediated by an alteration in mRNA decay but rather it is likely that GAPDH inhibits translation of AT1R by binding to its 3′-UTR.
Figure 4.
The effect of RNAi-mediated GAPDH knockdown on the expression of AT1R. HEK293 cells were cotransfected with luciferase vector and with 30 nM gene-specific GAPDH siRNA (black bars) or equal amount of control siRNA (white bars). (A) Upper panel: Luciferase activities of luciferase coding region only or fused with full length AT1R 3′-UTR or mutated full-length 3′-UTR del 9/11. Luminometer measurements for this and subsequent experiments are expressed as fold of promoter control without 3′-UTR in relative light units of firefly luciferase. Results represent the means ± SD of an average of three independent experiments performed as a triplicate for each construct. β-Tubulin was measured to control for protein loading. *P < 0.05 versus siControl-treated cells. Lower panel: Luciferase mRNA was quantified by quantitative PCR. All the results were normalized to β-actin expression. *P < 0.05 versus siControl transfected cells. (B) GAPDH suppresses AT1R 3′-UTR translation. The same transcripts as in Figure 1 were used. Increasing amounts of GAPDH was added to the reaction mixture (white bars 0 ng, black bars 50 ng and gray bars 150 ng). The translated product was evaluated by two methods. First, luciferase activity of the translation mixture was measured (upper panel). Second, lysate was separated on SDS–PAGE gel, blotted and detected on a membrane by streptavidin-HRP (lower panel). Experiment with and without AT1R 3′-UTR represent different exposures and therefore baseline difference between these two constructs cannot be compared. The results represent the means ± SD of an average of three independent experiments. *P < 0.05 versus control reaction with no GAPDH.
To test directly the role of translation as a molecular mechanism of GAPDH-induced decrease in AT1R expression, we performed in vitro translation. To further examine the molecular mechanism of GAPDH effects, luciferase construct with and without AT1R 3′-UTR was in vitro translated. Translation reactions were conducted in the presence of varying concentrations of purified GAPDH. The resulting products were measured for luciferase activity as well as separated on a SDS–PAGE gel and detected with streptavidin-HRP. In keeping with GAPDH's silencing effect on protein expression, GAPDH inhibited protein synthesis of a luciferase construct with AT1R 3′-UTR in a concentration dependent manner (Figure 4B). There was only a minor change in luciferase translation in the absence of 3′-UTR indicating that the GAPDH effect is mediated by AT1R 3′-UTR. Inclusion of BSA as a control protein did not have any effect on translation (data not shown). These results support the view that GAPDH functions as a translational repressor of AT1R.
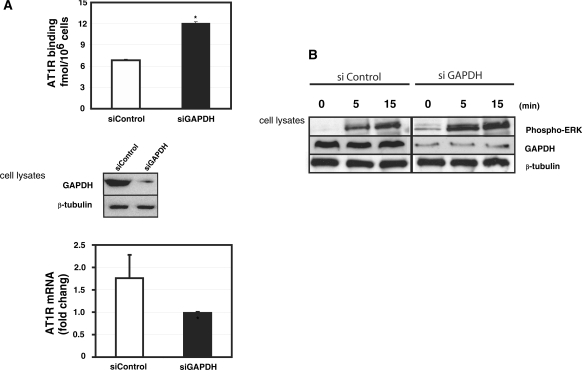
GAPDH silencing increases the expression of endogenous AT1R
To test the importance of GAPDH on the expression of endogenous AT1R in coronary artery VSMCs, we silenced GAPDH expression. To control for siRNA silencing efficiency, we measured GAPDH expression in the samples and observed an 8-fold decrease in GAPDH mRNA as compared to controls (data not shown). As shown in Figure 5A (upper panel), the use of siRNA targeting GAPDH increased AT1R protein expression as measured by ligand binding and thus GAPDH functional effects are similar in HEK293 cells and coronary artery VSMCs. GAPDH protein levels were significantly decreased by 72 h after transfection of a GAPDH-targeting siRNA. Similarly to HEK293 cells, siGAPDH modestly decreased AT1R 3′-UTR mRNA in coronary artery VSMCs, as measured by quantitative PCR analysis (Figure 5A, lower panel). GAPDH stabilizes AT1R mRNA in a similar way as it has been described to influence some other mRNAs (15). We tested whether increased AT1R expression would lead to enhanced MAPK phosphorylation by angiotensin II in coronary artery VSMC. AT1R stimulation by angiotensin II causes ERK phosphorylation. GAPDH-silenced cells showed stronger phosphorylation of ERK MAPKs in response to angiotensin II, Figure 5B. These results implicate GAPDH in the regulation of AT1R expression.
