Abstract
Retroviral replication proceeds through the formation of a provirus, an integrated DNA copy of the viral RNA genome. The linear cDNA product of reverse transcription is the integration substrate and two different integrase activities, 3′ processing and DNA strand transfer, are required for provirus formation. Integrase nicks the cDNA ends adjacent to phylogenetically-conserved CA dinucleotides during 3′ processing. After nuclear entry and locating a suitable chromatin acceptor site, integrase joins the recessed 3′-OHs to the 5′-phosphates of a double-stranded staggered cut in the DNA target. Integrase functions in the context of a large nucleoprotein complex, called the preintegration complex (PIC), and PICs are analyzed to determine levels of integrase 3′ processing and DNA strand transfer activities that occur during acute virus infection. Denatured cDNA end regions are monitored by indirect end-labeling to measure the extent of 3′ processing. Native PICs can efficiently integrate their viral cDNA into exogenously added target DNA in vitro, and Southern blotting or nested PCR assays are used to quantify the resultant DNA strand transfer activity. This study details HIV-1 infection, PIC extraction, partial purification, and quantitative analyses of integrase 3′ processing and DNA strand transfer activities.
Keywords: HIV-1, AIDS, integration, integrase, preintegration complex, 3′ processing activity, DNA strand transfer activity, real-time PCR
1. Introduction
The vast majority of animal viruses replicate efficiently while maintaining their genomes separate from those of their hosts. Retroviruses, by contrast, are the only animal viruses that must intertwine their genomes with their hosts. The integrated form of the virus, the provirus, is required for efficient gene expression and ensures for equal segregation of the genetic material to both daughter cells upon division.
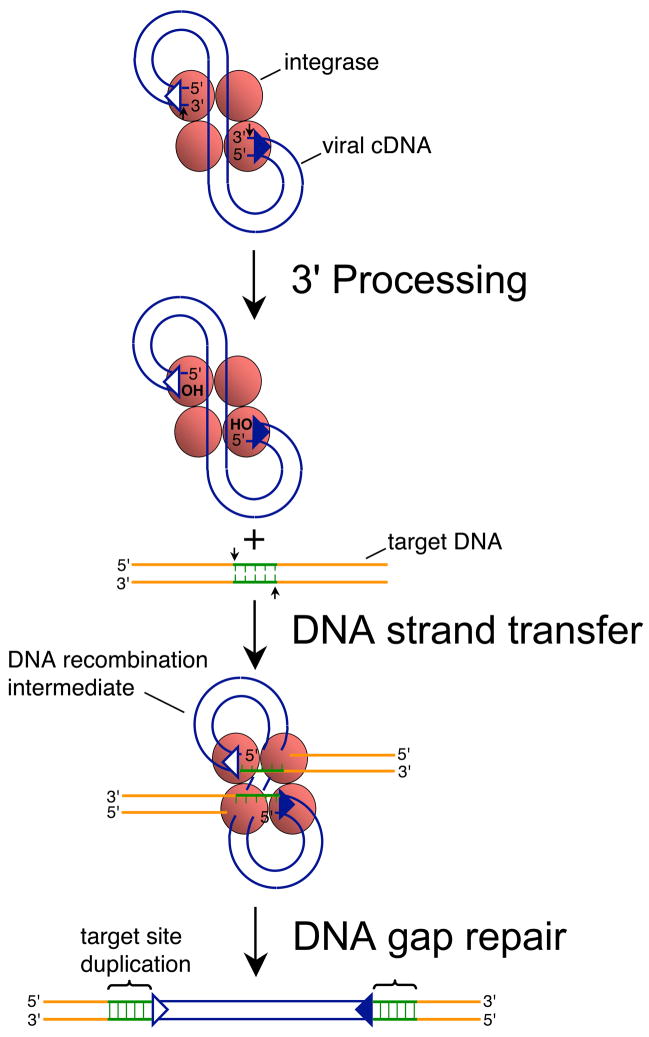
The retroviral genome is RNA. Retroviruses must therefore convert their genomes into double-stranded cDNA as a prelude to integration. The viral-encoded integrase enzyme then catalyzes two distinct endonucleolytic reactions en route to provirus formation. In the first reaction, which is called 3′ processing, integrase nicks the linear ends of the reverse transcript adjacent to the conserved sequence 5′-CA-3′, which in the majority of cases hydrolyzes a dinucleotide from each 3′ end [1–5] (Fig. 1). The second reaction, DNA strand transfer, links the viral and host DNAs together. Integrase uses the recessed CAOH ends to cleave the phosphodiester backbone of chromosomal DNA in a staggered fashion, which at the same time joins the viral 3′ ends to the resulting 5′-phosphates [6]. The DNA recombination intermediate, with free viral DNA 5′ ends abutting single-stranded gaps [1, 2, 5], is repaired by host cell enzymes to yield the provirus flanked by the duplicated sequence of the staggered cut made during DNA strand transfer (Fig. 1).
Fig. 1.
Mechanism of retroviral DNA integration. Reverse transcription yields linear cDNA with a copy of U3/R/U5 sequences, also known as the long terminal repeat (LTR), at each end. Integrase recognizes approximately 15–20 bp of each end region [39, 40]. The upstream recognition or attachment (att) site is therefore comprised of U3 sequences (open triangle) whereas the downstream att site is U5 (filled blue triangle). Integrase (red shaded ball) cleaves each att site adjacent to the invariant sequence 5′-CA-3′ during 3′ processing (marked by small vertical arrows), removing the dinucleotide GT from each end of HIV-1 cDNA [4, 6]. Integrase uses the oxygen nucleophiles at the exposed CAOH ends during DNA strand transfer to cut the target DNA at the small vertical arrows, which at the same time joins the viral ends to the resultant 5′-phosphates [6]. The DNA recombination intermediate, with unjoined viral DNA 5′ ends, is the structure formed in in vitro PIC integration assays [1, 2, 5]. DNA repair yields the integrated provirus flanked by the sequence duplication of the target DNA staggered cut (green lines), which is 5 bp for HIV-1 [41, 42]. Integrase functions as a multimer in vitro and in vivo, though the precise make-up of the active multimer during infection is unknown. It is depicted here as a tetramer for simplicity.
