Abstract
Pref-1 is an EGF-repeat containing transmembrane protein that produces a biologically active soluble form by TACE mediated cleavage. Although Pref-1 inhibition of adipogenesis has been well established, the specific target(s) of Pref-1 or the Pref-1 function in mesenchymal cell commitment/differentiation are not known. Here, we show that Sox9 downregulation is required for adipocyte differentiation and that Pref-1 inhibits adipocyte differentiation through upregulating Sox9 expression. Sox9 directly binds to the promoter regions of C/EBPβ and C/EBPδ to suppress their promoter activity, preventing adipocyte differentiation. Furthermore, we also show that by inducing Sox9, Pref-1 promotes chondrogenic induction of mesenchymal cells but prevents chondrocyte maturation as well as osteoblast differentiation, with supporting in vivo evidence in Pref-1 null and Pref-1 transgenic mice. Thus, Sox9 is a Pref-1 target and Pref-1 directs multipotent mesenchymal cells to the chondrogenic lineage but inhibits differentiation into adipocytes as well as osteoblasts and chondrocytes.
Keywords: Pref-1/dlk1, adipocyte differentiation, Sox9, chondrocyte, osteoblast
INTRODUCTION
Preadipocyte factor 1 (Pref-1) was originally cloned by us as a preadipocyte transmembrane protein that inhibits adipogenesis (Smas and Sul, 1993). The full-length form of Pref-1 (Pref-1A) is a protein of 385 amino acids. Pref-1 contains an extracellular domain with 6 EGF-like repeats, a juxtamembrane region, a single transmembrane domain and a short cytoplasmic tail (Smas et al., 1994). The extracellular domain of Pref-1 is cleaved from Pref-1 by A Disintegrin And Metalloproteinase 17 (ADAM17, TNF-α Converting Enzyme (TACE)) to generate the biologically active soluble Pref-1 (Mei et al., 2002; Smas et al., 1997); (Wang and Sul, 2006). Pref-1 mRNA and protein levels are high in 3T3-L1 preadipocytes but Pref-1 expression decreases during adipocyte differentiation, and is absent in mature adipocytes. Dexamethasone (DEX), a component of adipogenic agents routinely used for adipocyte differentiation, suppresses Pref-1 expression. Thus, Pref-1 is commonly used as a unique preadipocyte marker. Constitutive expression of Pref-1 inhibits adipocyte differentiation and absence of Pref-1 accelerates/increases the degree of adipocyte differentiation in cultured cells (Smas et al., 1999; Smas and Sul, 1993). Although the direct target of Pref-1 is not known, Pref-1 activates ERK/MAPK to inhibit adipocyte differentiation (Kim et al., 2007). The function of Pref-1 in adipogenesis in vivo has been firmly established. Pref-1 null mice display enhanced adipose tissue formation with higher expression of adipocyte markers, showing increased circulating lipid levels, a characteristic usually associated with obesity (Moon et al., 2002). Conversely, overexpression of Pref-1 in adipose tissue decreases adipose tissue mass and impairs adipocyte differentiation (Lee et al., 2003). In adults, Pref-1 expression is restricted mainly to preadipocytes of adipose tissue, although Pref-1 is detected in some neuroendocrine cell types and was identified as a gene (dlk1) expressed in neuroblastoma cells (Laborda et al., 1993). During embryonic development, however, Pref-1 is widely expressed in multiple embryonic tissues including liver, lung and developing vertebra. Thus, the soluble form of Pref-1 has been purified from fetal circulation and named as fetal antigen 1(FA1) (Bachmann et al., 1996). Pref-1 is coded by a paternally expressed imprinted gene and a majority of imprinted genes code for proteins that regulate fetal growth and organogenesis (da Rocha and Ferguson-Smith, 2004). Although Pref-1 function in embryonic development has not been studied, we observed defects in bone development in both Pref-1 null and Pref-1 transgenic mice (Lee et al., 2003; Moon et al., 2002).
Multipotent mesenchymal cells can commit and differentiate into various cell types, including adipocytes, chondrocytes and osteoblasts (Gesta et al., 2007). Adipogenesis occurs late in embryonic development and in postnatal periods. C/EBPα and PPARγ play critical roles in adipogenesis and induction of adipocyte markers including adipocyte fatty acid binding protein (aP2/FABP4) (Farmer, 2006; Gregoire et al., 1998; Rosen and MacDougald, 2006). Chondrogenesis and osteogenesis during skeletal development, however, occur in early embryonic stages. Chondrogenesis occurs from mesenchymal condensations and osteogenesis of some bones occurs directly from mesenchymal condensations by intramembranous ossification. Most bones, however, are formed by endochondral ossification: Mesenchymal condensations first become immature chondrocytes that undergo maturation to become hypertrophic, when some of the surrounding mesenchymal cells invade the zone of hypertrophic chondrocytes to become osteoblasts. Sox9 is required for mesenchymal condensation and for initiation of early chondrogenesis to generate immature chondrocytes that express cartilaginous matrix proteins, such as type II and IX collagens (Col), and aggrecan (Akiyama et al., 2004; Bi et al., 2001). Thus, Sox9 is expressed in all chondro-osteoprogenitor cells and in mesenchymal condensations and promotes chondrogenic commitment. During chondrocyte hypertrophy and early osteogenesis, Runx2 is upregulated and Col X and osteocalcin are induced (Takeda et al., 2001). However, Sox9 downregulation is a prerequisite for chondrocyte maturation and osteogenesis. Sox9, therefore, plays a critical role in skeletogenesis. Since Sox9 is found in chondro-osteoprogenitor cells and regulates skeletogenesis, it is possible that Sox9 may also affect differentiation of adipocytes that are derived from the common mesenchymal cells, but this possibility has not been examined before.
The present study was to elucidate the molecular target and effect on mesenchymal cell commitment/differentiation by Pref-1. Here, we show that Sox9 directly binds to C/EBPβ and C/EBPδ promoters to suppress their promoter activity, preventing adipocyte differentiation. Thus, constitutive Sox9 expression inhibits, and Sox9 knockdown enhances adipocyte differentiation. More importantly, Pref-1 inhibition of adipose conversion is by preventing Sox9 downregulation. Furthermore, by inducing Sox9, Pref-1 promotes chondrogenic induction of mesenchymal cells to generate immature chondrocytes, but Pref-1 inhibits chondrocyte maturation as well as osteoblast differentiation. Overall, by inducing Sox9, Pref-1 promotes the chondrogenic commitment of mesenchymal cells, but inhibits adipocyte, chondrocyte and osteoblast differentiation.
RESULTS
Sox9 downregulation is required for adipocyte differentiation and Pref-1 inhibition is through Sox9 regulation
During the search for Pref-1 targets by microarray analysis and other candidate approaches, we detected regulation of Sox9 expression by Pref-1. We found that treatment of Pref-1 null MEFs with Pref-1-hFc increased Sox9 expression rapidly by 4-fold within 1hr and this high level of expression was maintained for up to 4 hrs (Fig. 1A). A similar increase in Sox9 at the protein level is detected also. Pref-1 treatment also increased Sox9 expression in multipotent mesenchymal C3H10T1/2 cells as well as in 3T3-L1 preadipocytes (data not shown). Expression of other factors with documented effects on embryonic development such as Smad1 and Smad6 for TGFβ BMP family remained the same upon Pref-1 treatment. TNFα, a cytokine which can inhibit adipocyte differentiation also was ineffective in upregulating Sox9, suggesting that upregulation of Sox9 is Pref-1 specific. We previously have shown that Pref-1 inhibition of adipocyte differentiation is through Pref-1 mediated ERK1/2 activation. We therefore examined whether ERK1/2 phosphorylation is required for Pref-1 induction of Sox9 (Fig. 1B). As previously shown, Pref-1 treatment transiently increased ERK1/2 phosphorylation at 30 min of Pref-1 treatment in Pref-1 null MEFs but decreased to basal levels after 4 hrs. In these cells, we detected an increase in Sox9 protein level 30 min after Pref-1 treatment and the Sox9 level was further increased at 4 hr. Thus, ERK phosphorylation/activation by Pref-1 appears to precede Sox9 induction. When we treated these cells with MEK inhibitors, PD98059 or U0126, we did not observe any increase in Sox9 levels upon Pref-1 treatment. Similarly, siRNA mediated knockdown of ERK1/2 prevented the increase in Sox9 level by Pref-1 treatment. Overall, these results demonstrate that, by activating ERK1/2, Pref-1 induces Sox9 expression.
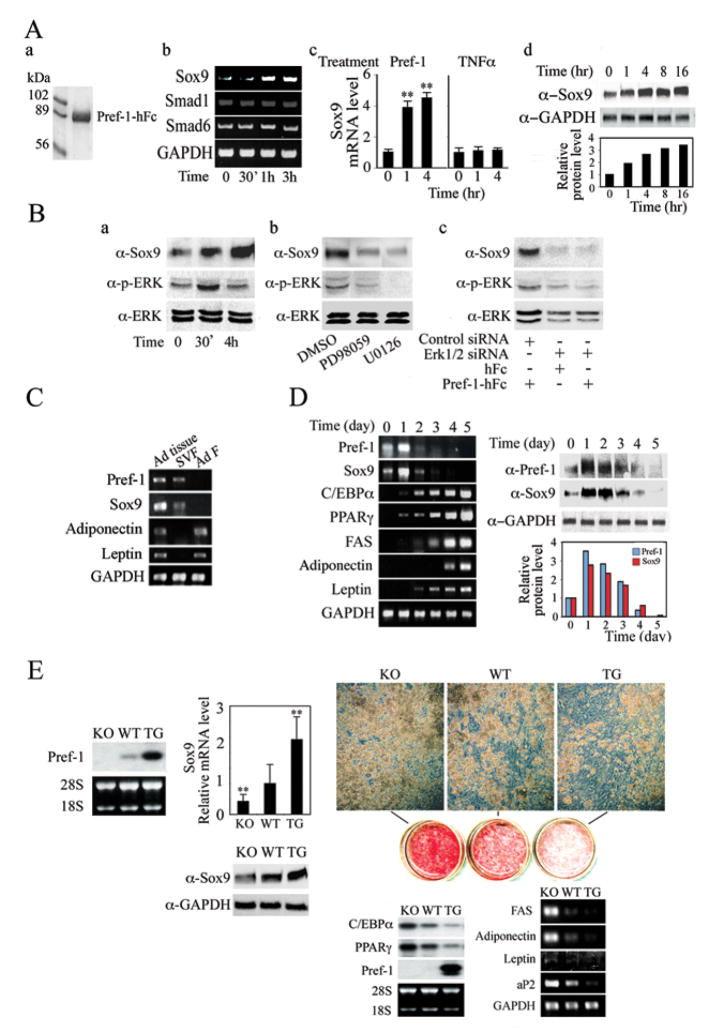
Figure 1.