Figure 5.
Role of GAPDH in the regulation of endogenous AT1R. (A) Upper panel: The effect of GAPDH silencing on AT1R expression in early passage (3–5) coronary artery VSMCs. VSMCs were transfected with 30 nM GAPDH siRNA or equal amount of control siRNA. AT1R expression was quantified by ligand binding. The western blot below demonstrates the effect of GAPDH siRNA on GAPDH expression. Lower panel: AT1R mRNA was quantified by RT–qPCR. All the results were normalized to β-actin expression. *P < 0.05 versus siControl transfected cells. (B) Activation of ERK MAPKs in human coronary artery VSMC by angiotensin II. Coronary artery VSMC were transfected either with siGAPDH or control siRNA. These cells were serum starved for 24 h before stimulation by 10−7 M angiotensin II. Cells were flash frozen at appropriate time points, proteins extracted and results analyzed by western blotting using anti-phospho-ERK antibody, GAPDH antibodies, and β-tubulin antibody.
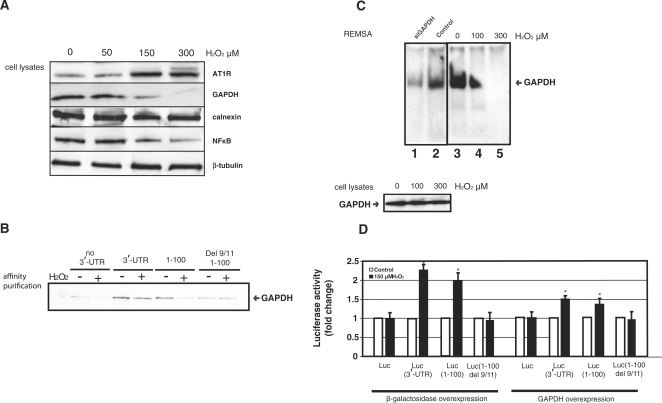
GAPDH mediates the effect of H2O2 on AT1R
Exposure of coronary artery VSMC to hydrogen peroxide caused a dose-dependent down-regulation of GAPDH while it increased AT1R expression (24,25), Figure 6A. Several lines of evidence support the role of GAPDH in the H2O2-induced regulation of AT1R. Affinity purification assay demonstrates that H2O2-treatment of cell lysates reduced GAPDH binding to both full-length AT1R 3′-UTR and 1–100 of 3′-UTR, Figure 6B. In Figure 6B we see a smaller reduction by H2O2 in full-length 3′-UTR expression (from ∼1.5- to 2-fold) whereas with 1–100 there is a larger reduction. The full length AT1R 3′-UTR may be subject to additional counteracting regulatory mechanisms. In contrast to intact cells, GAPDH protein level did not change in cell lysates exposed to H2O2. In Figure 6C, REMSA performed with H2O2-treated lysates showed decreased GAPDH binding to 3′-UTR of AT1R in a H2O2-concentration dependent manner. Thus, the reduction in GAPDH binding to AT1R 3′-UTR was not entirely due to reduced GAPDH expression but also due to decreased GAPDH-binding affinity.
Figure 6.
