Abstract
Herein we present a simple, cost-effective TopDown (TD) gene synthesis method that eliminates the interference between the polymerase chain reactions (PCR) assembly and amplification in one-step gene synthesis. The method involves two key steps: (i) design of outer primers and assembly oligonucleotide set with a melting temperature difference of >10°C and (ii) utilization of annealing temperatures to selectively control the efficiencies of oligonucleotide assembly and full-length template amplification. In addition, we have combined the proposed method with real-time PCR to analyze the step-wise efficiency and the kinetics of the gene synthesis process. Gel electrophoresis results are compared with real-time fluorescence signals to investigate the effects of oligonucleotide concentration, outer primer concentration, stringency of annealing temperature, and number of PCR cycles. Analysis of the experimental results has led to insights into the gene synthesis process. We further discuss the conditions for preventing the formation of spurious DNA products. The TD real-time gene synthesis method provides a simple and efficient method for assembling fairly long DNA sequence, and aids in optimizing gene synthesis conditions. To our knowledge, this is the first report that utilizes real-time PCR for gene synthesis.
INTRODUCTION
Synthetic gene with man-made DNA sequences has become a powerful molecular tool with broad applications on engineering proteins (1,2), artifical gene networks (3) and synthetic genomes (4–6). The DNA biomolecules are constructed by assembling pools of oligonucleotides into larger DNA using de novo PCR- (6,7) or ligase chain reaction (LCR)-based (4,8) synthesis methods. The most reported methods for constructing long DNA were based on the PCR process, which relied on the use of overlapped oligonucleotides to construct genes. Various PCR-based methods have been proposed in attempt to optimize the PCR process for long DNA sequences, and to enhance the accuracy of assembly. These methods include the thermodynamically balanced inside-out (TBIO) method (9), successive PCR (10), dual asymmetrical PCR (DA-PCR) (11), overlap extension PCR (OE-PCR) (12,13), PCR-based two-step DNA synthesis (10,14,15) and one-step gene synthesis (16). While various PCR approaches have been implemented, mysteriously the reported optimal synthesis conditions always coincided with an oligonucleotide concentration of 10–60 nM, an outer primer concentration of 200–800 nM, and a PCR cycle number of 20–35 (9,10,15–18). Genes assembled from unpurified, desalted oligonucleotides often resulted in full-length sequence with spurious products of higher molecular weights (9–12,16–19), which degraded the purity of the synthesized products. This abnormal event is usually neglected without explaination by authors. Although PCR-based gene synthesis has been widely implemented, there is a lack of capability in accurately predicting the gene synthesis. The development of an accurate model of gene synthesis would aid in understanding these phenomena and designing optimal reaction conditions.
Another issue associated with gene synthesis is the lack of a standard or universal method (20). Depending on the complexity of target genes, the synthetic genes are often constructed with a one-step or two-step overlapping process. For its simplicity, the one-step process is preferred for short DNAs (<500 bp), wherein the amplification primers are mixed with assembly oligonucleotides in a single PCR reaction. The melting temperatures (Tm) of these oligonucleotides are commonly designed with homologous Tm to balance the PCR assembly and amplification. Hence, the excess outer primers would preferentially anneal with extended oligonucleotides, resulting in a potentially large portion of amplification primers participating in the initial gene assembly cycles. This could deplete the deoxynucleotide triphosphates (dNTPs) and cease the PCR reaction (17,21). This competitive effect is more critical for DNA with high GC content or length (9,10), and is eliminated in the two-step PCR process whereby the amplification and assembly are performed separately with the extra cost of fresh PCR mixture and additional human effort. In the two-step process, a pool of short oligonucleotides is first assembled into long double-stranded DNA (dsDNA) (termed ‘template’) with the desired length and sequence information using the polymerase chain assembly (PCA). The quantity of the assembled template DNA is then amplified by the PCR step.
Herein we described a novel approach that combined the simplicity and cost effectiveness of the one-step process, with the assembly efficiency of the two-step process for the synthesis of relatively long genes. This method utilized software to design primers and oligonucleotides with distinct melting temperatures to minimize the competition between the PCA assembly and PCR amplification in the one-step gene synthesis. Figure 1 shows the concept of the proposed TopDown (TD) one-step gene assembly method. The outer primers (Tm ∼50°C) and inner oligonucleotides (Tm ∼65°C) were designed with a melting temperature difference (ΔTm) of ∼15°C. The overlapping gene synthesis was conducted in one PCR mixture with two annealing temperature segments for oligonucleotide assembly and full-length template amplification, respectively. A high annealing temperature (Tah) (matched to the Tm of oligonucleotides) was used for the first 20 cycles aimed at assembly. This was followed by a low annealing temperature (matched to the Tm of primers) to boost the full-length amplification. The outer primers were subjected to an elevated annealing condition (Tah – Tm = 15°C) during the early assembly process, which prevented mis-pairing among primers and oligonucleotides.
Figure 1.
Schematic illustration of TD one-step gene synthesis combining PCR assembly and amplification into a single stage with different annealing temperatures designed for assembly and amplification. Inner oligonucleotides and outer primers are designed with a melting temperature difference of >15°C to minimize potential interference during PCR.
