Summary
Homopolymeric stretches of deoxyadenosine nucleotides (A’s) on one strand of double stranded DNA, referred to as poly(dA:dT) tracts or A-tracts, are overabundant in eukaryotic genomes. They have unusual structural, dynamic, and mechanical properties, and may resist sharp bending. Such unusual material properties, together with their overabundance in eukaryotes, raised the possibility that poly(dA:dT) tracts might function in eukaryotes to influence the organization of nucleosomes at many genomic regions. Recent genome-wide studies strongly confirm these ideas and suggest that these tracts play major roles in chromatin organization and genome function. Here we review what is known about poly(dA:dT) tracts and how they work.
Functional importance of poly(dA:dT) tracts in vivo
Poly(dA:dT) tracts – homopolymeric stretches of deoxyadenosine nucleotides (A’s), often having lengths of 10–20 bp or even greater – are highly enriched in eukaryotic genomes but, intriguingly, not in prokaryotic genomes [1], suggesting that they may have a functional role unique to eukaryotic genomes. Indeed, studies of many individual genes showed that poly(dA:dT) tracts are important for transcriptional regulation [**2,**3,4–6], recombination [7], and blocking the spread of histone posttranslational modifications that are linked to transcriptional repression [8]. An early suggestion, inspired in part by in vitro studies described below, was that poly(dA:dT) tracts might function in vivo to facilitate gene activation by excluding nucleosomes [**2]. A recent genome-wide analysis showed further that poly(dA:dT) tracts are associated with and may cause nucleosome depletion at promoters, origins of DNA replication, and 3′ ends of genes; that genes whose promoters contain poly(dA:dT) tracts tend to exhibit less transcriptional noise; and that origins of replication that have poly(dA:dT) tracts in their vicinity tend to have a greater likelihood of utilization per round of DNA replication [**9].
The X-ray crystallographic structure of the nucleosome shows nucleosomal DNA to be highly distorted and sterically occluded, thereby hindering interaction of the nucleosomal DNA with other DNA binding proteins [10]. Thus, nucleosomal organization of DNA may have a generally repressive effect on DNA activity [11]. If nucleosomes were excluded from poly(dA:dT) tracts in vivo (and from their vicinity; see below), this nucleosome exclusion would facilitate access of other proteins to the DNA, helping to explain these functional roles of poly(dA:dT) tracts.
Poly(dA:dT) tracts and their flanking DNA are relatively depleted of nucleosomes in vivo
The possibility that poly(dA:dT) tracts might function in vivo to facilitate gene activation by excluding nucleosomes [**2] focused attention on the nucleosome organization around poly(dA:dT) tracts at many genes [**2,**3,4–7,12–16]. The results of these studies at individual loci were conflicting, in part because some were not carried out quantitatively. Certain assays for nucleosome occupancy can sensitively reveal the presence of nucleosome-free DNA even if a given sequence is in fact wrapped in nucleosomes across most of the cells in the population, while other assays have a converse sensitivity. Consequently, in a real situation, in which a poly(dA:dT) tract is wrapped in nucleosomes in only some fraction of the cells in the population, such analyses could report either nucleosome absence or nucleosome occupancy, depending simply on which kind of experiment was carried out. Thus, the real in vivo nucleosome occupancy over poly(dA:dT) tracts remained unclear.
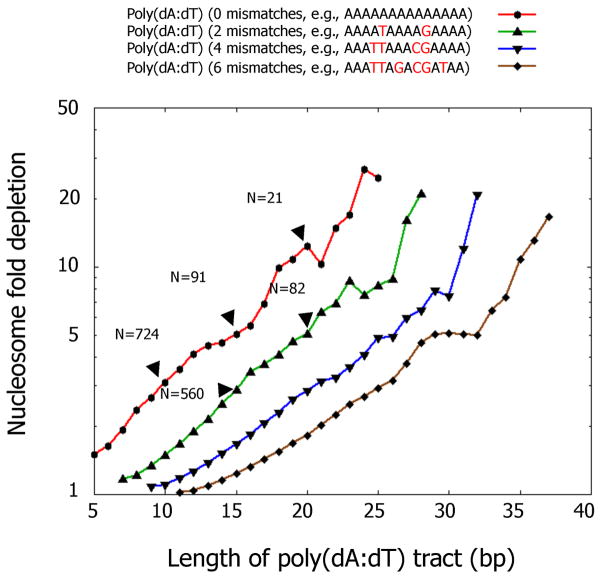
Recently, quantitative genome-wide analyses establish that poly(dA:dT) tracts are, on average, relatively depleted of nucleosomes in vivo [**9,**17,18–21]. These studies reveal that nucleosomes are depleted not just over perfect poly(dA:dT) tracts, but also over imperfect tracts containing multiple basepair substitutions or containing clusters of shorter tracts that alternate between strands [**9]. The magnitude of nucleosome depletion increases with both the length and the perfection of the poly(dA:dT) tracts (Figure 1). The fold depletion over a perfect or imperfect poly(dA:dT) tract can be predicted from the sequence itself, and can be surprisingly large. In the yeast genome, there are hundreds of poly(dA:dT) tracts with relative nucleosome depletions of 10-fold or greater [**9].
Figure 1. Nucleosome are relatively depleted over poly(dA:dT) tracts in vivo.