(A) Regulation of Sox9 expression by Pref-1. Coomassie staining of purified Pref-1-hFc (a). RT-PCR (b), RT-qPCR (c) and Western blotting (d) in Pref-1 null MEFs treated with 50 nM Pref-1-hFc or 50 ng/ml TNFα. Results are mean ± SEM; **, P<0.01 compared to that before treatment. (B) Western blotting of cells with Pref-1-hFc treatment (a), ERK1/2 inhibitor (b), and ERK1/2 siRNA transfection (c). (C) RT-PCR for adipose tissue (Ad tissue), stromal vascular fraction (SVF) and adipocyte fraction (Ad F). (D) Pref-1 regulates Sox9 expression during adipocyte differentiation. (E) Comparison of adipocyte differentiation of MEFs from Pref-1 null, wild-type and aP2-Pref-1 transgenic embryos. Northern blot (left panel), RT-qPCR, and Western blot analysis (middle panels) in MEFs at confluence. The value for wild-type MEFs was defined as 1. Results are mean ± SEM; **, P < 0.01 compared to wild-type MEFs. Microscopic morphology, Oil red O staining (right upper panels) and adipogenic transcription factors by Northern blotting and late adipocyte markers by RT-PCR 8 days after induction of adipocyte differentiation (right lower panels).
Expression of Sox9 was easily detected in adipose tissue (Fig. 1C). However, unlike adipocyte markers such as adiponectin and leptin, Sox9 mRNA was not detected in adipocyte fraction, but was found in stromal vascular fraction that contains preadipocytes as well as other cell types such as macrophages/endothelial cells. Interestingly, Sox9 mRNA levels were lower in adipose tissue from high fat diet-induced as well as genetically obese ob/ob mice (Suppl. Fig. 2). In this regard, mRNA levels of preadipocyte marker, Pref-1, were lower in adipose tissue from these obese mouse models whereas mRNA levels of various adipocyte markers such as C/EBPα, PPARγ and leptin were higher. We next examined Sox9 expression during MEF differentiation into adipocytes (Fig. 1D). As expected, upon treatment with adipogenic agents, DEX/MIX, there was a time dependent increase in expression levels of adipogenic transcription factors, C/EBPα and PPARγ as well as other adipocyte markers including FAS, adiponectin and leptin, as MEFs underwent adipocyte differentiation. As we have reported previously, unlike the decrease observed during 3T3-L1 adipocyte differentiation, during MEF differentiation Pref-1 expression displayed a transient increase at Day 1, but rapidly decreased and was not detectable at Day 5 after conversion into adipocytes. Interestingly, we found that the Sox9 expression also was transiently increased and then decreased during MEF differentiation, closely following the Pref-1 expression. Protein levels of Sox9 and Pref-1 showed a similar pattern although the decrease was slower than that observed for mRNA levels (Fig. 1D right panels). The difference in the time course of Pref-1 downregulation during adipocyte differentiation between MEFs and 3T3-L1 cells has been noticed previously (Kim et al., 2007; Smas et al., 1999). In this regard, unlike 3T3-L1 cells that are committed to adipocyte lineage, MEFs are multipotent mesenchymal cells and Pref-1 levels may increase during commitment to adipocyte lineage when these cells are treated with adipogenic agents. Likewise, Sox9 expression may be transiently induced in this culture condition. Regardless, the parallel patterns of Pref-1 and Sox9 expression support a notion that Pref-1 regulates Sox9 expression, thereby inhibiting adipocyte differentiation.
We next compared Sox9 expression in MEFs from Pref-1 null, wild-type and aP2-Pref-1 transgenic mice (Fig. 1E). Correlating with Pref-1 expression, Sox9 mRNA and protein levels were 50% lower in Pref-1 null MEFs than wild-type MEFs, while those in Pref-1 transgenic MEFs were approximately 2-fold higher than in wild-type MEFs. After treatment with adipognic agents, 80% of Pref-1 null MEFs differentiated into adipocytes, while 50% of wild-type MEFs, and 20% of Pref-1 transgenic MEFs differentiated as judged by the rounded adipocyte morphology. Oil red O staining for lipid accumulation displayed corresponding differences in the degree of adipocyte differentiation. Likewise, following differentiation, mRNA levels of C/EBPα and PPARγ in Pref-1 null MEFs were 2-fold higher than those in wild-type MEFs, and levels in Pref-1 transgenic MEFs were 65–70% lower than in wild-type MEFs, reflecting the differences in adipocyte differentiation. As expected, Pref-1 expression was not detected in Pref-1 null MEFs and was completely suppressed in wild-type MEFs during adipocyte differentiation. On the other hand, Pref-1 transgenic MEFs showed a high level of Pref-1 expression due to inhibition of differentiation in addition to expression from the transgene. Overall, these results show that Pref-1 and Sox9 expression changes in a parallel fashion during differentiation and is inversely correlated with the degree of differentiation of MEFs into adipocytes.
We next employed soluble Pref-1-hFc in Pref-1 null MEFs to test whether inhibition of adipocyte differentiation by Pref-1 accompanies changes in Sox9 expression (Fig. 2A). Pref-1-hFc caused a marked decrease in adipocyte differentiation as judged by Oil red O staining and expression of adipocyte markers including adiponectin and leptin. In control hFc treated cells, C/EBPα and PPARγ levels increased by 10- and 5-fold, respectively, while in Pref-1-hFc treated cells, C/EBPα levels increased by only 1.8-fold and PPARγ expression did not change. We found that Sox9 expression was drastically decreased by 90% during adipocyte differentiation in control hFc treated cells, whereas in Pref-1-hFc treated cells, Sox9 expression did not change and remained at levels seen prior to treatment with adipogenic agents. We also examined Pref-1 and Sox9 expression during differentiation of wild-type MEFs into adipocytes (Suppl. Fig. 3). Pref-1 and Sox9 mRNA levels increased approximately 3-fold at Day 1 (after DEX/MIX treatment), and then decreased precipitously to very low levels upon differentiation. In cells treated with Pref-1-hFc, the expression levels also decreased, but slowly and not below those in preadipocytes. Overall, these results show that, during Pref-1 inhibition of adipocyte differentiation of MEFs, Pref-1 prevents the decrease in Sox9 expression that normally occurs during adipose conversion.
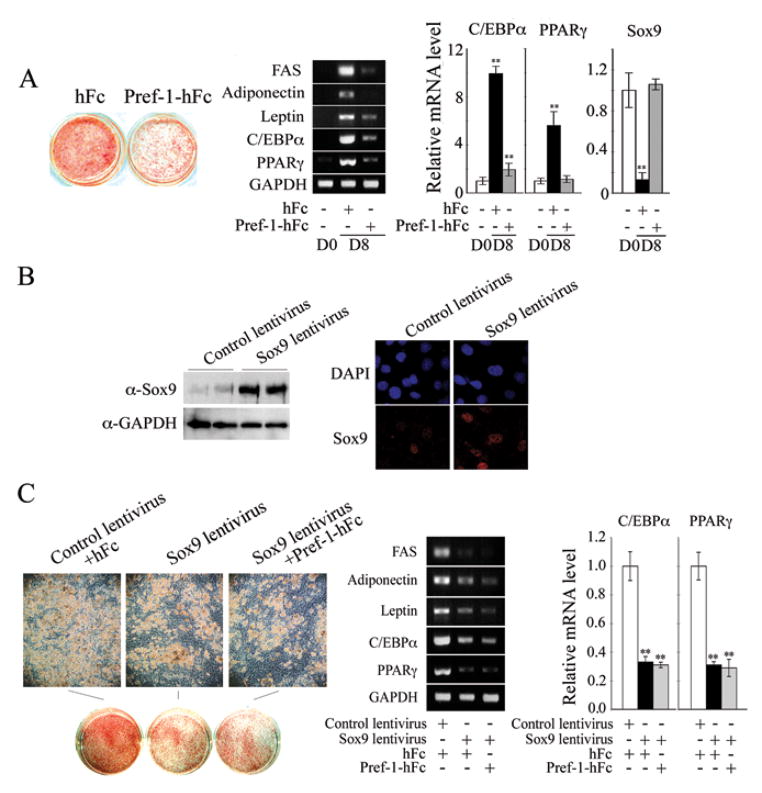
Figure 2.
(A) Pref-1 null MEFs were subjected to adipocyte differentiation in the presence of Pref-1-hFc or control hFc for 8 days. Oil red O staining (left panel), RT-PCR (middle panel) and RT-qPCR (right panel). (B) Expression level of Sox9 was verified by Western blotting (left panel) and by immunofluorescence 48 hrs after infection (right panel). (C) Pref-1 null MEFs overexpressing Sox9 by lentivirus infection were subjected to adipocyte differentiation in the presence of Pref-1-hFc or control hFc. Results are mean ± SEM; **, P < 0.01 compared to differentiated cells infected with control lentivirus or in the presence of hFc.