We further combined the TD one-step gene synthesis with real-time PCR to investigate the gene synthesis process. LCGreen I and SYBR Green I intercalation dyes were examined for real-time gene synthesis. Gel electrophoresis results were compared with the real-time fluorescence signals to study the effects of the oligonucleotide concentration, outer primer concentration, stringency of annealing temperature, and number of PCR cycles. Performing real-time gene synthesis provided a novel approach to explore these factors in a systematic study, which revealed unique information not available from gel electrophoresis results (16). An analytical model was further developed, which uncovered the mystery of coincident optimal conditions, and the formation of undesired, spurious DNA products. This real-time method provided new insights into gene synthesis and a universal method for gene synthesis.
MATERIALS AND METHODS
Design of oligonucleotides for gene synthesis
Gene sequence for the promoter of human calcium-binding protein A4 (S100A4, 752 bp; chr1:1503312036-1503311284) (22) was selected for synthesis via assembly PCR. Oligonucleotides were derived by a custom-developed program, which first divided the given sequence into oligonucleotides of approximately equal lengths by markers, and computed the average and deviation in melting temperatures among the overlapping regions using the nearest-neighbor model with SantaLucia's thermodynamic parameter (23), corrected with salt and oligonucleotide concentrations. Next, the oligonucleotide lengths were adjusted through shifting the marker positions to minimize the deviations in the overall overlapping melting temperature. The summary of oligonucleotide set is shown in Table 1 with the detail information provided in Supplementary Table S1.
Table 1.
Data of oligonucleotide set
| Gene | Length (bp) | Average Tm (min, max) (°C) | Number of oligonucleotides | Overlap length (nt) | Oligonucleotide length (nt) |
|---|---|---|---|---|---|
| S100A4 | 752 | 66.0 (61.1, 69.8) | 30 | 19–33 | 19, 41–66 |
TopDown one-step real-time gene synthesis
TopDown one-step process was optimized using real-time PCR conducted with Roche's LightCycler 1.5 real-time thermal cycling machine with a temperature transition of 20°C/s. Real-time gene synthesis was conducted with 20 µl of reaction mixture including 1× PCR buffer (Novagen), 1 µl of 0.25× to 5× SYBR Green I (1× = 1/20 000 dilution; Invitrogen) or LCGreen I (Idaho Technology Inc.), 4 mM of MgSO4, 1 mM each of dNTP (Stratagene), 500 µg/ml of bovine serum albumin (BSA), 5–80 nM of oligonucleotides, 60 nM to 1 µM of forward and reverse primers and 1 U of KOD Hot Start (Novagen). The PCR were conducted under the following conditions: 2 min of initial denaturation at 95°C; 20 cycles of 95°C for 5 s, 58–70°C for 10 s, 72°C for 30 s; followed by 20 cycles of 95°C for 5 s, 49°C for 10 s, 72°C for 30 s; and final extension at 72°C for 10 min. Desalted oligonucleotides were purchased from Research Biolabs (Singapore) and Proligo (Singapore) without additional purification.
One-step and two-step PCR-based gene synthesis
Conventional gene synthesis via PCR was performed either as a one-step process, combining PCR assembly and amplification into a single stage, or as a two-step process with separate stages for assembly and amplification. All PCR reactions, whether for assembly or amplification, were run in standard 0.2 ml PCR tubes with a commercial thermal cycler (DNA Engine PTC-200, Bio-Rad) using the same oligonucleotides set and out primers as in the non-competitive one-step PCR. The one-step process was performed with 50 µl of reaction mixture including 1× PCR buffer (Novagen), 4 mM of MgSO4, 1 mM each of dNTP (Stratagene), 500 µg/ml of BSA, 10 nM of oligonucleotides, 400 nM of forward and reverse primers and 1 U of KOD Hot Start (Novagen). The one-step PCR was conducted under the following conditions: 2 min initial denaturation at 95°C; 30 cycles of 95°C for 5 s, 58°C for 10 s, 72°C for 30 s; and final extension at 72°C for 10 min. The PCR protocol of the two-step process was essentially the same as that for one-step process except for the concentration of oligonucleotides and annealing temperature. For PCR assembly, 10 nM of oligonucleotides were used without the forward and reverse primers. For gene amplification, 2 µl of the assembled product was diluted in 25 µl of amplification reaction mixture with primers concentration of 400 nM each, and an annealing temperature of 49°C was employed. The PCR conditions of these three gene syntheses are summarized in Table 2.
Table 2.
PCR conditions for one-step, two-step and TD one-step gene syntheses
| Method | PCAa |
PCR |
||
|---|---|---|---|---|
| Annealing temperature | Number of cycles | Annealing temperature | Number of cycles | |
| One-step | 58°Cb | 30 | – | – |
| Two-step | 58°C | 30 | 49°Cc | 30 |
| TD one-step | 58–70°C | 20 | 49°Cc | 20 |
aAverage melting temperature of oligonucleotide set: 66.0°C.
bMelting temperature of outer primers: 59.4°C and 63.4°C.
cMelting temperature of outer primers: 49.4°C and 50.9°C.
Agarose gel electrophoresis
The synthesized products were analyzed by 1.5% agarose gel (NuSieve® GTG®, Cambrex Corporation), stained with ethidium bromide (Bio-Rad Laboratories) or SYBR Green (Invitrogen), and visualized using Typhoon 9410 variable imager (Amersham Biosciences). Gel electrophoreses were performed at 100 V for 45 min with 100 bp ladder (New England) and 5 μl of DNA samples.