Shown is the combined nucleosome fold depletion over all poly(dA:dT) tracts of length k, for k = 5,6,7,…, and for tracts with exactly 0, 2, 4, or 6 base substitutions. Each graph is trimmed at a length K at which there are less than 10 such tracts in the S. cerevisiae genome, and the fold depletion at this final point is computed over all elements whose length is at least K. The number of underlying elements at various points in the graph is indicated (N). Figure adapted from ref. [**9].
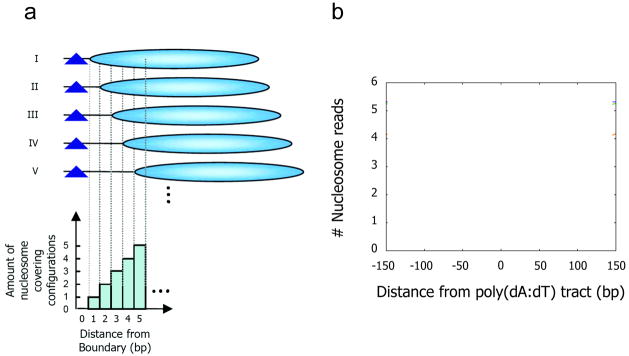
The nucleosome depletion over poly(dA:dT) tracts extends for considerable distances also into the flanking DNA on both sides of the poly(dA:dT) tract. The depletion is maximal over the poly(dA:dT) tract, but (on average) remains significant over much greater distances, ±100–150 bp (Figure 2), comparable to the length of the nucleosomal DNA itself. This longer-range nucleosome excluding behavior arises as a consequence of configurational statistics: there are a smaller number of configurations in which a nucleosome can be located nearby to a nucleosome excluding region, compared to regions that are far from such constraints [**22].
Figure 2. Poly(dA:dT) tracts create larger nucleosome-depleted regions.
(a) Shown is a simple example focusing only on the immediate neighborhood of the boundary. All (five) possible nucleosome configurations are illustrated, in which a nucleosome (cyan ovals) can be placed within five basepairs of the boundary (blue triangle). The number and set of nucleosome configurations occupying each of the five basepairs immediately adjacent to the boundary are shown in the graph below. If all configurations are equally likely, then basepairs closer to the poly(dA:dT) tract will exhibit lower nucleosome occupancy simply because fewer nucleosome configurations cover those basepairs [22]. (b) Schematic showing that nucleosome depletion caused by a poly(dA:dT) tract is maximal over the tract itself, but extends for considerable distances in either direction. Thus specific factor DNA binding sites located nearby to a poly(dA:dT) tract will have relatively enhanced accessibility compared to factor sites located far from a poly(dA:dT) tract, facilitating binding of the factor. Panel (a) adapted from ref. [**9].
In summary, nucleosomes are, on average, strongly depleted from poly(dA:dT) tracts in vivo, and this depletion extends for considerable distances into the flanking DNA. Since nucleosomes occlude their wrapped DNA from interacting with many other proteins, decreased nucleosome occupancy over such an extended DNA region will have the effect of increasing the accessibility of all of the DNA in that region – both the poly(dA:dT) tract itself and its flanking DNA – to other DNA binding proteins. Such enhanced DNA accessibility could explain many of the in vivo functions that have been associated with poly(dA:dT) tracts.
Nucleosome depletion over poly(dA:dT) tracts results chiefly from the tracts’ intrinsically lower nucleosome affinity
What causes the dramatic nucleosome depletion over poly(dA:dT) tracts? The simplest hypothesis, that the nucleosome depletion is due to competition with another protein that binds specifically to poly(dA:dT) tracts, is ruled out. To date, a single protein in S. cerevisiae, called Datin (Dat1p), which recognizes poly(dA:dT) tracts of length 9 bp or greater, has been identified [23]. Datin may be the only DNA binding protein in S. cerevisiae that binds poly(dA:dT) tracts, since yeast cell extracts in a Datin deletion do not exhibit any detectable protein binding to poly(dA:dT) tracts. Many of the studies that revealed functional roles for poly(dA:dT) tracts specifically tested the role of Datin binding by deleting the DAT1 gene [**3,5–7,15,23]. However, Datin binding was found to be important for transcriptional activation in only one case [4]. These studies prove that Datin binding is not responsible for the transcription activating function of most poly(dA:dT) tracts or for the nucleosome depletion over the poly(dA:dT) tracts.
Another possibility is that the binding of transcription factors to sites near the poly(dA:dT) tracts causes nucleosome depletion over poly(dA:dT) tracts. Indeed, such an effect is to be expected on thermodynamic grounds [**22]; the question is the relative significance of this effect. If transcription factor binding to sites flanking poly(dA:dT) tracts were a dominant cause of nucleosome depletion over the poly(dA:dT) tracts themselves, then one would expect similar nucleosome depletion over factor binding sites regardless of whether or not they are close to poly(dA:dT) tracts. However, this is not the case: nucleosomes are strongly depleted over factor binding sites that are near poly(dA:dT) tracts, but only weakly depleted over factor sites that are not near poly(dA:dT) tracts [**9]. Conversely, the extent of nucleosome depletion over poly(dA:dT) tracts is similar regardless of whether the poly(dA:dT) tracts are near to binding sites for transcription factors, or not. Thus, binding by transcription factors is not the major cause of nucleosome depletion over poly(dA:dT) tracts in vivo.
