Abstract
Homologue pairing mediates both recombination and segregation of chromosomes at meiosis I. The recognition of nucleic-acid-sequence homology within the somatic nucleus has an impact on DNA repair and epigenetic control of gene expression. Here we investigate interchromosomal interactions using a non-invasive technique that allows tagging and visualization of DNA sequences in vegetative and meiotic live yeast cells. In non-meiotic cells, chromosomes are ordered in the nucleus, but preferential pairing between homologues is not observed. Association of tagged chromosomal domains occurs irrespective of their genomic location, with some preference for similar chromosomal positions. Here we describe a new phenomenon that promotes associations between sequence-identical ectopic tags with a tandem-repeat structure. These associations, termed interchromosome trans-associations, may underlie epigenetic phenomena.
The pairing of homologous chromosomes during meiosis I is well characterized. Such pairing is required for proper recombination and chromosome segregation, and is controlled by a specialized meiotic molecular machine. In addition to meiotic pairing, a variety of interactions between DNA sequences in the non-meiotic (somatic) nucleus has been described. Recombinational DNA repair and epigenetic control of gene expression are the two prominent processes that might benefit from allelic and ectopic interchromosomal associations1–3. These phenomena require physical association between two interacting DNA molecules, the genomic location of which has only a limited effect on this process3,4. In contrast, homologue pairing during meiosis exhibits a strong allelic preference between the two interacting sequences5. Is there some continuity between somatic and meiotic homologue pairing? The definite answer to this question is complicated by inter-species differences, lack of non-invasive methods of investigation, and disparity between different chromosomes and even distinct chromosomal domains. In S. cerevisiae and Schizosaccharomyces pombe, homologue pairing has been reported to occur as early as the preceding mitotic cellular division6–9. In contrast, maize and human chromosomes seem not to be paired premeiotically10,11. In addition, the mechanism of interchromosome pairing in somatic cells remains elusive. A possible explanation for the phenomenon of somatic pairing is that such interactions provide the basis of meiotic pairing9. Indeed, some data indicate that both meiotic and non-meiotic (somatic) pairing may originate from the similar, unstable, side-to-side (paranemic) interactions that occur between homologous chromosomes7,12. However, this hypothesis is challenged by the complete disruption of pre-existing homologue associations during premeiotic S phase, before the re-establishment of pairing at the onset of meiotic prophase I7. In view of this contradiction, this hypothesis has recently been re-examined13.
In the yeast S. cerevisiae, the presence of ectopic (non-allelic) mitotic recombination provides some evidence that pairing between DNA sequences occurs transiently but regularly in mitotically propagating cells3. On the other hand, several nuclear features might constrain relative chromosome positions in the nucleus, including, for example, clustering of centromeric regions at the nuclear periphery14,15, opposite to telomeres, which in turn tend to form multiple clusters16.
Here we report the use of a non-invasive tagging technique, chromosomal green fluorescent protein (GFP) tagging17, to monitor allelic and ectopic interactions between the tagged stretches of DNA. This technique allows visualization of specific segments of the genome in the nuclei of intact mitotic and meiotic cells. We used two independent tagging systems based on tandem-repeat arrays, the lactose (lacO)17 and tetracycline (tetO)18 operators.
Our results indicate that two different types of interaction occur in the S. cerevisiae nucleus. The first class occurs mainly during meiosis and is based on the allelic positions of the two interacting DNA tags. This type, referred to as ‘meiotic pairing’, allows interaction of even nonhomologous tags in the allelic position, probably because of the stable allelic interaction between the flanking sequences. The second class of interaction preferentially occurs in mitotically propagating cells, and is absolutely dependent on the sequence homology between the tags themselves, not of the flanking sequences. We refer to this class as ‘flanking-independent’ interaction or ‘trans-association’, as it occurs with equal frequency between allelic and ectopic tags.
Results
Design of chromosomal tags
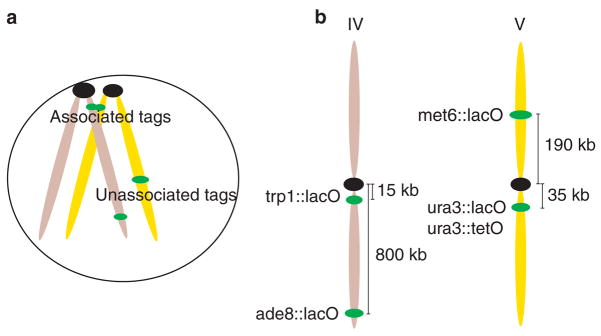
Our aim was to detect and characterize chromosome interactions in vivo. Traditional methods to visualize chromosome pairing in yeast are based on nuclear spreads and fluorescent in situ hybridization (FISH) analysis7,9. In these experiments the nuclear content is fixed, deproteinized and dispersed over an area up to ten times greater than the diameter of an intact nucleus. As a result, the three-dimensional information is lost and the spacing between chromatin domains is distorted. Recently, indirect techniques using site-specific recombination have been used to evaluate somatic-homologue pairing13. We wished to monitor interchromosome interactions directly, without perturbing cell physiology and changing relative distances within the nucleus. We therefore used a method to visualize specific DNA sequences in living yeast cells by tagging chromosomes with GFP17 (see Table 1 for strain genotypes). This method exploits the intrinsic fluorescence of GFP and the highly specific binding of the bacterial lacI repressor to the lacO DNA sequence. Initially, we integrated a tandem array of 256 lacO copies17 at different genomic locations (Fig. 1) in haploid yeast, which we crossed to form diploid strains with the following tag positions: the trp1 locus (15 kilobases (kb) from CEN4), the ura3 locus (35 kb from CEN5), the met6 locus (190 kb from CEN5) or the ade8 locus (800 kb from CEN4). We used an alternative tagging system, tetracycline (tetO)-operator18 arrays, to evaluate the contribution of sequence homology in the tag interactions. We integrated tandem arrays of 336 copies of tetO at the same loci as some of the lacO tags. In each case, direct visualization of the array was mediated by engineering strains expressing GFP-fused lacI or tet repressors, respectively. To confirm that tagging did not confer significant lethality, we monitored colony formation in tagged strains after fluorescence microscopy, and observed no significant reduction in viability (see Supplementary Information).
Table 1.