RESULTS
Performance of TD one-step gene synthesis
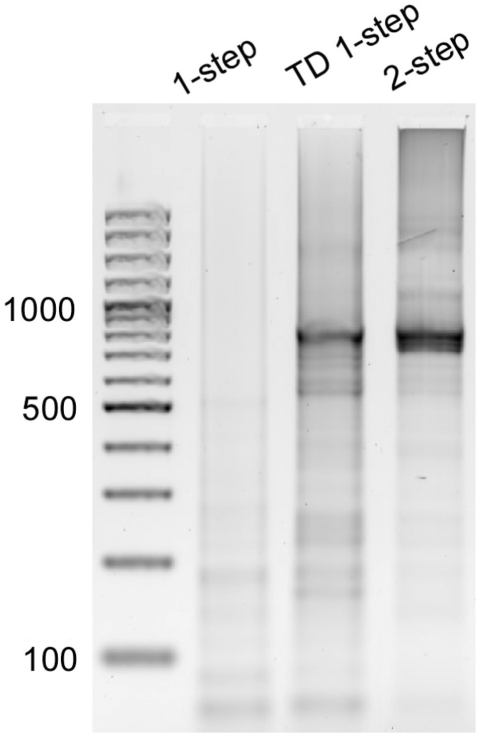
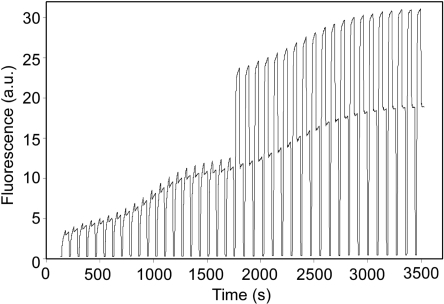
Successful gene synthesis was achieved using TD one-step and two-step processes, while no obvious full-length gene product was obtained in one-step PCR process as shown by gel electrophoresis (Figure 2). The TD one-step process was conducted with an annealing temperature (Tah) of 67°C (average Tm of oligonucleotides = 66°C) for the first 20 cycles, followed by an annealing temperature of 49°C (average Tm of primers = 50.1°C) for another 20 cycles. The continuous fluorescence monitoring revealed the efficiency of the gene synthesis process (Figure 3). Unlike the exponential nature of PCR amplification, the assembly efficiency was more likely linear in nature.
Figure 2.
Agarose gel electrophoresis results of one-step (30 cycles), TD one-step (40 cycles) and two-step (PCA: 30 cycles; PCR: 30 cycles) gene syntheses. The TD one-step process is conducted with 20 cycles with an annealing temperature of 67°C, followed by 20 cycles with an annealing temperature of 49°C. The concentrations of oligonucleotides and outer primers are 10 nM and 400 nM, respectively.
Figure 3.
Continuous fluorescence monitoring of real-time gene synthesis with 1× LCGreen I. The first 20 cycles are conducted with an annealing temperature of 67°C, and the next 20 cycles are conducted with an annealing temperature of 49°C. The concentrations of oligonucleotides and outer primers are 10 nM and 400 nM, respectively.
Two intercalating fluorescent dyes (SYBR Green I and LCGreen I) were investigated for real-time gene synthesis (Supplementary Figure S1). The LCGreen I (24) was more suitable for studying the real-time gene synthesis, which has a fluorescence spectrum similar to the commonly adopted SYBR Green I in real-time PCR. The SYBR Green I would bind preferentially to long DNA fragments (25,26), and could redistribute from short DNA fragments to long DNA fragments during thermal cycling (27). This would make it difficult to analyze the fluorescence signal since the PCA mixture would contain various lengths of dsDNA.
The initial DNA quantity (∼6 pmol; 10 nM × 20 µl × 30 oligonucleotides) in PCA mixture was much larger (by >6 orders of magnitude) than that in standard PCR amplification (<106 copies of template DNA) (28). The real-time PCR conditions were adjusted for this factor. The optimal concentration of LCGreen I was studied and increased from 1× for standard PCR to 2× (Supplementary Figure S1). The dNTPs concentration was adjusted from 0.2 mM each for standard PCR to 1 mM each to prevent the depletion of dNTPs. The Mg2+ ion (MgSO4) concentration has been empirically optimized (at 4 mM) based on the concentration of dNTP, which could chelate with Mg2+ and affect the polymerase activity (29,30) (Supplementary Figure S2). The manufacturer's recommended Mg2+ ion concentration was 1.5 mM for standard PCR with 0.2 mM of dNTPs each.
Analysis of real-time gene synthesis
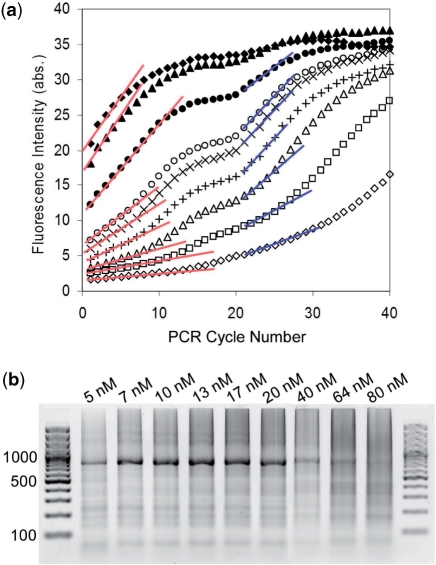
Mechanistically, gene synthesis took place in several phases, as revealed by the variation in slopes with the number of PCR cycles (Figure 4). This phenomenon was remarkable with an oligonucleotide concentration of 10–20 nM. In the early cycles of PCA, most annealing between paired oligonucleotides formed an extendable duplex, which could undergo extension by polymerase (phase 1; cycles less than 7). The fluorescence signal revealed a linear increment of DNA length extension with each cycle. In contrast to that reported by Wu et al. (16) and Lee et al. (21), the assembly efficiency increased with further PCR cycles (phase 2; cycles ∼7–14). Our hypothesis was that the assembly process switched in favor of full-length template amplification as the full-length fragments emerged, and was promoted by the excess outer primers. The PCA reaction then reached the first plateau (phase 3; cycles 15–20) whereby the outer primers priming was limited by the elevated annealing conditions (Tah – Tm = 15°C). At cycle 21, the annealing temperature was reduced to 49°C to match with the Tm of primers (phase 4; cycles ∼21–29). The exponential amplification was boosted, and caused a sudden jump in fluorescence signals. Finally, the process reached the second plateau, presumably due to the depletion of outer primers or non-specific products annealing (phase 5). The plateau stages were delayed or completely missing for low oligonucleotide concentration (<7 nM) due to its low assembly efficiency.