Integrase proteins purified from a variety of recombinant sources display 3′ processing and DNA strand transfer activities in vitro. These simplified biochemical assays were central for dissecting critical aspects of enzyme function, as well as establishing high throughput screens for the identification of integrase inhibitors (refer to chapters by Grandgenett, Merkel, and Grobler within this Methods issue). During infection, integrase functions in the context of the preintegration complex (PIC), a relatively large nucleoprotein structure that is derived from the core of the incoming virion [7–9]. Following extraction, levels of integrase 3′ processing activity can be quantified by digesting the purified viral cDNA with restriction endonucleases to yield approximate 100 bp end regions, and analyzing the resulting structures after denaturation for dinucleotide loss from the U5 plus- and U3 minus-strands [1–5, 9] (Fig. 2). Native PICs moreover support DNA strand transfer activity in vitro [1, 2, 5, 10–12]. These seminal studies revealed several unknown mechanistic aspects of retroviral integration, including that (i) it occurred in the context of a higher-order nucleoprotein complex, (ii) proceeded in the absence of an added high-energy cofactor like ATP, and (iii) the immediate precursor was linear cDNA and not circular DNA forms that also arise during infection. PIC analyses remain central to investigations of integrase catalytic function in the context of the infected cell. This paper details biochemical analyses of HIV-1 PICs.
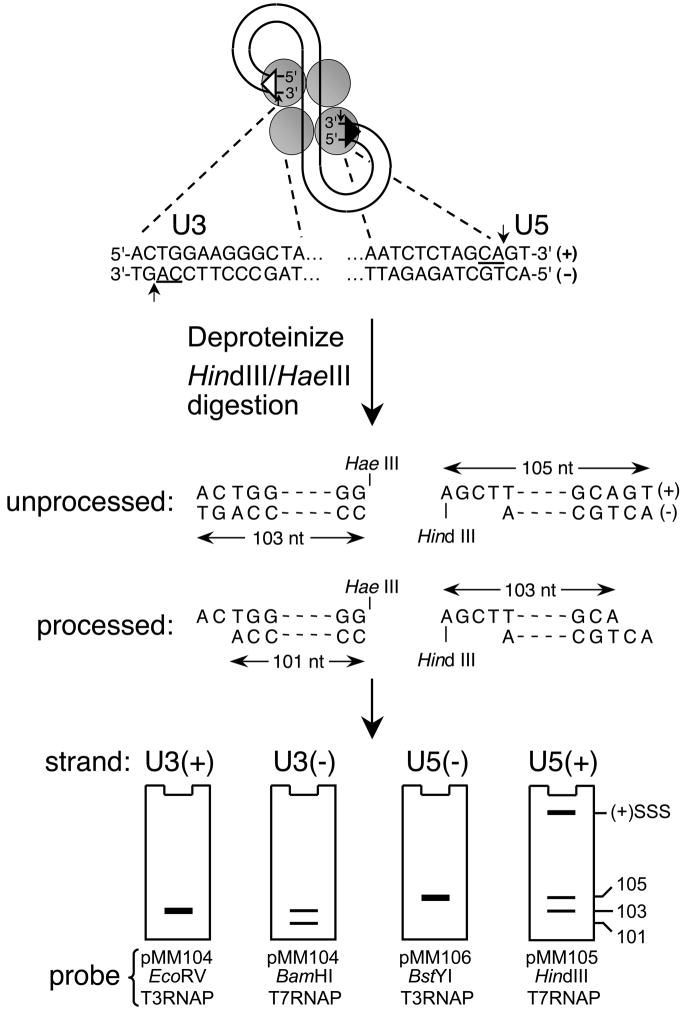
Fig. 2.
Indirect end-labeling of HIV-1 cDNA end structures. The terminal 13 bp of the U3 and U5 DNA att sites are shown beneath the HIV-1 PIC, with the phosphodiester bonds cleaved during 3′ processing and invariant CA dinucleotides marked by vertical arrows and underlines, respectively. The viral DNA ends are isolated as approximate 100 bp regions upon digestion with HindIII and HaeIII. The initial U3 minus-strand is 103 nucleotides, whereas integrase processing yields a 101 nt product. The unprocessed U5 plus-strand is 105 bases, with 3′ processing generating a shortened 103 nt strand. The U3 plus- and U5 minus-strands by contrast remain unchanged during 3′ processing. The various end structures are visualized by indirect end-labeling using riboprobes generated from the indicated linearized plasmid DNA templates, with the cartoon gels depicting the approximate mobilities of the different strands/3′ processing reaction products. (+)SSS, plus-strand strong-stop DNA; RNAP, RNA polymerase.
2. Description of methods
The original PIC assay utilized a genetic approach to quantify the level of in vitro integration [10]. Extracts were prepared from cells infected with Moloney murine leukemia virus (MLV) modified to contain the bacterial SupF gene, and purified bacteriophage lambda gtWES DNA was added as a surrogate to cell chromosomes. Following in vitro integration, purified DNA was reacted with phage lambda packaging extract, and the resulting phage were plated on two different indicator cells. Bacteria that naturally complemented the nonsense codons in lambda gtWES yielded overall phage titer, whereas phage that had acquired SupF via MLV integration gained the ability to plate on suppressor-minus cells. The fraction of suppressor-minus colonies thus defined the frequency of MLV DNA integration. Importantly, the resulting proviruses lost 2 bp from each end of viral DNA and were flanked by a 4 bp duplication of lambda DNA sequence at the site of integration. The advent of this in vitro assay that recapitulated the known genetics of MLV integration in one fell swoop redefined the retroviral integration field.
Though quantitative, the Brown et al. [10] assay was laborious and moreover scored integration indirectly through phage lambda plating efficiencies. Deciphering viral cDNA end structures either before or after integration were therefore not approachable using the genetic screen. Direct physical assays were needed, and Fujiwara and Mizuuchi spearheaded the use of indirect end-labeling to analyze viral DNA end structures as well as to monitor the formation of the in vitro DNA recombination intermediate [1]. Native PICs were reacted with linearized bacteriophage φX174 target DNA, and purified DNAs were separated by agarose gel electrophoresis. Blotting with a virus-specific probe yielded two linear DNA species: the faster migrating cDNA substrate represented unreacted PICs, whereas the more slowly migrating band identified the MLV DNA recombination intermediate (Fig. 3). PIC activity could therefore be defined as the fraction of viral cDNA converted into integration reaction product. Monitoring integrase 3′ processing and DNA strand transfer activities by indirect end-labeling was quickly adapted to other viral systems such as HIV-1 [4, 11, 12] and Rous sarcoma virus [5].