Yeast Strains
| Strain | Relevant genotype | Integrated plasmids | Source |
|---|---|---|---|
| MJL1026-2a | MATα ura3 lys2 ho::LYS2 leu2-Δ trp1::hisG | – | M. Lichten |
| MJL1025-9b | MATa ura3 lys2 ho::LYS2 leu2-Δ trp1::hisG | – | M. Lichten |
| K7022 |
MATa ade2 his3 trp1 leu2::(LEU2 tetR:GFP) ura3::(URA3 3×112tetO) |
– |
K. Nasmyth |
| LA120 | MATa/MATα ura3/ura3::(URA3 pDMC1:ECFP) his3 lys2 leu2 trp1 | pLA673 | This study |
| LA123 |
MATa/MATα ura3::(URA3 pDMC1:ECFP)/ura3::(URA3 lacI-GFP) trp1::(TRP1 256lacO)/trp1::(TRP1 256lacO) his3 leu2 |
pLA673, pLA671, pAS399 | This study |
| LA124 |
MATa/MATα ura3::(URA3 256lacO)/ura3::(URA3 pDMC1:ECFP) leu2::(LEU2 lacI:GFP) trp1/trp1::(TRP1 256lacO) his3 |
pAFS135, pLA673, pAS399 | This study |
| LA126 |
MATa/MATα ura3::(URA3 3×112tetO)/ura3::(URA3 lacI:GFP) his3 leu2::(LEU2 tetR:GFP) trp1/trp1::(TRP1 256lacO) ade2/ADE2 |
pLA671, pAS399 | This study |
| LA127 |
MATa/MATα his3::(HIS3 lacI:GFP) URA3/ura3::(URA3 256lacO) MET6/met6::(TRP1 URA3 256lacO) leu2 trp1 |
pAFS135, pAS584, pAS399 | This study |
| LA128 |
MATa/MATα ura3::(URA3 256lacO)/ura3::(URA3 lacI:GFP) ADE8/ade8::(LEU2 256lacO) his3 leu2 trp1 |
pLA675, pLA673, pAS399, pLA101 | This study |
| LA129 |
MATa/MATα ade2/ADE2 his3/his3::(HIS3 lacI:GFP) ura3/ura3::(URA3 3×112tetO) leu2/leu2::(LEU2 tetR:GFP) MET6/met6::(TRP1 URA3 256 lacO) |
pAFS135, pAS584, pAS399 | This study |
| LA130 |
MATa/MATα ade2/ADE2 his3/his3::(HIS3 lacI:GFP) ura3/ura3::(URA3 3×112tetO) leu2/leu2::(LEU2 tetR:GFP) ADE8/ade8::(LEU2 256lacO) |
pLA675, pLA673 pAS399, pLA101 |
This study |
Figure 1. Interchromosome-association assay and tag positions.
a, Schematic diagram of the assay. lacO arrays were introduced in different genomic positions and the associations between these loci were recorded. Association of lacO repeats is visible as a single fluorescent dot. b, Positions of the tagged loci: two centromere-linked sites (ura3 and trp1) and two distal sites (met6 and ade8). Chromosome numbers are shown at the top.
Visualization of homologue pairing in meiotic cells
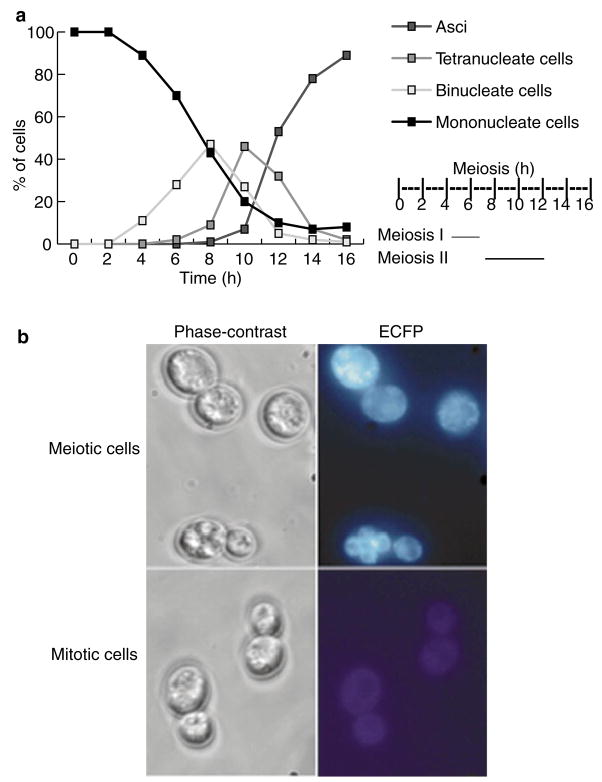
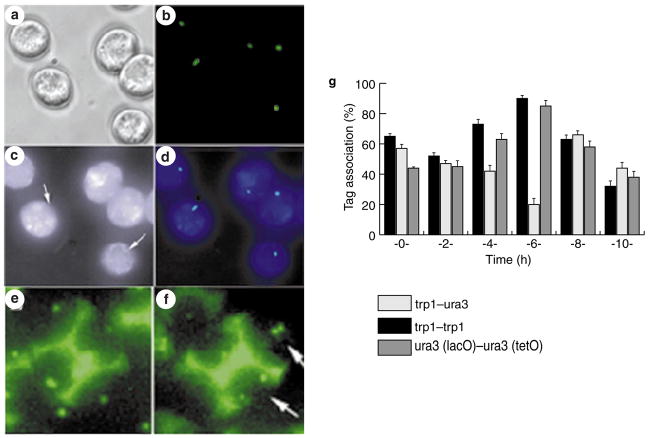
To test the reliability of the assay, we first examined its potential in meiosis. We developed a new approach using a cytoplasmic reporter of meiosis by placing a cyan fluorescent protein with a nuclear-export signal (ECFP-NES) under the control of a meiosis-specific promoter, pDMC1. Using this in vivo marker we were able to exclude cells that did not enter meiosis from our analysis. ECFP-positive cells (cyan-coloured) appeared 2–3 h after transfer to sporulation-inducing media, the time at which DMC1 transcription is induced19 (Fig. 2). The first binuclear cells appeared 4 h after induction of meiosis at 30 °C, and reached their highest levels at 8 h. Cells with four nuclei (meiosis II) appeared at 7 h and peaked at 10 h. Mature asci appeared at 10 h and by 16 h reached 89%, demonstrating efficient sporulation (Fig. 2). We compared the association of tags during meiosis between strain LA123, containing two lacO arrays at allelic positions (trp1 loci), and strain LA124, containing two lacO arrays at non allelic locations (trp1 and ura3 loci; Fig. 1). In LA123 cells, we detected association levels of up to 90 ± 3% after 6 h, whereas the levels for LA124 cells were down to 14 ± 5% (Fig. 3g). The concurrent increase in allelic and reduction in non-allelic associations after 6 h (Figs 2a and 3g), which was coincident with pachytene (as determined by chromosome spreading), demonstrates that the lacO-pairing assay is both sensitive and specific. We further tested the system using a strain, LA125, that was engineered to contain different DNA tags at allelic positions in both chromosome V homologues (ura3::lacO; ura3::tetO). The results were similar to those for LA123 cells (Fig. 3g), confirming that the pairing during meiosis is governed by a genome-wide bias that favours interaction between homologous chromosomes over ectopic interactions.