Figure 4.
The oligonucleotide concentration is critical in the successful gene synthesis. S100A4 (752 bp) is synthesized with various oligonucleotide concentrations ranging from 5 nM to 80 nM, and annealing temperatures of 67°C for the first 20 cycles and 49°C for the next 20 cycles. (a) Fluorescence as a function of PCR cycle number for oligonucleotide concentrations of 5 nM (unfilled diamond), 7 nM (unfilled square), 10 nM (unfilled triangle), 13 nM (+), 17 nM (×), 20 nM (unfilled circle), 40 nM (filled circle), 64 nM (filled triangle) and 80 nM (filled diamond). The slopes of fluorescence increment in the early cycles and cycles #21 indicate the efficiencies of the assembly and amplification processes. (b) The corresponding agarose gel electrophoresis results.
For gene synthesis with >64 nM of oligonucleotides, the PCR process reached the plateau within 15 cycles. Additional cycles would most likely favor non-specific PCR, and lead to the generation of spurious bands and the buildup of high molecular weight products in gel electrophoresis (Figure 4b), as observed in most reported gene synthesis results (9–12,16–19). The consistent gel results and real-time PCR curves suggested that the optimal oligonucleotide concentration was 10–20 nM for TD gene synthesis, which coincided with that of both the one-step (16,17) and two-step (18) processes.
We further investigated the effect of primer by varying primer concentration from 60 nM to 1 µM while keeping the oligonucleotide concentration at 10 nM. The highest full-length quantity was obtained with 400 nM of primers (Figure 5), which was consistent with observations in one-step (16) and two-step (18) processes. Assembly efficiencies, depicted by the slopes of fluorescence increment, were indifferent in the early cycles (less than cycle 7), even though the primer concentration was varied by 16-fold (inset in Figure 5a). This demonstrated the non-interference feature of the TD process, wherein the outer primers did not intervene with the assembly process. The assembly efficiencies started to deviate at around cycle 8 as the full-length products emerged, in favor of full-length template amplification. Unlike the oligonucleotide concentration, which dominated the assembly reaction and critically influenced the success of gene synthesis, the primer concentration was less critical. It presumably controlled the late amplification process and the quantity of desired DNA. The optimal PCR cycles depended on the initial oligonucleotide concentration and target gene length. This was clearly demonstrated by the experiment on oligonucleotide concentration (Figure 4). As oligonucleotide concentration increased from 20 nM to 80 nM, the full-length band gradually disappeared and became widened.
Figure 5.
S100A4 (752 bp) is successfully synthesized with various primer concentrations ranging from 60 nM to 1 µM, as indicated by the sharp, narrow gel band of the desired length. (a) Fluorescence as a function of PCR cycle number for outer primer concentrations of 60 nM (unfilled diamond), 120 nM (unfilled square), 200 nM (unfilled triangle), 300 nM (×), 400 nM (+) and 1 µM (unfilled circle). The inset shows the fluorescence signal of the first 20 cycles. (b) The corresponding agarose gel electrophoresis results.
The overlapping assembly was a parallel process. Relatively few PCR cycles were needed to complete the assembly. The theoretical minimum number of cycles (x) in order to construct a dsDNA molecule of length (L) from uniform oligonucleotide length (n) and overlapping size (s) is given by:
Theoretically, six PCA cycles were sufficient for assembling S100A4 (752 bp) from a pool of 40 nt oligonucleotides with an overlap of 20 nt. To determine whether excess cycling was necessary for gene assembly, we used the optimal condition determined in previous experiments with various PCA cycles of 6–20, followed by 20 amplification cycles. Gene synthesis was fairly efficient. Indeed, full-length assembly was achieved within 11 PCA cycles (Figure 6).
Figure 6.
S100A4 is synthesized with various assembly cycles (6–20 cycles), followed by another 20 cycles for amplification. Agarose gel electrophoresis results indicate that full-length assembly is achieved within 11 cycles.
The gene synthesis was insensitive to the variation in assembly annealing temperature (Tah) from 58°C to 70°C, as visualized in both gel results and fluorescence signals (Figure 7). The fluorescence intensity curves were indiscriminant to the annealing temperatures during the assembly phase (first 13 cycles), and began to deviate only after the first phase (see inset in Figure 7a). The indifference in fluorescence intensity during the first 13 cycles implied that the primers did not intervene with the assembly reaction. The primers were designed with an average Tm of 50.9°C, which meant that the primers encountered an annealing stringent of 7.1–19.9°C (Tah – Tm) during the PCA process. This suggested that we could potentially reduce the melting temperature window (ΔTm of primers and oligonucleotides) to 7.1°C, and ensure the non-competitive feature of TD gene synthesis method. Interestingly, a higher yield of the desired DNA was obtained with a stringent annealing temperature (>67°C) higher than the average Tm of oligonucleotides (66°C).