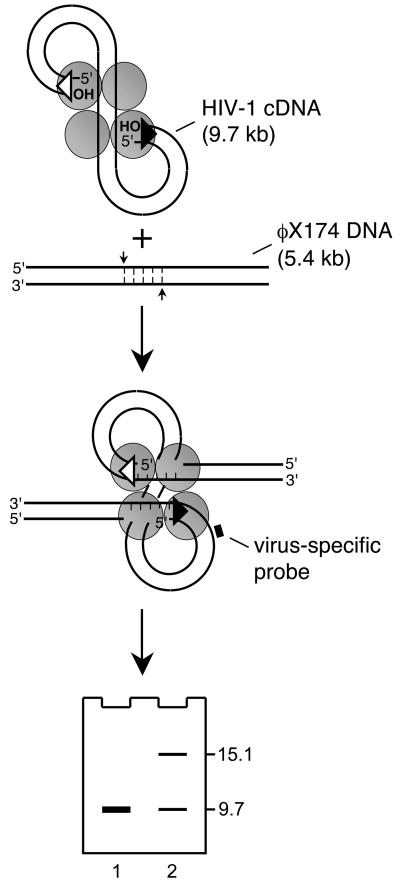
Fig. 3.
Southern blot assay for PIC function. Integration of 9.7 kb HIV-1 cDNA into linearized 5.4 kb φX174 yields a 15.1 kb DNA recombination intermediate. Reaction mixtures following deproteinization are fractionated by agarose gel electrophoresis. After Southern blotting with a virus-specific probe, DNA strand transfer activity is defined as the percent of cDNA substrate converted into reaction product (lane 2; φX174 DNA was omitted from the integration reaction analyzed in lane 1). Though integration occurs essentially throughout the φX174 genome, the resulting population of recombinants displays an electrophoretic mobility consistent with linear double-stranded 15.1 kb DNA.
Indirect end-labeling remains the industry-standard for quantifying integrase activities in the context of viral infection. Considering the utilization of defined target DNA molecules in vitro, DNA strand transfer reaction products can also be discerned via PCR amplification, and a number of assay designs have been described [13–18]. The main advantage of PCR over Southern blotting is that less input PIC material is in general required for a positive readout, an important consideration for high throughput approaches. One drawback of PCR is that the detection of the DNA recombination product is indirect. In other words, as compared to Southern blotting, the complete DNA recombination intermediate is not visualized. Most PCR assays also amplify just one of the two viral-target DNA junctions. This is expected in the vast majority of cases to represent the concerted integration of both viral DNA ends into opposing target DNA strands with defined spacing (Fig. 1, DNA recombination intermediate), though it should be noted that under certain conditions, for example when one of the viral DNA ends is mutated, the mutant end can insert independent of integrase function [19].
2.1 Step-by-step protocols
PICs are extracted from acutely-infected cells during the peak of reverse transcription, which can vary among different retroviruses (cells infected with HIV-1 are generally lysed 5–8 h post-infection, whereas MLV infections proceed overnight).
2.1.1 HIV-1 infection
Relatively high multiplicities of infection are essential for PIC analyses. Additional parameters of cell culture, some of which are undefined, can greatly affect the level of in vitro DNA strand transfer activity. In our experience it is essential to keep cell lines in optimal working order. Maintain the concentrations of suspension cell cultures at 0.1–1.0 × 106/ml unless stated otherwise. Do not let adherent 293T cells reach confluency, and limit the amount of time these cells are exposed to trypsin during subculture. Grow all flasks broad side-down in a 5% CO2 humidified incubator to maximize gas exchange. Test different lots of fetal bovine serum (FBS) for those that yield optimal virus production from transiently-transfected 293T cells, and secure as many bottles of the optimal lot as feasible.
SupT1 [20] and C8166-45 [21] T-cells support efficient HIV-1 reverse transcription and are therefore preferred target cells [22]. Infections are initiated by incubating uninfected cells with chronically-infected virus-producer cells or cell-free virus derived via transient transfection [22]. Identify individual lots of cell lines that yield efficient levels of reverse transcription and in vitro PIC integration activity, and carefully expand these to make additional freezer vials. We routinely thaw fresh cells and expand these to sufficient numbers for each experiment.
2.1.1.1 Infection by co-culture
MOLTIIIB cells, which are chronically infected with the IIIB strain of HIV-1 [12], as well as uninfected SupT1 and C8166-45 cells, are grown in RPMI 1640 media supplemented to contain 10% v/v heat-inactivated FBS, 100 IU of penicillin G sodium, and 0.1 mg of streptomycin sulfate per ml. MOLTIIIB cells are treated in one of two ways the day before infection. First, incubate the cells to be used in the co-culture at 4 × 105/ml with 10 ng/ml phorbol 12-myristate 13-acetate (PMA; Sigma-Aldrich catalog number P1585) to stimulate virus production [12, 22]. Plate a second lot of cells at 106/ml (without PMA) to prepare virus-conditioned media. Twenty-four hours later, co-culture 4 × 107 SupT1 cells with 4 × 106 PMA-stimulated MOLTIIIB cells in a final volume of 20 ml of virus-conditioned media for 5 h. The majority of cells should fuse to form giant multi-nucleated syncytia during this time.
2.1.1.2 Cell-free virus infection
High-titer virus is made by transfecting 293T cells with pNL4-3 [23] or pNL43/XmaI, a somewhat smaller derivative lacking ~1.1 kb of flanking human DNA [24]. Grow 293T cells in Dulbecco’s modified Eagles medium (DMEM) containing 10% FBS, 100 IU of penicillin G sodium, and 0.1 mg of streptomycin sulfate per ml (DMEM-FBS). A number of transfection techniques yield virus of sufficient titer. Calcium-phosphate co-precipitation is desirable due to its relatively low cost. It is important to treat resulting virus-containing supernatants with DNase to degrade the bulk of the plasmid DNA that remains after transfection. This step is more efficient with lipid-based as compared to calcium-phosphate transfection reagents. We therefore generate virus by transfection with Fugene 6 as recommended by the manufacturer (Roche Applied Science catalog number 11814443001) for experiments that examine reverse transcription and/or integration by PCR. For indirect end-labeling studies, virus generated by the following calcium-phosphate co-precipitation method suffices.
Seed 3.3 × 106 293T cells in 10 ml DMEM-FBS per 10 cm tissue culture dish 24 h prior to transfection (if need be, adjust the cell number such that 60–70% confluency is attained by the time of transfection).
Add 0.5 ml of 2x HEPES-buffered saline (2x HBS; 280 mM NaCl, 50 mM HEPES-NaOH pH 7.1, 1.5 mM Na2HPO4) to the bottom of a 15 ml conical tube1. In a separate tube, mix 20 μg of plasmid DNA in a final volume of 450 μl sterile water, add 50 μl 2.5 M CaCl2, and mix thoroughly.