(A) Effect of H2O2 on AT1R expression in human coronary artery VSMC. These VSMC were exposed to varying concentrations of H2O2 for 15 h. Western blots were probed with appropriate antibodies. (B) GAPDH binding to AT1R 3′-UTR was evaluated by affinity purification using the 3′-UTR 0–847 as a probe (same as in Figure 2B and E) and western blotting of the isolated proteins by GAPDH antibodies. GAPDH expression was evaluated in total cellular lysates prepared from HEK293 cells exposed to 150 μM H2O2 for 15 h (lower panel, right). β-Tubulin was measured to control for protein loading. (C) H2O2 treatment reduces GAPDH binding to AT1R. Exposure of protein lysates to H2O2 does not influence protein levels but decreases GAPDH binding to 3′-UTR in a dose-dependent manner. Left side of the REMSA has a control blot (lanes 1–2) and the right side has the experiment (lanes 3–5) in which protein lysate was exposed to H2O2 for 1 h. (D) GAPDH mediates H2O2-induced effect on the posttranscriptional regulation of AT1R. The same luciferase constructs as in Figure 4A were used. HEK293 cells were transfected with either β-galactosidase (left panel) or GAPDH expression vector (right panel) together with luciferase constructs with (3′-UTR, 1–100) or without GAPDH-binding site (no 3′-UTR, 1–100 del 9/11). Cells were exposed to 150 μM H2O2 for 15 h (black bar) or to vehicle (white bar), lysed and assayed for luciferase activities. Values were normalized to the activity of untreated luciferase construct. The results represent the means ± SD of an average of three independent experiments performed in triplicate for each construct. *P < 0.05 versus AT1R without 3′-UTR.
To determine if GAPDH interaction with 3′-UTR was necessary for the H2O2-induced increase in AT1R expression, we tested the effect of deleting the GAPDH-binding site in the reporter gene. HEK293 cells were transfected with luciferase construct either with or without AT1R 3′-UTR. Similarly to endogenous AT1R, H2O2 treatment decreased luciferase-AT1R 3′-UTR activity. Deletion of GAPDH-binding site in 3′-UTR led to an attenuated response to hydrogen peroxide (Figure 6D). We transiently transfected GAPDH to reverse H2O2-induced decrease in GAPDH expression and indeed found that GAPDH overexpression attenuated this response. Taken together, these results indicate that decreased GAPDH binding to AT1R 3′-UTR in response to H2O2 increased AT1R expression.
DISCUSSION
We found that GAPDH bound the AT1R mRNA and affected its translation, that H2O2 treatment decreased (GAPDH-AT1 mRNA) complexes. GAPDH is an abundant protein that is located mainly but not exclusively within the cytoplasm of cells. It is essential for glycolysis, but recent evidence suggests it may have several other important cellular functions. GAPDH binds RNA through NAD-binding region, however, not much is known about the sequence specificity of GAPDH binding (17). Our data suggest that GAPDH effects on AT1R expression are governed by cis-element in AT1R mRNA and mediated by a direct GAPDH–mRNA interaction. Gel shift shows that the interaction of GAPDH with AT1R 3′-UTR is direct and requires neither miRNA nor other proteins. GAPDH binding is specific based on competition experiment in gel shift as well as deletion and mutagenesis analysis of the sequence requirements for GAPDH binding within 3′-UTR. Deletion of bases at 9 and 11 leads to a loss of GAPDH binding to AT1R 3′-UTR. Although there appears to be much flexibility in the RNA-binding protein binding sites, single mutations have been shown to decrease protein binding to mRNA (40). It is likely that an alteration in the secondary structure of AT1R by two single-base deletions removes essential structural element of the GAPDH-binding site.