Figure 7.
S100A4 (752 bp) synthesized with various assembly annealing temperatures ranging from 58°C to 70°C for the first 20 cycles, followed by an annealing temperature of 49°C for the next 20 cycles. (a) Fluorescence as a function of PCR cycle number for annealing temperatures of 58°C (unfilled diamond), 60°C (unfilled square), 62°C (unfilled triangle), 65°C (×), 67°C (+) and 70°C (unfilled circle). The inset shows the middle 15 cycles (no. 13–27). (b) The corresponding agarose gel electrophoresis results. Higher synthesis yield is obtained with a stringent assembly annealing temperature (>67°C).
DISCUSSION
We have presented the TD gene synthesis method, which offered a simple, rapid and low-cost method for synthesizing fairly long DNA (752 bp) with only one PCR step. The primer interference problem present in one-step process has been eliminated by designing primers and assembly oligonucleotide set with melting temperature variation (ΔTm) of >15°C, and performing Tm-matched PCR to selectively control the efficiencies of oligonucleotide assembly and full-length template amplification. Our data suggested that the TD process might also work well with a relaxed ΔTm of 7.1°C. The TD synthesis conditions have been empirically optimized using real-time PCR with LCGreen I dye. It was noted that the popular SYBR Green I dye was not suitable for real-time gene synthesis. Other intercalating dyes such as LCGreen (25) and SYTO9 (31) might also work, but required further optimization in dye concentration.
Based on the data presented herein, we have developed an analytical model that described the TD one-step gene synthesis process clearly. The TD synthesis occurred in five phases in term of PCR cycles: (i) linear assembly, (ii) emerging amplification, (iii) first plateau, (iv) exponential amplification and (v) non-specific amplification. Our experiments have demonstrated that TD one-step gene synthesis was fairly efficient, as compared to the conventional one-step and two-step processes. We found the assembly process automatically switched to preferential full-length amplification as the full-length template emerged. This greatly improved the assembly efficiency of the PCA process as compared to the conventional one-step and two-step processes.
We found that the quality and quantity of PCR-based gene synthesis were influenced by several factors, including annealing temperature, concentration of oligonucleotides, concentration of monomers and number of PCR cycles. The fluorescence curve (10 nM curve in Figure 4) suggested the assembly and amplification processes reached the plateau at around cycle 15 and cycle 35, respectively. This implied that the optimal PCR cycles were 30 cycles: 15 cycles each for assembly and amplification reactions. Furthermore, after the PCR reached the plateau, the amplification efficiency of PCR decreased. Additional PCR cycling would favor the non-specific annealing of the full-length product to assembled random-fragments or the full-length product itself. Both extendable and unextendable pairings could occur. Products annealed in the 3′ recessed configuration could be extended to higher molecular weight DNA and be randomly terminated during the later cycles. In contrast, products annealed with 3′ ends protruded would reduce the amount of full-length DNA, and generate DNA with lower molecular weight. Both of these types of non-specific annealing would result in diminished full-length gel band (32), as observed in gel electrophoresis. These abnormal products with incorrect DNA sequences would potentially complicate the enzymatic cleavage or consensus shuffling error correction process (33,34) when consecutive PCRs were adopted for gene re-assembly or DNA re-amplification.
Although the gel results (Figure 1) indicated that the two-step process was superior to the TD one-step process, we could argue that the optimized TD approach could eventually outperform the two-step process in providing better purity of synthetic products, with the understanding of gene synthesis mechanism. The two-step method suffered the same rapid decrease on extension efficiency as the one-step process during the assembly process (PCA), which created a lot of intermediate DNAs with lower molecular weight besides the full-length products. The non-specific annealing and extension would most likely also occur during PCA, considering the observed fairly efficient full-length assembly (<10 cycles) (Figure 5). These higher molecular weight products could also contain primers’ binding sites, and be exponentially amplified at the next amplification step (PCR) as the full-length products (32,35). These incorrect products would complicate the error filtering process (33,36), requiring excess MutS enzyme. We could improve the TD process by having the PCR amplification tailored after the emergence of full-length DNA to avoid excess PCR.
With the help of our model, insights into the coincidental optimal conditions reported for various PCR-based gene synthesis processes were attained (Table 3). The optimal conditions were universal with an oligonucleotide concentration of 10–60 nM and a primer concentration of 200–800 nM, as reported by a distinct gel band of full-length products. We have demonstrated that the success of gene synthesis relied on the assembly efficiency of PCA, which was in turn dominated by the oligonucleotide concentration and number of PCR cycles (Figures 4 and 6). The fluorescence signals indicated that an oligonucleotide concentration of 10–60 nM provided optimal assembly efficiency with full-length products. It was noteworthy that the optimal number of PCR cycles should be adjusted according to the oligonucleotide concentration and gene length to avoid excessive PCR. Successful gene synthesis (752 bp) was also achieved with an oligonucleotide concentration of <5 nM in 40 PCR cycles. However, it might not be reliable for genes with a high GC content or a high length due to the comparatively low assembly efficiency. The primer concentration was less critical; it controlled the late amplification process and the quantity of desired DNA (Figure 5). The optimal primer concentration balanced the quantity and purity of the desired DNA, avoiding the formation of spurious DNA products.