A remaining alternative is that poly(dA:dT) tracts are relatively nucleosome-depleted in vivo because the tracts themselves intrinsically disfavor nucleosome formation. This possibility is supported by both in vivo and in vitro studies at specific genes. The most important of the in vivo experiments include: In the yeast HIS3 promoter, a poly(dA:dT) tract, but not a Gal4 protein binding site, can induce transcription by bacteriophage T7 RNA polymerase, suggesting that the poly(dA:dT) tract acts by exclusion of a repressive nucleosome and not by inducing interactions with the basal transcriptional machinery [14]. At HIS3, RPS28a, and BAR1, replacing the poly(dA:dT) tract by poly (dC:dG) resulted in similar transcriptional induction, with longer poly(dA:dT) tracts resulting in greater transcriptional induction, consistent with increasing nucleosome exclusion but less-so with a role for a sequence-specific DNA binding protein [**3,5]. One apparent contradictory result suggested that a poly(dA:dT) tract in the DED1 promoter cannot function only through its nucleosome exclusion effects [24]. However, since that poly(dA:dT) tract has 7 non-A nucleotides (in a 38 bp-long tract), it may well contain a binding site for an additional site-specific DNA binding activity; and in any case, that finding does not contradict a possible additional role for an intrinsic nucleosome disfavoring activity for that imperfect poly(dA:dT) tract in vivo. Thus, the consensus of studies of specific genes support a role for poly(dA:dT) tracts in causing a relative nucleosome depletion in vivo, with the resulting nucleosome depletion facilitating the binding of factors to specific DNA target sites.
Similarly, studies in vitro establish that poly(dA:dT) tracts intrinsically disfavor nucleosome organization. First, however, contrary to some claims, poly(dA:dT) tracts are capable of being incorporated into nucleosomes [25–29]. Any remaining question about this was definitively settled by the determination of a high resolution X-ray crystallographic structure of a nucleosome containing a 16 bp-long perfect poly(dA:dT) tract [30]. Small structural differences caused by the poly(dA:dT) tract are detectable, but the overall wrapping of the nucleosome DNA is essentially unchanged. It follows that any effects of poly(dA:dT) tracts on nucleosome occupancy or affinity will necessarily be quantitative in nature, not absolute. Only studies that are sensitive to quantitative differences in occupancy or affinity can shed light on such questions; and early such quantitative studies showed that poly(dA:dT) tracts do indeed disfavor nucleosome formation [31,32], with a magnitude that increases with the length of the poly(dA:dT) tract [25,26]. More recent studies confirm that even a relatively short (16 bp) poly(dA:dT) tract significantly decreases nucleosome affinity [33]; many copies of a 4–6 bp poly(dA:dT) tract greatly reduced affinity [34]; and poly(dA:dT) tract-containing DNAs present in several different yeast promoters disfavor nucleosome incorporation [**9] by an amount comparable to that of other non-natural DNAs that were selected in vitro for their ability to resist nucleosome formation [35]. These findings were strongly confirmed and extended in a recent genome-wide analysis [**36]: the distribution of nucleosomes reconstituted on genomic DNA in vitro closely resembled the in vivo nucleosome distribution, with significant depletion of nucleosomes from over poly(dA:dT) tracts.
In summary, nucleosomes are on average significantly depleted from poly(dA:dT) tracts in vivo. This depletion in most cases is not due to competition with Datin binding specifically to the poly(dA:dT) tracts; and, while competition with other proteins binding to specific target sites located nearby the poly(dA:dT) tracts can contribute to the observed nucleosome depletion, this is not the dominant cause. Instead, the observed nucleosome depletion is due chiefly to nucleosomes intrinsically disfavoring occupancy over the poly(dA:dT) tracts; and this disfavoring is quantitative, not absolute.
Poly(dA:dT) tracts have unusual structural and dynamic properties
At the level of detailed molecular structure and mechanics, why is it that nucleosomes intrinsically disfavor wrapping poly(dA:dT) tracts relative to most other DNA sequences? The answer is not known definitively; but a growing body of studies points to a unique cooperative structure of poly(dA:dT) tracts, which in turn is associated with, and possibly due to, a unique hydration structure of the poly(dA:dT) tracts. Deforming this unique poly(dA:dT) tract structure by forcing it into a nucleosome conformation may be much more energetically costly than are comparable deformations of generic DNA sequences.
An early hypothesis was that AA dinucleotide steps might be intrinsically stiff compared to other dinucleotide steps, and thus poly(dA:dT) tracts might have an exaggerated stiffness, maximally-disfavoring the deformations required for nucleosome formation. However, analyses of newer, larger, databases of X-ray crystallographic structures of DNA and protein-DNA complexes, in which the variance among configurations in independent structures may serve as a proxy for basepair step flexibility [37], and recent molecular mechanics calculations [38,39], do not support a high intrinsic stiffness of the AA dinucleotide step. Thus, any intrinsic resistance of poly(dA:dT) tracts to nucleosome formation is not attributable to special mechanics of AA dinucleotides.
Several lines of evidence suggest instead that the structural, dynamic, and mechanical properties of poly(dA:dT) tracts may differ fundamentally from the corresponding properties of individual AA dinucleotides. Compared to generic sequence DNA, poly(dA:dT) has a shorter helical repeat [40,41], a narrow minor groove, a distinct spine of hydration within the minor grove, and maximal overlap of the bases separately within each strand [42,43]. The crystallographic studies [42,43] further suggested that poly(dA:dT) tracts also exhibit an unusual hydrogen bonding pattern (“bifurcated H-bonds”), in which amino groups on A bases formed hydrogen bonds both with their Watson-Crick partner and also to the O4 atom of an adjacent T base on the opposite strand. Such cross-strand H-bonds could potentially stiffen the DNA; however subsequent higher resolution studies show the shortest (presumably, tightest) such bonds to be at the long limit for a significant H-bond [**44], so whether such bonds truly exist, and how much they might contribute to special properties of poly(dA:dT) tracts, is unclear.