Figure 2. Time course of sporulation.
a, Synchronous meiotic time course (0–16 h after induction), as analysed by DAPI staining. The timing of meiosis I & II was calculated from the percentage of cells with one, two or four nuclei at the indicated times. b, Phase-contrast (left panels) and fluorescent (right panels) images of sporulating and vegetative LA120 cells with integrated pDMC1:ECFP–NES. Sporulating cells express ECFP–NES (blue), whereas vegetatively growing cells show no ECFP–NES expression.
Figure 3. In vivo chromosome association in meiosis.
a–d, Meiotic cells 6 h after transfer to sporulation media (SPM), carrying green fluorescent protein (GFP) tags at the trp1 loci in both homologues. a, Phase-contrast image; b, GFP chromosome tags shown in green; c, ECFP–NES expressed under control of the DMC1 promoter; arrows show the location of the nucleus; d, Colour overlay of b and c, showing GFP in green and ECFP in blue. e, f, Tetrads from a strain with both homologues tagged at the trp1 locus (e) or at one trp1 and one ura3 locus (f). Arrows indicate the segregation of GFP tags (green). g, Synchronous meiotic time-course analysis of interchromosome interactions 0–10 h after transfer to SPM. Trans-association between lacO–lacO (black bars, allelic; open bars, non-allelic) and lacO–tetO (grey bars) tags located at centromere-proximal sites (ura3 and trp1). Error bars represent s.d. Homologue pairing is detected at 4–6 h, at which time a reduction in non-allelic and an increase of allelic pairing is observed.
Somatic interchromosome association is dependent on several components
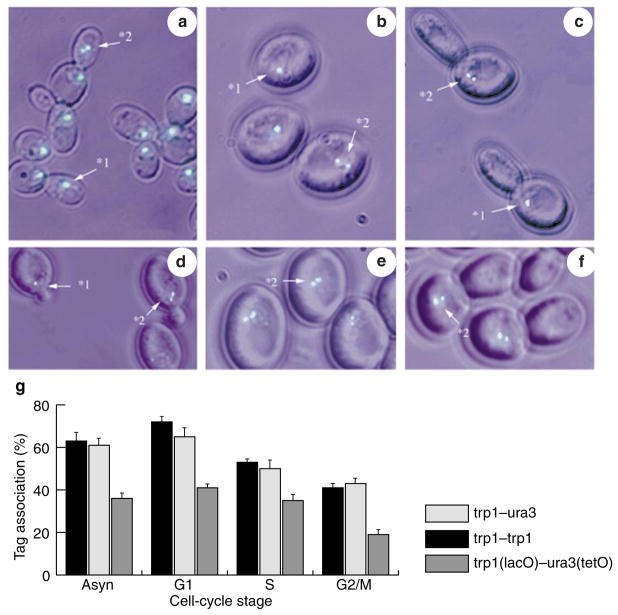
We used the same chromosomal tags to evaluate interchromosomal interaction in live, mitotically dividing (vegetative or somatic) cells. LA123 and LA124 cells exhibited marked differences with respect to their meiotic counterparts. In asynchronous culture, chromosomes in both strains showed high association levels of 63 ± 5% for LA123 and 61 ± 4% for LA124, as measured using lacO tags (Fig. 4). Remarkably, moving the tags to non-homologous locations had no effect on their association frequency, indicating that the underlying association mechanism may be distinct from meiotic pairing. To gain insight into the mechanism of this phenomenon, we investigated the components that contribute to the relatively high rate of ectopic interchromosomal associations.
Figure 4. In vivo association of GFP chromosome tags during the mitotic cycle.
a–f, Examples of yeast somatic cells at different stages of cell cycle, carrying lacO and tetO green fluorescent protein (GFP) tags. Images are colour overlays, showing GFP in green and phase contrast in cyan. a, Exponentially growing cells, with two trp1(lacO) GFP tags. b, Elutriated G1 cells, with two trp1(lacO) GFP tags. c, Cells sequentially treated with hydroxyurea and nocodazole, with trp1(lacO) and ura3(lacO) GFP tags. d, Elutriated S-phase cells, with ura3(lacO) and trp1(lacO) GFP tags. e, Elutriated G1 cells, with ura3(lacO) and met6(lacO) GFP tags. f, Cells sequentially treated with hydroxyurea and nocodazole, with trp1(lacO) and ura3(tetO) GFP tags. Numbers with arrows indicate how signals were scored: 1, associated; 2, separated. g, Interchromosome associations in somatic cells. Exponentially growing (Asyn) and synchronous (G1, S and G2/M) cultures were evaluated. Culture synchrony was achieved by elutriation (G1), or by elutriation followed by treatment with hydroxyurea (S), or nocodazole (G2/M). Allelic (black bars), non-allelic (open bars) and non-homologous lacO–tetO (grey bars, control) interactions are shown.
Using tag-association values for LA126 cells (trp1::lacO-ura3::tetO), we determined the importance of DNA-tag-sequence homology in the associations, which can be calculated as the difference between the association values for trp1::lacO-ura3::tetO (LA126) and trp1::lacO-ura3::lacO (LA124) cells (Fig. 4g, asyn). Over one-third of the interactions seen in LA124 cells may have originated from the DNA-sequence homology between tags. This indicates the possible existence of a genome-wide sequence-homology search mechanism that provides an opportunity for the association of short stretches of identical DNA sequences located on different chromosomes.
The polarized arrangement of interphase chromosomes in which centromeres are clustered on the opposite pole to telomeres, known as the Rabl configuration, has been clearly documented in S. cerevisiae14,15. Centromere clustering itself could mimic interchromosome interactions or could promote interaction between centromere-proximal sequences on different chromosomes even on non-homologous chromosomes, provided that they are equidistant from the corresponding centromeres. As both the trp1 and ura3 loci are centromere-proximal, this may account for some fraction of the observed associations. To evaluate the contribution of centromere clustering to trp1 and ura3 interactions, we used the microtubule-depolymerizing agent nocodazole, which disrupts spindle-pole-body-dependent clustering in fixed cells9,14. Chromosomal tags in both LA123 and LA124 cells showed similar association levels in the range of 40–42% (Fig. 4g, G2/M bars), showing that clustering of centromeres in vivo is responsible for some of the ectopic associations but not all. Nocodazole treatment of LA126 cells exhibits half fewer associations than identical treatment of LA123 and LA124 cells (Fig. 4g, G2/M), which further supports the idea that there exists a search mechanism for DNA-sequence homology. Cells arrested with hydroxyurea in S phase and treated with nocodazole exhibited identical reductions in tag association (data not shown); thus, it is microtubule depolymerization, and not mitotic arrest itself, that is the cause of the loss of centromere clustering in response to nocodazole treatment.