Next, we tested the role of Sox9 in adipocyte differentiation of Pref-1 null MEFs by transfecting Sox9 lentivirus. The high levels of Sox9 in nucleus after lentivirus infection was first verified (Fig. 2B) and Sox9 expression driven by the CMV promoter would remain the same during differentiation. Upon treatment of Pref-1 null MEFs with adipogenic agents, more than 80% of cells infected with control empty lentivirus differentiated into adipocytes, whereas only 40% of cells infected with Sox9 lentivirus differentiated, as judged by morphology and expression of the adipocyte markers FAS, adiponectin and leptin, and adipogenic transcription factors. Thus, Sox9 lentivirus infected cells had 70% lower C/EBPα and PPARγ levels compared to empty lentivirus infected cells after adipose conversion (Fig. 2C). We conclude that constitutive overexpression of Sox9 inhibits adipocyte differentiation. Furthermore, Sox9 lentivirus infected cells treated with Pref-1-hFc showed a similar degree of adipocyte differentiation to that observed in cells infected with Sox9 lentivirus alone. This observation indicates that Pref-1 and Sox9 did not have additive effects on adipocyte differentiation, suggesting a common underlying mechanism for inhibition of adipocyte differentiation.
We next employed 3T3-L1 cells to test the role of Pref-1 and Sox9 in adipocyte differentiation. As expected, during 3T3-L1 differentiation, various adipocyte markers such as FAS, adiponectin and leptin increased late during differentiation, while the increase in C/EBPα and PPARγ expression was detected somewhat earlier and continued to increase up to day 5 (Fig. 3Aa). Expression of C/EBPβ and C/EBPδ increased even earlier by 16 and 10-fold respectively, 24 hr after the addition of adipogenic agents (Fig. 3Ab). On the other hand, both Pref-1 and Sox9 expression decreased in parallel by 50% 12 hr after the addition of adipogenic agents, and further decreased by 80 to 90% by Day 2 (Fig. 3Ac). The decrease in Pref-1 at the protein level appears to precede that of Sox9 (Fig. 3c bottom panel). We have previously reported that, of the two adipogenic agents, DEX but not MIX suppresses Pref-1 expression. Similarly, we found that DEX but not MIX effectively decreased Sox9 mRNA levels by 70% after 12 hrs of treatment (Fig. 3Ad). We conclude that suppression of Pref-1 and Sox9 is an early event that precedes C/EBPα and PPARγ induction but better coincides, and in fact, precedes C/EBPβ C/EBPδ induction.
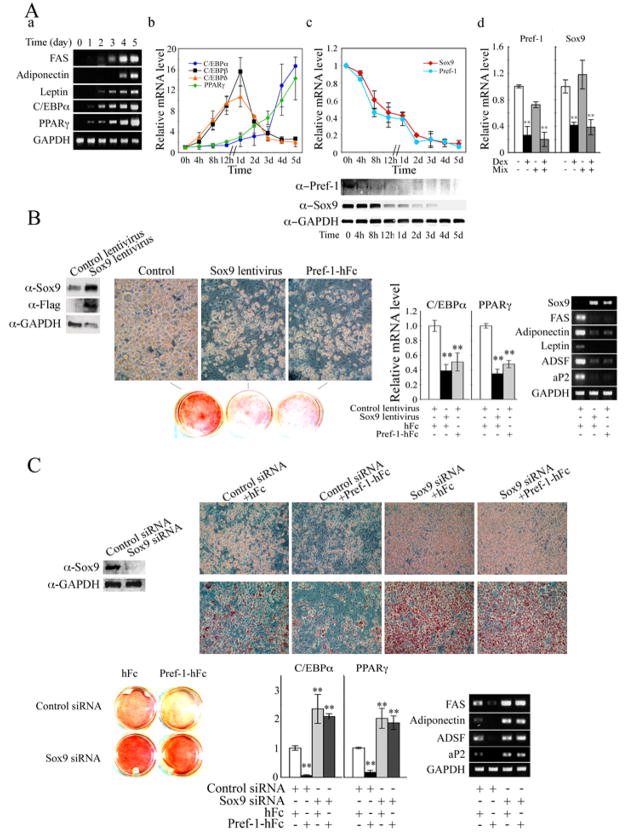
Figure 3.
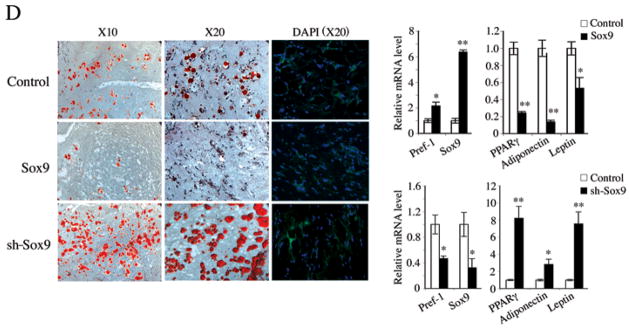
Pref-1 inhibits 3T3-L1 adipocyte differentiation through Sox9. (A) RT-PCR (a) and RT-qPCR (b and c) for various adipocyte markers during 3T3-L1 adipocyte differentiation. RT-qPCR after 12 h treatment (d). (B) Sox9 protein level after 48 h of infection (left panel). Morphology, Oil red O staining (middle panel), and RT-qPCR and RT-PCR for adipocyte markers (right panels). (C) Sox9 knockdown enhances adipocyte differentiation. Western blotting after 48 h siRNA transfection (left panel). Morphology, Oil red O staining and adipocyte marker expression after adipocyte differentiation. (D) 3T3-F442A-GFP preadipocytes stably overexpressing or with knockdown of Sox9, or control cells (3×107 cells mixed with Matrigel matrix (BD) per site) were injected subcutaneously into the back of SCID mice. The resulting tissues were dissected for Oil red O and DAPI staining and RT-qPCR (n = 4/group).
We next tested if Sox9 can inhibit 3T3-L1 adipocyte differentiation as was the case for MEF differentiation into adipocytes. Both Pref-1 hFc treatment or constitutive ovexpression of Pref-1 by 4-fold by Pref-1 lentivirus infection attenuated adipocyte differentiation as judged by cell morphology, Oil red O staining and expression of adipocyte markers FAS, adiponectin, leptin ADSF/resistin, and aP2, as well as the adipogenic transcription factors C/EBPα and PPARγ (Fig. 3B). Conversely, knockdown of Sox9 by 80% at the protein level by Sox9 siRNA transfection resulted in enhanced adipocyte differentiation compared to the control cells as judged by adipocyte morphology, Oil red O staining and expression of adipocyte markers (Fig. 3C). Furthermore, in Sox9 siRNA transfected cells, Pref-1-hFc treatment could not bring about inhibition of differentiation as observed by the same degree of adipocyte differentiation as in Sox9 siRNA transfected cells treated with control hFc. These data clearly show that Sox9 inhibits adipocyte differentiation and provide evidence that Pref-1 inhibition of adipocyte differentiation is through Sox9.
We previously demonstrated Pref-1 inhibition of adipogenesis in vivo by generating mice lacking Pref-1 as well as mice expressing soluble Pref-1 in adipose tissue. Sox9 ablation in mice causes embryonic lethality and thus could not be used to study adipogenesis that occurs mainly late after birth. In testing the Sox9 effect in vivo, we generated 3T3-F442A-GFP cells that either stably overexpress Sox9 by 6-fold or have knockdown of Sox9 by 65%. These cells were implanted subcutaneously into immunodeficient SCID mice. Two weeks after implantation, we determined lipid accumulation and adipocyte marker expression. As shown in Figure 3D, control cells formed a substantial number of adipocytes. On the other hand, the adipocytes that arose from the implanted cells overexpressing Sox9 formed fewer adipocytes. They showed lower Oil red O staining, higher expression of Pref-1 and lower expression of adipocyte markers such as PPARγ, adiponectin and leptin. Conversely, implanted cells expressing Sox9 shRNA formed a higher number of adipocytes as detected by higher lipid staining and by lower expression of Pref-1, and higher expression of PPARγ, adiponectin and leptin. These results clearly demonstrate the inhibitory role of Sox9 in adipogenesis in vivo.
Sox9 binds C/EBPβ and C/EBPδ promoters to suppress promoter activity in mediating Pref-1 inhibition of adipogenesis
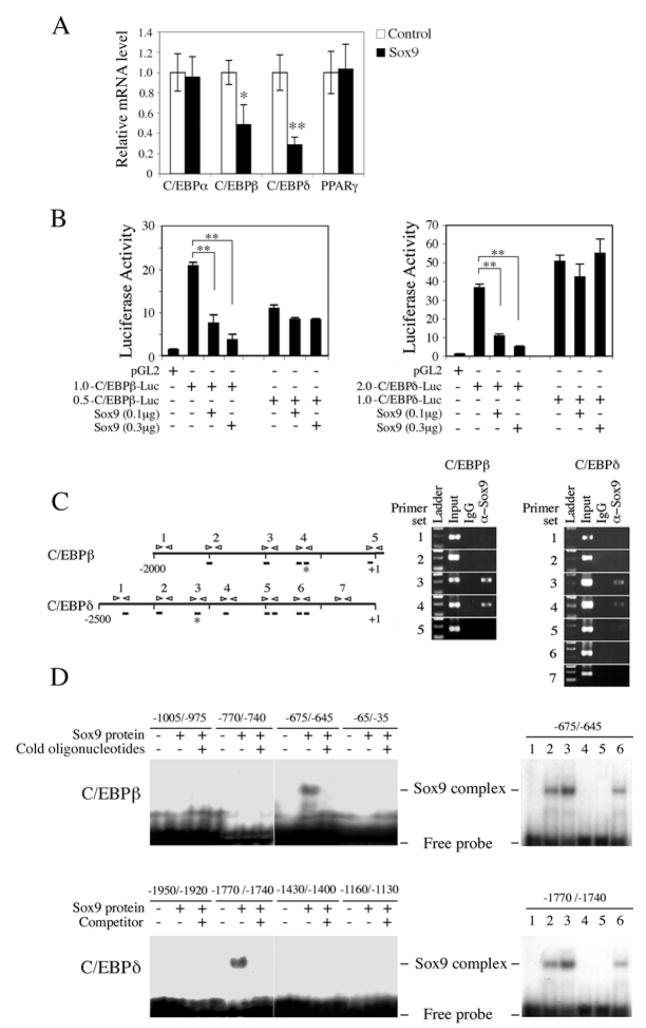
Since the decrease in Pref-1 and Sox9 expression upon treatment with adipogenic agents precedes the early induction of C/EBPβ and C/EBPδ (Fig. 3A), we hypothesized that Sox9 may regulate C/EBPβ and C/EBPδ expression to affect adipocyte differentiation. As shown in Figure 4A, overexpression of Sox9 did not affect C/EBPα and PPARγ mRNA levels but significantly suppressed C/EBPβ and C/EBPδ expression by 55% and 70%, respectively. Furthermore, cotransfection of Sox9 with 1.0 kb-C/EBPβ-Luc or 2.0 kb- C/EBPδ-Luc suppressed promoter activities by 80 or 85%, respectively (Fig. 4B). However, cotransfection of Sox9 with the shorter promoter-reporter constructs, 0.5 kb-C/EBPβ-Luc or 1.0 kb- C/EBPδ-Luc, did not affect the promoter activities. These data suggest that repression of C/EBPβ and C/EBPδ genes by Sox9 might be through Sox9 binding between −1.0 and −0.5 kb of the C/EBPβ promoter and between −2.0 and −1.0 kb of the C/EBPδ promoter.