AT1R expression is modulated by the level of GAPDH binding to AT1R mRNA. In an in vitro assay GAPDH dose-dependently inhibits AT1R translation. Deletion of GAPDH-binding site from the 3′-UTR increases reporter gene activity consistent with the loss of GAPDH-mediated translation suppression. Similar effect was achieved by reducing GAPDH expression. GAPDH silencing reduced GAPDH binding to AT1R mRNA and increased AT1R expression. The reversal of the GAPDH-mediated translation inhibition by deletion of GAPDH-binding site within 3′-UTR, further argues for the exquisite specificity of GAPDH's interaction with a defined 3′-UTR sequence of the AT1R mRNA to cause translation inhibition. GAPDH silencing and 3′-UTR mutation del 9/11 increase AT1R expression by decreasing GAPDH binding to 3′-UTR, therefore providing a strong argument for a direct link between GAPDH and AT1R expression.
As shown in this report, H2O2 treatment decreased GAPDH binding to AT1R mRNA, thereby increasing AT1R expression. Accordingly, loss of GAPDH-binding site within 3′-UTR resulted in a loss of H2O2 response, while overexpression of GAPDH attenuated H2O2 response. Similarly to GAPDH silencing experiments, decrease in GAPDH expression by oxidative stress led to decreased GAPDH binding to AT1R mRNA. This appears to be due to reduced GAPDH expression and change in GAPDH structure due to oxidative stress (41). The oxidative stress responsive element and GAPDH-binding site are the same as 3′-UTR mutant lacking both GAPDH binding and oxidative stress-response, providing a further argument for a link between GAPDH and AT1R regulation. Decrease in GAPDH binding to AT1R 3′-UTR by RNA silencing, deleting GAPDH-binding site, or by oxidative stress led to increased AT1R expression providing a strong argument for the role of GAPDH in the posttranscriptional control of AT1R. In addition to oxidative stress, GAPDH may regulate the basal expression of AT1R. Interestingly, there is a 15-fold difference in GAPDH mRNA in baseline expression between highest and lowest expressing tissue (42). Thus, the level of GAPDH-induced transcript-specific translational suppression of AT1R varies from tissue to tissue. Transcript-specific translational suppression has been described for a wide variety of transcripts including β-adrenergic receptor (43), cox-2 (44) and tumor necrosis factor α (45), for example. Although there are several proteins that inhibit translation by binding to the 3′-UTR including TIA-1 and Ago1, the molecular mechanism of translation inhibition by 3′-UTR-binding proteins remains unclear.
Computational analysis of GAPDH-binding mRNAs led to elucidation of AU-rich motif present in GAPDH target mRNAs. GAPDH-binding motif found in AT1R mRNA can also be found in ∼3% of transcripts suggesting that it has a more general role in cellular mRNA regulation. Two well-characterized RBPs, HuR and TIAR, have a large number target mRNAs and it could be that RBP have fairly broad target specificity (39,40). Depending on target mRNA, GAPDH either regulates mRNA stability or translation (46). It appears to be common for RBPs to have mRNA-dependent functional effects. In many instances, the same RNA-binding protein mediates both RNA destabilization and translational silencing (47,48). A well-characterized The Embryonic Lethal Abnormal Vision (ELAV)-like protein HuR regulates both mRNA stability and translation of the mRNAs it binds.
In summary, we identified human AT1R 3′-UTR as a potent negative regulator of mRNA expression through inhibition of translation and destabilization of mRNA. Posttranscriptional regulation of AT1R is a complex interplay of positive regulators such as p100 (14) and negative regulators such as GAPDH, calreticulin (6) and AUF1 (12). Translational suppression of AT1R mRNA by converting it to an untranslatable or less translatable form by interaction with GAPDH reduces the AT1R mRNA translation. The ability to regulate the expression of AT1R by altering translation efficiency would be a rapid and efficient way to control receptor expression, especially in response to oxidative stress in kidneys, blood vessels and the heart.