Add the DNA–calcium mixture drop-wise to the 2x HBS while simultaneously introducing bubbles via continuous pipetting of air through the solution. Allow the DNA to precipitate for 30 min at room temperature.
Add the precipitate drop-wise to the plate. Incubate the DNA–cell mixture for 14–17 h.
Rinse the cells twice with 4 ml serum-free DMEM. Add 8 ml DMEM-FBS–10 mM HEPES pH 7.6, and culture the cells for 24 h.
Collect virus-containing cell supernatant, pellet cell debris by centrifugation at 300g (1,200 rpm in a Sorvall RT 6000D centrifuge equipped with H1000B rotor) for 10 min, and filter the resulting supernatant by gravity through a 0.45 μm filter. Keep a 100 μl aliquot for determination of virus yield, which can be measured using an Alliance HIV-1 p24 Antigen ELISA Kit (PerkinElmer catalog number NEK050001KT) or exogenous reverse transcriptase (RT) assay [25]. Yields should be 4–10 × 107 32P RT-cpm/ml, which equate to ~3–10 μg p24/ml.
Treat the remaining virus supernatant with 40 U/ml Turbo DNase (Ambion catalog number AM2238) for 1 h at 37°C prior to infection.
Infect 3 × 107 C8166-45 T cells with 15 ml of DNase-treated virus for 7 h at 37°C. Unlike the co-culture method described in the previous section, this technique does not yield efficient syncytia formation.
SupT1 cells (4 × 107) can be substituted for C8166-45 T cells, though SupT1 cells should be infected by spinoculation due to their somewhat lower level of susceptibility [22]. Distribute the 15 ml cell-virus mixture evenly among a 6-well tissue culture plate, and centrifuge at 480g (1,800 rpm in the H1000B rotor) for 2 h at room temperature. Incubate the cells an additional 5 h at 37°C after the spin.
2.1.2 Cell lysis and PIC preparation
Cells lysed under non-denaturing conditions are separated into cytoplasmic and nuclear fractions.
1. Harvest cells at 300g for 10 min, and wash the cell pellet twice with buffer K−/− (20 mM HEPES pH 7.6, 150 mM KCl, 5 mM MgCl2) at room temperature. Remove residual buffer using a micropipettor.
2. Resuspend the pellet in 1 ml ice-cold buffer K+/+ (20 mM HEPES pH 7.6, 150 mM KCl, 5 mM MgCl2, 1 mM DTT, 20 μg/ml aprotinin, 0.025% w/v digitonin)2 by pipetting. Transfer to a 1.5 ml microcentrifuge tube and rock on a Nutator platform for 10 min at room temperature.
3. Separate the lysate into cytoplasmic (supernatant) and nuclear (pellet) fractions by centrifugation at 1,500g for 4 min at 4°C. All further manipulations are performed on ice using chilled buffers unless stated otherwise.
2.1.2.1 Cytoplasmic PIC preparation
4. Transfer the supernatant to a fresh microcentrifuge tube and centrifuge at 19,000g for 1 min at 4°C.
5. Transfer the supernatant to a fresh tube, add RNase A (QIAGEN catalog number 19101) to 20 μg/ml, and incubate for 30 min at room temperature to degrade cellular mRNA. This is the crude, cytoplasmic PIC fraction. Place on ice or freeze for later use.
6. PICs can be stored at −80°C for approximately 6 months without significant loss of integration activity. Add sucrose (60% w/v in Buffer K−/−) to a final concentration of 7%, flash freeze in liquid N2, and place at −80C. Thaw on ice as needed.
2.1.2.2 Preparation of nuclear PICs
7. Resuspend the nuclear pellet from step 3 in 1 ml buffer K+/− (20 mM HEPES pH 7.6, 150 mM KCl, 5 mM MgCl2, 1 mM DTT, 20 μg/ml aprotinin, 20 μg/ml RNase A)2, and transfer to a 5 ml polypropylene tube (Falcon catalog number 2063) for lysis.
8. Lyse the nuclei by ball bearing homogenization. We use a mounted PRO 250 unit (PRO Scientific) at a power setting that is sufficient for lysis but at the same time minimizes sample heating and frothing. Homogenize for 30 sec followed by 30 sec of cooling. Monitor lysis by light microscopy using samples diluted 1:10 in buffer K+/−. In our experience, 10 cycles of 30 sec homogenization–30 sec cooling lyses 80–90% of nuclei [26].
9. Transfer the lysate to a new microcentrifuge tube, spin at 19,000g for 3 min at 4°C, and transfer the supernatant to a fresh tube. This is the crude nuclear PIC fraction. Place on ice or flash freeze as described in step 6 for storage at −80°C.
2.1.3 Partial PIC purification by spin column chromatography
HIV-1 PICs can be partially purified by gel filtration chromatography using fast performance liquid handling devices [9, 11, 12] or centrifugation [22, 27]. We prefer Sepharose CL-4B (Sigma-Aldrich catalog number CL4B200) spin columns assembled in Bio-Rad Econo-Pac columns (Fig. 4).
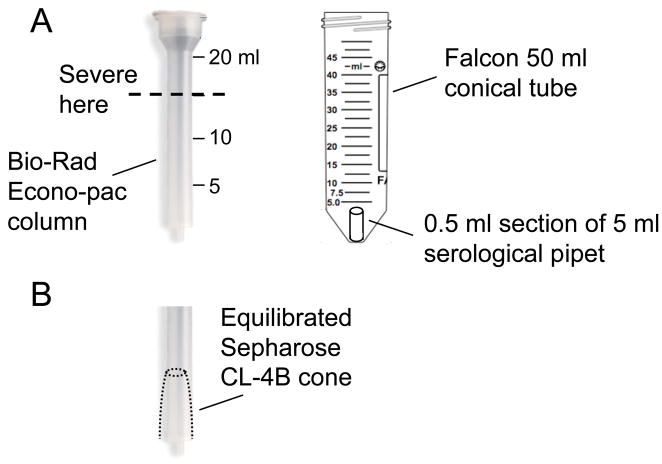
Fig. 4.
Partial PIC purification by spin column chromatography. (A) To accommodate the Bio-Rad Econo-Pac column in an appropriate centrifuge, it may be necessary to severe off the top as indicated. An approximate 0.5 ml section of a 5 ml serological pipet is used to prop the column up from the bottom of a 50 ml conical tube during centrifugation. (B) The spun bed should appear as a contracted cone with flattened top.