Figure 4.
Sox9 directly binds C/EBPβ and C/EBPδ promoters, and suppresses their transcription. (A) RT-qPCR for various transcription factors 72 h after Sox9 transfection into 3T3-L1 cells. *, P<0.05 and **, P<0.01 compared to cells transfected with empty vector. (B) C/EBPb and C/EBPd promoter activity after co-transfection with Sox9 into 293FT cells. pRL-SV40 was used as internal control. Results are mean ± SEM; **, P<0.01 (C) Numbering denotes primer sets in the C/EBPβ and C/EBPδ promoter regions used for ChIP (left panel). ChIP for C/EBPβ and C/EBPδ using 3T3-L1 cells (right panel). Bar; sites with core consensus Sox9 binding sequence, Star; actual Sox9 binding sites. (D) EMSA for Sox9 binding sites. In vitro translated Sox9 protein and 32P labeled oligonucleotides were used for EMSA. Probe only (lane 1); 1 μl (lane 2) and 2 μl (lane 3) of Sox9 translation reaction with probe; 1 μl of Sox9 with 100 × wild-type competitor (lane 4); 1 μl of translation reaction of empty pcDNA3.1 vector with probe (lane 5); 1 μl of Sox9 with 100 × mutant competitor (lane 6).
Examination of C/EBPβ and C/EBPδ promoter sequences revealed the presence of, respectively, 5 and 8 core consensus binding sites for Sox9 (Fig. 4C). ChIP analysis detected evidence for Sox9 binding when specific primer pairs 3 and 4 for C/EBPβ and specific primer pairs 3 and 4 for C/EBPβ but not other primer sets, were used. Furthermore, EMSA using in vitro translated Sox9 protein and oligonucleotides corresponding to the various Sox9 consensus sites showed that Sox9 can bind the −675 – −645 of the C/EBPβ promoter and −1770 – −1740 of C/EBPβ promoter (Fig. 4D). Excess cold oligonucleotides prevented the signals (left panels), whereas the oligonucleotides containing mutations of the core Sox9 binding CAAT sequence did not affect the binding (right panels). Overall, these results demonstrate that Sox9 directly binds its sites in the C/EBPβ and C/EBPδ promoter regions to suppress promoter activity in mediating Pref-1 inhibition of adipocyte differentiation.
Pref-1 inhibits osteoblast differentiation by maintaining Sox9 expression
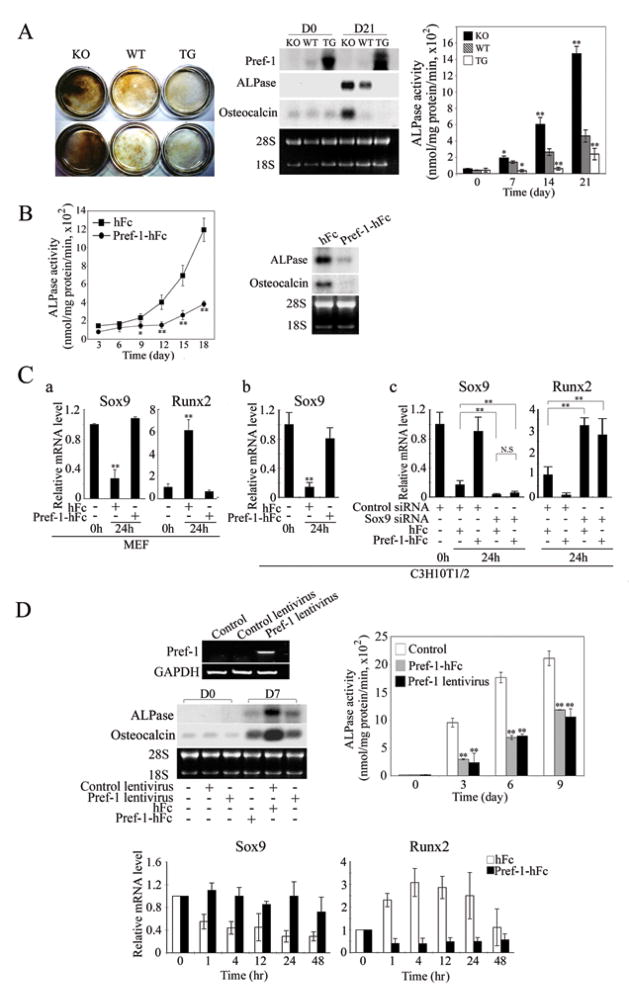
Since we previously observed bone malformation in Pref-1 null and Pref-1 transgenic mice, we tested the effect of Pref-1 on osteoblast differentiation. First, we compared osteoblast differentiation of MEFs from Pref-1 null, wild-type and Pref-1 transgenic embryos. After 21-days of osteogenic induction, the intensity of von Kossa staining was 3-fold higher in Pref-1 null MEFs than in wild-type MEFs, whereas the staining was barely detected in Pref-1 transgenic cells (Fig. 5A left panel). As predicted, Northern blotting showed an absence of Pref-1 in Pref-1 null cells, while, in wild-type cells, endogenous Pref-1 was detected at a low level, which was completely suppressed after differentiation. In contrast, Pref-1 expression was high in Pref-1 transgenic cells, and the levels remained high after treatment with osteogenic agents. After 21 days of osteoblast differentiation, expression of ALPase and osteocalcin, respective early and late osteogenic markers, increased drastically in Pref-1 null cells, to levels exceeding those in wild-type cells, whereas expression in Pref-1 transgenic cells was very low and did not change. Similarly, ALPase activity in Pref-1 null cells was significantly higher than that in wild-type cells after 7-days and the difference was more marked as differentiation proceeded. Thus, after 21 days of differentiation, ALPase activity in Pref-1 null cells increased by 15-fold, a level 3-fold higher than that in wild-type cells. The increase in ALPase activity was only 2-fold in Pref-1 transgenic cells, 60% lower than the 5-fold increase detected in wild-type cells. These data suggest that overexpression of Pref-1 inhibits, whereas lack of Pref-1 enhances, osteoblast differentiation in vitro. The inhibitory effect of Pref-1 on osteoblast differentiation was further tested by using Pref-1-hFc. As expected, the ALPase activity in Pref-1 null cells treated with control hFc increased drastically and reached a 12-fold increase at Day 18 (Fig. 5B). In contrast, ALPase activity in cells treated with Pref-1-hFc increased in a delayed fashion and was only 4-fold higher at Day 18. ALPase and osteocalcin mRNA levels at Day 14 were markedly lower in cells treated with Pref-1-hFc, showing Pref-1 inhibition of osteoblast differentiation (Fig. 5B).
Figure 5.
Pref-1 inhibits osteoblast differentiation by preventing the decrease in Sox9 expression. (A) Von Kossa staining of MEFs after 21-days of osteoblast differentiation (left panel). Northern blot analysis (middle panel) and ALPase activity (right panel). Results are means ± SEM; *, P<0.05 and **, P<0.01 compared to wild-type cells at indicated time points. (B) ALPase activities (left panel) and Northern blotting after 18-days of osteoblast differentiation (right panel). (C) RT-qPCR for Pref-1 null MEFs and C3H10T1/2 cells cultured for in osteogenic media (a and b). siRNA transfected C3H10T1/2 cells cultured in osteogenic media (c). For Sox9 expression, the value of untreated cells was defined as 1. For Runx2, due to its undetectable level in untreated cells, the value of control siRNA transfected cells after 24 hr treatment was defined as 1. (D) RT-PCR after 48 h lentivirus infection in MC3T3 cells. Northern blot analysis from confluent MC3T3 cells (D0) and 7-days osteogenic treatment (D7), and ALPase activity, **, P<0.01 compared to cells infected with control lentivirus and cultured with hFc at indicated time points. RT-qPCR during osteogenic differentiation (bottom panel).
