Abstract
Conditional gene expression systems have developed into essential tools for the study of gene functions. However, their utility is often limited by the difficulty of identifying clonal cell lines, in which transgene control can be realized to its full potential. Here, we describe HeLa cell lines, in which we have identified—by functional analysis—genomic loci, from which the expression of transgenes can be tightly controlled via tetracycline-regulated expression. These loci can be re-targeted by recombinase-mediated cassette exchange. Upon exchange of the gene of interest, the resulting cell line exhibits the qualitative and quantitative properties of controlled transgene expression characteristic for the parent cell line. Moreover, by using an appropriate promoter, these cell lines express the tetracycline controlled transcription activator rtTA2-M2 uniformly throughout the entire cell population. The potential of this approach for functional genomics is highlighted by utilizing one of our master cell lines for the efficient microRNA-mediated knockdown of the endogenous human lamin A/C gene.
INTRODUCTION
Our understanding of gene functions has greatly benefited from approaches that permit to predictably activate or deactivate the expression of individual genes and to monitor subsequent phenotypic changes. In this context, tetracycline controlled transcription activation is the most widely applied principle (1–3). Indeed, the ‘Tet System’ was shown to function not only within a broad spectrum of cultured cells, but also in whole organisms from fungi to non-human primates (4,5). Salient features of the system are reversibility, tightness of control, a wide regulation window as well as quantitative control of gene expression in incremental steps.
For optimal function of Tet regulation, two main prerequisites have to be met, which sometimes are not trivial to establish experimentally. First, the target cell has to constitutively produce appropriate concentrations of one of the tetracycline controlled transcription activators tTA (1) or rtTA (2,6) uniformly throughout the cell population. Second, the tTA/rtTA responsive RNA polymerase II promoter, Ptet, has to be integrated in the target cell's genome in such a way that the highly specific interaction between Ptet and tTA or rtTA is not perturbed by the local chromosomal context. Possible interferences with its desired expression characteristics can for example be caused by transcriptional enhancers or silencers in the vicinity of Ptet or by obstructing chromatin structures surrounding the integrated Ptet-controlled transcription unit (1,7).
Here, we address the challenge of predictably placing Ptet-controlled transcription units into a genomic site where the full potential of Tet regulation can be exploited. We describe the identification of a chromosomal locus in a novel rtTA2-M2 expressing HeLa cell line, where a Ptet-directed transcription unit is virtually inactive in the absence of doxycycline (dox), but activated over more than four orders of magnitude in its presence. This functionally defined ‘silent but activatable’ (s/a) locus (7–9) can be directly targeted via FLP recombinase-mediated cassette exchange (RMCE) (10). RMCE empowers us to efficiently insert any gene of interest into the s/a locus and to control its expression, mirroring the regulation of expression of the parental transgene. Moreover, by expressing the transactivator under control of the human elongation factor 1 alpha promoter (EF1α), a uniform production of rtTA2-M2 is warranted throughout the entire population of cells.
The possibility to readily place controllable transcription units into pre-characterized genomic loci of otherwise isogenic cell lines as described herein will significantly contribute to the study of gene functions under highly defined conditions. We exemplify this principle by precisely controlling the concentrations of the intermediate filament lamin A/C and the nuclear pore protein Pom121, by Tet-regulated RNA interference.
MATERIALS AND METHODS
Plasmid constructs
The S2f-lMCg-F3 vector was derived from the retroviral SIN-vector S2f-lMCg (9) by exchanging the F5 Flp-recombinase recognition site for the mutated F3 site (10) (Figure 1A). The plasmid pE11.F3.M.F was derived from pCMV.MCS.pA.FRTN1ampFRT (a generous gift from G. Schütz, DKFZ, Heidelberg) by flanking the multiple cloning site (MCS) with heterospecific Flp recognition sites F3 and FRT. For cloning of pE11.F3.htk.F., the hygTK fusion gene was released by XhoI and HindIII from p.F3.HygTK.F (11) and inserted into XhoI/HindIII cut pE11.F3.M.F plasmid.
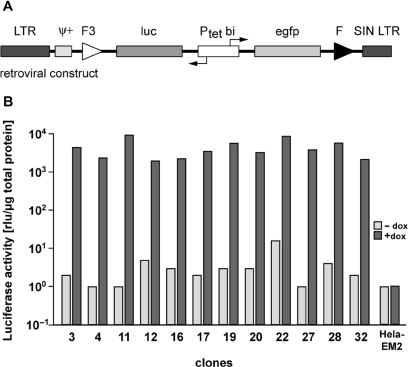
Figure 1.
Generation of HeLa cell lines with highly regulated gene expression by retroviral transduction. (A) Schematic outline of the proviral MuLV-based S2f-lMCg-F3 vector used for stable transduction of HeLa-EM2 cells. The bidirectional tetracycline-inducible promoter (Ptetbi) controls the bicistronic expression of the reporter genes luc (luciferase) and egfp (enhanced green fluorescent protein). The reporter unit is flanked by a wild type (F, downstream) and a mutant (F3, upstream) Flp-recombinase target site. The viral packaging signal Ψ+ and the 5′LTR and 3′SIN-LTR (self-inactivating long terminal repeats) are retroviral elements, the latter defining the borders of the integrated provirus. (B) Independent HeLa cell clones show silent, but highly activatable expression of luciferase. Clonal HeLa-EM2 cell lines derived from transduction with the S2f-lMCg-F3 retrovirus were kept in the absence or presence of dox (1 µg/ml). Luciferase activity in all extracts was measured and normalized to the protein content. Clone HeLa-EM2-11 was chosen for further analysis.
The RMCE vectors depicted in Figures 5 and 6 are based on the plasmid pBI4 (12). pBI4 contains two MCSs up- and downstream of the bidirectional tetracycline-inducible promoter (Ptet-bi), which enable the insertion of two transcription units in opposing directions. To construct pBI.F3.M.F, the bidirectional expression cassette of pBI4 was flanked by two heterospecific Flp recognition sites F3 and FRT, synthesized as complementary oligonucleotides. Subsequently, the cDNAs for d1EGFP (13), mCherry (cherry) (14), luciferase and nuclear localized β-galactosidase were inserted in the MCS of pBI.F3.M.F, thereby constructing the recombination vectors pd1gfpPtetcherry and plucPtetlacZ.