Table 3.
Some reported optimal gene synthesis conditions
| Name of synthesized DNA | Length (bp) | Oligos (nM) | Primer (nM) | Number of PCR cycles | Method |
|---|---|---|---|---|---|
| PDK1 gene | 1712 | 20 | 200 | 25–35/25–35 | Two-step PCR (9) |
| PDK1 gene | 1712 | 40–200 | 200 | 25/25 | TBIO (9) |
| Various DNAs | 470–1200 | 40 | 200 | 20/15 | Two-step PCR (12) |
| vip3aI gene | 2370 | 30 | 600 | 25/25 | Two-step PCR (10) |
| Various DNAs | 209–936 | 10–25 | 400–800 | 25 | One-step PCR (16) |
| Pur operon | 12 000 | 60 | 600 | 25/25 | PCR-based (15) |
| Various DNAs | 327–993 | 10–25 | 500 | 35–45 | One-step PCR (17) |
| GFP segment | 752 | 5–15 | 400 | 30/30 | Two-step PCR (18) |
As the synthesis yield was insensitive to the assembly annealing temperature (Figure 7), we suggested performing the assembly with an annealing temperature slightly higher than the average Tm of oligonucleotides. This provided several advantages in (i) eliminating potential competition between the assembly and amplification reactions, (ii) minimizing the possibility of truncated oligonucleotides (n – 1) participating in the assembly process and the resulting errors, (iii) providing an stringent annealing condition to reduce the potential of forming secondary structures and (iv) increasing the specialization of oligonucleotides hybridization as in Touchdown PCR (37). All of these would prevent the generation of faulty sequence, especially for gene with high GC contents.
Besides providing a tool for optimizing the gene synthesis conditions, the real-time method would potentially provide other functions for characterizing the synthesis products via analysis of DNA-melting curve. Presumably, successful gene synthesis would yield a product with a single, sharp melting peak, while incomplete synthesis would result in a broad melting curve. The integrated area of the melting peak in the negative derivative of the fluorescence with respect to temperature (-dF/dT versus T) would give the quantity of the desired full-length product (38). In combination with DNA-melting simulation software (31,39), the purity and quantity of assembled products could be estimated with the use of melting curve analysis, which would eliminate the necessity for agarose gel electrophoresis. This approach could be integrated with microfluidic gene synthesis (17,18,40) to develop automated gene synthesis.
Another important factor for successful gene synthesis was the polymerase enzyme. The performance of various polymerases has been studied and compared for the gene synthesis process (2,16,41–43). The KOD series of polymerases were suggested for gene synthesis (2,16,41). We have observed that the KOD Hot Start outperformed the Taq and Pfu polymerases (data not shown). No obvious full-length gene product was obtained with Taq or Pfu for the TD method.
Herein we have presented a simple, efficient and cost-effective TD one-step gene synthesis method that combines the advantages of one-step and two-step gene synthesis processes. We recommend conducting TD synthesis with the following conditions: (i) design primers (Tm = 50–55°C) and inner oligonucleotide (Tm ∼65°C) with distinct melting temperature (ΔTm > 8°C); (ii) 2× LCGreen I, 4 mM of MgSO4, 1 mM each of dNTP, 500 µg/ml of BSA, 10 nM of oligonucleotides, 400 nM of forward and reverse primers and 1 U of KOD Hot Start, (iii) 2 min of initial denaturation at 95°C; 15 cycles of 95°C for 5 s, 67–70°C (according to the Tm of inner oligonucleotides) for 30 s and 72°C for 30 s; followed by 15 cycles of 95°C for 5 s, 50–55°C (according to the Tm of outer primers) for 30 s and 72°C for 30 s.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Institute of Bioengineering and Nanotechnology (Biomedical Research Council, Agency for Science, Technology and Research, Singapore). Funding for open access charge: Institute of Bioengineering and Nanotechnology.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors acknowledge Drs Shu Wang and Jérôme Boulaire for their helpful suggestions on S100A4.