We next examined the effect of Pref-1 on Sox9 expression early in osteoblast differentiation of MEFs (Fig. 5C). Treatment with osteogenic agents drastically decreased Sox9 expression after 24 hrs in Pref-1 null MEFs (Fig. 5Ca). Importantly, Pref-1-hFc prevented this decrease. Expression of Runx2, a transcription factor required for initiation of osteogenesis as well as chondrocyte maturation, on the other hand, increased markedly, and this increase was also prevented by Pref-1-hFc treatment. Similar to the effects observed in MEFs, Sox9 expression also decreased drastically in C3H10T1/2 cells upon treatment with osteogenic agents, which was prevented by Pref-1-hFc treatment (Fig. 5Cb). Unlike in MEFs and MC3T3 cells, however, we could not detect Runx2 expression in C3H10T1/2 cells. After 24 hrs of osteogenic treatment, Runx2 was clearly detected and Pref-1-hFc treatment prevented this Runx2 induction (data not shown).
To further test the Pref-1 effect on Sox9 expression during osteogenesis, we transfected Sox9 siRNA into C3H10T1/2 cells. As expected, Sox9 siRNA transfection decreased Sox9 levels markedly (Fig. 5Cc). After 24 hrs of treatment with osteogenic agents, Pref-1-hFc prevented the drastic decrease in Sox9 expression observed in control siRNA transfected cells. Expression of Runx2 was 3-fold higher in Sox9 siRNA transfected cells compared to control cells. More importantly, Pref-1 suppressed Runx2 expression by 90% in control siRNA transfected cells treated with osteogenic agents, whereas Pref-1 was unable to suppress Runx2 expression in Sox9 siRNA transfected cells (Fig. 5Cc). These results show that Pref-1 inhibits osteoblast differentiation by preventing Sox9 downregulation. We also detected the Pref-1 effect by comparing osteoblast differentiation of primary osteogenic cells from wild-type, Pref-1 null and Pref-1 transgenic E18.5 limbs and ribs (Suppl. Fig. 4). Overall, these results clearly show Pref-1 inhibits osteoblast differentiation through Sox9 regulation.
The effect of Pref-1 on osteoblast differentiation was also tested in preosteoblastic MC3T3 cells (Fig. 5D). Pref-1 expression was not detectable in MC3T3 cells (Fig. 5D top, left panel). Osteocalcin and ALPase expression was very low in confluent cells, but increased markedly after 7 days of treatment with osteogenic agents. These increases were blunted in cells infected with Pref-1 lentivirus or upon Pref-1-hFc treatment. Similarly, ALPase activity rapidly increased at Day 3 and further increased at Days 6 and 9 after osteogenic stimulation (Fig. 5D top, right panel). The increase in ALPase activity was delayed and significantly blunted at all time points in cells infected with Pref-1 lentivirus or upon Pref-1-hFc treatment. We also examined the effect of Pref-1-hFc on expression of Sox9 early during osteogenesis in these cells (Fig. 5D bottom panels). During early osteogenesis, expression of Sox9 was rapidly down-regulated by 50% within 1 hr after osteogenic stimulation in control hFc treated cells, and Sox9 levels further decreased up to 2 days after treatment, whereas expression of Runx2 was rapidly induced reaching a maximum of 3-fold at 4 hrs, that gradually decreased. In contrast, the Sox9 expression level was maintained in cells treated with Pref-1-hFc, while Runx2 expression did not increase in these cells upon treatment with osteogenic agents. Overall, These results show that Pref-1 prevents Sox9 downregulation and thereby inhibits mesenchymal cell differentiation into osteoblasts.
Pref-1 directs multipotent mesenchymal cells to chondrogenic commitment but inhibits maturation of chondrocytes by inducing Sox9 expression
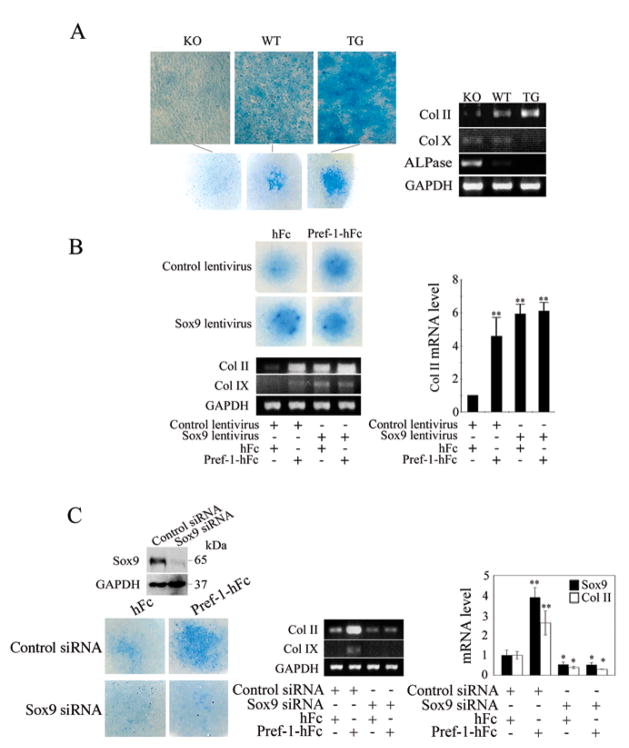
Because Pref-1 inhibits osteoblast differentiation and osteoblasts and chondrocytes are from common mesenchymal progenitor cells, we hypothesized that Pref-1 may regulate chondrocyte formation also (Gaur et al., 2005; Lengner et al., 2004). We, therefore, compared MEFs from Pref-1 null, wild-type and Pref-1 overexpressing transgenic embryos in micromass culture for chondrogenesis. Alcian blue staining showed the effect of Pref-1 on chondrogenesis (Fig. 6A left panel): Pref-1 null cells showed the weakest staining, while wild-type cells showed a 4-fold higher intensity of staining, which was even higher by 8-fold in transgenic cells. We used Col II and Col IX as immature chondrocyte markers and Col X and ALPase as hypertrophic chondrocyte markers. After 14 days of culture in the presence of chondrogenic agents, wild-type cells showed 4.3-fold higher Col II levels, while Pref-1 transgenic cells displayed 6.7-fold higher Col II expression compared to Pref-1 null cells (Fig. 6A right panel). On the contrary, Col X and ALPase expression was highest in Pref-1 null cells. Upon treatment with chondrogenic agents, ALPase expression decreased by 4.7-fold in wild-type cells but was not detected in Pref-1 transgenic cells. Col X expression was also lower in Pref-1 transgenic cells. These results show that Pref-1 promotes mesenchymal cell commitment to the chondrogenic lineage but inhibits chondrocyte maturation in vitro.
Figure 6.
Pref-1 directs MEF commitment to chondrogenic lineage through Sox9 regulation. (A) Micromass culture of MEFs. Alcian blue staining (left panel) and RT-PCR for chondrogenic markers (right panel) after 14 days of culture. (B) Pref-1 null MEFs infected with Sox9 lentivirus after 10 days micromass culture. **, P<0.01 compared to differentiated cells infected with control empty lentivirus and in the presence of control hFc. (C) Knockdown of Sox9 blocks chondrogenic induction by Pref-1. Alcian blue staining for Pref-1 null MEFs transfected with siRNA after micromass culture. Western blotting after 48 h transfection (left top panel). Results are mean ± SEM; *, P<0.05 and **, P<0.01 compared to cells transfected with control siRNA and treated with hFc.
Next, to test the effect of Pref-1 and Sox9 on chondrogensis, we infected Pref-1 null MEFs with Sox9 lentivirus or treated Pref-1 null MEFs with Pref-1-hFc and subjected them to micromass culture (Fig 6B). We found that Pref-1-hFc treatment or lentivirus mediated Sox9 overexpression in MEFs caused increased immature chondrocytes as judged by alcian blue staining and Col II and Col IX expression. Furthermore, overexpression of Sox9 along with Pref-1-hFc treatment did not produce additive effects, suggesting that Pref-1 and Sox9 may affect chondrogenesis by the same mechanism. Similar results were obtained from C3H10T1/2 cells subjected to chondrogenesis (data not shown). To further examine the role of Sox9 in mediating the Pref-1 effect on chondrogenesis, we treated the Pref-1 null MEFs transfected with Sox9 siRNA with soluble Pref-1-hFc (Fig. 6C). As judged by alcian blue staining and chondrogenic marker expression, we found that Pref-1-hFc treatment did not have an effect on chondrogenesis in cells transfected with Sox9 siRNA, compared to control cells treated with Pref-1 that had increased immature chondrocytes. Overall, these data from MEFs and C3H10T1/2 cells are consistent with the notion of Sox9 mediating the Pref-1 effect, and clearly demonstrate that, like Sox9, Pref-1 can promote chondrogenic induction of uncommitted mesenchymal cells in micromass culture.
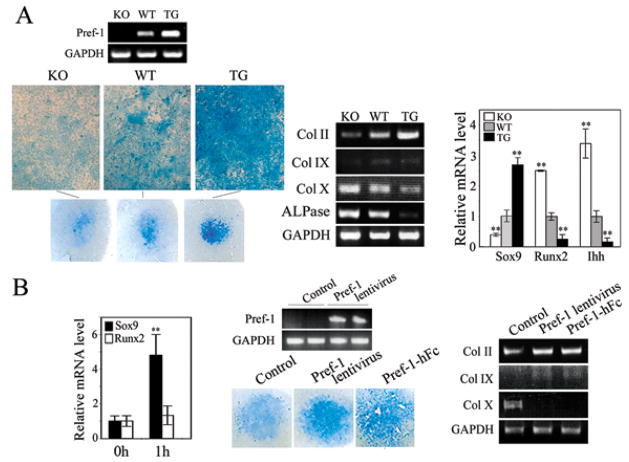
We also tested the involvement of Sox9 and Pref-1 in chondrocyte maturation by employing committed chondrogenic cells from skinned tails from newborn mice in micromass culture (Fig. 7A and B). After 14 days of culture, compare to wild-type cells, Pref-1 null cells displayed a lower intensity of alcian blue staining with lower expression of Col II and Col IX, but higher expression of Col X and ALPase. In contrast, Pref-1 transgenice cells displayed stronger alcian blue staining with higher Col II and Col IX, but lower Col X and ALPase expression (Fig. 7A). In addition, Sox9 expression was lower, while Runx2 was higher in Pref-1 null cells than in wild-type cells, and Pref-1 transgenic cells showed higher Sox9 expression but lower Runx2 expression. We also found that expression of the prehypertrophic marker, Ihh, was higher in Pref-1 null cells, but was lower in Pref-1 transgenic cells than in wild-type cells. These observations demonstrate that Pref-1 increases immature chondrocytes but inhibits maturation of chondrocytes. Furthermore, Pref-1 lentivirus infection or Pref-1-hFc treatment increased immature chondrocytes as judged by alcian blue staining and chondrogenic marker expression with increased Col II and Col IX expression but decreased Col X expression. These observations provide further evidence that Pref-1 promotes chondrogenic induction/early chondrocyte differentiation, but inhibits chondrocyte maturation. Thus, Pref-1 maintains the chondrocytes in an immature state and prevents their differentiation into mature chondrocytes in vitro. When we tested the effect of Pref-1 on Sox9 and Runx2 expression in Pref-1 null primary chondrogenic cells (Fig. 7B, left panel), Sox9 expression increased by 4.6-fold after 1 hr of Pref-1-hFc treatment. On the other hand, we did not detect any significant changes in Runx2 expression at this time point, further suggesting Pref-1 function is through Sox9 induction.