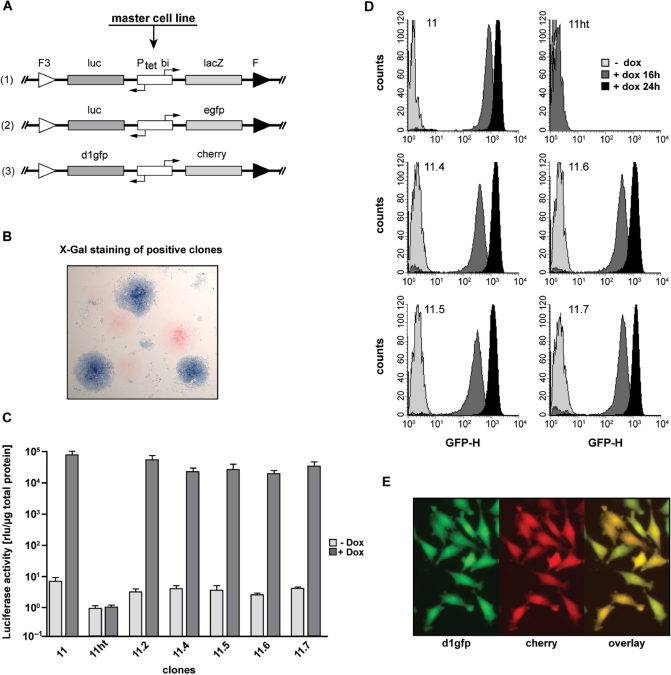
Figure 5.
Stable, uniform and reproducible transgene expression in retargeted HeLa EM2-11 clones. (A) Schematic layout of the targeting vectors used for RMCE. The bidirectional tetracycline inducible expression cassettes luc/lacZ (1), luc/egfp (2) or d1gfp/mcherry (3) were used as substrate for Flp-mediated recombination in HeLa-EM2-11ht to obtain stable single-copy cell lines. (B) Retargeting of the hygtk cassette is very efficient. Recombination efficiency of the retargeting (construct A-1) was determined by in-situ β-galactosidase staining. Stable clones obtained after the RMCE selection procedure were induced with 200 ng/ml dox for 2 days before they were fixed and β-galactosidase activity was visualized by staining with X-Gal. More than 50% of the clones derived by RMCE from master target cell line EM2-11ht show successful targeting. (C) The master cell line HeLa EM2-11ht retargeted by the original luc/egfp reporter (construct A-2) recovers original induction characteristics of the parental EM2-11 line. Luciferase activity (measured in cell lysates and normalized to the total protein content) was induced in the original clone EM2-11 and RMCE derived daughter lines (EM2-11.2, 4, 5, 6 and 7) with 200 ng/ml of dox for 16 h. The bars represent the mean values and the standard deviations of three independent measurements. (D) Histogram plots of EGFP expression in clones measured in C. The expression of EGFP was measured in the absence (light gray) and the presence of 200 ng/ml of dox after 16 (dark gray) and 24 h (black). Each plot represents 10 000 cells. (E) Coexpression of both fluorescent proteins d1gfp and mcherry in individual cells of a given clone (construct A-3) was detected by fluorescence microscopy.
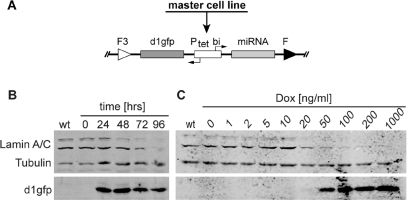
Figure 6.
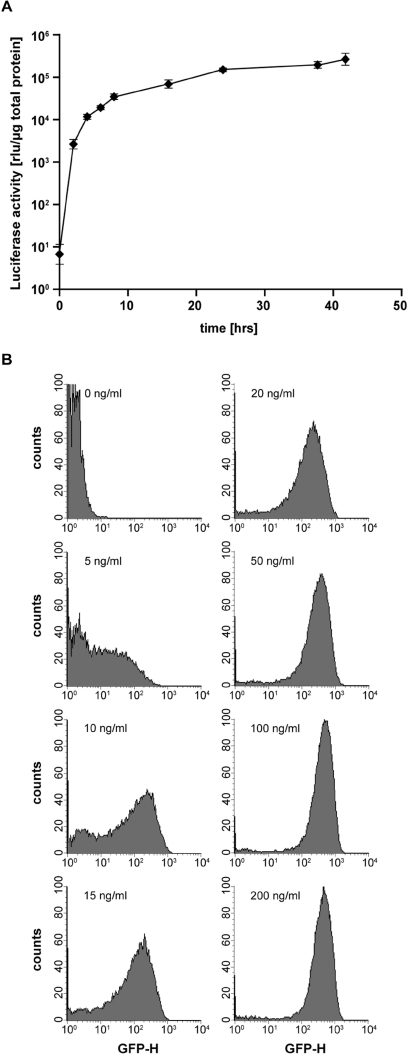
Conditional downregulation of endogenous lamin A/C by tetracycline controlled expression of miRNA. (A) Schematic representation of the Tet-regulated microRNA (miRNA) construct pd1gfpPtetmiR after site-directed genomic integration via RMCE. The bidirectional expression cassette contains a lamin A/C specific miRNA and a destabilized green fluorescent protein (d1gfp). (B) Western blot analysis of the lamin A/C knock-down after miRNA expression over time. Clonal miRNA harboring cells derived from the master cell line EM2-11ht by RMCE were induced with 200 ng/ml dox for the time indicated. Tubulin expression served as a loading control. After 96 h, lamin A/C expression is undetectable (C) Dose response analysis of lamin A/C knock-down in stable pd1gfpPtetmiR containing HeLa EM2-11 cells. Cells were cultured with increasing concentrations of dox for 96 h before collection. The expression of miRNA is mirrored by the appearance of the coexpressed reporter protein d1gfp. Significant reduction of lamin expression is visible with 20 ng/ml of dox.
For RNA interference, we used an optimized microRNA design (miRNA3) described in detail in Berger et al. (manuscript submitted). The targeting sequence for lamin A/C (GenBank acc. no X03444) corresponds to positions 608–628 relative to the start codon (15) and for human Pom121 to positions 1161–1181, respectively. The miRNA sequence was interposed as a synthetic double-stranded oligonucleotide in pBI.F3M.F facing the d1EGFP cDNA in the second expression unit, thereby creating the targeting plasmid pd1gfpPtetmiR.