Sepharose CL-4B is sold as a slurry in 20% ethanol. Equilibrate beads by transferring 20 ml packed volume to a 50 ml conical tube, spinning at 300g for 5 min at room temperature, decanting the supernatant, and thoroughly mixing the beads with 20 ml H2O. Repeat. Then wash the beads three times with buffer K−/−. Store unused beads at 4°C in the presence of 0.02% w/v Na azide. Wash the azide from the stored beads prior to spin column preparation.
Prepare a spin column with the equilibrated beads. The Econo-Pac column (catalog number 732–1010) is too tall for a Sorvall 6000D centrifuge. To accommodate, cut the top off at approximately the 15 ml mark (Fig. 4A). This is most easily done by heating the blade of a utility knife in a flame to melt the plastic during cutting. During centrifugation, prop the column up from the bottom of a 50 ml Falcon conical tube holder using an approximate 0.5 ml section of a plastic 5 ml serological pipet prepared using the heated knife blade (Fig. 4A). The spin column volume is determined by the volume of extract to be purified. A 3 ml wet bed volume is used for 0.5 ml input sample, whereas 7 ml and 12 ml bed volumes are used for 1.0 ml and 1.5 ml samples, respectively. Spin the poured column at 200g (1,000 rpm in the H1000B rotor) for 2 min at 4°C. Carefully inspect the resultant packed bed– it should resemble a cone shape with flat top (Fig. 4B). If the top of the bed is cracked, dispose and start again. To equilibrate the column, add the appropriate volume of buffer (K+/+ if purifying cytoplasmic extract; K+/− for nuclear extract) drop-wise to the top of the packed bed, taking care to avoid the inner sides of the plastic cylinder. Centrifuge at 200g for 2 min at 4°C. Discard the flow-through, and repeat except increase the spin to 830g (2,000 rpm in the H1000B rotor). Discard the flow-through.
Carefully load the cell extract to the center of the equilibrated, packed bed. Place a fresh microcentrifuge tube in the 50 ml tube holder, and collect the flow-through during centrifugation at 830g for 2 min at 4°C. Place the purified sample on ice.
Mix 18 μl of the input and spin column-purified samples with 2 μl 1% w/v Na dodecyl sulfate (SDS), and determine protein concentration using the Bio-Rad Dc Protein Assay Kit (catalog number 500–0116). Due to the relatively large PIC size, the spin column removes 80–90% of total protein without significantly affecting the yield of HIV-1 cDNA [22].
2.1.4 Quantitation of integrase 3′ processing activity by indirect end-labeling
Purified viral DNA cleaved with restriction endonucleases is denatured, fractioned through sequencing gels, and transferred to a solid support to visualize the end structures via strand-specific riboprobes [1–5, 9, 26, 28].
Add proteinase K, SDS, and EDTA to 1 ml of PIC cytoplasmic extract (section 2.1.2.1, step 5) to the final concentrations of 0.5 mg/ml, 0.5%, and 8 mM, respectively. Deproteinize by heating overnight at 56°C.
Add an equal volume of phenol, mix thoroughly, and separate the aqueous and organic phases by centrifugation at 16,000g for 2 min at room temperature. Transfer the aqueous phase to a fresh tube, and repeat. Transfer the resultant aqueous phase to a fresh tube, and an equal volume of a 1:1 mixture of phenol/chloroform, mix thoroughly, and separate the phases by centrifugation. Transfer the resultant supernatant to a fresh tube, and perform a final extraction with chloroform. Transfer the supernatant to a fresh tube, add 1 μl linear polyacrylamide (LPA; Sigma-Aldrich catalog number 56575) and Na acetate to the final concentration of 0.3 M. Add 2.5 volumes of 100% ethanol, vortex, and pellet the DNA by centrifugation at 16,000g for 10 min at room temperature. Rinse the pellet with 80% ethanol, and lyophilize.
Digest the DNA in 50 μl using 20 U each of HaeIII and HindIII under conditions recommended by the manufacturer (New England Biolabs).
Add RNase A to the final concentration of 10 μg/ml, and incubate for 30 min at 37°C. Extract the reaction with phenol/chloroform, and recover the DNA by ethanol precipitation. Resuspend the dried pellet in 6 μl TE buffer (10 mM Tris pH 8.0, 1 mM EDTA).
Heat the DNA at 68°C for 30 min. Add 6 μl of sequencing stop buffer (95% formamide, 20 mM EDTA, 0.05% w/v bromophenol blue [BB], 0.05% w/v xylene cyanol [XC]). Heat the mixture at 68°C for another 30 min, and then boil for 3 min.
Electrophorese 6 μl of the sample at 60 W through a 6% denaturing polyacrylamide (sequencing) gel until the bromophenol blue dye reaches the bottom of the gel. In a separate set of lanes, fractionate the dideoxy sequencing reaction generated from M13 mp18 DNA using the −40 universal primer (Sequenase Version 2.0 DNA Sequencing Kit, USB Corporation catalog number 70770).
Transfer the DNA containing region of the gel to Duralon-UV membrane (Stratagene) using a TransBlot SD Cell (Bio-Rad) in 0.3X TBE buffer (27 mM Tris, 27 mM borate, 0.6 mM EDTA) at 50 mA for 1 h.
Remove the membrane, rinse briefly in 10X SSC (1.5 M NaCl, 0.15 M Na citrate), air dry, and bake at 80°C for 2 h under vacuum. Expose to UV light using the autocrosslink option on the UV Stratalinker 2400 (Stratagene).
Prepare 32P-labeled strand specific riboprobes using Promega’s Riboprobe System kits. The probe that detects the 103 nucleotide (nt) non-processed U3 plus-strand is transcribed from EcoRV-digested pMM104 plasmid DNA [9, 28] using bacteriophage T3 RNA polymerase, whereas the unprocessed and processed U3 minus-strands are visualized using the T7 RNA polymerase probe generated after BamHI digestion (Fig. 2). For the U5 end, the non-processed 105 nt minus-strand is detected using the T3 RNA polymerase probe transcribed from BstYI-digested pMM106, while the unprocessed and processed plus-strands are developed via the T7 RNA polymerase transcript from HindIII-cut pMM105 [9, 28] (Fig. 2). Determine the specific activities of the various riboprobes by counting 0.5 μl in a liquid scintillation counter.