Moreover, the unusual structural properties of poly(dA:dT) grow in cooperatively with length of the poly(dA:dT) tract, and are accompanied by unusual dynamic properties. Hydroxyl radical footprinting studies reveal a progressive decrease in reactivity with increasing distance inside poly(dA:dT) tracts, for tracts of length 4 bp or greater (also for A2T2), implying the existence of a distinct, length-dependent, structural state for the poly(dA:dT) tract [45]. The detailed cleavage pattern further suggested that the minor groove width decreased progressively with distance inside the poly(dA:dT) tract, a conclusion that is strongly upheld in atomic resolution crystallographic structures of A2T2- and A3T3-containing DNAs [**44,46,47]. Correspondingly, NMR measurements of imino proton exchange rates reveal extraordinarily long basepair lifetimes (high lifetimes) for T residues located 4 or more nucleotides inside a poly(dA:dT) tract of length 4 bp or greater (again, also for A2T2 and A3T3 tracts), with the basepair lifetimes increasing with depth inside the tract [48–50]. These results imply that not only do the poly(dA:dT) tracts possess unusual length-dependent structures, but these structures have corresponding unusual dynamics, which could well translate into unusual mechanical properties – including, potentially, into a relatively great resistance to the bending and twisting deformations that are characteristic of DNA in the nucleosome [10]. Other evidence for cooperative formation of a distinctive DNA structure with increasing length of a poly(dA:dT) tract includes an abrupt change in gel mobility for tracts of length 4 bp or greater [51]; a remarkable cooperative premelting transition in DNAs having several poly(dA:dT) tracts of length 5 bp [52]; and structural discontinuities including local DNA bending within poly(dA:dT) tract and at the two ends where the tract connects to arbitrary DNA sequence [53,54].
In summary, there is overwhelming evidence from diverse experiments that poly(dA:dT) tracts of lengths of 4 bp or greater adopt a novel cooperative state whose structures, dynamics, and thermodynamic properties differ fundamentally from those of generic sequence DNA. But why does this state disfavor nucleosome incorporation?
The simplest possibility, mentioned above in connection with the results of NMR studies, is that the unique length-dependent structure of poly(dA:dT) tracts might be uniquely resistant to the deformation(s) required for nucleosome formation. Imposing such deformations on a poly(dA:dT) tract by incorporating it into a nucleosome would then incur a particularly large cost in free energy, producing a nucleosome with an intrinsically reduced stability [30], or, equivalently, causing the nucleosome to preferentially occupy locations on the DNA that exclude the poly(dA:dT) tract, as observed in the many in vitro nucleosome reconstitution studies mentioned above.
Structural studies provide further evidence that is suggestive of such a picture. The atomic resolution crystallographic studies of DNAs containing the poly(dA:dT) tracts A2T2 and A3T3 [**44,46] and NMR solution studies [55] reveal highly ordered spines of hydration, at least four water-layers deep for A3T3 (Figure 3), restricted to the narrowed minor groove regions of the tracts themselves. In certain cases specific high occupancy and/or long lifetime bound cations can also be detected [46,56]. The existence of such highly ordered waters (and localized cations when present) strongly suggests that they must be held in place by favorable energetic interactions. Similarly, the long bound-state lifetimes of these localized waters are analogous to those of water molecules in the interiors of globular proteins, which are integral parts of the proteins’ structure [55], further suggestive of a net favorable energetic interaction. The extensive H-bonding of these waters with both DNA and each other, including between the successive water layers, would be expected to contribute to a length-dependent cooperative formation of the poly(dA:dT) tract’s special structure.
Figure 3. Narrow minor groove and multilayer spine of hydration in a poly(dA:dT) tract.