We further evaluated cell-cycle-dependent association of chromosome tags by analysing interactions at different stages of the cell cycle (Fig. 4g). The results show that association levels are indeed cell-cycle-dependent. Associations were at their highest level at the G1 stage (observed for elutriated unbudded cells) in all tag configurations (Fig. 4g). Extensive homologue association during G1 has previously been reported7,9. However, comparing association values for strains LA123, LA124 and LA126 (Figs 1b and 4g) shows that association between heterologous DNA tags (lacO and tetO) are also higher (Fig. 4g). Thus, the increased G1 interaction values are neither due to homologue pairing nor to ‘sequence-homology searches’, but rather represent an inherent property of this phase of the cell cycle. It is plausible that the absence of sister chromatids increases the potential of loci interaction. Cells arrested during S phase (with hydroxyurea) also exhibited a 20% decrease in association compared with G1 cells (Fig. 4g). This decrease in association may reflect changes in chromatin structure as a result of initiation of DNA replication20. Association between GFP tags was reduced by a further 10% when hydroxyurea arrest was followed by nocodazole treatment (Fig. 4c, f).
Our data regarding the somatic association between DNA tags indicate that clustering of centromeres polarizes chromosomes in vivo, promoting some associations between all loci at similar distances from centromeres. In addition, there is a mechanism that acts during the entire cell cycle that is able to associate loci of similar DNA-sequence composition.
Trans-association is independent of genomic location
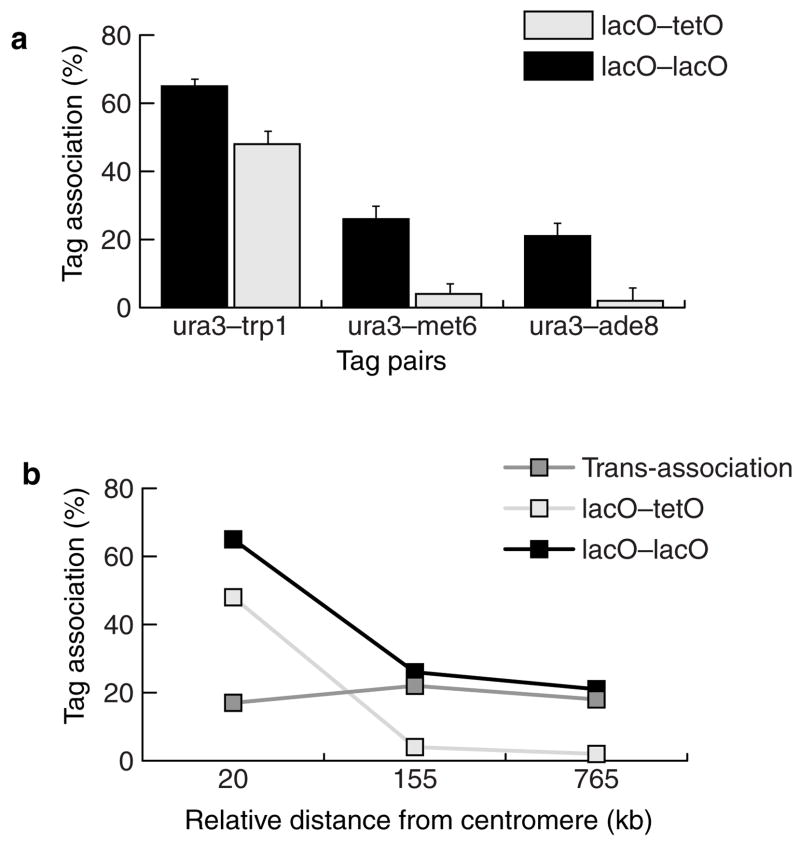
We defined trans-association as association between two given identical sequences, corrected for the sequence-homology-independent interaction. Subtracting the rate of lacO–tetO association from the corresponding rate of lacO–lacO association under the same experimental conditions and tag positions gives a measure of trans-association. As an example, the trans-association value for the trp1–ura3 pair would be the result of subtracting trp1::lacO–ura3::tetO interactions (48 ± 2%) from trp1::lacO–ura3::lacO interactions (69 ± 3%). Thus the trans-association value for this pair is 69 – 48 = 21%. Using this simple approach, we further investigated the properties of the ‘sequence-homology search’ phenomenon manifested in trans-association. We reasoned that if trans-association is mechanistically related to epigenetic and DNA-repair interactions, identical-sequence tags located at different physical distances from the centromeres would be expected to exhibit similar trans-association rates. We engineered four strains to contain either one lacO tag or one tetO tag at the ura3 locus and another lacO tag at the met6 locus (LA127, LA129), or at the ade8 locus (LA128, LA130; Table 1, Fig. 1b). We plotted the results against the relative distances of the tags from the centromeres (Fig. 5b). We included the association levels of LA124 cells (trp1::lacO–ura3::lacO) in the analysis to allow plotting of trans-association against tag-pair distances in relation to their corresponding centromeres (Fig. 5b). We obtained the trans-association component by subtracting the G1 lacO–tetO interaction values (strains LA126, LA129 and LA130) from the interactions of the corresponding lacO–lacO tags (strains LA124, LA127 and LA128). All the tag configurations yielded similar trans-association values (~20%), clearly demonstrating that trans-association of tags does not depend on their genomic location or on the sequences flanking them, but depends primarily on their DNA-sequence homology.
Figure 5. Trans-association of chromosomal tags.
a, Trans-association between lacO–lacO (black bars) and lacO–tetO (open bars) tags located at different centromere-to-locus distances. The tagged loci are ura3 (35 kb from CEN), trp1 (15 kb), met6 (190 kb) and ade8 (800 kb). lacO–tetO associations were used as a control to identify the subset of lacO–lacO associations that is due to sequence identity. b, Association between lacO–lacO (black points) repeats, lacO–tetO (open points) and trans-association (grey points), plotted as a function of relative centromere distance (subtraction of centromeric distances of two loci in each pair). Trans-association values were obtained by subtracting lacO–tetO associations from lacO–lacO.