Denature tRNA (10 mg/ml, Ambion catalog number AM7119) at 68°C for 5 min. Rinse the membrane in H2O for 2 min, and transfer it to a hybridization container. Pre-hybridize using QuikHyb hybridization solution (Stratagene catalog number 201220) containing 300 μg/ml denatured tRNA for minimally 1 h.
Calculate the volume of riboprobe needed to attain a final concentration of 2.0–5.0 × 106 cpm/ml within the utilized volume of QuikHyb solution. In a fresh microcentrifuge tube, add denatured tRNA to the final concentration of 300 μg/ml to the riboprobe, and heat at 68°C for 5 min. Add 0.5 ml preheated QuikHyb solution, and transfer the mixture to the prehybridization solution. Hybridize at 68°C for 1–4 h. Perform separate hybridization reactions for each viral DNA end strand (Fig. 2, bottom)
Wash the blot for 15 min in 500 ml of 2X SSC–0.1% SDS at room temperature on an orbital shaker. Repeat. Then wash for 10 min in 500 ml of 0.1X SSC–0.1% SDS at 60°C with agitation. Repeat the final wash, blot the membrane dry, wrap in saran wrap, and expose to X-ray film or phosphor screen. Read the M13 sequencing ladder to identify the region of the blot that contains 100–105 base DNAs. Integrase 3′ processing activity is quantified as the percent of the U3 minus- and U5 plus-strands converted into their 2 base shortened reaction products (Fig. 2).
2.1.5 Southern blot assay for PIC function
Assay 0.5 ml of crude or Sepharose CL-4B-purified PIC preparation for determination of DNA strand transfer activity.
Pre-linearize φX174 target DNA by digestion with XhoI. Extract with phenol/chloroform, and resuspend the ethanol precipitated DNA at 1 mg/ml in TE.1 buffer (10 mM Tris pH 8.0, 0.1 mM EDTA).
Add the linearized target DNA to the final concentration of 3 μg/ml to one set of samples, and omit the DNA from a second set as minus-target DNA control reactions. Incubate at 37°C for 45 min. Stop the reactions by adding proteinase K, SDS, and EDTA to the concentrates noted in step 1 of the previous section. Extract the denatured proteins and recover the DNA by ethanol precipitation as described in step 2 of the prior section. Resuspend the DNA in 5 μl TE.1 buffer.
Add 1 μl of 6X gel loading buffer (40% w/v sucrose, 0.25% BB, 0.25% XC), and electrophorese at 18 V for 15 h through a 0.6% agarose (I.D.NA, Cambrex catalog number 50170)–TAE (40 mM Tris-acetate pH 7.6, 1 mM EDTA) gel. Include 250 pg of BamHI-linearized pNL4-3 DNA [23] in a separate lane to control for hybridization sensitivity. Circulate the tank buffer during electrophoresis.
Stain the gel with 0.5 μg/ml ethidium bromide for 10 min in TAE buffer. Destain for 15 min using H2O. Record the electrophoresis pattern by photography under UV illumination.
Prepare the gel for transfer: Denature it for 15 min in 0.24 N HCl, neutralize for 30 min in 0.6 M NaCl–0.4 N NaOH, and equilibrate for 30 min in 1.5 M NaCl–0.5 M Tris pH 7.5.
Transfer the gel to GeneScreen Plus hybridization membrane (PerkinElmer catalog number NEF994001PK). A variety of techniques can be utilized. We favor the Stratagene PosiBlot 30-30 Pressure Blotter; consult http://www.stratagene.com/produts/displayproduct.aspx?pid =291 or reference [18] for detailed instructions. See [29] or [30] for detailed capillary-based transfer methods.
After transfer, rinse the membrane with 2X SSC. Blot it dry, wrap in saran wrap, and crosslink the DNA as described in step 8 of the previous section. Unwrap the membrane and wash it thoroughly with H2O to remove excess salt. Prehybridize the membrane in QuikHyb solution at 65°C.
Prepare the 32P-labeled riboprobe from HindIII-digested pSP73/XH plasmid [18] using T7 RNA polymerase. Hybridize the membrane at 65°C for 1 h in QuikHyb solution containing 0.1 mg salmon sperm DNA (Stratagene catalog number 201190) and 2.0 × 106 cpm of riboprobe per ml.
Wash the membrane with 100 ml 2X SSC–0.5% SDS at 65°C for 10 min with agitation, and then repeat. Wash the membrane twice with 250 ml 0.5X SSC–0.1% SDS at room temperature with agitation, followed by two washes with 250 ml 0.3X SSC-0.1% SDS at room temperature with agitation. Blot the membrane dry, warp it in saran wrap, and expose to phosphor screen or X-ray film. The 14.9 kb linearized plasmid control should be readily discerned after overnight exposure. DNA strand transfer activity is quantified as the percent of 9.7 kb viral cDNA converted into the 15.1 kb reaction product (diagrammed as 50% in Fig. 3).
2.1.6 Quantitation of PIC activity via nested PCR
Our assay quantifies the extent of U5 att site integration into circular pTZ18U/PL plasmid DNA (Fig. 5). An initial exponential PCR amplifies HIV-plasmid DNA junctions, whereas the second real-time or kinetic PCR quantifies the resulting levels of HIV-1 DNA sequences [16, 18]. The revamped method here incorporates the AE3014 first round chimera primer comprised of bacteriophage λ and HIV-1 R sequences (Fig. 5). This design, adopted from Brussel and Sonigo [31] who used it to measure integration into human cellular DNA, increases overall signal-to-noise by incorporating λ DNA-specific primer AE3013 in the nested quantitative (Q) PCR. See Table 1 for primer and TaqMan probe sequences.
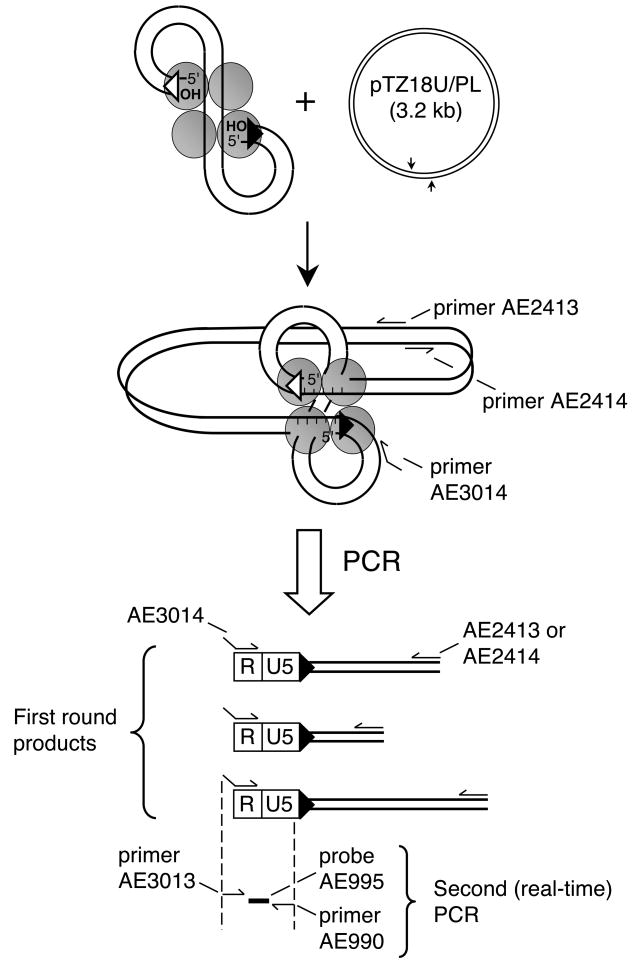
Fig. 5.