Generating the HeLa-EM2 cell line
HeLa-EM2 cells are HeLa cells stably expressing an improved version of the reverse tetracycline controlled transactivator gene coding for rtTA2S-M2 (6) under the control of the human EF1α promoter. The expression vector was derived from pEF1 (16). Its MCS was deleted by 5′-HindIII/NotI-3′ digestion and substituted with a 5′-HindIII/NotI-3′- fragment of the pBluescript polylinker (Stratagene, La Jolla, CA); sequence: 5′ AAGCTTGATATCGAATTCCTGCAGCCCGGGGGATCCACTAGTTCTAGAGCGGCCGC 3′. This modification facilitated the insertion of a 5′-EcoRI/BamHI-3′ fragment from pUHrT62-1 (6), containing the rtTA2S-M2 open-reading frame. The resulting expression vector, pEF-M2neo, was introduced in HeLa cells using the Roti-Fect liposome formulation (Carl Roth, Karlsruhe, Germany) according to the manufacturer's protocol. Several growing colonies were selected and tested for the expression of the M2 reverse tetracycline controlled transactivator by transfection with the luciferase reporter plasmid pUHC13-3 as described earlier (1). Clone EM2 showed the best regulation properties, with low basal luciferase expression levels and induction factors in the range of three to four orders of magnitude in transient experiments.
Retroviral transduction
The retroviral infectious particles were produced by 293T cells as described before (9). Several rounds of fluorescence activated cell sorting (FACS) in the presence and absence of 200 ng/ml dox were used for the isolation of individual gfp-luc positive clones such as HeLa-EM2-11.
Flow cytometric analysis
For flow cytometric analysis (FACS), cells were prepared as described before (9) and sorted using a FACSAria flow cytometer. Data acquisition and analysis were performed with the CellQuest program. Workstation and software were both from BD Biosciences, Germany.
Isolation of highly inducible HeLa-EM2 clones
HeLa-EM2 cells were infected with infectious particles at a MOI of 0.03–0.05 to ensure single-copy integration of the S2f-lMCg-F3 retrovirus in the majority of transduced cells. For induction of EGFP expression, the cells were treated with dox (200 ng/ml) for 16 h. About 0.8% of the cells were EGFP-positive as monitored at the single-cell level using FACS. The EGFP positive cells were collected in a six-well dish (ON-sort) and cultured for 8 days in the absence of dox. Subsequent FACS demonstrated that most of the cells remained EGFP-negative in the absence of dox. These cells were collected (OFF-sort) while cells expressing EGFP in the absence of inductor were discarded. After another 8 days of cultivation in dox-containing medium, cell samples were again analyzed by FACS (Supplementary Figure 1). At this point, single EGFP positive cells were isolated (ON-sort) for clonal expansion.
Flp RMCE
To replace the gfp-luc expression unit in HeLa-EM2-11 cells by the hygtk positive/negative selection cassette, Flp-mediated RMCE was used. HeLa-EM2-11 cells were seeded in six-well plates at a density of 2.5 × 105 cells 24 h before transfection. Two micrograms of the targeting plasmid pE11.F3.htk.F (17) together with 2 µg of pCAGGS-FLPe-IRESpuro (18) (Gene Bridges, Heidelberg, Germany) were used for transfection with Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol. Twelve hours post transfection, the cells were detached with trypsin/EDTA, transferred into 100 mm Petri-dishes and cultured in DMEM supplemented with hygromycin B (Invitrogen, 300 µg/ml). Hygromycin-resistant colonies became visible after 8–10 days of selection. The master cell line HeLa-EM2-11ht was established from an EGFP negative clone.
Targeting the master cell line HeLa EM2-11ht
HeLa-EM2-11ht cells were seeded and transfected as described earlier. The targeting plasmids were pd1gfpPtetmiR, pgfpPtetcherry or pd1gfpPtetlacZ, respectively. Twelve hours after transfection, the cells were transferred to 100 mm Petri dishes and incubated with 5 µg/ml puromycin (Sigma-Aldrich) for 36 h. Subsequently, the medium was exchanged for DMEM supplemented with 30–50 µM ganciclovir (Sigma-Aldrich) for negative selection. After 8–10 days, single clones were selected, expanded and characterized.
Luciferase assay
The preparation of cell lysates and the determination of luciferase activity were described earlier (1). Total protein concentration in the lysates was measured at 595 nm using Coomassie Brilliant Blue G-250 (BioRad Laboratories).
Western blotting
Cell lysates were obtained from subconfluent, PBS washed cell cultures after incubation with sample buffer (2 mM MgCl2, 4% SDS, 140 mM Tris base pH 8, 50 mM dithiothreitol, 5 M urea and a spatula tip of bromphenol blue) for 10 min at 24°C. Benzonase nuclease (Sigma-Aldrich) was added fresh at a concentration of 5 U/100 µl of sample buffer. Equal concentrations of total protein (corresponding to 5 × 104 cells) per lane were separated on 10% polyacrylamide Laemli gels and transferred to nitrocellulose membranes. The membranes were incubated with primary antibodies mouse anti-lamin A/C (19), rabbit anti-EGFP and mouse anti-α-tubulin (Sigma-Aldrich) for 3 h. Goat anti-mouse and goat anti-rabbit conjugated to infrared fluorophores IRDye 800 (LI-COR Biosciences) were used as secondary antibodies. Detection was performed on a LI-COR Odyssey imaging system.
Immunofluorescent analysis and microscopy
Cells growing on coverslips were briefly washed with pre-warmed PBS, fixed for 2 min in 3% pre-warmed paraformaldehyde, again washed in PBS and quenched with 50 mM NH4Cl in PBS for 5 min. About 0.1% Triton X-100 in PBS was used to permeabilize cells followed by a block for 30 min with 1% BSA, 10% goat serum and 0.1% Triton X-100 in PBS. The primary antibodies mouse anti-lamin A/C (a gift from K. Weber, MPI for Biophysical Chemistry, Goettingen, Germany), anti-Pom121 (20) and Nup107 (21) were diluted in block solution (1:500, 1:30 000 and 1:1500, respectively) and applied to the cells for 1 h. Unbound antibodies were washed off with PBS before Alexa conjugated secondary antibodies (Invitrogen, diluted 1:500 in block solution) were applied for 30 min. Following incubation, samples were washed repeatedly by PBS before and after post-fixation with 2% paraformaldehyde for 3 min and mounted with Vectashield (Vector Laboratories). Confocal laser scanning microscopy was performed with a Leica TCS SP5 laser-scanning microscope. The laser lines 405, 488, 561 and 633 nm were used for excitation and all pictures were taken with a Leica TCX PL APO 63x NA 1.4 oil objective.
β-Galactosidase staining
Ganciclovir resistant clones were incubated with β-galactosidase substrate 4-bromo-3-chloro-2-indolyl-β-galactosidase (X-gal) for 2 h at 37°C as described (22). A 0.1% Nuclear Fast Red-solution (Certistain, MERCK) in 5% aqueous aluminium sulfate solution was used for counterstaining.