REFERENCES
- 1.Ramachandran N, Hainsworth E, Bhullar B, Eisenstein S, Rosen B, Lau AY, Walter JC, Labaer J. Self-assembling protein microarrays. Science. 2004;305:86–90. doi: 10.1126/science.1097639. [DOI] [PubMed] [Google Scholar]
- 2.Cox JC, Lape J, Sayed MA, Hellinga HW. Protein fabrication automation. Protein Sci. 2007;16:379–390. doi: 10.1110/ps.062591607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sprinzak D, Elowitz MB. Reconstruction of genetic circuits. Nature. 2005;438:443–448. doi: 10.1038/nature04335. [DOI] [PubMed] [Google Scholar]
- 4.Smith HO, Hutchison CA, III, Pfannkoch C, Venter JC. Generating a synthetic genome by whole genome assembly: ΦX174 bacteriophage from synthetic oligonucleotides. Proc. Natl Acad. Sci. USA. 2003;100:15440–15445. doi: 10.1073/pnas.2237126100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gibson DG, Benders GA, Andrews-Pfannkoch C, Denisova EA, Baden-Tillson H, Zaveri J, Stockwell TB, Brownley A, Thomas DW, Algire MA, et al. Complete chemical synthesis, assembly, and cloning of a Mycoplasma genitalium genome. Science. 2008;319:1215–1220. doi: 10.1126/science.1151721. [DOI] [PubMed] [Google Scholar]
- 6.Cello J, Paul AV, Wimmer E. Chemical synthesis of poliovirus cDNA: Generation of infectious virus in the absence of natural template. Science. 2002;297:1016–1018. doi: 10.1126/science.1072266. [DOI] [PubMed] [Google Scholar]
- 7.Kodumal SJ, Patel KG, Reid R, Menzella HG, Welch M, Santi DV. Total synthesis of long DNA sequences: Synthesis of a contiguous 32-kb polyketide synthase gene cluster. Proc. Natl Acad. Sci. USA. 2004;101:15573–15578. doi: 10.1073/pnas.0406911101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Au LC, Yang FY, Yang WJ, Lo SH, Kao CF. Gene synthesis by a LCR-based approach: High-level production of leptin-L54 using synthetic gene in Escherichia coli. Biochem. Biophys. Res. Commun. 1998;248:200–203. doi: 10.1006/bbrc.1998.8929. [DOI] [PubMed] [Google Scholar]
- 9.Gao X, Yo P, Keith A, Ragan TJ, Harris TK. Thermodynamically balanced inside-out (TBIO) PCR-based gene synthesis: A novel method of primer design for high-fidelity assembly of longer gene sequences. Nucleic Acids Res. 2003;31:e143. doi: 10.1093/nar/gng143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xiong A-S, Yao Q-H, Peng R-H, Li X, Fan H-Q, Cheng Z-M, Li Y. A simple, rapid, high-fidelity and cost-effective PCR-based two-step DNA synthesis method for long gene sequences. Nucleic Acids Res. 2004;32:e98. doi: 10.1093/nar/gnh094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sandhu GS, Aleff RA, Kline BC. Dual asymmetric PCR: One-step construction of synthetic genes. Biotechniques. 1992;12:14–16. [PubMed] [Google Scholar]
- 12.Toung L, Dong Q. Two-step total gene synthesis method. Nucleic Acids Res. 2004;32:e59. doi: 10.1093/nar/gnh058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Prodromou C, Pearl L. Recursive PCR: A novel technique for total gene synthesis. Protein Eng. 1992;5:827–829. doi: 10.1093/protein/5.8.827. [DOI] [PubMed] [Google Scholar]
- 14.Stemmer WP, Crameri A, Ha KD, Brennan TM, Heyneker HL. Single-step assembly of a gene and entire plasmid from large numbers of oligodeoxyribonucleotides. Gene. 1995;164:49–53. doi: 10.1016/0378-1119(95)00511-4. [DOI] [PubMed] [Google Scholar]
- 15.Xiong A-S, Yao Q-H, Peng R-H, Duan H, Li X, Fan H-Q, Cheng Z-M, Li Y. PCR-based accurate synthesis of long DNA sequences. Nat. Protoc. 2006;1:791–797. doi: 10.1038/nprot.2006.103. [DOI] [PubMed] [Google Scholar]
- 16.Wu G, Wolf JB, Ibrahim AF, Vadasz S, Gunasinghe M, Freeland SJ. Simplified gene synthesis: A one-step approach to PCR-based gene construction. J. Biotech. 2006;124:496–503. doi: 10.1016/j.jbiotec.2006.01.015. [DOI] [PubMed] [Google Scholar]
- 17.Kong DS, Carr PA, Chen L, Zhang S, Jacobson JM. Parallel gene synthesis in a microfluidic device. Nucleic Acids Res. 2007;35:e61. doi: 10.1093/nar/gkm121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang MC, Ye H, Kuan YK, Li M-H, Ying JY. Integrated two-step gene synthesis in a microfluidic device. Lab. Chip. 2009;9:276–285. doi: 10.1039/b807688j. [DOI] [PubMed] [Google Scholar]
- 19.Hoover DM, Lubkowski J. DNAWorks: an automated method for designing oligonucleotides for PCR-based gene synthesis. Nucleic Acids Res. 2002;30:e43. doi: 10.1093/nar/30.10.e43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu G, Dress L, Freeland SJ. Optimal encoding rules for synthetic genes: the need for a community effort. Mol. Syst. Biol. 2007;3:1–5. doi: 10.1038/msb4100176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee JY, Lim H-W, Yoo S-I, Zhang B-T, Park TH. Efficient initial pool generation for weighted graph problems using parallel overlap assembly. Lect. Notes Comp. Sci. 2005;3384:215–223. [Google Scholar]
- 22.Saleem M, Kweon M-H, Johnson JJ, Adhami VM, Elcheva I, Khan N, Hafeez BB, Bhat KMR, Sarfaraz S, Reagan-Shaw S, et al. S100A4 accelerates tumorigenesis and invasion of human prostate cancer through the transcriptional regulation of matrix metalloproteinase 9. Proc. Natl Acad. Sci. USA. 2007;103:14825–14830. doi: 10.1073/pnas.0606747103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.SantaLucia J, Jr, Hicks D. The thermodynamics of DNA structural motifs. Annu. Rev. Biophys. Biomol. Struct. 2004;33:415–440. doi: 10.1146/annurev.biophys.32.110601.141800. [DOI] [PubMed] [Google Scholar]