PCR assay for PIC function. Integration of HIV-1 cDNA (9.7 kb) into circular 3.2 kb pTZ18U/PL yields a 12.9 kb circular DNA recombination intermediate. Following deproteinization, reaction mixtures are subjected to two PCR rounds to specifically amplify newly-formed HIV-target DNA junctions. Primers AE2413 and AE2414 anneal to opposite strands of pTZ18U/PL in tail-to-tail fashion [16], whereas primer AE3014 is a hybrid composed of lambda and HIV-1 R DNA sequences. First round reaction products are a population of fragments with a fixed viral R end abutting variably-lengthened pTZ18U/PL sequences. The level of HIV-1-specific DNA in this population is quantified in the second real-time PCR using primers AE3013 and AE990 with probe AE995.
Table 1.
Oligonucleotides for PCR-based integration assay
| AE2413 | 5′-GTTGTTCCAGTTTGGAACAAGAGTC-3′ |
| AE2414 | 5′-ACTCAACCCTATCTCGGTCTATT-3′ |
| AE3014 | 5′-ATGCCACGTAAGCGAAACTCTGGCTAACTAGGGAACCCACTG-3′ |
| AE3013 | 5′-ATGCCACGTAAGCGAAACTC-3′ |
| AE990 | 5′-CTGACTAAAAGGGTCTGAGG-3′ |
| AE995 | 5′-[FAM]TTAAGCCTCAATAAAGCTTGCCTTGAGTGC[TAMRA]-3′ |
| MH531 | 5′-TGTGTGCCCGTCTGTTGTGT-3′ |
| MH532 | 5′-GAGTCCTGCGTCGAGAGAGC-3′ |
| LRT-P | 5′-[FAM]CAGTGGCGCCCGAACAGGGA[TAMRA]3′ |
FAM, 6-carboxyfluorescein; TAMRA, 6-carboxytetramethylrhodamine
Add pTZ18U/PL to 0.2 ml of PIC extract to the final concentration of 3 μg/ml, and incubate at 37°C for 45 min. Omit pTZ18U/PL from a second set of tubes for minus-target DNA control reactions. Stop the reaction and process the sample as described in step 2 of the previous section, except dissolve the purified DNA in 50 μl TE.1 buffer.
Since integration will occur at numerous locations within the pTZ18U/PL plasmid, molecular clones are not used to construct the PCR standard curve. Instead, construct an end-point dilution series of the reaction expected to yield the greatest level of DNA strand transfer activity. For example, if the experiment analyzed the activity of an integrase mutant virus, dilute the integration reaction that was conducted with wild-type PICs. If the effects of small molecules were investigated, dilute the sample extracted from cells infected in the absence of inhibitor [16].
-
Construct first round PCR samples:
5 μl DNA sample
5 μl 10X HotStarTaq buffer
1 μl 10 mM dNTP mix
0.5 μM (final concentration) primer AE2413
0.5 μM primer AE2414
0.5 μM primer AE3014
0.25 μl HotStarTaq polymerase (QIAGEN catalog number 203203)
→ H2O to 50 μl
The following two control PCRs are essential. The first substitutes TE.1 buffer for the integration reaction sample. The second omits target DNA primers AE2413 and AE2414 from the different integration reaction samples to account for potential HIV-1 cDNA carry over into the second round PCR. Master mixes containing all reaction components except DNA can be assembled on ice. Aliquot 45 μl into separate PCR tubes, and add 5 μl DNA or buffer control sample.
Incubate the reactions in a thermal cycler at 95°C for 5 min, followed by 23 cycles of 94°C for 30 sec, 58°C for 30 sec, and 72°C for 4 min. End with a 10 min extension at 72°C.
-
Dilute the first round reactions 1:200 in H2O, and assay by Q-PCR in duplicate:
5 μl diluted DNA sample
15 μl QuantiTect Probe PCR Mix (QIAGEN catalog number 204341)
0.3 μM primer AE3013
0.3 μM primer AE990
0.1 μM probe AE995
→ H2O to 30 μl
Include duplicate no DNA (water only) controls. A master mix lacking DNA and assembled on ice can be aliquoted to empty plate wells.
Incubate the reactions in a real-time thermal cycler for 15 min at 95°C, followed by 40 cycles of 94°C for 15 sec, 58°C for 30 sec, and 72°C for 30 sec.
-
It is important to normalize the resulting DNA strand transfer reaction products to the total levels of HIV-1 cDNA in the different samples. Q-PCR with primers that anneal to the U5 and gag regions of HIV-1 measure late reverse transcription (LRT) products that require the second template switch for their formation [32]. Construct a standard curve by diluting pNL4-3 five fold from 106 copies/μl to 64 copies/μl. Assay reactions in duplicate:
2 μl integration reaction or standard curve DNA
15 μl QuantiTect Probe PCR Mix
0.3 μM primer MH531
0.3 μM primer MH532
0.1 μM probe LRT-P
→ H2O to 30 μl
Include water-only control reactions. Cycle as described in the previous step.
DNA strand transfer activities are determined by comparing cycle threshold (Ct) values of unknown samples to those obtained with the end-point diluted integration standard curve. If Ct values above background are detected from the control reactions that lacked target DNA primers AE2413 and AE2414, subtract these from the values obtained in the presence of the primers. The adjusted Ct values are then normalized to the LRT levels within the different DNA samples. The assay is optimal when LRT values exceed 10,000 copies/μl of integration reaction, which equates to approximately 50 pg of HIV-1 cDNA/0.5 ml of PIC extract. As high multiplicity infections yield approximately 250 pg, the nested PCR assay for the determination of integrase DNA strand transfer activity is approximately 5 to 10-fold more sensitive than the Southern blotting assay.