Locus identification and verification
Linear amplification-mediated PCR (LAM-PCR) was performed with genomic DNA of clone EM2-11 by GATC-Biotech, Konstanz, Germany, as described previously (23). The genomic sequence flanking the 3′ end of the integrated DNA was amplified, sequenced and identified using NCBIs Human Genome BLAST as a specific region within LOC642354 on chromosome 5. The result of the LAM PCR was verified by conventional PCR using HeLa-EM2-11 DNA as control template. The 5′ interfaces of the retroviral insertion was amplified with primers FP-chr5.upstream (5′TAAGTCCCAAAGGAGTGTTCTAACCAGAGCTTGTG3′) in combination with a primer localized within the retroviral packaging signal Psi (5′AGGTAACCCAACGTCTCTTCTTGACATCTACCGAC 3′). The 3′ interface was amplified with the primers EGFP (5′AAGACCCCAACGAGAAGCGCGATCACAT3′) and RP-chr5.downstream (5′GTGAATGGGGGCCAGGGAAGGAGAGACGTG3′). As a control, the rtTA-M2 sequence was amplified using the primers FP-rtTA (5′CCA TGT CTA GACTGG ACA AGA3′) and RP-rtTA (5′CTC CAG GCC ACA TAT GAT TAG3′) (Figure 3).
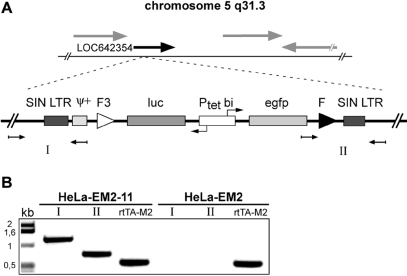
Figure 3.
Identification of the retroviral integration site in clone HeLa-EM2-11. (A) Schematic representation of the retroviral construct integrated as single copy on chromosome 5, region q31.3. The provirus is integrated within an intronic sequence of the hypothetical gene LOC642354 (black arrow). Neighboring genes are indicated by gray arrows. The integration site was first identified using LAM-PCR (see ‘Materials and Methods’ section for details). (B) Verification of the LAM-PCR results. Genomic DNA was extracted from EM2-11 cells as well as non-infected HeLa-EM2 cells and used as templates for PCR. Primers designed to anneal specifically within gene LOC642354 up- and downstream of the integration site were combined with internal primers for the amplification of the products I and II, indicated in (A). An rtTA2S-M2 PCR product served as a positive control for the PCR-reaction.
RESULTS
Generation of the HeLa-EM2 cell line
Synthetic transcription factors of heterologous expression systems such as the transactivators used in the Tet system should ideally be produced uniformly throughout a stable cell population. To this end, we placed the reverse tetracycline controlled transactivator rtTA2-M2 (6) under control of the human EF1α promoter. This promoter has been characterized to be efficient for homogenous transgene expression and was used as a control element for Tet transactivators previously (24,25). A corresponding expression vector was used to generate stable HeLa cell clones. These were initially characterized by introducing suitable Tet-responsive reporter genes, both in transient and stable transfection experiments. One of the cell lines, HeLa-EM2, showing a high dynamic range of Tet-responsiveness without signs of mosaic regulation and its derivatives were used throughout this study (data not shown).
Identification of a silent but activatable (s/a) genomic locus in the HeLa–EM2 cell line
The identification of transgene insertion sites fulfilling the criteria for s/a loci was implemented such that these genomic loci could be retargeted in subsequent experiments. To this end, HeLa-EM2 cells were transduced with self-inactivating retroviruses S2f-lMGg-F/F3 at low MOI to favor single integration of the bidirectional tTA/rtTA dependent promoter Ptet-bi3 that simultaneously controls the expression of luciferase and EGFP (9). This bidirectional transcription unit is flanked by a wild type (F) and a mutated, non-compatible (F3) FRT site (Figure 1A) allowing directed cassette exchange via Flp recombinase (26) as outlined below.
Transduced cells were cultured in presence of dox to induce expression of EGFP, which was exploited for monitoring and separating off the most intensely GFP producing cell population by FACS. The population of cells obtained, representing the leading edge of the ‘activated’ GFP peak, was grown in absence of dox for 1 week to allow for degradation of GFP. Cells giving no or a marginal fluorescence signal were harvested, expanded and taken through a second round of cell sorting in the presence and absence of dox. The selected cell population was again induced with dox and single GFP producing cells were isolated via FACS (see Supplementary Figure S1). Clonal cell populations were expanded and examined individually for luciferase activity in the presence and absence of dox. The results from 12 selected clones are shown in Figure 1B. In the majority of clones, luciferase activity can be induced over more than three orders of magnitude to very high levels. Moreover, while 11 of 12 clones analyzed show luciferase levels <10 rlu/µg protein, for three of these clones (4,11,27) reporter measurements in absence of dox was actually indistinguishable from that of the luc-negative parental cell line. Thus, we conclude that according to our isolation procedure, clones with highly dynamic tet-controlled expression can be routinely isolated. Based on these findings, several clones were picked and characterized in more detail. Clone 11 (Figure 1B), henceforth designated as cell line HeLa-EM2-11, was chosen for all further experiments reported herein. In this line, the expression of luciferase can be regulated following exposure with dox over a range of about four orders of magnitude (Figure 2A). The kinetics of luciferase induction was fast (Figure 2A), in line with previous findings (2). Remarkably, the apparent luciferase background measured in the un-induced state did not exceed 10 rlu/µg of protein, comparable to control extracts from HeLa cells without luciferase transgene (Figures 1B and 2A). To examine the induction characteristics of HeLa-EM2-11 cells at the single cell level, we analyzed the induction of EGFP at different dox concentrations via FACS. As shown in Figure 2B, full induction is reached at dox concentrations of 50–100 ng/ml. Importantly, the induced state prevails throughout the entire population of cells. This demonstrates both the even expression of rtTA2-M2 governed by the EF1α promoter as well as the accessibility of the reporter unit in all cells of the clonal population.
Figure 2.
Induction kinetics of luciferase expression in HeLa-EM2-11 cells. (A) HeLa-EM2-11 cells were incubated with 200 ng/ml of dox and harvested as indicated. Luciferase activity was measured and normalized to the protein concentration. The data represent the mean values and the standard deviations of four independent measurements. (B) Low concentrations of dox effectively induce EGFP gene expression in s/a loci. The histogram plots delineate the flow cytometric analysis of GFP expression after induction with increasing concentrations of dox for 16 h. Each plot represents 15 000 cells.
Our data indicate that, in HeLa-EM2-11 cells, the bidirectional expression unit is indeed integrated in a genomic locus, which has all the characteristics of a s/a locus: tight control in the absence of dox and high, non-variegating inducibility when dox is administered.