Figure 7.
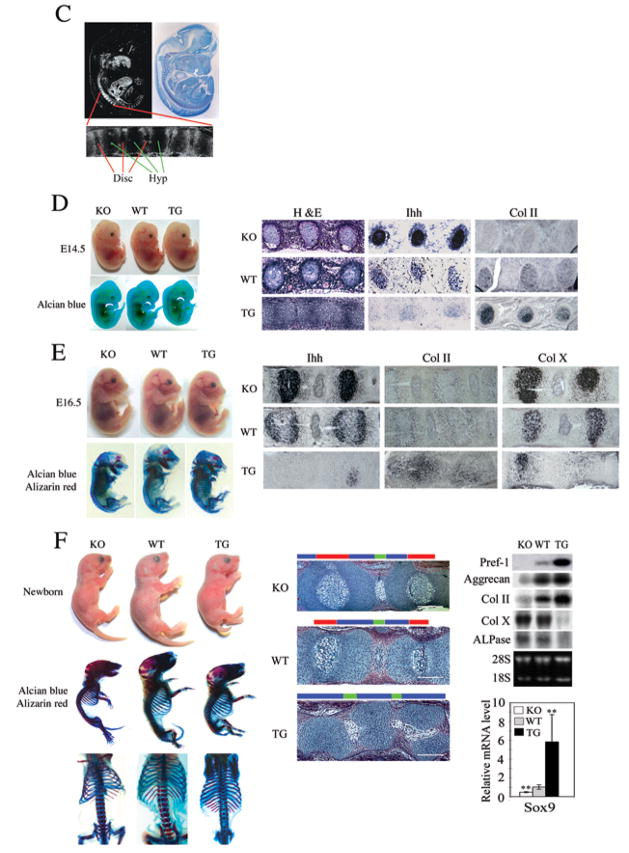
Pref-1 promotes chondrogenic induction but inhibits chondrocyte maturation through upregulation of Sox9. (A) Micromass culture of cells from newborn tails. Alcian blue staining and RT-PCR and RT-qPCR for chondrogenic markers. **, P<0.01 compared to wild-type. (B) RT-qPCR after 1 hr treatment with 50 nM Pref-1-hFc (left panel), **, P<0.01 compared to untreated cells. RT-PCR in cells 48 hrs after infection. (C) In situ hybridization of E13.5 embryos with 33P labeled Pref-1 anti-sense probe (left panel) and bright field view with hematoxylin staining (left panel). Thoracic-abdominal vertebra region is enlarged (bottom). Disc, spinal disc; Hyp, hypertrophic zone. Embryos and mice, and their skeleton staining (alcian blue for cartilage and alizarin red for bone) in various developmental stages (D–F). Thoracic-abdominal vertebra was used for in situ hybridization with digoxigenin labeled anti-sense probe. H&E staining of 4–5th coccyges from newborn mice (F, middle panel). Color bars represent the length of different histological area. Red bar, zones of hypertrophic chondrocytes; blue bar, the area of resting and proliferating chondrocytes and prehypertrophic chondrocytes; green bar, spinal disc. White bar = 200 μm. Northern blotting and RT-qRCR for skinned tails of newborn mice. **, P<0.01 compare to wild-type. (G and H) Role of Pref-1 in MSC commitment and differentiation. (G) H&E staining (upper panel) of 5th coccyges of 1 wk-old mice and higher magnitude of bone marrow region (right). RT-qPCR for femur bone marrow from 12 week-old mice (n = 5) (lower panel). Results are mean ± SEM; **, P<0.01 compare to wild-type. (H) RT-qPCR of human primary MSC after 2 days adipogenic (upper panel) or chondrogenic treatment (lower panel). *, P<0.05 and **, P<0.01.
As mentioned, in adults Pref-1 expression is mainly restricted to preadipocytes as well as some neuroendocrine cell types, but Pref-1 is widely expressed during embryonic development with prominent expression in developing vertebra as shown by in situ hybridization at day E13.5 (Fig. 7C). In vertebra, Pref-1 expression was restricted to the periphery of vertebral bodies but was undetectable in hypertrophic zones (Fig. 7C, bottom panel), suggesting that Pref-1 functions in immature chondrocytes but not in hypertrophic chondrocytes. To demonstrate the effects of Pref-1 on chondrogenesis in vivo, we examined embryonic developmental stages by comparing Pref-1 null, wild-type and Pref-1 transgenic embryos. As shown in Figure 7D, we did not observe drastic malformation at E14.5 when mesenchymal condensations start to commit to the chondrocyte linage and in part to become prehypertrophic chondrocytes. But, the prehypertrophic marker, Ihh, was strongly expressed in Pref-1 null vertebral bodies compared to wild-type, whereas it was barely detectable in Pref-1 overexpressing vertebral bodies. In contrast, expression of the immature chondrocyte marker, Col II, was lower in Pref-1 null embryos, but was strongly expressed in Pref-1 overexpressing ones. At E16.5 stage when the central vertebral bodies normally become hypertrophic chondrocytes, both Pref-1 null and Pref-1 overexpressing embryos showed more visible skeletal malformation (Fig. 7E). Compared to wild-type, Pref-1 null embryos were smaller with higher expression of the prehypertrophic and hypertrophic chondrocyte markers, Ihh and Col X, respectively. Pref-1 transgenic embryos were also smaller with shorter kinky tails, and showed higher expression of the immature chondrocyte marker Col II, and markedly lower expression of the prehypertrophic and hypertrophic markers, Ihh and Col X.
Phenotypes were more severe in the newborn stage (Fig. 7F). Histological staining of the vertebral region showed that, compared to wild-type mice, Pref-1 null mice had a 40% increase in the hypertrophic zone, whereas the hypertrophic zone was absent in Pref-1 transgenic mice (Fig. 7F). Similarly, we detected a 40% increase in the hypertrophic zone in Pref-1 null long bones (E18.5 femurs) compared to wild-type mice, whereas we found a 25–30% decrease in the hypertrophic zone in Pref-1 transgenic mice (data not shown). Conversely, the zone of immature chondrocytes was 30% lower in Pref-1 null mice, while Pref-1 transgenic mice had only immature chondrocytes without an obvious hypertrophic zone. As expected, by Northern blot analysis, Pref-1 was not detected in Pref-1 null mice but was markedly higher in Pref-1 transgenic mice. Similarly, Sox9 expression was significantly lower in Pref-1 null mice compared to wild-type mice, but was 6-fold higher in Pref-1 transgenic mice. Examination of chondrocyte/osteoblast marker expression also indicated that the levels of immature chondrocyte markers including Col II and aggrecan were lower in Pref-1 null mice but higher in Pref-1 transgenic mice compared to wild-type mice. In contrast, expression of mature chondrocyte marker Col X and hypertrophic/early osteogenic marker ALPase was higher in Pref-1 null mice but barely detectable in Pref-1 transgenic mice. Overall, these data show that Pref-1 brings about chondrogenic induction but inhibits chondrocyte maturation.
Effect of Pref-1 on bone marrow stromal cell fate
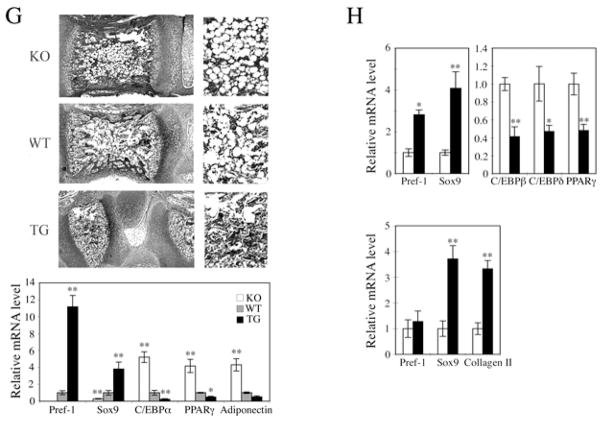
Since bone marrow mesenchymal stem cells (MSC) can commit and differentiate into adipocytes as well as chondrocytes and Pref-1 is detected in bone marrow, we next examined the presence of adipocytes in bone marrow of 1 wk-old vertebral bodies from Pref-1 null, wild-type and Pref-1 transgenic mice. H&E staining showed that rounded adipose features were higher in Pref-1 null than in wild-type vertebra and they were barely detectable in Pref-1 overexpressing vertebra (Fig. 7G upper panel). Expression of adipocyte markers in bone marrow from 12 wk-old femur bone was inversely correlated with Pref-1 expression: C/EBPα, PPARγ and adiponectin levels were markedly higher in Pref-1 null bone marrow, but were very low in Pref-1 transgenic ones. In contrast, Sox9 expression levels were very low in Pref-1 null bone marrow but were markedly higher in Pref-1 transgenic bone marrow (Fig. 7G lower panel). Overall, these observations indicate a potential in vivo effect of Pref-1 on MSC differentiation into adipocytes.