FUNDING
The Finnish Academy, the Finnish Foundation for Cardiovascular Research, the Sigrid Juselius Foundation (to K.K and J.L.), Finnish Medical Foundation and Instrumentarium Research Foundation (to J.L.). Funding for open access charge: Finnish Heart Foundation.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We thank Ms Susanna Tverin, Ms Saara Nyqvist, Ms Tuula Soppela and Gunilla Rönnholm for skilful technical assistance.
REFERENCES
- 1.Dzau V. The cardiovascular continuum and renin-angiotensin-aldosterone system blockade. J. Hypertens. Suppl. 2005;23:S9–S17. [PubMed] [Google Scholar]
- 2.Horiuchi M, Akishita M, Dzau VJ. Recent progress in angiotensin II type 2 receptor research in the cardiovascular system. Hypertension. 1999;33:613–621. doi: 10.1161/01.hyp.33.2.613. [DOI] [PubMed] [Google Scholar]
- 3.Lehtonen JY, Daviet L, Nahmias C, Horiuchi M, Dzau VJ. Analysis of functional domains of angiotensin II type 2 receptor involved in apoptosis. Mol. Endocrinol. 1999;13:1051–1060. doi: 10.1210/mend.13.7.0303. [DOI] [PubMed] [Google Scholar]
- 4.Daviet L, Lehtonen JY, Tamura K, Griese DP, Horiuchi M, Dzau VJ. Cloning and characterization of ATRAP, a novel protein that interacts with the angiotensin II type 1 receptor. J. Biol. Chem. 1999;274:17058–17062. doi: 10.1074/jbc.274.24.17058. [DOI] [PubMed] [Google Scholar]
- 5.Elton TS, Martin MM. Angiotensin II type 1 receptor gene regulation: transcriptional and posttranscriptional mechanisms. Hypertension. 2007;49:953–961. doi: 10.1161/HYPERTENSIONAHA.106.070565. [DOI] [PubMed] [Google Scholar]
- 6.Nickenig G, Michaelsen F, Muller C, Berger A, Vogel T, Sachinidis A, Vetter H, Bohm M. Destabilization of AT(1) receptor mRNA by calreticulin. Circ. Res. 2002;90:53–58. doi: 10.1161/hh0102.102503. [DOI] [PubMed] [Google Scholar]
- 7.Nickenig G, Murphy TJ. Down-regulation by growth factors of vascular smooth muscle angiotensin receptor gene expression. Mol. Pharmacol. 1994;46:653–659. [PubMed] [Google Scholar]
- 8.Nickenig G, Murphy TJ. Enhanced angiotensin receptor type 1 mRNA degradation and induction of polyribosomal mRNA binding proteins by angiotensin II in vascular smooth muscle cells. Mol. Pharmacol. 1996;50:743–751. [PubMed] [Google Scholar]
- 9.Nickenig G, Sachinidis A, Michaelsen F, Bohm M, Seewald S, Vetter H. Upregulation of vascular angiotensin II receptor gene expression by low-density lipoprotein in vascular smooth muscle cells. Circulation. 1997;95:473–478. doi: 10.1161/01.cir.95.2.473. [DOI] [PubMed] [Google Scholar]
- 10.Nickenig G, Roling J, Strehlow K, Schnabel P, Bohm M. Insulin induces upregulation of vascular AT1 receptor gene expression by posttranscriptional mechanisms. Circulation. 1998;98:2453–2460. doi: 10.1161/01.cir.98.22.2453. [DOI] [PubMed] [Google Scholar]
- 11.Nickenig G, Bohm M. Interaction between insulin and AT1 receptor. Relevance for hypertension and arteriosclerosis. Basic Res. Cardiol. 1998;93(Suppl. 2):135–139. doi: 10.1007/s003950050240. [DOI] [PubMed] [Google Scholar]
- 12.Pende A, Giacche M, Castigliola L, Contini L, Passerone G, Patrone M, Port JD, Lotti G. Characterization of the binding of the RNA-binding protein AUF1 to the human AT(1) receptor mRNA. Biochem. Biophys. Res. Commun. 1999;266:609–614. doi: 10.1006/bbrc.1999.1862. [DOI] [PubMed] [Google Scholar]