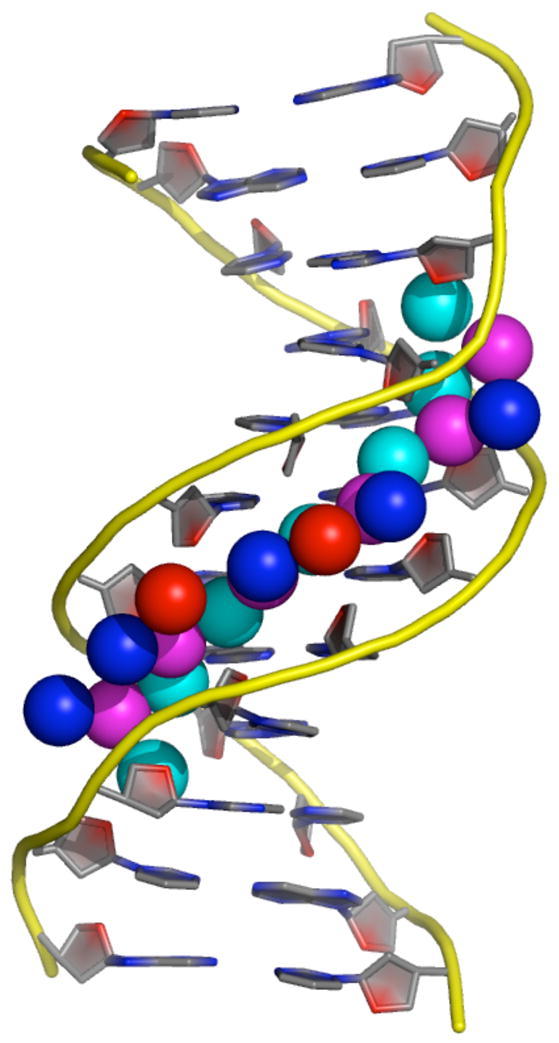
Shown is a representation of the atomic resolution X-ray crystallographic structure of [d(CGCAAATTTGCG)]2 [**44]. The DNA backbones are shown as yellow curves, with the bases shown in a partial-charge-coded stick representation. The narrow minor groove of the A3T3 stretch has many high occupancy water molecules, 4 layers deep, shown here as spheres, color coded according to their layer from innermost to outermost as cyan, purple, blue, and red, respectively. The multiple layers, extensive hydrogen bonding, and high occupancy of these waters all suggest that they may have strongly favorable energetic interactions with themselves and the DNA. Figure kindly provided by Prof. L.D. Williams (Georgia Tech.).
The DNA structural deformations required for nucleosome formation could likely disrupt such energetically favorable water- or specific cation-DNA interactions, thereby causing the poly(dA:dT) tracts to have a decreased affinity (less-negative free energy change) for nucleosome formation. Consistent with this view, a model for the sequence-dependent free energy of DNA wrapping in nucleosomes suggests that the curvature-dependent DNA hydration changes coupled to sharp DNA bending plays a significant role in the energetics of nucleosome formation [57].
All of these facts – together with the experimentally proven intrinsic resistance of poly(dA:dT) tracts to nucleosome incorporation in vitro – suggest that poly(dA:dT) tracts intrinsically resist the structural deformations required for nucleosome formation, relative to generic DNA sequences. But is this true? Taken at face value, the available literature does not support this conclusion: some studies suggest that poly(dA:dT) tracts are not more-resistant to bending and twisting, but less-so, than are other simple sequences [58], while other studies suggest stiffnesses that are within the normal range [59,60]. However, these experiments are somewhat indirect, moreover, they monitor DNA flexibility in situations in which the DNA is rather less distorted than is DNA in nucleosomes. Thus, the requirements of nucleosome organization would greatly exaggerate the effects of what might otherwise be only small differences in the mechanics of differing DNA sequences, such that the differences are not detectable with presently available methods.
Conclusions
In summary, poly(dA:dT) tracts strongly resist incorporation into nucleosomes in vitro, and, if incorporated into nucleosomes, reduce the stability of those nucleosomes. This intrinsic resistance to incorporation into nucleosomes may be due to an intrinsic resistance of the poly(dA:dT) tracts to adopting the substantially distorted structures required by the nucleosome, although idea this remains unproven. Whatever the physical mechanism for their preferential avoidance of nucleosome incorporation, poly(dA:dT) tracts are dominant determinants of the in vivo nucleosome organization of eukaryotic genomes, and strongly influence genome function, including by controlling the accessibility of other nearby specific DNA target sites to their cognate regulatory proteins. Better ways of analyzing the sequence-dependent mechanical properties of DNA are plainly needed.
Supplementary Material
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Dechering KJ, Cuelenaere K, Konings RN, Leunissen JA. Distinct frequency-distributions of homopolymeric DNA tracts in different genomes. Nucleic Acids Research. 1998;26:4056–4062. doi: 10.1093/nar/26.17.4056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2**.Struhl K. Naturally occurring poly(dA-dT) sequences are upstream promoter elements for constitutive transcription in yeast. Proc Natl Acad Sci USA. 1985;82:8419–8423. doi: 10.1073/pnas.82.24.8419. This paper was the first to report that poly(dA:dT) elements in a gene’s promoter facilitate transcription in vivo, and the first to connect this observation with a possible mechanism, suggesting that poly(dA:dT) elements could act by depleting repressive nucleosomes from the promoter. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3**.Iyer V, Struhl K. Poly(dA:dT), a ubiquitous promoter element that stimulates transcription via its intrinsic DNA structure. EMBO J. 1995;14:2570–2579. doi: 10.1002/j.1460-2075.1995.tb07255.x. This paper was the first to provide concrete evidence in support of the model in which poly(dA:dT) elements act by depleting repressive nucleosomes from a gene’s promoter, thereby effectively enhancing transcription. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moreira JM, Remacle JE, Kielland-Brandt MC, Holmberg S. Datin, a yeast poly(dA:dT)-binding protein, behaves as an activator of the wild-type ILV1 promoter and interacts synergistically with Reb1p. Mol Gen Genet. 1998;258:95–103. doi: 10.1007/s004380050711. [DOI] [PubMed] [Google Scholar]