Somatic interchromosome associations are distinct from meiotic pairing
Chromosome interactions in the mitotic nucleus are often described in the literature as somatic pairing, a term that implies the association of entire chromosomes with their homologous partners, as seen in meiotic prophase I. To test whether meiotic pairing and somatic interactions share a common mechanism, we investigated interchromosome tag interactions during premeiotic stages. From the time S. cerevisiae cells are transferred to sporulation medium (t = 0), they initiate the developmental programme in response to nutrient limitation21. We used LA123 and LA124 cells that were grown in presporulation media, and were thus synchronized at premeiotic G1 (ref. 7) to analyse this early stage of meiosis (Fig. 3g). Association levels were very similar to those observed for somatic G1 cells of the same strain (Figs 3g and 4g). Similarly, no differences in the level of association between pre-meiotic and mitotic S phases (Fig. 3g, 2h) were observed. Only at t = 6–8 h, which is coincident with meiotic pachytene, was a meiotic-specific mode of tag association clearly seen (Figs 2a and 3g). We further investigated this change in tag-association levels (at t = 6 h) using a strain, LA125, that was engineered to contain tags of different DNA sequences at allelic positions in both chromosome V homologues (ura3::lacO; ura3::tetO). During pachytene the allelic but heterologous tags behaved as if they were homologous loci (LA123; Fig. 3g), thus confirming that at t = 6 h there is a marked change in the mode of tag association. This change is manifested in switching from a DNA-tag-sequence bias in mitotic and pre-meiotic nuclei to a genome-wide bias that favours interaction between homologous chromosomes, that is, allelic loci, over non-homologous chromosomes. Thus, meiotic pairing is established in the course of execution of the meiotic programme and is not a consequence of the pre-existing phenomenon of somatic pairing. Our analysis of two modes of adhesion between DNA sequences in the nucleus of S. cerevisiae demonstrates they are both separated in time and distinct in nature (Fig. 6).
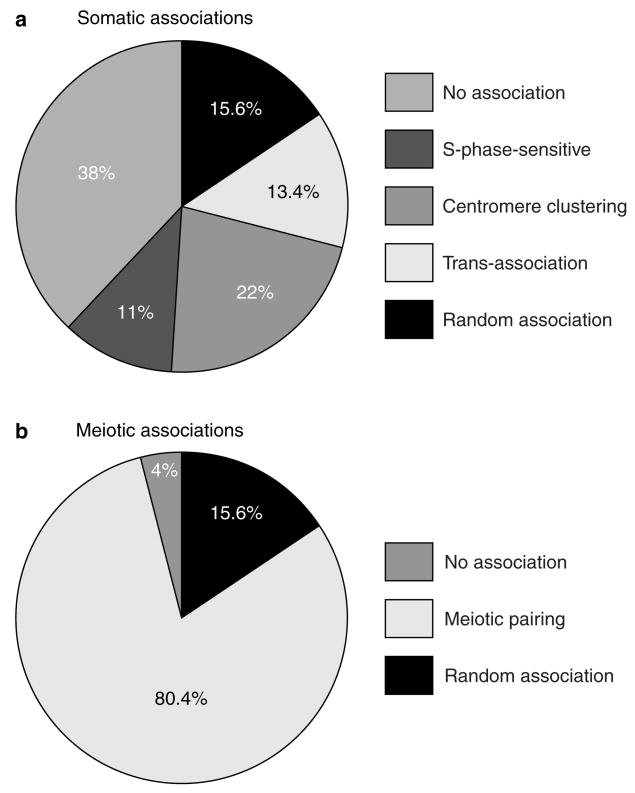
Figure 6. Dissection of somatic interchromosomal trans-association and meiotic homologue pairing.
a, Factors that contribute to somatic interchromosomal associations averaged for lacO–lacO interactions at allelic (trp1–trp1), and non-allelic centromere-linked (trp1–ura3) sites. Percentages were calculated as follows: random association, average lacO–tetO stochastic-collision values from ura3–trp1, ura3–met6 and ura3–ade8 associations (Fig. 5a); S-phase sensitive, subtraction of S values from G2/M values (Fig. 4g); centromere clustering, subtraction of G2/M values from G1 values (Fig. 4g); trans-association, average values obtained by subtracting lacO–tetO values from lacO–lacO values (Fig. 4g); no association, subtraction of G1 values from 100% (Fig. 4g). Some of the categories may overlap to some degree; for instance, both centromere clustering and trans-association may have an S-phase-sensitive component. b, Comparison of homologue and non-homologue interactions for meiotic cells. Percentages were calculated as follows: random, same values assumed as for a; no association, subtraction of lacO–lacO allelic pairing at t = 6 h from 100% (Fig. 3g); meiotic pairing, subtraction of random values from values for lacO–lacO allelic sites at t = 6 h (Fig. 3g).
Discussion
The aim of this study was to characterize somatic interchromosome interactions using live-cell imaging, and to compare them to meiotic chromosome-pairing in meiosis I. Using S. cerevisiae as a model organism, we found that the somatic interactions that are described in the literature by the collective term ‘somatic pairing’ are in fact composed of several organizational factors, including centromere clustering, cell-cycle-dependent alterations, and a new phenomenon, termed ‘trans-association’, that allows short stretches of homologous DNA sequences to find each other in the nucleus (Fig. 6). Dissection of somatic pairing and analysis of meiotic cells with the same set of tags have shown that the mode of interchromosome association is markedly different between vegetatively growing cells and those at meiosis I (Fig. 6).
What other processes could be dependent on somatic interchromosome associations based on sequence homology (trans-association)? Several normal cellular processes in somatic cells involve ectopic association of DNA segments. Such processes include recombinational repair22–25, annealing of broken DNA molecules26 and epigenetic regulation of gene expression27,28. In S. cerevisiae, the high level of spontaneous mitotic recombination has been proposed to correspond to recombinational repair of DNA damage. Rates of recombination between sequences located on allelic and non-allelic positions at similar distances from their corresponding centromeres are essentially the same3, but change substantially when the loci in the non-allelic situation are at different distances from the centromere (>370 kb difference, ref. 29). Our results show that ectopic recombination, although sensitive to chromosome latitude, occurs at lower frequencies between sites with greater locus-to-centromere distances. Recently, similar results have been obtained using an assay for Cre/loxP site-specific recombination13. Overall, these observations and the results presented here are consistent with the the idea that recombinational repair depends on trans-association mechanisms. In a study focused on the repair of double-strand breaks (DSBs) through single-strand annealing26, repair of DSBs was equally represented by two different mechanisms, involving reciprocal translocations or intrachromosomal deletions. This indicates that the broken ends of mitotic chromosomes may be free to search the entire genome for appropriate partners, which is consistent with our findings regarding the interchromosomal trans-association phenomenon.