- 13.Zhang W, Wagner BJ, Ehrenman K, Schaefer AW, DeMaria CT, Crater D, et al. Purification, characterization, and cDNA cloning of an AU-rich element RNA-binding protein, AUF1. Mol. Cell Biol. 1993;13:7652–7665. doi: 10.1128/mcb.13.12.7652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Paukku K, Kalkkinen N, Silvennoinen O, Kontula KK, Lehtonen JY. p100 increases AT1R expression through interaction with AT1R 3′-UTR. Nucleic Acids Res. 2008;36:4474–4487. doi: 10.1093/nar/gkn411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bonafe N, Gilmore-Hebert M, Folk NL, Azodi M, Zhou Y, Chambers SK. Glyceraldehyde-3-phosphate dehydrogenase binds to the AU-Rich 3′ untranslated region of colony-stimulating factor-1 (CSF-1) messenger RNA in human ovarian cancer cells: possible role in CSF-1 posttranscriptional regulation and tumor phenotype. Cancer Res. 2005;65:3762–3771. doi: 10.1158/0008-5472.CAN-04-3954. [DOI] [PubMed] [Google Scholar]
- 16.Dollenmaier G, Weitz M. Interaction of glyceraldehyde-3-phosphate dehydrogenase with secondary and tertiary RNA structural elements of the hepatitis A virus 3′ translated and non-translated regions. J. Gen. Virol. 2003;84:403–414. doi: 10.1099/vir.0.18501-0. [DOI] [PubMed] [Google Scholar]
- 17.Nagy E, Rigby WF. Glyceraldehyde-3-phosphate dehydrogenase selectively binds AU-rich RNA in the NAD(+)-binding region (Rossmann fold) J. Biol. Chem. 1995;270:2755–2763. doi: 10.1074/jbc.270.6.2755. [DOI] [PubMed] [Google Scholar]
- 18.Sirover MA. New insights into an old protein: the functional diversity of mammalian glyceraldehyde-3-phosphate dehydrogenase. Biochim. Biophys. Acta. 1999;1432:159–184. doi: 10.1016/s0167-4838(99)00119-3. [DOI] [PubMed] [Google Scholar]
- 19.Sirover MA. New nuclear functions of the glycolytic protein, glyceraldehyde-3-phosphate dehydrogenase, in mammalian cells. J. Cell Biochem. 2005;95:45–52. doi: 10.1002/jcb.20399. [DOI] [PubMed] [Google Scholar]
- 20.Hara MR, Agrawal N, Kim SF, Cascio MB, Fujimuro M, Ozeki Y, et al. S-nitrosylated GAPDH initiates apoptotic cell death by nuclear translocation following Siah1 binding. Nat. Cell Biol. 2005;7:665–674. doi: 10.1038/ncb1268. [DOI] [PubMed] [Google Scholar]
- 21.Hara MR, Cascio MB, Sawa A. GAPDH as a sensor of NO stress. Biochim. Biophys. Acta. 2006;1762:502–509. doi: 10.1016/j.bbadis.2006.01.012. [DOI] [PubMed] [Google Scholar]
- 22.Barbini L, Rodriguez J, Dominguez F, Vega F. Glyceraldehyde-3-phosphate dehydrogenase exerts different biologic activities in apoptotic and proliferating hepatocytes according to its subcellular localization. Mol. Cell Biochem. 2007;300:19–28. doi: 10.1007/s11010-006-9341-1. [DOI] [PubMed] [Google Scholar]
- 23.Andrade J, Pearce ST, Zhao H, Barroso M. Interactions among p22, glyceraldehyde-3-phosphate dehydrogenase and microtubules. Biochem. J. 2004;384:327–336. doi: 10.1042/BJ20040622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sukhanov S, Higashi Y, Shai SY, Itabe H, Ono K, Parthasarathy S, Delafontaine P. Novel effect of oxidized low-density lipoprotein: cellular ATP depletion via downregulation of glyceraldehyde-3-phosphate dehydrogenase. Circ. Res. 2006;99:191–200. doi: 10.1161/01.RES.0000232319.02303.8c. [DOI] [PubMed] [Google Scholar]