We next used the human MSC in culture to test if Pref-1 affects the fate of these cells during adipogenic and chondrogenic commitment/differentiation (Fig. 7H). RT-qPCR showed that 2 days after initiation of adipocyte differentiation, Sox9 as well as endogenous Pref-1 expression levels were significantly higher in Pref-1-hFc treated cells compared to control hFc treated cells (Fig. 8, upper panel). On the other hand, expression levels of adipocyte markers, C/EBPβ, C/EBPδ and PPARγ were lower in Pref-1-hFc treated cells. Upon treatment with chondrogenic agents of MSC for 2 days, Sox9 and Col II expression levels were significantly higher in Pref-1-hFc treated cells compared to control hFc cells, demonstrating promotion to the chondrogenic lineage by Pref-1 (lower panel). Overall, these results from MSC in culture further confirm that Pref-1 promotes commitment to the chondrocyte-lineage but inhibits adipogenesis.
DISCUSSION
We previously reported Pref-1 inhibition of adipocyte differentiation by both in vitro and in vivo approaches. In fact, we originally identified Pref-1 as a protein found in preadipocytes but not in adipocytes. During adipocyte differentiation, Pref-1 is extinguished. Constitutive expression of Pref-1 inhibits 3T3-L1 preadipocyte differentiation into adipocytes, and Pref-1 knockdown causes enhanced differentiation. The significance of Pref-1 function in adipogenesis has been documented in vivo in Pref-1 null and Pref-1 overexpressing transgenic mice. Although expression of the two key adipogenic transcription factors C/EBPα and PPARγ clearly is suppressed by Pref-1 treatment, the proximal downstream molecule that mediates Pref-1 inhibition has not been defined. It has been reported previously that HES-1 expression is induced by Pref-1 treatment in thymocytes (Kaneta et al., 2000). However, we could not detect any changes in HES-1 expression by Pref-1 (unpublished results). In the present study, we show that Sox9 is a target of Pref-1 and Pref-1 increases Sox9 expression rapidly in various cell types including multipotent mesenchymal cells. Although a receptor for Pref-1 has not been identified, we have previously shown that Pref-1 activates the MEK/ERK pathway. The fact that Sox9 is known to be induced by ERK activation (Murakami et al., 2000) and that inhibition of Pref-1-mediated ERK activation prevents Sox9 induction, further supports the notion of Sox9 as a downstream target of Pref-1. In this regard, although Sox9 may not be the only target for Pref-1, we found that Pref-1 expression parallels Sox9 expression during adipocyte differentiation of MEFs and 3T3-L1 cells as well as in vivo adipogenesis.
Although the role of Sox9 in chondro- and osteogenesis is well understood, its role in adipocyte commitment/differentiation has not been studied. Here, we show that overexpressing Sox9 constitutively or maintaining Sox9 levels by treating the cells with Pref-1-hFc causes inhibition of adipocyte differentiation. We conclude that Sox9 suppression is required before induction of the adipogenic transcription factors PPARγ and C/EBPα for adipocyte differentiation. Furthermore, downregulation of Sox9 coincides with the induction of C/EBPβ C/EBPδ that occurs earlier than C/EBPα and PPARγ induction. Suppression of Pref-1 decreases Sox9 expression that, in turn, allows C/EBPβ and C/EBPδ induction early at the start of adipocyte differentiation. A synthetic glucocorticoid DEX, routinely used for adipocyte differentiation suppresses Pref-1 expression. We also found that DEX suppresses Sox9 expression also. We thus predicted that Sox9 might function through regulating C/EBPβ and/or C/EBPδ during adipocyte differentiation. In this regard, glucocorticoids have been reported to induce C/EBPδ expression. In the present study, we detected suppression of C/EBPδ and C/EBPβ upon Sox9 overexpression. Furthermore, cotransfection of Sox9 resulted in decreased C/EBPβ and C/EBPδ promoter activity and EMSA and ChIP analysis revealed the binding sites of Sox9 at −675 – −645 of C/EBPβ and −1770 – −1740 of C/EBPδ promoter sequences. Here, we also demonstrate in vivo the role of Sox9 in adipogenesis by subcutaneous implanting (Fig. 3D). The present study demonstrates that Sox9 is downregulated during adipocyte differentiation and Sox9 inhibits adipocyte differentiation. Our study also shows that the function of Sox9 is due to maintain cells in the preadipocyte stage by directly binding to the C/EBPβ and C/EBPδ promoter regions to suppress transcription.
Pref-1 is expressed in multipotent mesenchymal stem cells that can differentiate into several cell types, including chondrocytes, osteoblasts and adipocytes. Although the role of Pref-1 in adipogenesis has been extensively studied (Lee et al., 2003; Mei et al., 2002; Moon et al., 2002; Smas et al., 1998), Pref-1 effects on chondrocyte commitment and maturation have not been studied before. Here, we employed MEFs that have the ability to undergo commitment/differentiation into various mesenchymal lineages including chondrocytes, osteoblasts and adipocytes. Sox9 and Pref-1 have similar but not additive effects on MEF commitment and early differentiation into immature chondrocytes. We also employed bone marrow stromal cells to demonstrate that Pref-1 inhibits adipogenesis, while promoting chondrogenic commitment. Examination of the bone marrow clearly shows that Pref-1 expression is inversely correlated with adipose morphology in vivo. Moreover, chondrogenic induction by Pref-1 in micromass culture is abolished in the absence of Sox9, evidence that Sox9 mediates the Pref-1 effect of promoting chondrogenic induction. It is well recognized that Sox9 plays an essential role in recruiting mesenchymal cells to undergo chondrogenic commitment and early chondrogenesis but inhibits maturation to hypertrophic chondrocytes (Akiyama et al., 2004). Sox9 function in chondrogenesis has been documented by loss- and gain- of function studies in vitro and in vivo (de Crombrugghe et al., 2001). Inactivation of Sox9 before mesenchymal condensation in mice results in complete prevention of condensation and cartilage formation. Thus, Sox9 deficiency and Sox9 overexpression in mice results in smaller skeletons as seen in Pref-1 null and Pref-1 transgenic mice. Pref-1 null mice appear to have, albeit less severely, phenotypes of skeletal development resembling phenotypes found when Sox9 is knockeddown or mutated, while the phenotypes of Pref-1 transgenic mice have similarities to the Sox9 transgenic mouse model. In this regard, deficiencies in some of the known growth factors/hormones that affect skeletogenesis, at least in part, through Sox9 regulation also have similar phenotypes (Karaplis et al., 1994; Liu et al., 2002). Regardless, these apparent phenotypic similarities of in vivo gain- and loss- of function further support the notion that Pref-1 effects are through regulation of Sox9 expression.
With our present study demonstrating the role of Pref-1 on chondrogenesis, the Pref-1 effect on bone formation could be an indirect one. However, the Pref-1 effect on MEFs as well as preosteoblastic MC3T3 cells that can undergo osteoblast differentiation in vitro indicates a direct inhibitory effect of Pref-1 on osteoblast differentiation. This idea is also supported by a previous report of Pref-1 leading to inhibition of osteoblast differentiation of human bone marrow stromal cells (Abdallah et al., 2004). More importantly, we demonstrate here that Pref-1 prevents Sox9 downregulation and thereby inhibits osteoblast differentiation and the expression of the osteogenic transcription factor Runx2. Progenitor cells in early mesenchymal condensations have dual differentiation potentials as they co-express Sox9 and Runx2. Runx2 has been shown to accelerate chondrocyte maturation to become hypertrophic chondrocytes and to promote osteogenesis (Takeda et al., 2001). Runx2 appears to exert its function only when Sox9 is downregulated in hypertrophic chondrocytes or in osteoblasts. We conclude that Pref-1 inhibits osteoblast differentiation by preventing downregulation of Sox9.
In conclusion, we show here that Sox9 downregulation is required for adipocyte differentiation. Sox9 suppresses C/EBPβ/C/EBPδ expression by directly binding to its sites in the promoter regions of these genes. Importantly, we demonstrate that Pref-1 inhibition of adipogenesis is through maintaining Sox9 expression. We also show that by inducing Sox9, Pref-1 promotes chondrogenic induction of mesenchymal cells but prevents chondrocyte maturation as well as osteoblast differentiation, with supporting in vivo evidence in Pref-1 null and Pref-1 transgenic mice. Overall, Sox9 is a Pref-1 target and Pref-1 directs multipotent mesenchymal cells to the chondrogenic lineage but inhibits differentiation into adipocytes as well as osteoblasts and chondrocytes.
MATERIALS AND METHODS
Lentivirus constructs and plasmid and animals
The lentiviral construct was made by subcloning Sox9 into pLenti6/V5-D-TOPO vector. Sox9 was also subcloned into pcDNA3.1. 1.0-C/EBPβ-luc, 0.5-C/EBPβ-luc, 2.0-C/EBPδ-luc and 1.0-C/EBPδ-luc were generated by inserting 1.0 and 0.5 kb C/EBPβ and 2.0 and 1.0 kb C/EBPδ 5′ flanking regions into the pGL2 basic vector (Promega). Sox9 shRNA plasmid was purchased from SABiosciences. Generation of Pref-1 null and Pref-1 transgenic mice were described previously (Lee et al., 2003; Moon et al., 2002). Scid mice were purchased from the Jackson laboratory.
Cells and siRNA transfection
MC3T3 cells were cultured in α-MEM (Invitrogen) containing 10% FBS. MEFs were prepared from E13.5 embryos as described previously (Wang and Sul, 2006). Human derived marrow stromal cells (MSC) and MSC media were purchased from Allcells. Sox9 target-specific siRNA (sc-36534) (Santa Cruz) and the control siRNA (sc-36869) were transfected into cells using transfection reagent (Santa Cruz).