Why is it that in non-meiotic yeast cells, small repetitive tags are reproducibly found in association with each other, but a stable pairing between the non-repetitive homologous chromosomes is not formed? At present, neither the mechanism of these associations nor the reason for their transient nature is understood. Interaction between lacI repressor molecules can be excluded, as the tetramerisation domain30 is deleted in the constructs used. We can, however, hypothesize that the repeated arrangement of DNA sequences (as in the tags used here) can promote and stabilize tag interactions, thus invoking a parallel with a variety of epigenetic phenomena. Recent discoveries have linked the recognition of nucleic-acid-sequence homology to the targeting of DNA methylation, chromatin remodelling and RNA turnover27,28. The interaction between repeated DNA sequences has been shown to trigger the formation and transmission of inactive genetic states. These include phenomena such as transvection31, position-effect variegation32 and post-transcriptional gene silencing33–35. In Neurospora crassa and Ascobolus immersus, phenomena known as repeat-induced point mutation (RIP, ref. 36) and methylation induced premeiotically (MIP, ref. 37) also result in pairing-dependent modification of DNA. The key element in these and other epigenetic phenomena is the recognition of homology between nucleic-acid sequences.
We have identified, through dissection of somatic association into its components (Fig. 6), a new element that drives a quarter of these interactions, in addition to previously known, but not well-characterized, centromere clustering. This phenomenon has a bias towards short, homologous sequences, and is independent of the genomic position of the interacting sequences. Such features make trans-association a conceptually different mechanism from meiotic pairing. The former is able to associate ectopic homologous DNA segments in non-homologous chromosomes (and within the same chromosome), whereas the latter must favour allelic interaction between homologous chromosomes. Therefore, a sweeping description of interchromosomal associations in budding yeast, such as ‘somatic pairing’, seems inaccurate, as this trans-association does not reflect the specific pairing of each allele between the two homologues, and is not a prelude to meiotic pairing. The functional significance of trans-association must be looked for in recombinational repair and/or epigenetic control of gene expression.
Methods
Yeast cultures, strains and cell-cycle methods
All strains used were isogenic with SK1. Haploid strains were used for integration of plasmid constructs. tetO tags were derived from K7022 by backcrossing five times to an SK1 strain; genotypes are shown in Table 1. Yeast transformations were carried out using the lithium-acetate method38. For all experiments, yeast were grown to mid-log phase at 23 °C or 30 °C in YPD (10 g l−1 yeast extract, 20 g l−1 BactoPeptone, 20 g l−1 dextrose, supplemented with 50 μg l−1 adenine). Complete synthetic medium lacking tryptophan, histidine and uracil (CSM-TRP-HIS-URA) was supplemented with 50 μg l−1 adenine, 50 μg l−1 leucine and 6.5 g l−1 sodium citrate. For experiments with synchronous cultures, cells were arrested with 15 μg l−1 nocodazole for 2 h, with 0.1 M hydroxyurea for 3 h, or with 0.1 M hydroxyurea for 2 h followed by nocodazole treatment for 1 h in the presence of 0.1 M hydroxyurea. Cultures were synchronized in G1 phase by centrifugal elutriation. Cells grown in YPD to ~107 cells ml−1 were loaded into a Beckman JE-5.0 elutriation chamber at a flow rate of 100 ml min−1 and centriguged at 4,000 r.p.m. in a Beckman Avanti J-20I centrifuge. Unbudded cells began eluting at 135 ml min−1 and 4,000 r.p.m. Samples (150 ml) were collected, pelleted and resuspended in either 15 μg l−1 nocodazole/YPD or 0.1 M hydroxyurea/YPD. After incubation at 23 °C for 2 h, nocodazole was added to hydroxyurea-arrested cells to a concentration of 15 μg l−1. Three hours after elutriation, aliquots from both cultures were briefly centrifuged, resuspended in 50 μl of the corresponding medium and visualized under the microscope. The effect of centrifugation on the 3D location of GFP tags was evaluated by comparing the association frequencies of trp1–ura3 tags in asynchronous cultures with and without centrifugation. Similar results were obtained for both situations (59% of tags were associated after centrifugation, compared to 61% when cells were not centrifuged). As microscopy was carried out at 23 °C, and in some experiments cells were grown at 30 °C, the effect of temperature was also tested on the same tag pair. Association frequency was identical at 23 °C and 30 °C. Fluorescence-activated cells sorting (FACS) analysis or DAPI staining of nuclear contents (data not shown) confirmed the synchrony of the cultures. For microscopy, cells were briefly centrifuged at 3,500 r.p.m. and resuspended in 50 μl of adequate medium. For meiotic experiments, YPD-grown cells were transferred to presporulation SPS media (5 g l−1 yeast extract, 10 g l−1 BactoPeptone, 1.7 g l−1 yeast nitrogen base, 10 g l−1 potassium acetate, 5 g l−1 ammonium sulphate and 0.5 g l−1 potassium biphthalate, pH 5.5) at 30 °C for 18 h. Cells were transferred to sporulation media (SPM, 10 g l−1 potassium acetate with 50 μg l−1 leucine). Meiotic time-course experiments were also carried out at 30 °C. Aliquots of sporulating cells were taken every 30 min, briefly centrifuged at 3,500 r.p.m. and resuspended in 50 μl SPM before visualization under the microscope. The timing of meiotic events was calibrated using ECFP–NES and nuclear morphology. The meiotic data presented in this study were collected from ECFP-positive cells.