- 5.Lascaris RF, Groot E, Hoen PB, Mager WH, Planta RJ. Different roles for abf1p and a T-rich promoter element in nucleosome organization of the yeast RPS28A gene. Nucleic Acids Research. 2000;28:1390–1396. doi: 10.1093/nar/28.6.1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Koch KA, Thiele DJ. Functional analysis of a homopolymeric (dA-dT) element that provides nucleosomal access to yeast and mammalian transcription factors. J Biol Chem. 1999;274:23752–23760. doi: 10.1074/jbc.274.34.23752. [DOI] [PubMed] [Google Scholar]
- 7.Schultes NP, Szostak JW. A poly(dA. dT) tract is a component of the recombination initiation site at the ARG4 locus in Saccharomyces cerevisiae. Molecular and Cellular Biology. 1991;11:322–328. doi: 10.1128/mcb.11.1.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bi X, Yu Q, Sandmeier JJ, Zou Y. Formation of boundaries of transcriptionally silent chromatin by nucleosome-excluding structures. Molecular and Cellular Biology. 2004;24:2118–2131. doi: 10.1128/MCB.24.5.2118-2131.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9**.Field Y, Kaplan N, Fondufe-Mittendorf Y, Moore IK, Sharon E, Lubling Y, Widom J, Segal E. Distinct modes of regulation by chromatin encoded through nucleosome positioning signals. PLoS Comput Biol. 2008;4:e1000216. doi: 10.1371/journal.pcbi.1000216. This paper is one of several that reports that nucleosomes are relatively depleted over poly(dA:dT) elements in vivo, and it associates nucleosome depletion with functional roles, including, at promoters, with decreased transcriptional noise; and at origins of DNA replication, with increased probability of origin firing per round of replication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Richmond T, Davey C. The structure of DNA in the nucleosome core. Nature. 2003;423:145–150. doi: 10.1038/nature01595. [DOI] [PubMed] [Google Scholar]
- 11.Kornberg RD, Lorch Y. Twenty-five years of the nucleosome, fundamental particle of the eukaryote chromosome. Cell. 1999;98:285–294. doi: 10.1016/s0092-8674(00)81958-3. [DOI] [PubMed] [Google Scholar]
- 12.Verdone L, Camilloni G, Di Mauro E, Caserta M. Chromatin remodeling during Saccharomyces cerevisiae ADH2 gene activation. Molecular and Cellular Biology. 1996;16:1978–1988. doi: 10.1128/mcb.16.5.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Struhl K. Constitutive and inducible Saccharomyces cerevisiae promoters: evidence for two distinct molecular mechanisms. Molecular and Cellular Biology. 1986;6:3847–3853. doi: 10.1128/mcb.6.11.3847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen W, Tabor S, Struhl K. Distinguishing between mechanisms of eukaryotic transcriptional activation with bacteriophage T7 RNA polymerase. Cell. 1987;50:1047–1055. doi: 10.1016/0092-8674(87)90171-1. [DOI] [PubMed] [Google Scholar]
- 15.Suter B, Schnappauf G, Thoma F. Poly(dA. dT) sequences exist as rigid DNA structures in nucleosome-free yeast promoters in vivo. Nucleic Acids Research. 2000;28:4083–4089. doi: 10.1093/nar/28.21.4083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shimizu M, Mori T, Sakurai T, Shindo H. Destabilization of nucleosomes by an unusual DNA conformation adopted by poly(dA) small middle dotpoly(dT) tracts in vivo. EMBO J. 2000;19:3358–3365. doi: 10.1093/emboj/19.13.3358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17**.Yuan GC, Liu YJ, Dion MF, Slack MD, Wu LF, Altschuler SJ, Rando O. Genome-scale identification of nucleosome positions in S. cerevisiae. Science. 2005;309:626–630. doi: 10.1126/science.1112178. This paper reports the first genome-wide analysis of nucleosome positioning in vivo, and showed that on average poly(dA:dT) tracts are relatively nucleosome depleted. [DOI] [PubMed] [Google Scholar]
- 18.Lee W, Tillo D, Bray N, Morse R, Davis R, Hughes T, Nislow C. A high-resolution atlas of nucleosome occupancy in yeast. Nat Genet. 2007;39:1235–1244. doi: 10.1038/ng2117. [DOI] [PubMed] [Google Scholar]
- 19.Whitehouse I, Rando O, Delrow J, Tsukiyama T. Chromatin remodelling at promoters suppresses antisense transcription. Nature. 2007;450:1031–1035. doi: 10.1038/nature06391. [DOI] [PubMed] [Google Scholar]
- 20.Mavrich T, Ioshikhes I, Venters BJ, Jiang C, Tomsho L, Qi J, Schuster SC, Albert I, Pugh B. A barrier nucleosome model for statistical positioning of nucleosomes throughout the yeast genome. Genome Research. 2008;18:1073–1083. doi: 10.1101/gr.078261.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ozsolak F, Song JS, Liu XS, Fisher DE. High-throughput mapping of the chromatin structure of human promoters. Nat Biotechnol. 2007;25:244–248. doi: 10.1038/nbt1279. [DOI] [PubMed] [Google Scholar]