HeLa-EM2-11 cells contain a single copy of the bidirectional transcription unit
Given the low MOI of the transduction experiment and the expression characteristics of luciferase and GFP, it seemed unlikely that HeLa-EM2-11 cells contain more than one copy of the bidirectional transcription unit. However, to avoid any ambiguity when targeting the locus via RMCE, it was important to prove that indeed only a single integration event had taken place. We therefore identified the genomic site where the SIN vector had integrated by LAM-PCR (23). A single site within the long arm of chromosome 5 was revealed and sequence analysis delineated the insertion at locus 5q31.3. Sequence analysis showed furthermore that the integration had occurred at a putative intronic sequence of a hypothetical gene (LOC642354). These findings were verified by sequence analysis of the junctions between the insert and the flanking genomic regions (Figure 3). Together, our results show that HeLa-EM2-11 cells indeed contain a single integrate of the bidirectional transcription unit and that no deletion or rearrangement had occurred at the site during the insertion process of the SIN vector. It will be interesting to obtain a more comprehensive picture of integration sites displaying s/a characteristics by advanced sequence analysis methods (27). Such an experimental approach might reveal interesting insights in the nature of genomic loci displaying such expedient features for genetic engineering.
Targeted exchange of the bidirectional transcription unit in HeLa-EM2-11 cells and generation of master cell line HeLa-EM2-11ht
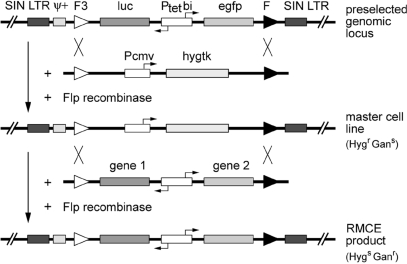
The quantitative data obtained so far met our definition of a s/a locus, where the activity of a gene of interest can be predictably regulated via dox. The inclusion of the heterospecific FRT sites allows for cassette exchange by RMCE. The expression of Flp recombinase does not result in the deletion of DNA sequences between those sites, but rather drives recombination and cassette exchange in trans, when an incoming DNA is equipped with a corresponding pair of recombination sites (28) (see Figure 4 for outline). In a first step, we aimed at the exchange of the bidirectional transcription unit in HeLa-EM2-11 cells for a cassette encoding the hygtk fusion protein controlled by the constitutive CMV promoter (Figure 4). The resulting master cell line would be resistant to hygromycin and sensitive to ganciclovir, providing excellent selection criteria for subsequent replacements of the hygtk cassette by various Tet-controlled expression units (Figure 4).
Figure 4.
Experimental flow in Flp-mediated RMCE: generation of HeLa-EM2-11ht and subsequent restoration of the reporter unit. In order to use the preselected genomic locus in the HeLa-EM2-11 cell line for RMCE, the bidirectional luc/egfp reporter cassette is first replaced by the selection marker gene hygtk, which confers resistance to hygromycin (Hygr) and sensitivity to ganciclovir (Gans) (top). Cassette exchange occurs by double reciprocal crossover between identical pairs of Flp-recombinase target sites after transfection with the circular hygtk plasmid and the Flp-recombinase expression vector. The resulting clone (HeLa-EM2-11ht) is used as master cell line for a second round of RMCE, in which the gene of interest under control of Ptetbi is introduced in the respective locus as surrogate for the hygtk cassette. Cells that have successfully undergone RMCE are selected based on their resistance to ganciclovir in this step (bottom).
HeLa-EM2-11 cells were cotransfected with the circular plasmids pE11.F3.htk.F (17) and pCAGGS-FLPe-IRESpuro (18), and stable hygromycin resistant clones were derived as described in the Methods section. All clones analyzed were sensitive to ganciclovir and had lost their potential to produce luciferase and EGFP upon addition of dox, thus indicating that the bidirectional expression unit encoding luciferase and EGFP was replaced by the hygtk cassette. To verify the anticipated function of this hygtk master cell line, we initially set out to reintroduce the original bidirectional transcription unit encoding luciferase and EGFP in several of our hygtk positive clones. The outcome of this reconstruction experiment was identical for all clones examined: the hygtk cassette could be readily exchanged again for the original bidirectional transcription unit, as will be detailed below. One of the ‘hygtk clones’ was therefore selected as master cell line and designated HeLa-EM2-11ht. All experiments described in the following were carried out with this master cell line.
Insertion of bidirectional Tet-controlled transcription units into master cell line HeLa-EM2-11ht—quantitative aspects
For HeLa-EM2-11ht to serve as a master cell line for all future experiments two quantitative parameters were of particular importance to us: (i) the efficiency of the replacement reaction in this cell line and (ii) the reproducibility of controlled expression, when the s/a locus is repeatedly targeted. To this end, we generated the three bidirectional transcription units shown in Figure 5A, which were inserted in plasmids tailor-made for RMCE.
In a first experiment, we examined the efficiency of insertion by cotransfecting HeLa-EM2-11ht cells with plucPtetlacZ (Figure 5A) and the Flp recombinase encoding plasmid pCAGGS-FLPe-IRESpuro. After negative selection by ganciclovir for 10 days, we exposed the resulting colonies to dox for 2 days. Subsequent staining for β-galactosidase activity revealed that 50–70% of the colonies were homogeneously stained as shown in Figure 5B. This high efficiency of cassette exchange allows readily identification and isolation of desired cell lines.
In a second experiment, we replaced the hygtk cassette in HeLa-EM2-11ht cells by exactly the same bidirectional gfp/luc transcription unit (Figure 5A) present in the parental cell line HeLa-EM2-11 following the selection approach described earlier. From a large number of clones obtained after the selection procedure, five were randomly selected and analyzed for luciferase activity in the presence and absence of dox. Like the parental cell line HeLa-EM2-11, all clones analyzed showed extremely low background activities (<10 rlu/µg protein) in the non-induced state. However, in the presence of dox, more than 104-fold induction of luciferase activity was measured in each clone tested (Figure 5C).
These data show that the cell lines derived from the master cell line HeLa-EM2-11ht exhibit the same regulation potential (>104-fold) as the original parental cell line while maintaining the low background. If at all, only marginal differences exist between the individual clones. Examining the induction of EGFP in various clones via FACS corroborates this finding. The analysis depicted in Figure 5D demonstrates again that upon induction with dox, the entire population of cells is uniformly producing EGFP.
Finally, by integrating a bidirectional unit, where the expression of two fluorescent proteins, namely d1GFP (13) and mCherry (14), are co-regulated via dox, we show that both proteins are simultaneously produced in individual cells (Figure 5E).
Together, our results demonstrate that the genomic locus identified in cell line HeLa-EM2-11 and brought to use in the master cell line HeLa-EM2-11ht can be efficiently targeted by site-specific recombination, giving rise to stable cell lines with highly predictable expression properties.