- 25.Li D, Saldeen T, Romeo F, Mehta JL. Oxidized LDL upregulates angiotensin II type 1 receptor expression in cultured human coronary artery endothelial cells: the potential role of transcription factor NF-kappaB. Circulation. 2000;102:1970–1976. doi: 10.1161/01.cir.102.16.1970. [DOI] [PubMed] [Google Scholar]
- 26.Grifoni SC, Gannon KP, Stec DE, Drummond HA. ENaC proteins contribute to VSMC migration. Am. J. Physiol. Heart. Circ. Physiol. 2006;291:H3076–H3086. doi: 10.1152/ajpheart.00333.2006. [DOI] [PubMed] [Google Scholar]
- 27.Fan X, Roy E, Zhu L, Murphy TC, Kozlowski M, Nanes MS, et al. Nitric oxide donors inhibit luciferase expression in a promoter-independent fashion. J. Biol. Chem. 2003;278:10232–10238. doi: 10.1074/jbc.M209911200. [DOI] [PubMed] [Google Scholar]
- 28.Masuda H, Fukabori Y, Nakano K, Shimizu N, Yamanaka H. Expression of bone morphogenetic protein-7 (BMP-7) in human prostate. Prostate. 2004;59:101–106. doi: 10.1002/pros.20030. [DOI] [PubMed] [Google Scholar]
- 29.Minami M, Daimon Y, Mori K, Takashima H, Nakajima T, Itoh Y, Okanoue T. Hepatitis B-virus-related insertional mutagenesis in chronic hepatitis B patients as an early drastic genetic change leading to hepatocarcinogenesis. Oncogene. 2005;24:4340–4348. doi: 10.1038/sj.onc.1208628. [DOI] [PubMed] [Google Scholar]
- 30.Shevchenko A, Wilm M, Vorm O, Mann M. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Biochem. 1996;68:850–858. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- 31.Peritz T, Zeng F, Kannanayakal TJ, Kilk K, Eiriksdottir E, Langel U, Eberwine J. Immunoprecipitation of mRNA-protein complexes. Nat. Protoc. 2006;1:577–580. doi: 10.1038/nprot.2006.82. [DOI] [PubMed] [Google Scholar]
- 32.Team RDC. R Foundation for Statistical Computing. Vienna, Austria: 2007. R: A Language and Environment for Statistical Computing. [Google Scholar]
- 33.Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, et al. Bioconductor: Open software development for computational biology and bioinformatics. Genome Biology. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dai M, Wang P, Boyd AD, Kostov G, Athey B, Jones EG, Bunney WE, Myers RM, Speed TP, Akil H, et al. Evolving gene/transcript definitions significantly alter the interpretation of GeneChip data. Nucleic Acids Res. 2005;33:e175. doi: 10.1093/nar/gni179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Irizarry R, Bolstad B, Collin F, Cope L, Hobbs B, Speed T. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 2003;33:e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hubbard T, Andrews D, Caccamo M, Cameron G, Chen Y, Clamp M, Clarke L, Coates F, Cox T, Cunningham F, et al. Ensemble 2005. Nucleic Acids Res. 2006;33:D447–D453. doi: 10.1093/nar/gki138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yao Z, Weinberg Z, Ruzzo W. CMfinder – A covariance model based RNA motif finding algorithm. Bioinformatics. 2006;22:445–452. doi: 10.1093/bioinformatics/btk008. [DOI] [PubMed] [Google Scholar]