- 22**.Kornberg RD, Stryer L. Statistical distributions of nucleosomes: nonrandom locations by a stochastic mechanism. Nucleic Acids Research. 1988;16:6677–6690. doi: 10.1093/nar/16.14.6677. This paper explains how any short stretch of DNA that nucleosomes preferentially avoid will create a larger nucleosome depletion centered on the avoided stretch but extending considerable distances in either direction. The argument imagined a DNA target site occupied by a bound protein, but applies generally. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Winter E, Varshavsky A. A DNA binding protein that recognizes oligo(dA). oligo(dT) tracts. EMBO J. 1989;8:1867–1877. doi: 10.1002/j.1460-2075.1989.tb03583.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lue NF, Buchman AR, Kornberg RD. Activation of yeast RNA polymerase II transcription by a thymidine-rich upstream element in vitro. Proc Natl Acad Sci USA. 1989;86:486–490. doi: 10.1073/pnas.86.2.486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kunkel GR, Martinson HG. Nucleosomes will not form on double-stranded RNa or over poly(dA). poly(dT) tracts in recombinant DNA. Nucleic Acids Research. 1981;9:6869–6888. doi: 10.1093/nar/9.24.6869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Prunell A. Nucleosome reconstitution on plasmid-inserted poly(dA). poly(dT) EMBO J. 1982;1:173–179. doi: 10.1002/j.1460-2075.1982.tb01143.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schieferstein U, Thoma F. Modulation of cyclobutane pyrimidine dimer formation in a positioned nucleosome containing poly(dA. dT) tracts. Biochemistry. 1996;35:7705–7714. doi: 10.1021/bi953011r. [DOI] [PubMed] [Google Scholar]
- 28.Losa R, Omari S, Thoma F. Poly(dA). poly(dT) rich sequences are not sufficient to exclude nucleosome formation in a constitutive yeast promoter. Nucleic Acids Research. 1990;18:3495–3502. doi: 10.1093/nar/18.12.3495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fox KR. Wrapping of genomic polydA. polydT tracts around nucleosome core particles. Nucleic Acids Research. 1992;20:1235–1242. doi: 10.1093/nar/20.6.1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bao Y, White CL, Luger K. Nucleosome core particles containing a poly(dA. dT) sequence element exhibit a locally distorted DNA structure. Journal of Molecular Biology. 2006;361:617–624. doi: 10.1016/j.jmb.2006.06.051. [DOI] [PubMed] [Google Scholar]
- 31.Rhodes D. Nucleosome cores reconstituted from poly (dA-dT) and the octamer of histones. Nucleic Acids Research. 1979;6:1805–1816. doi: 10.1093/nar/6.5.1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Simpson RT, Künzler P. Cromatin and core particles formed from the inner histones and synthetic polydeoxyribonucleotides of defined sequence. Nucleic Acids Research. 1979;6:1387–1415. doi: 10.1093/nar/6.4.1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Anderson JD, Widom J. Poly(dA-dT) promoter elements increase the equilibrium accessibility of nucleosomal DNA target sites. Molecular and Cellular Biology. 2001;21:3830–3839. doi: 10.1128/MCB.21.11.3830-3839.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Scipioni A, Pisano S, Anselmi C, Savino M, De Santis P. Dual role of sequence-dependent DNA curvature in nucleosome stability: the critical test of highly bent Crithidia fasciculata DNA tract. Biophys Chem. 2004;107:7–17. doi: 10.1016/S0301-4622(03)00214-X. [DOI] [PubMed] [Google Scholar]
- 35.Thåström A, Lowary PT, Widlund HR, Cao H, Kubista M, Widom J. Sequence motifs and free energies of selected natural and non-natural nucleosome positioning DNA sequences. Journal of Molecular Biology. 1999;288:213–229. doi: 10.1006/jmbi.1999.2686. [DOI] [PubMed] [Google Scholar]
- 36**.Kaplan N, Moore IK, Fondufe-Mittendorf Y, Gossett AJ, Tillo D, Field Y, Leproust EM, Hughes TR, Lieb JD, Widom J, et al. The DNA-encoded nucleosome organization of a eukaryotic genome. Nature. 2008 doi: 10.1038/nature07667. This paper analyzes the genome-wide distribution of nucleosomes that were reconstituted in a purified system in vitro, and shows the characteristic depletion of nucleosomes over poly(dA:dT) tracts in vivo to be recapitulated in vitro. This observation proves that the depletion is due at least in part to an intrinsic unfavorable property of the poly(dA:dT) tract–nucleosome interaction. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Olson WK, Gorin AA, Lu XJ, Hock LM, Zhurkin VB. DNA sequence-dependent deformability deduced from protein-DNA crystal complexes. Proc Natl Acad Sci USA. 1998;95:11163–11168. doi: 10.1073/pnas.95.19.11163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gardiner E, Hunter CA, Packer MJ, Palmer DS. Sequence-dependent DNA Structure: A Database of Octamer Structural Parameters. Journal of Molecular Biology. 2003 doi: 10.1016/j.jmb.2003.08.006. [DOI] [PubMed] [Google Scholar]
- 39.Dixit SB, Beveridge DL, Case DA, Cheatham TE, Giudice E, Lankas F, Lavery R, Maddocks JH, Osman R, Sklenar H, et al. Molecular dynamics simulations of the 136 unique tetranucleotide sequences of DNA oligonucleotides. II: sequence context effects on the dynamical structures of the 10 unique dinucleotide steps. Biophysical Journal. 2005;89:3721–3740. doi: 10.1529/biophysj.105.067397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Peck LJ, Wang JC. Sequence dependence of the helical repeat of DNA in solution. Nature. 1981;292:375–378. doi: 10.1038/292375a0. [DOI] [PubMed] [Google Scholar]