Chromatin Immunoprecipitation (ChIP) and Electrophoretic mobility shift assay (EMSA)
For ChIP analysis, 1×107 3T3-L1 cells were cross-linked with 1% formaldehyde before nuclei preparation. After sonication, DNA sizes were 0.3–0.9 kb. 10% of the total volume was used as input, the rest was divided into two fractions: one for control IgG and the second for 5 μg of Sox9 antibody immunoprecipitation and purified DNA was used in PCR reactions.
Sox9 protein were in vitro translated using Promega TNT® Translation Systems and used in EMSA. Thirty bp sense and antisense oligonucleotides containing potential Sox9 core binding sequence CAAT were labeled by γ 32P-dCTP. Mutant competitors were the oligonucleotides mutated CAAT to CCAT. Samples were separated by 5% native polyacrylamide gel electrophoresis before exposure to film.
Statistical analysis
Data are expressed as the mean ± SEM. The statistical differences in mean values were assessed by Student’s t test. All experiments are repeated at least twice and representative data are shown. All scanned images were quantified by NIH image software.
Supplementary Material
Acknowledgments
This work was supported in part by the National Institutes of Health grant, DK50828, to HSS. We thank Dr. Benoit de Crombrugghe (University of Texas M. D. Anderson Cancer Center, Houston) and Dr. Ormond A. MacDougald (University of Michigan Medical School, Ann Arbor) for Sox9 plasmid and for MC3T3 cells. We thank Roger Wong for technical help and Drs. Mary Ann Williams, Robin Duncan and Maryam Ahmadian for careful reading of the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abdallah BM, Jensen CH, Gutierrez G, Leslie RG, Jensen TG, Kassem M. Regulation of human skeletal stem cells differentiation by Dlk1/Pref-1. J Bone Miner Res. 2004;19:841–852. doi: 10.1359/JBMR.040118. [DOI] [PubMed] [Google Scholar]
- Akiyama H, Lyons JP, Mori-Akiyama Y, Yang X, Zhang R, Zhang Z, Deng JM, Taketo MM, Nakamura T, Behringer RR, et al. Interactions between Sox9 and beta-catenin control chondrocyte differentiation. Genes Dev. 2004;18:1072–1087. doi: 10.1101/gad.1171104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachmann E, Krogh TN, Hojrup P, Skjodt K, Teisner B. Mouse fetal antigen 1 (mFA1), the circulating gene product of mdlk, pref-1 and SCP-1: isolation, characterization and biology. J Reprod Fertil. 1996;107:279–285. doi: 10.1530/jrf.0.1070279. [DOI] [PubMed] [Google Scholar]
- Bi W, Huang W, Whitworth DJ, Deng JM, Zhang Z, Behringer RR, de Crombrugghe B. Haploinsufficiency of Sox9 results in defective cartilage primordia and premature skeletal mineralization. Proc Natl Acad Sci U S A. 2001;98:6698–6703. doi: 10.1073/pnas.111092198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Rocha ST, Ferguson-Smith AC. Genomic imprinting. Curr Biol. 2004;14:R646–649. doi: 10.1016/j.cub.2004.08.007. [DOI] [PubMed] [Google Scholar]
- de Crombrugghe B, Lefebvre V, Nakashima K. Regulatory mechanisms in the pathways of cartilage and bone formation. Curr Opin Cell Biol. 2001;13:721–727. doi: 10.1016/s0955-0674(00)00276-3. [DOI] [PubMed] [Google Scholar]
- Farmer SR. Transcriptional control of adipocyte formation. Cell Metab. 2006;4:263–273. doi: 10.1016/j.cmet.2006.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaur T, Lengner CJ, Hovhannisyan H, Bhat RA, Bodine PV, Komm BS, Javed A, van Wijnen AJ, Stein JL, Stein GS, Lian JB. Canonical WNT signaling promotes osteogenesis by directly stimulating Runx2 gene expression. J Biol Chem. 2005;280:33132–33140. doi: 10.1074/jbc.M500608200. [DOI] [PubMed] [Google Scholar]
- Gesta S, Tseng YH, Kahn CR. Developmental origin of fat: tracking obesity to its source. Cell. 2007;131:242–256. doi: 10.1016/j.cell.2007.10.004. [DOI] [PubMed] [Google Scholar]
- Gregoire FM, Smas CM, Sul HS. Understanding adipocyte differentiation. Physiol Rev. 1998;78:783–809. doi: 10.1152/physrev.1998.78.3.783. [DOI] [PubMed] [Google Scholar]
- Kaneta M, Osawa M, Sudo K, Nakauchi H, Farr AG, Takahama Y. A role for pref-1 and HES-1 in thymocyte development. J Immunol. 2000;164:256–264. doi: 10.4049/jimmunol.164.1.256. [DOI] [PubMed] [Google Scholar]
- Karaplis AC, Luz A, Glowacki J, Bronson RT, Tybulewicz VL, Kronenberg HM, Mulligan RC. Lethal skeletal dysplasia from targeted disruption of the parathyroid hormone-related peptide gene. Genes Dev. 1994;8:277–289. doi: 10.1101/gad.8.3.277. [DOI] [PubMed] [Google Scholar]
- Kim KA, Kim JH, Wang Y, Sul HS. Pref-1 (preadipocyte factor 1) activates the MEK/extracellular signal-regulated kinase pathway to inhibit adipocyte differentiation. Mol Cell Biol. 2007;27:2294–2308. doi: 10.1128/MCB.02207-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laborda J, Sausville EA, Hoffman T, Notario V. dlk, a putative mammalian homeotic gene differentially expressed in small cell lung carcinoma and neuroendocrine tumor cell line. J Biol Chem. 1993;268:3817–3820. [PubMed] [Google Scholar]
- Lee K, Villena JA, Moon YS, Kim KH, Lee S, Kang C, Sul HS. Inhibition of adipogenesis and development of glucose intolerance by soluble preadipocyte factor-1 (Pref-1) J Clin Invest. 2003;111:453–461. doi: 10.1172/JCI15924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengner CJ, Lepper C, van Wijnen AJ, Stein JL, Stein GS, Lian JB. Primary mouse embryonic fibroblasts: a model of mesenchymal cartilage formation. J Cell Physiol. 2004;200:327–333. doi: 10.1002/jcp.20118. [DOI] [PubMed] [Google Scholar]
- Liu Z, Xu J, Colvin JS, Ornitz DM. Coordination of chondrogenesis and osteogenesis by fibroblast growth factor 18. Genes Dev. 2002;16:859–869. doi: 10.1101/gad.965602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mei B, Zhao L, Chen L, Sul HS. Only the large soluble form of preadipocyte factor-1 (Pref-1), but not the small soluble and membrane forms, inhibits adipocyte differentiation: role of alternative splicing. Biochem J. 2002;364:137–144. doi: 10.1042/bj3640137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon YS, Smas CM, Lee K, Villena JA, Kim KH, Yun EJ, Sul HS. Mice lacking paternally expressed Pref-1/Dlk1 display growth retardation and accelerated adiposity. Mol Cell Biol. 2002;22:5585–5592. doi: 10.1128/MCB.22.15.5585-5592.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami S, Kan M, McKeehan WL, de Crombrugghe B. Up-regulation of the chondrogenic Sox9 gene by fibroblast growth factors is mediated by the mitogen-activated protein kinase pathway. Proc Natl Acad Sci U S A. 2000;97:1113–1118. doi: 10.1073/pnas.97.3.1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen ED, MacDougald OA. Adipocyte differentiation from the inside out. Nat Rev Mol Cell Biol. 2006;7:885–896. doi: 10.1038/nrm2066. [DOI] [PubMed] [Google Scholar]
- Smas CM, Chen L, Sul HS. Cleavage of membrane-associated pref-1 generates a soluble inhibitor of adipocyte differentiation. Mol Cell Biol. 1997;17:977–988. doi: 10.1128/mcb.17.2.977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smas CM, Chen L, Zhao L, Latasa MJ, Sul HS. Transcriptional repression of pref-1 by glucocorticoids promotes 3T3-L1 adipocyte differentiation. J Biol Chem. 1999;274:12632–12641. doi: 10.1074/jbc.274.18.12632. [DOI] [PubMed] [Google Scholar]
- Smas CM, Green D, Sul HS. Structural characterization and alternate splicing of the gene encoding the preadipocyte EGF-like protein pref-1. Biochemistry. 1994;33:9257–9265. doi: 10.1021/bi00197a029. [DOI] [PubMed] [Google Scholar]
- Smas CM, Kachinskas D, Liu CM, Xie X, Dircks LK, Sul HS. Transcriptional control of the pref-1 gene in 3T3-L1 adipocyte differentiation. Sequence requirement for differentiation-dependent suppression. J Biol Chem. 1998;273:31751–31758. doi: 10.1074/jbc.273.48.31751. [DOI] [PubMed] [Google Scholar]
- Smas CM, Sul HS. Pref-1, a protein containing EGF-like repeats, inhibits adipocyte differentiation. Cell. 1993;73:725–734. doi: 10.1016/0092-8674(93)90252-l. [DOI] [PubMed] [Google Scholar]
- Takeda S, Bonnamy JP, Owen MJ, Ducy P, Karsenty G. Continuous expression of Cbfa1 in nonhypertrophic chondrocytes uncovers its ability to induce hypertrophic chondrocyte differentiation and partially rescues Cbfa1-deficient mice. Genes Dev. 2001;15:467–481. doi: 10.1101/gad.845101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Sul HS. Ectodomain shedding of preadipocyte factor 1 (Pref-1) by tumor necrosis factor alpha converting enzyme (TACE) and Inhibition of Adipocyte Differentiation. Mol Cell Biol. 2006;26:5421–5435. doi: 10.1128/MCB.02437-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.