- 24.Herrmann MG, Durtschi JD, Bromley LK, Wittwer CT, Voelkerding KV. Amplicon DNA melting analysis for mutation scanning and genotyping: Cross-platform comparison of instruments and dyes. Clin. Chem. 2006;52:494–503. doi: 10.1373/clinchem.2005.063438. [DOI] [PubMed] [Google Scholar]
- 25.Wittwer CT, Reed GH, Gundry CN, Vandersteen JG, Pryor RJ. High-resolution genotyping by amplicon melting analysis using LCGreen. Clin. Chem. 2003;49:853–860. doi: 10.1373/49.6.853. [DOI] [PubMed] [Google Scholar]
- 26.Giglio S, Monis PT, Saint CP. Demonstration of preferential binding of SYBR Green I to specific DNA fragments in real-time multiplex PCR. Nucleic Acids Res. 2003;31:e136. doi: 10.1093/nar/gng135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Varga A, James D. Real-time PCR and SYBR Green I melting curve analysis for the identification of plum pox virus strains C, EA, and W: effect of amplicon size, melt rate, and dye translocation. J. Virol. Methods. 2006;132:146–153. doi: 10.1016/j.jviromet.2005.10.004. [DOI] [PubMed] [Google Scholar]
- 28.Wittwer CT, Herrmann MG, Moss AA, Rasmussen RP. Continuous fluorescence monitoring of rapid cycle DNA amplification. Biotechniques. 1997;22:130–138. doi: 10.2144/97221bi01. [DOI] [PubMed] [Google Scholar]
- 29.Ely JJ, Reeves-Daniel A, Campbell ML, Kohler S, Stone WH. Influence of magnesium ion concentration and PCR amplification conditions on cross-species PCR. Biotechniques. 1998;25:38–40. doi: 10.2144/98251bm07. [DOI] [PubMed] [Google Scholar]
- 30.von Ahsen N, Wittwer CT, Schütz E. Oligonucleotide melting temperatures under PCR conditions: nearest-neighbor corrections for Mg2+, deoxynucleotide triphosphate, and dimethyl sulfoxide concentrations with comparison to alternative empirical formulas. Clin. Chem. 2001;47:1956–1961. [PubMed] [Google Scholar]
- 31.Rasmussen JP, Saint CP, Monis PT. Use of DNA melting simulation software for in silico diagnostic assay design: Targeting regions with complex melting curves and confirmation by real-time PCR using intercalating dyes. BMC Bioinformatics. 2007;8:107–118. doi: 10.1186/1471-2105-8-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Luo R, Zhang D. Partial strands synthesizing leads to inevitable aborting and complicated products in consecutive polymerase chain reactions (PCRs) Sci. China Ser. C-Life Sci. 2007;50:548. doi: 10.1007/s11427-007-0043-z. [DOI] [PubMed] [Google Scholar]
- 33.Binkowski BF, Richmond KE, Kaysen J, Sussman MR, Belshaw PJ. Correcting errors in synthetic DNA through consensus shuffling. Nucleic Acids Res. 2005;33:e55. doi: 10.1093/nar/gni053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fuhrmann M, Oertel W, Berthold P, Hegemann P. Removal of mismatched bases from synthetic genes by enzymatic mismatch cleavage. Nucleic Acids Res. 2005;33:e58. doi: 10.1093/nar/gni058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bell DA, DeMarini DM. Excessive cycling converts PCR products to random-length higher molecular weight fragments. Nucleic Acids Res. 1991;19:5079. doi: 10.1093/nar/19.18.5079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Carr PA, Park JS, Lee YJ, Yu T, Zhang S, Jacobson JM. Protein-mediated error correction for de novo DNA synthesis. Nucleic Acids Res. 2004;32:e162. doi: 10.1093/nar/gnh160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Don RH, Cox PT, Wainwright BJ, Baker K, Mattick JS. ‘Touchdown’ PCR to circumvent spurious priming during gene amplification. Nucleic Acids Res. 1991;19:4008. doi: 10.1093/nar/19.14.4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ririe KM, Rasmussen RP, Wittwer CT. Product differentiation by analysis of DNA melting curves during the polymerase chain reaction. Anal. Biochem. 1997;245:154–160. doi: 10.1006/abio.1996.9916. [DOI] [PubMed] [Google Scholar]
- 39.Blake RD, Bizzaro JW, Blake JD, Day GR, Delcourt SG, Knowles J, Marx KA, SantaLucia J., Jr Statistical mechanical simulation of polymeric DNA melting with MELTSIM. Bioinformatics. 1999;15:370–375. doi: 10.1093/bioinformatics/15.5.370. [DOI] [PubMed] [Google Scholar]
- 40.Neuzil P, Pipper J, Hsieh TM. Disposable real-time microPCR device: Lab-on-a-chip at a low cost. Mol. BioSyst. 2006;2:292–298. doi: 10.1039/b605957k. [DOI] [PubMed] [Google Scholar]
- 41.Mamedov TG, Padhye NV, Viljoen H, Subramanian A. Rational de novo gene synthesis by rapid polymerase chain assembly (PCA) and expression of endothelial protein-C and thrombin receptor genes. J. Biotech. 2007;131:379–387. doi: 10.1016/j.jbiotec.2007.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Arezi B, Xing W, Sorge JA, Hogrefe HH. Amplification efficiency of thermostable DNA polymerase. Anal. Biochem. 2003;321:226–235. doi: 10.1016/s0003-2697(03)00465-2. [DOI] [PubMed] [Google Scholar]
- 43.Cherry J, Nieuwenhuijsen BW, Kaftan EJ, Kennedy JD, Chanda PK. A modified method for PCR-directed gene synthesis from large number of overlapping oligodeoxyribonucleotides. J. Biochem. Biophys. Methods. 2008;70:820–822. doi: 10.1016/j.jprot.2007.12.009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.