Controlled expression of micro-RNA
Our system obviously offers itself not only for the overexpression of genes, but also for the controlled interference with the function of endogenous genes via micro-RNA (miRNA). To exemplify this approach, we developed a Ptet-controlled miRNA construct (Berger et al., manuscript submitted), which targets the mRNA of lamin A and C.
Lamin A and C are two abundant proteins, which localize at the nuclear membrane. The A and C variants are encoded by a single gene and originate by alternative splicing of the respective transcript. Our miRNA was directed against a sequence, which is shared by both mRNAs (15). Upon insertion of the miRNA sequence in the exchange vector, lamin-specific miRNA was co-regulated with destabilized d1GFP (Figure 6A). HeLa-EM2-11ht cells were co-transfected with the respective plasmids, and clones were selected as described earlier. Several stable clones were cultured in the presence of dox to induce the miRNA as well as the synthesis of d1GFP. In all 19 examined clones, we observed a strong knock-down of endogenous lamin by immunofluorescence with lamin A/C specific monoclonal antibody (19) (data not shown). One of the clones was analyzed in more detail. It was expanded in the presence of dox, and cells were harvested after 24, 48, 72 and 96 h. Cell extracts were analyzed for lamin A/C and d1GFP by Western blots and as shown in Figure 6B, lamin begins to disappear after 48 h and is hardly visible after 96 h. In parallel, d1GFP is induced demonstrating its coexpression with the lamin-specific miRNA. A dose-response experiment shows that above 50 ng/ml of dox, the production of both, miRNA and d1GFP, are effectively induced.
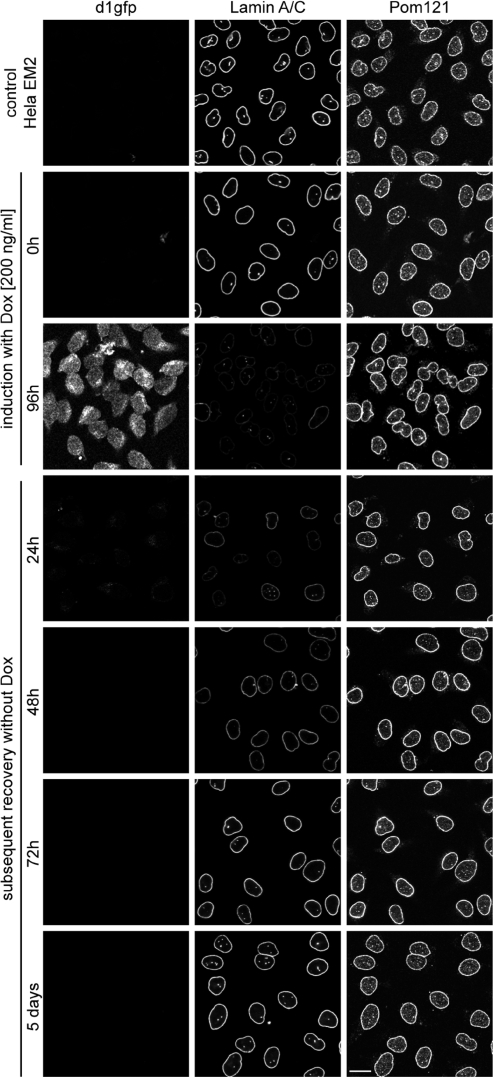
To track the shutdown of lamin A/C synthesis and the induction of d1GFP at the cellular level, we monitored the cells by immunofluorescence with a lamin-specific antibody and by GFP fluorescence, respectively. As shown in Figure 7, lamin becomes almost undetectable within 96 h while d1GFP can be observed. Removal of dox leads to full recovery of lamin staining after a period of 72 h. Note the uniformity in changes of lamin concentration in response to dox-mediated miRNA regulation. Similar results were obtained with several other clones (data not shown), underscoring the high degree of predictability of this experimental approach. In a series of continuative experiments, we were able to reduce the weak, but still detectable lamin expression by the simultaneous expression of multiple miRNAs directed against different positions in the lamin mRNA. In these experiments, the knock-down of lamin A/C could be significantly enhanced by the additive action of two and four miRNAs, respectively, arrayed in a tandem configuration (Supplementary Figure S3). Additionally, we elucidated the possibility of the simultaneous inactivation of two genes by constructing a tandem array with miRNAs directed against lamin A/C and Pom121 (20), both proteins located at the inner membrane of the nuclear envelope. Here, we demonstrate that the combination of two different miRNAs in a polycistronic cluster is as effective as the single-expression constructs (Supplementary Figure S4).
Figure 7.
Fast and efficient reversal of the lamin A/C knock-down. Conditional knock-down of lamin A/C is demonstrated by immunofluorescence staining of stable HeLa EM2-11 cells harboring a single-copy miRNA expression cassette. Cells were seeded on cover slips and induced with 200 ng/ml of dox for 96 h. Subsequently, dox was removed from the growth medium, and cells were imaged at the time points indicated on the left side. Anti-Pom121 staining served as a positive control visualizing nuclear pore complexes within the nuclear envelope. Scale bar 20 μm.
DISCUSSION
The generation of cultured cells capable of expressing transgenes of interest stably and in a well defined and comparable manner is generally marred by a considerable clone-to-clone variability both in absolute expression levels and expression characteristics of the transgene. Copy number and spatial arrangement of the transgenes as well as its epigenetic modifications can be responsible for these unpredictable effects. The latter can result in partial or complete gene silencing, which usually strongly depends on the chromosomal integration site. The development of the recombinase-mediated cassette exchange, RMCE, was a major advance in handling these so-called position effects (28). Targeting pre-defined genomic loci has been shown to significantly reduce and simplify procedures for identifying a desired cell line and, importantly, it facilitates comparative analyses of different transgenes expressed in an otherwise identical genetic background (29–31). Our goal was to utilize this method for further expanding the scope of application for the tetracycline controlled gene expression technology including inducible and reversible gene suppression by RNAi.
To this end, we set out to identify genomic loci that are optimal recipients of Tet-responsive transcription units, in the sense that they qualify as s/a loci (7–9): transcriptionally silent under non-induced conditions but highly active upon induction. Once identified, these loci can be retargeted by suitably engineered exchange vectors. Others have recently succeeded in combining Tet regulation and site-specific recombinases for the same purpose. While achieving a comparable stringency of transgene regulation as shown by us (about 104-fold), their strategies are technically more complex, inherently increasing the risk of failure. Brough and colleagues (32) require two recombination steps to transfer efficient inducibility to the transgene of choice, while Wong et al. (33) had to include an additional trans-acting silencer in their experimental design. We could avoid these complications by using a strategy of identifying s/a loci, which relies exclusively on quantitative expression characteristics of the Tet-controlled reporter genes introduced. Additionally, this screening procedure allowed us to omit the introduction of an antibiotic resistance gene for the generation of stable clones, a procedure which has been previously validated (34). Our results show that this approach of stable cell line generation can be used for the identification of genomic loci, which not only support uniform, persistent and bidirectional expression of two transgenes in the presence of dox, but also do not affect minimal promoters such as Ptet in the ‘off’ state, i.e. in the absence of dox. Thus, the s/a loci identified are well suitable recipients for transgenes of choice by RMCE.