- 41.Rhodes D, Klug A. Helical periodicity of DNA determined by enzyme digestion. Nature. 1980;286:573–578. doi: 10.1038/286573a0. [DOI] [PubMed] [Google Scholar]
- 42.Nelson HC, Finch JT, Luisi BF, Klug A. The structure of an oligo(dA). oligo(dT) tract and its biological implications. Nature. 1987;330:221–226. doi: 10.1038/330221a0. [DOI] [PubMed] [Google Scholar]
- 43.DiGabriele AD, Steitz TA. A DNA dodecamer containing an adenine tract crystallizes in a unique lattice and exhibits a new bend. Journal of Molecular Biology. 1993;231:1024–1039. doi: 10.1006/jmbi.1993.1349. [DOI] [PubMed] [Google Scholar]
- 44**.Woods KK, Maehigashi T, Howerton SB, Sines CC, Tannenbaum S, Williams LD. High-resolution structure of an extended A-tract: [d(CGCAAATTTGCG)]2. J Am Chem Soc. 2004;126:15330–15331. doi: 10.1021/ja045207x. This atomic resolution crystallographic structure of a poly(dA:dT) tract dispells myths about the nature of these tracts that had been suggested from earlier studies, and reveals an exceptionally prominent multilayered hydration structure that may explain the cooperative formation and special properties of poly(dA:dT) tracts. [DOI] [PubMed] [Google Scholar]
- 45.Burkhoff AM, Tullius TD. The unusual conformation adopted by the adenine tracts in kinetoplast DNA. Cell. 1987;48:935–943. doi: 10.1016/0092-8674(87)90702-1. [DOI] [PubMed] [Google Scholar]
- 46.Tereshko V, Minasov G, Egli M. A “hydrat-ion” spine in a B-DNA minor groove. J Am Chem Soc. 1999;121:3590–3595. [Google Scholar]
- 47.Hizver J, Rozenberg H, Frolow F, Rabinovich D, Shakked Z. DNA bending by an adenine--thymine tract and its role in gene regulation. Proc Natl Acad Sci USA. 2001;98:8490–8495. doi: 10.1073/pnas.151247298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Leroy JL, Charretier E, Kochoyan M, Guéron M. Evidence from base-pair kinetics for two types of adenine tract structures in solution: their relation to DNA curvature. Biochemistry. 1988;27:8894–8898. doi: 10.1021/bi00425a004. [DOI] [PubMed] [Google Scholar]
- 49.Wärmländer S, Sen A, Leijon M. Imino proton exchange in DNA catalyzed by ammonia and trimethylamine: evidence for a secondary long-lived open state of the base pair. Biochemistry. 2000;39:607–615. doi: 10.1021/bi991863b. [DOI] [PubMed] [Google Scholar]
- 50.Moe JG, Folta-Stogniew E, Russu IM. Energetics of base pair opening in a DNA dodecamer containing an A3T3 tract. Nucleic Acids Research. 1995;23:1984–1989. doi: 10.1093/nar/23.11.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Haran TE, Crothers DM. Cooperativity in A-tract structure and bending properties of composite TnAn blocks. Biochemistry. 1989;28:2763–2767. doi: 10.1021/bi00433a003. [DOI] [PubMed] [Google Scholar]
- 52.Chan SS, Breslauer KJ, Austin RH, Hogan ME. Thermodynamics and premelting conformational changes of phased (dA)5 tracts. Biochemistry. 1993;32:11776–11784. doi: 10.1021/bi00095a005. [DOI] [PubMed] [Google Scholar]
- 53.Barbic A, Zimmer DP, Crothers DM. Structural origins of adenine-tract bending. Proc Natl Acad Sci USA. 2003;100:2369–2373. doi: 10.1073/pnas.0437877100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stefl R, Wu H, Ravindranathan S, Sklenár V, Feigon J. DNA A-tract bending in three dimensions: solving the dA4T4 vs. dT4A4 conundrum. Proc Natl Acad Sci USA. 2004;101:1177–1182. doi: 10.1073/pnas.0308143100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liepinsh E, Otting G, Wüthrich K. NMR observation of individual molecules of hydration water bound to DNA duplexes: direct evidence for a spine of hydration water present in aqueous solution. Nucleic Acids Research. 1992;20:6549–6553. doi: 10.1093/nar/20.24.6549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hud NV, Sklenár V, Feigon J. Localization of ammonium ions in the minor groove of DNA duplexes in solution and the origin of DNA A-tract bending. Journal of Molecular Biology. 1999;286:651–660. doi: 10.1006/jmbi.1998.2513. [DOI] [PubMed] [Google Scholar]
- 57.Anselmi C, Bocchinfuso G, De Santis P, Savino M, Scipioni A. Dual role of DNA intrinsic curvature and flexibility in determining nucleosome stability. Journal of Molecular Biology. 1999;286:1293–1301. doi: 10.1006/jmbi.1998.2575. [DOI] [PubMed] [Google Scholar]
- 58.Hogan M, LeGrange J, Austin B. Dependence of DNA helix flexibility on base composition. Nature. 1983;304:752–754. doi: 10.1038/304752a0. [DOI] [PubMed] [Google Scholar]
- 59.Podtelezhnikov AA, Mao C, Seeman NC, Vologodskii A. Multimerization-cyclization of DNA fragments as a method of conformational analysis. Biophysical Journal. 2000;79:2692–2704. doi: 10.1016/S0006-3495(00)76507-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang Y, Xi Z, Hegde RS, Shakked Z, Crothers DM. Predicting indirect readout effects in protein-DNA interactions. Proc Natl Acad Sci USA. 2004;101:8337–8341. doi: 10.1073/pnas.0402319101. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.