Plasmids
All plasmids were propagated in E. coli strain TOP10 in medium containing 100 μg ml−1 ampicillin. The lacO GFP-based chromosome-tagging system was designed by modification of vectors pAFS59 and pAFS135 (ref. 17). Plasmid pAS399, containing the lacO repeat array, was created by ligating the BamHI–SalI fragment of pAFS59 into the BamHI and XhoI sites of pRS404 (ref. 39). Plasmid pLA671, containing the GFP:lacI:NLS fragment from pAFS135, was cloned as a polymerase chain reaction (PCR) product (primers: 5′-GCAAAGCTCGTCTAGATGAGTAAAGGAGAAGAACTTTTCAC and 5′-TGGCGGCCGCCTCGAGTTAGGCAACCTTTCTCTTCTTCTTTGGT) into the XbaI and XhoI sites of plasmid pRS406 (ref. 39). Plasmid pLA672 was created by ligation of the XbaI–XhoI fragment from pLA671 into the same sites of p416GPD (ref. 40), thereby placing the GFP:lacI:NLS fusion under the control of the GPD promoter. To place pAS399 at the met6 locus, pAS584 (containing a truncation of the MET6 gene and a URA3 marker) was used. pAS399 (ref. 41) was targeted to the β-lactamase gene of integrated pAS584. To place the lacO repeats at the ade8 locus, an internal ade8 PCR fragment (primers: 5′-GGGTTAACTAGAGCTCCGGACAATAATATCCCCACAAAGGTTTGT and 5′-TTGTGAAGCTGCTGTAAAACCTTATATGTAGCTTC) was cloned into the SacI–XhoI sites of pAF59, generating pLA101. To express the ECFP specifically in meiotic cells, pLA673 was constructed by cloning the DMC1 promoter (primers: 5′-CGCATGATATGGTACCCTGGAAGCGCCATTTTATAGCAAG and 5′-CAGTTCCTGTAAGCTTCATATTTGTTCAAATGTTTCCTGAT) and ECFP fused to a NES (primers: 5′-GCTTAAGCTTAATGAATTAGCCTTGAAATTAGCAGGTCTTGATATCAACAAGACAGTGAGCAAGGGCGAGGA and 5′-CGCGTGATCACAGTCGGGAAACCTGTCGTGCCAGCTGCATTAATGAATCGGCCAACTTGTACAGCTCGTCCA) into the BamHI–Asp718 sites of pRS406 (ref. 39).
The 256 lacO repeat was introduced into yeast by integration of pAS399 pAS584 (ref. 41) into either the ura3 locus (StuI site), which is located in the left arm of chromosome V, 35 kb from CEN5, the trp1 locus (PmlI site), which is located in the right arm of chromosome IV, 13 kb from CEN4, or the ampicillin fragment of pAS584 (ref. 41), which was previously integrated (BglII-mediated) at the met6 locus (ScaI site), 190 kb from CEN5. lacO was also integrated at the ade8 locus by integration of pLA101 (XhoI site). Expression of lacI–GFP was achieved either by integration of pLA671 in the ura3 locus (StuI site) or by transformation with pLA672. ECFP–NES was expressed by integration of pLA673 into the ura3 locus (StuI-targeted). In all cases plasmid integration was confirmed by PCR.
Microscopy
Live cells were imaged on a wide-field Zeiss AxioVert 135M microscope using a 100× 1.4 oil-immersion lens. The viability of strains containing tags was comparable to that of untagged strains after imaging. Fluorescence was visualized with standard excitation filters and a long-pass emission filter. Images were acquired with the MicroMax cooled charge-coupled device (CDD) camera (Princeton Instruments, Trenton, New Jersey) using IP-Lab software (Scanalytics, Fairfax, Virginia). Cells were imaged from 10–15 optical frames of 0.1–0.2 μm for each z section. Data was analysed on maximum-intensity projections of collected z sections; unclear cases were resolved by 3D analysis.
Chromosome pairing was evaluated by calculating the frequencies with which allelic and non-allelic GFP tags were associated, that is, appeared as a single fluorescent dot rather than two independent dots in a three-dimensional image (Fig. 1). Association frequencies for each category were calculated from at least 800 nuclei.
Supplementary Material
Acknowledgments
We thank M. Lichten, A. Straight, A. Murray, D. Koshland and K. Nasmyth for research materials, and A. Wolffe, E. Ballestar, M. Dasso, O. Cohen-Fix, L. Freeman, F. Urnov, C. P. Lichtestein and A. R. Leitch for ideas and comments on the manuscript. L.A-A. was supported by the NICHD Intramural Research Training Award and in part by the BBSRC.
References
- 1.Csink AK, Henikoff S. Genetic modification of heterochromatic association and nuclear organization in Drosophila. Nature. 1996;381:529–531. doi: 10.1038/381529a0. [DOI] [PubMed] [Google Scholar]
- 2.Dorer DR, Henikoff S. Transgene repeat arrays interact with distant heterochromatin and cause silencing in cis and trans. Genetics. 1997;147:1181–1190. doi: 10.1093/genetics/147.3.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lichten M, Haber JE. Position effects in ectopic and allelic mitotic recombination in Saccharomyces cerevisiae. Genetics. 1989;123:261–268. doi: 10.1093/genetics/123.2.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kassis JA, VanSickle EP, Sensabaugh SM. A fragment of engrailed regulatory DNA can mediate transvection of the white gene in Drosophila. Genetics. 1991;128:751–761. doi: 10.1093/genetics/128.4.751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Haber JE, et al. Physical monitoring of meiotic and mitotic recombination in yeast. Prog Nucleic Acids Res Mol Biol. 1988;35:209–259. [PubMed] [Google Scholar]
- 6.Loidl J, Klein F, Scherthan H. Homologous pairing is reduced but not abolished in asynaptic mutants of yeast. J Cell Biol. 1994;125:1191–1200. doi: 10.1083/jcb.125.6.1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weiner BM, Kleckner N. Chromosome pairing via multiple interstitial interactions before and during meiosis in yeast. Cell. 1994;77:977–991. doi: 10.1016/0092-8674(94)90438-3. [DOI] [PubMed] [Google Scholar]
- 8.Scherthan H, Bahler J, Kohli J. Dynamics of chromosome organization and pairing during meiotic prophase in fission yeast. J Cell Biol. 1994;127:273–285. doi: 10.1083/jcb.127.2.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Burgess SM, Kleckner N, Weiner BM. Somatic pairing of homologs in budding yeast: existence and modulation. Genes Dev. 1999;13:1627–1641. doi: 10.1101/gad.13.12.1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bass HW, Marshall WF, Sedat JW, Agard DA, Cande WZ. Telomeres cluster de novo before the initiation of synapsis: a three-dimensional spatial analysis of telomere positions before and during meiotic prophase. J Cell Biol. 1997;137:5–18. doi: 10.1083/jcb.137.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Scherthan H, et al. Centromere and telomere movements during early meiotic prophase of mouse and man are associated with the onset of chromosome pairing. J Cell Biol. 1996;134:1109–1125. doi: 10.1083/jcb.134.5.1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kleckner N, Weiner BM. Potential advantages of unstable interactions for pairing of chromosomes in meiotic, somatic, and premeiotic cells. Cold Spring Harb Symp Quant Biol. 1993;58:553–565. doi: 10.1101/sqb.1993.058.01.062. [DOI] [PubMed] [Google Scholar]