To fully test the capacity of s/a loci to support the desired expression parameters, we exchanged the reporter used for clone isolation by a positive/negative selection marker, solely to facilitate selection and monitoring of subsequent exchange reactions. The potential of the resulting master clones has been scrutinized in two different experimental setups.
First, we re-introduced the original reporter genes to examine the reproducibility of expression features between independently isolated but completely isogenic cell lines. We could show that there is very little clone-to-clone variability, an essential step in making transgene expression in stable cell lines more predictable and ensuring better comparability of cell lines in which different transgenes are to be analyzed in parallel. In addition, these clones showed quantitative parameters of transgene control closely resembling, if not identical, to the parental clone in which the respective s/a locus was identified. These experiments were not only carried out with HeLa EM-11 clone, described here in detail, but also with clones HeLa EM-17 (data not shown) and HeLa EM-22 (Supplementary Figure S2). With all three clones, we obtained fully comparable results, which indicate to us that the loci identified are epigenetically stable over time.
Second, we introduced Ptet-controlled miRNA directed against endogenous cellular targets. Wang et al. (35) recently published another approach allowing the recombinase-mediated integration of inducible miRNA transcription units in a predetermined chromosomal locus. It remains unclear, though, how effectively this locus will support regulated transcription in quantitative terms, as it relied on a pre-existing recipient cell line. In our example, lamin A/C and Pom121 could be reversibly knocked down close to the detection limits, a process which occurred synchronously throughout the entire cell population. By experience, the performance of newly established, regulatable miRNA constructs is largely empirical and problems with inefficient and poorly controllable knock-down phenotypes are frequently encountered. These may either be due to insufficient control over the miRNA transcription unit or the poor efficacy of the miRNA/target sequence interaction. With the latter still difficult to predict, it is even more important to rely on predetermined integration sites, for which Polymerase II transcription control parameters have been established beforehand.
In addition, we could also limit miRNA interference to a defined time window, shown by the complete reversibility of dox-dependent knock-down of target genes, and we succeeded to combine miRNAs in a polycistronic configuration, which enabled the knock-down of two different proteins at the same time. Furthermore, the controlled expression of miRNA in incremental steps as implemented here by properly adjusting the dox concentration may allow for the precise adjustment of protein concentrations and the controlled perturbation of equilibria in complex biological systems.
Taken together, the features of the conditional RNA interference system introduced here will greatly increase the flexibility of the experimental design in knock-down experiments and strengthen the confidence in the interpretation of resulting phenotypes in cell based experimental systems.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
FP6 EU grant LSHG-CT-2003-503259 Integrated Technologies for In-Vivo Molecular Imaging, the University of Heidelberg [Frontier Grant to D.B., S.B. and K.S.]; Federal State Baden-Württemberg [collaborative Grant ‘Network Aging Research’ to D.B. and K.S.]. Funding for open access charge: Deutsche Forschungsgemeinschaft (SFB 636).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Brigitte Pesold and Julia Lenz for technical assistance and G. Schütz and K. Weber for generously providing materials
REFERENCES
- 1.Gossen M, Bujard H. Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc. Natl Acad. Sci. USA. 1992;89:5547–5551. doi: 10.1073/pnas.89.12.5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gossen M, Freundlieb S, Bender G, Muller G, Hillen W, Bujard H. Transcriptional activation by tetracyclines in mammalian cells. Science. 1995;268:1766–1769. doi: 10.1126/science.7792603. [DOI] [PubMed] [Google Scholar]
- 3.Gossen M, Bujard H. Studying gene function in eukaryotes by conditional gene inactivation. Annu. Rev. Genet. 2002;36:153–173. doi: 10.1146/annurev.genet.36.041002.120114. [DOI] [PubMed] [Google Scholar]
- 4.Park YN, Morschhauser J. Tetracycline-inducible gene expression and gene deletion in Candida albicans. Eukaryotic Cell. 2005;4:1328–1342. doi: 10.1128/EC.4.8.1328-1342.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chenuaud P, Larcher T, Rabinowitz JE, Provost N, Joussemet B, Bujard H, Samulski RJ, Favre D, Moullier P. Optimal design of a single recombinant adeno-associated virus derived from serotypes 1 and 2 to achieve more tightly regulated transgene expression from nonhuman primate muscle. Mol. Ther. 2004;9:410–418. doi: 10.1016/j.ymthe.2003.12.015. [DOI] [PubMed] [Google Scholar]
- 6.Urlinger S, Baron U, Thellmann M, Hasan MT, Bujard H, Hillen W. Exploring the sequence space for tetracycline-dependent transcriptional activators: novel mutations yield expanded range and sensitivity. Proc. Natl Acad. Sci. USA. 2000;97:7963–7968. doi: 10.1073/pnas.130192197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baron U, Bujard H. Tet repressor-based system for regulated gene expression in eukaryotic cells: principles and advances. Methods Enzymol. 2000;327:401–421. doi: 10.1016/s0076-6879(00)27292-3. [DOI] [PubMed] [Google Scholar]
- 8.Schonig K, Schwenk F, Rajewsky K, Bujard H. Stringent doxycycline dependent control of CRE recombinase in vivo. Nucleic Acids Res. 2002;30:e134. doi: 10.1093/nar/gnf134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Loew R, Vigna E, Lindemann D, Naldini L, Bujard H. Retroviral vectors containing Tet-controlled bidirectional transcription units for simultaneous regulation of two gene activities. J. Mol. Genet. Med. 2006;2:107–118. doi: 10.4172/1747-0862.1000021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schlake T, Bode J. Use of mutated FLP recognition target (FRT) sites for the exchange of expression cassettes at defined chromosomal loci. Biochemistry. 1994;33:12746–12751. doi: 10.1021/bi00209a003. [DOI] [PubMed] [Google Scholar]