- 38.Akishita M, Ito M, Lehtonen JY, Daviet L, Dzau VJ, Horiuchi M. Expression of the AT2 receptor developmentally programs extracellular signal-regulated kinase activity and influences fetal vascular growth. J. Clin. Invest. 1999;103:63–71. doi: 10.1172/JCI5182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lopez de Silanes I, Zhan M, Lal A, Yang X, Gorospe M. Identification of a target RNA motif for RNA-binding protein HuR. Proc. Natl Acad. Sci. USA. 2004;101:2987–2992. doi: 10.1073/pnas.0306453101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim HS, Kuwano Y, Zhan M, Pullman M, Jr., Mazan-Mamczarz K, Li H, Kedersha N, Anderson P, Wilce MC, Gorospe M, et al. A: Elucidation of a C-rich signature motif in target mRNAs of RNA-binding protein TIAR. Mol. Cell Biol. 2007;27:6806–6817. doi: 10.1128/MCB.01036-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nakajima H, Amano W, Fujita A, Fukuhara A, Azuma Y-T, Hata F, Inui T, Takeuchi T. The Active Site Cysteine of the Proapoptotic Protein Glyceraldehyde-3-phosphate Dehydrogenase Is Essential in Oxidative Stress-induced Aggregation and Cell Death. J. Biol. Chem. 2007;282:26562–26574. doi: 10.1074/jbc.M704199200. [DOI] [PubMed] [Google Scholar]
- 42.Barber RD, Harmer DW, Coleman RA, Clark BJ. GAPDH as a housekeeping gene: analysis of GAPDH mRNA expression in a panel of 72 human tissues. Physiol. Genomics. 2005;21:389–395. doi: 10.1152/physiolgenomics.00025.2005. [DOI] [PubMed] [Google Scholar]
- 43.Kandasamy K, Joseph K, Subramaniam K, Raymond JR, Tholanikunnel BG. Translational control of beta2-adrenergic receptor mRNA by T-cell-restricted intracellular antigen-related protein. J. Biol. Chem. 2005;280:1931–1943. doi: 10.1074/jbc.M405937200. [DOI] [PubMed] [Google Scholar]
- 44.Sureban SM, Murmu N, Rodriguez P, May R, Maheshwari R, Dieckgraefe BK, et al. Functional antagonism between RNA binding proteins HuR and CUGBP2 determines the fate of COX-2 mRNA translation. Gastroenterology. 2007;132:1055–1065. doi: 10.1053/j.gastro.2006.12.031. [DOI] [PubMed] [Google Scholar]
- 45.Garnon J, Lachance C, Di Marco S, Hel Z, Marion D, Ruiz MC, et al. Fragile X-related protein FXR1P regulates proinflammatory cytokine tumor necrosis factor expression at the post-transcriptional level. J. Biol. Chem. 2005;280:5750–5763. doi: 10.1074/jbc.M401988200. [DOI] [PubMed] [Google Scholar]
- 46.Atasoy U, Watson J, Patel D, Keene JD. ELAV protein HuA (HuR) can redistribute between nucleus and cytoplasm and is upregulated during serum stimulation and T cell activation. J. Cell Sci. 1998;111(Pt 21):3145–3156. doi: 10.1242/jcs.111.21.3145. [DOI] [PubMed] [Google Scholar]
- 47.Leandersson K, Riesbeck K, Andersson T. Wnt-5a mRNA translation is suppressed by the Elav-like protein HuR in human breast epithelial cells. Nucleic Acids Res. 2006;34:3988–3999. doi: 10.1093/nar/gkl571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Piecyk M, Wax S, Beck AR, Kedersha N, Gupta M, Maritim B, Chen S, Gueydan C, Kruys V, Streuli M, et al. TIA-1 is a translational silencer that selectively regulates the expression of TNF-alpha. EMBO J. 2000;19:4154–4163. doi: 10.1093/emboj/19.15.4154. [DOI] [PMC free article] [PubMed] [Google Scholar]