- 13.Burgess SM, Kleckner N. Collisions between yeast chromosomal loci in vivo are governed by three layers of organisation. Genes Dev. 1999;13:1871–1883. doi: 10.1101/gad.13.14.1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guacci V, Hogan E, Koshland D. Centromere position in budding yeast: evidence for anaphase A. Mol Biol Cell. 1997;8:957–972. doi: 10.1091/mbc.8.6.957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jin Q, Trelles-Sticken E, Scherthan H, Loidl J. Yeast nuclei display prominent centromere clustering that is reduced in nondividing cells and in meiotic prophase. J Cell Biol. 1998;141:21–29. doi: 10.1083/jcb.141.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gotta M, et al. The clustering of telomeres and colocalization with Rap1, Sir3, and Sir4 proteins in wild-type Saccharomyces cerevisiae. J Cell Biol. 1996;134:1349–1363. doi: 10.1083/jcb.134.6.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Straight AF, Belmont AS, Robinett CC, Murray AW. GFP tagging of budding yeast chromosomes reveals that protein–protein interactions can mediate sister chromatid cohesion. Curr Biol. 1996;6:1599–1608. doi: 10.1016/s0960-9822(02)70783-5. [DOI] [PubMed] [Google Scholar]
- 18.Michaelis C, Ciosk R, Nasmyth K. Cohesins: chromosomal proteins that prevent premature separation of sister chromatids. Cell. 1997;91:35–45. doi: 10.1016/s0092-8674(01)80007-6. [DOI] [PubMed] [Google Scholar]
- 19.Bishop DK, Park D, Xu L, Kleckner N. DMC1: a meiosis-specific yeast homolog of E. coli recA required for recombination, synaptonemal complex formation, and cell cycle progression. Cell. 1992;69:439–456. doi: 10.1016/0092-8674(92)90446-j. [DOI] [PubMed] [Google Scholar]
- 20.Li G, Sudlow G, Belmont AS. Interphase cell cycle dynamics of a late-replicating, heterochromatic homogeneously staining region: precise choreography of condensation/decondensation and nuclear positioning. J Cell Biol. 1998;140:975–989. doi: 10.1083/jcb.140.5.975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mitchell AP. Control of meiotic gene expression in Saccharomyces cerevisiae. Microbiol Rev. 1994;58:56–70. doi: 10.1128/mr.58.1.56-70.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haber JE, Ray BL, Kolb JM, White CI. Rapid kinetics of mismatch repair of heteroduplex DNA that is formed during recombination in yeast. Proc Natl Acad Sci USA. 1993;90:3363–3367. doi: 10.1073/pnas.90.8.3363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jinks-Robertson S, Michelitch M, Ramcharan S. Substrate length requirements for efficient mitotic recombination in Saccharomyces cerevisiae. Mol Cell Biol. 1993;13:3937–3950. doi: 10.1128/mcb.13.7.3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Inbar O, Kupiec M. Homology search and choice of homologous partner during mitotic recombination. Mol Cell Biol. 1999;19:4134–4142. doi: 10.1128/mcb.19.6.4134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Melamed C, Kupeiec M. Effect of donor copy number on the rate of gene conversion in the yeast Saccharomyces cerevisiae. Mol Gen Genet. 1992;235:97–103. doi: 10.1007/BF00286186. [DOI] [PubMed] [Google Scholar]
- 26.Haber JE, Leung W. Lack of chromosome territoriality in yeast: promiscuous rejoining of broken chromosome ends. Proc Natl Acad Sci USA. 1996;93:13949–13954. doi: 10.1073/pnas.93.24.13949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wolffe A, Matzke MA. Epigenetics: regulation through repression. Science. 1999;286:481–486. doi: 10.1126/science.286.5439.481. [DOI] [PubMed] [Google Scholar]
- 28.Henikoff S. Nuclear organization and gene expression: homologous pairing and long-range interactions. Curr Opin Cell Biol. 1997;9:388–395. doi: 10.1016/s0955-0674(97)80012-9. [DOI] [PubMed] [Google Scholar]
- 29.Jinks-Robertson S, Petes TD. Chromosomal trans-locations generated by high-frequency meiotic recombination between repeated yeast genes. Genetics. 1986;114:731–752. doi: 10.1093/genetics/114.3.731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lewis M, et al. Crystal structure of the lactose operon repressor and its complexes with DNA and inducer. Science. 1996;271:1247–1254. doi: 10.1126/science.271.5253.1247. [DOI] [PubMed] [Google Scholar]
- 31.Lewis EB. The theory and application of a new method of detecting chromosomal rearrangements in Drosophila melanogaster. Am Nat. 1954;88:225–239. [Google Scholar]
- 32.Judd BH. Transvection: allelic cross talk. Cell. 1988;53:841–843. doi: 10.1016/s0092-8674(88)90209-7. [DOI] [PubMed] [Google Scholar]
- 33.Wassenegger M, Pelissier T. A model for RNA-mediated gene silencing in higher plants. Plant Mol Biol. 1998;37:349–362. doi: 10.1023/a:1005946720438. [DOI] [PubMed] [Google Scholar]
- 34.Cogoni C, Macino G. Isolation of quelling-defective (qde) mutants impaired in posttranscriptional transgene-induced gene silencing in Neurospora crassa. Proc Natl Acad Sci USA. 1997;94:10233–10238. doi: 10.1073/pnas.94.19.10233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tabara H, Grishok A, Mello CC. RNAi in C. elegans: soaking in the genome sequence. Science. 1998;282:430–431. doi: 10.1126/science.282.5388.430. [DOI] [PubMed] [Google Scholar]
- 36.Selker EU. Epigenetic phenomena in filamentous fungi: useful paradigms or repeat-induced confusion? Trends Genet. 1997;13:296–301. doi: 10.1016/s0168-9525(97)01201-8. [DOI] [PubMed] [Google Scholar]
- 37.Rossignol JL, Faugeron G. MIP: an epigenetic gene silencing process in Ascobolus immersus. Curr Top Microbiol Immunol. 1995;197:179–191. doi: 10.1007/978-3-642-79145-1_12. [DOI] [PubMed] [Google Scholar]
- 38.Ito H, Fukuda Y, Murata K, Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983;153:163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mumberg D, Muller R, Funk M. Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene. 1995;156:119–122. doi: 10.1016/0378-1119(95)00037-7. [DOI] [PubMed] [Google Scholar]
- 41.Freeman L, Aragón-Alcaide L, Strunnikov A. The condesin complex governs chromosome condensation and mitotic transmission of rDNA. J Cell Biol. 2000;149:811–824. doi: 10.1083/jcb.149.4.811. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.