- 11.Seibler J, Bode J. Double-reciprocal crossover mediated by FLP-recombinase: a concept and an assay. Biochemistry. 1997;36:1740–1747. doi: 10.1021/bi962443e. [DOI] [PubMed] [Google Scholar]
- 12.Baron U, Freundlieb S, Gossen M, Bujard H. Co-regulation of two gene activities by tetracycline via a bidirectional promoter. Nucleic Acids Res. 1995;23:3605–3606. doi: 10.1093/nar/23.17.3605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li X, Zhao X, Fang Y, Jiang X, Duong T, Fan C, Huang CC, Kain SR. Generation of destabilized green fluorescent protein as a transcription reporter. J. Biol. Chem. 1998;273:34970–34975. doi: 10.1074/jbc.273.52.34970. [DOI] [PubMed] [Google Scholar]
- 14.Shaner NC, Campbell RE, Steinbach PA, Giepmans BN, Palmer AE, Tsien RY. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat. Biotechnol. 2004;22:1567–1572. doi: 10.1038/nbt1037. [DOI] [PubMed] [Google Scholar]
- 15.Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 16.Strathdee CA, McLeod MR, Underhill TM. Dominant positive and negative selection using luciferase, green fluorescent protein and β-galactosidase reporter gene fusions. BioTechniques. 2000;28:210–212. doi: 10.2144/00282bm04. 214. [DOI] [PubMed] [Google Scholar]
- 17.Kentner D. 2004. Modifikation eines BAC-Vektors durch Rekombination. Diploma thesis. University of Heidelberg. [Google Scholar]
- 18.Schaft J, Ashery-Padan R, van der Hoeven F, Gruss P, Stewart AF. Efficient FLP recombination in mouse ES cells and oocytes. Genesis. 2001;31:6–10. doi: 10.1002/gene.1076. [DOI] [PubMed] [Google Scholar]
- 19.Rober RA, Gieseler RK, Peters JH, Weber K, Osborn M. Induction of nuclear lamins A/C in macrophages in in vitro cultures of rat bone marrow precursor cells and human blood monocytes, and in macrophages elicited in vivo by thioglycollate stimulation. Exp. Cell Res. 1990;190:185–194. doi: 10.1016/0014-4827(90)90184-c. [DOI] [PubMed] [Google Scholar]
- 20.Stavru F, Nautrup-Pedersen G, Cordes VC, Gorlich D. Nuclear pore complex assembly and maintenance in POM121- and gp210-deficient cells. J. Cell Biol. 2006;173:477–483. doi: 10.1083/jcb.200601002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hase ME, Cordes VC. Direct interaction with nup153 mediates binding of Tpr to the periphery of the nuclear pore complex. Mol. Biol. Cell. 2003;14:1923–1940. doi: 10.1091/mbc.E02-09-0620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sanes JR, Rubenstein JL, Nicolas JF. Use of a recombinant retrovirus to study post-implantation cell lineage in mouse embryos. EMBO J. 1986;5:3133–3142. doi: 10.1002/j.1460-2075.1986.tb04620.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schmidt M, Schwarzwaelder K, Bartholomae C, Zaoui K, Ball C, Pilz I, Braun S, Glimm H, von Kalle C. High-resolution insertion-site analysis by linear amplification-mediated PCR (LAM-PCR) Nat. Methods. 2007;4:1051–1057. doi: 10.1038/nmeth1103. [DOI] [PubMed] [Google Scholar]
- 24.Kim DW, Uetsuki T, Kaziro Y, Yamaguchi N, Sugano S. Use of the human elongation factor 1 α promoter as a versatile and efficient expression system. Gene. 1990;91:217–223. doi: 10.1016/0378-1119(90)90091-5. [DOI] [PubMed] [Google Scholar]
- 25.Gopalkrishnan RV, Christiansen KA, Goldstein NI, DePinho RA, Fisher PB. Use of the human EF-1α promoter for expression can significantly increase success in establishing stable cell lines with consistent expression: a study using the tetracycline-inducible system in human cancer cells. Nucleic Acids Res. 1999;27:4775–4782. doi: 10.1093/nar/27.24.4775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Baer A, Bode J. Coping with kinetic and thermodynamic barriers: RMCE, an efficient strategy for the targeted integration of transgenes. Curr. Opin. Biotechnol. 2001;12:473–480. doi: 10.1016/s0958-1669(00)00248-2. [DOI] [PubMed] [Google Scholar]
- 27.Wang GP, Ciuffi A, Leipzig J, Berry CC, Bushman FD. HIV integration site selection: analysis by massively parallel pyrosequencing reveals association with epigenetic modifications. Genome Res. 2007;17:1186–1194. doi: 10.1101/gr.6286907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bode J, Schlake T, Iber M, Schubeler D, Seibler J, Snezhkov E, Nikolaev L. The transgeneticist's toolbox: novel methods for the targeted modification of eukaryotic genomes. Biol. Chem. 2000;381:801–813. doi: 10.1515/BC.2000.103. [DOI] [PubMed] [Google Scholar]
- 29.Fukushige S, Sauer B. Genomic targeting with a positive-selection lox integration vector allows highly reproducible gene expression in mammalian cells. Proc. Natl Acad. Sci. USA. 1992;89:7905–7909. doi: 10.1073/pnas.89.17.7905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schubeler D, Maass K, Bode J. Retargeting of retroviral integration sites for the predictable expression of transgenes and the analysis of cis-acting sequences. Biochemistry. 1998;37:11907–11914. doi: 10.1021/bi9807052. [DOI] [PubMed] [Google Scholar]
- 31.Feng YQ, Seibler J, Alami R, Eisen A, Westerman KA, Leboulch P, Fiering S, Bouhassira EE. Site-specific chromosomal integration in mammalian cells: highly efficient CRE recombinase-mediated cassette exchange. J. Mol. Biol. 1999;292:779–785. doi: 10.1006/jmbi.1999.3113. [DOI] [PubMed] [Google Scholar]
- 32.Brough R, Papanastasiou AM, Porter AC. Stringent and reproducible tetracycline-regulated transgene expression by site-specific insertion at chromosomal loci with pre-characterised induction characteristics. BMC Mol. Biol. 2007;8:30. doi: 10.1186/1471-2199-8-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wong ET, Kolman JL, Li YC, Mesner LD, Hillen W, Berens C, Wahl GM. Reproducible doxycycline-inducible transgene expression at specific loci generated by Cre-recombinase mediated cassette exchange. Nucleic Acids Res. 2005;33:e147. doi: 10.1093/nar/gni145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kaufman WL, Kocman I, Agrawal V, Rahn HP, Besser D, Gossen M. Homogeneity and persistence of transgene expression by omitting antibiotic selection in cell line isolation. Nucleic Acids Res. 2008;36:e111. doi: 10.1093/nar/gkn508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang J, Theunissen TW, Orkin SH. Site-directed, virus-free, and inducible RNAi in embryonic stem cells. Proc. Natl Acad. Sci. USA. 2007;104:20850–20855. doi: 10.1073/pnas.0710565105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.