3. Concluding Remarks
Many assays exist to measure levels of HIV-1 integrase catalytic function, and most of these utilize purified, recombinant protein. It however remains essential to quantify the levels of integrase activities in the context of virus infection. For example, indirect end-labeling analyses helped to pinpoint the functionality of an important pre-clinical class of HIV-1 integrase inhibitor to the DNA strand transfer step of integration [33]. Raltegravir has more recently been approved for clinical use [34], and PIC analyses are predicted to remain central to investigations into mechanisms of drug resistance. Indirect end-labeling and nested PCR analyses of complexes isolated from lens epithelium-derived growth factor (LEDGF) knockout cells were moreover crucial to ascertain that this critical lentiviral integrase binding protein functioned downstream of PIC formation to affect the frequency and distribution of HIV-1 integration [26]. PIC analyses will therefore remain a critical research tool as studies of host cell factors that potentially affect integrase function and HIV-1 integration expand [35–38].
Acknowledgments
We thank Michael Miller for technical advice and generous gifts of reagents. This work was supported by NIH grants AI039394 and AI052014 to A.E.
Footnotes
Sterilize biochemical reagents by filtration through 0.22 μm filters.
Make buffers K+/+ and K+/− fresh for each experiment. Digitonin stock solution (5% w/v in dimethyl sulfoxide) is stored at 4°C.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Fujiwara T, Mizuuchi K. Cell. 1988;54:497–504. doi: 10.1016/0092-8674(88)90071-2. [DOI] [PubMed] [Google Scholar]
- 2.Brown PO, Bowerman B, Varmus HE, Bishop JM. Proc Natl Acad Sci USA. 1989;86:2525–2529. doi: 10.1073/pnas.86.8.2525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Roth MJ, Schwartzberg PL, Goff SP. Cell. 1989;58:47–54. doi: 10.1016/0092-8674(89)90401-7. [DOI] [PubMed] [Google Scholar]
- 4.Pauza C. Virology. 1990;179:886–889. doi: 10.1016/0042-6822(90)90161-j. [DOI] [PubMed] [Google Scholar]
- 5.Lee YM, Coffin JM. Mol Cell Biol. 1991;11:1419–1430. doi: 10.1128/mcb.11.3.1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Engelman A, Mizuuchi K, Craigie R. Cell. 1991;67:1211–1221. doi: 10.1016/0092-8674(91)90297-c. [DOI] [PubMed] [Google Scholar]
- 7.Bowerman B, Brown PO, Bishop JM, Varmus HE. Genes Dev. 1989;3:469–478. doi: 10.1101/gad.3.4.469. [DOI] [PubMed] [Google Scholar]
- 8.Bukrinsky M, Sharova N, McDonald T, Pushkarskaya T, Tarpley W, Stevenson M. Proc Natl Acad Sci USA. 1993;90:6125–6129. doi: 10.1073/pnas.90.13.6125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Miller M, Farnet C, Bushman F. J Virol. 1997;71:5382–5390. doi: 10.1128/jvi.71.7.5382-5390.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brown PO, Bowerman B, Varmus HE, Bishop JM. Cell. 1987;49:347–356. doi: 10.1016/0092-8674(87)90287-x. [DOI] [PubMed] [Google Scholar]
- 11.Ellison V, Abrams H, Roe T, Lifson J, Brown P. J Virol. 1990;64:2711–2715. doi: 10.1128/jvi.64.6.2711-2715.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Farnet CM, Haseltine WA. Proc Natl Acad Sci USA. 1990;87:4164–4168. doi: 10.1073/pnas.87.11.4164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bukrinsky M, Sharova N, Dempsey M, Stanwick T, Bukrinskaya A, Haggerty S, Stevenson M. Proc Natl Acad Sci USA. 1992;89:6580–6584. doi: 10.1073/pnas.89.14.6580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hansen MS, Smith GJ, 3, Kafri T, Molteni V, Siegel JS, Bushman FD. Nat Biotechnol. 1999;17:578–582. doi: 10.1038/9886. [DOI] [PubMed] [Google Scholar]
- 15.Brooun A, Richman DD, Kornbluth RS. J Biol Chem. 2001;276:46946–46952. doi: 10.1074/jbc.M108000200. [DOI] [PubMed] [Google Scholar]
- 16.Lu R, Vandegraaff N, Cherepanov P, Engelman A. J Virol. 2005;79:12584–12591. doi: 10.1128/JVI.79.19.12584-12591.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dismuke DJ, Aiken C. J Virol. 2006;80:3712–3720. doi: 10.1128/JVI.80.8.3712-3720.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Engelman A. Methods Mol Biol. 2009;485:135–149. doi: 10.1007/978-1-59745-170-3_10. [DOI] [PubMed] [Google Scholar]
- 19.Oh J, Chang KW, Hughes SH. J Virol. 2006;80:451–459. doi: 10.1128/JVI.80.1.451-459.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smith SD, Shatsky M, Cohen PS, Warnke R, Link MP, Glader BE. Cancer Res. 1984;44:5657–5660. [PubMed] [Google Scholar]
- 21.Salahuddin SZ, Markham PD, Wong-Staal F, Franchini G, Kalyanaraman VS, Gallo RC. Virology. 1983;129:51–64. doi: 10.1016/0042-6822(83)90395-1. [DOI] [PubMed] [Google Scholar]
- 22.Chen H, Wei SQ, Engelman A. J Biol Chem. 1999;274:17358–17364. doi: 10.1074/jbc.274.24.17358. [DOI] [PubMed] [Google Scholar]
- 23.Adachi A, Gendelman HE, Koenig S, Folks T, Willey R, Rabson A, Martin MA. J Virol. 1986;59:284–291. doi: 10.1128/jvi.59.2.284-291.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brown HEV, Chen H, Engelman A. J Virol. 1999;73:9011–9020. doi: 10.1128/jvi.73.11.9011-9020.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Engelman A, Englund G, Orenstein JM, Martin MA, Craigie R. J Virol. 1995;69:2729–2736. doi: 10.1128/jvi.69.5.2729-2736.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shun MC, Raghavendra NK, Vandegraaff N, Daigle JE, Hughes S, Kellam P, Cherepanov P, Engelman A. Genes Dev. 2007;21:1767–1778. doi: 10.1101/gad.1565107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Farnet CM, Bushman FD. Cell. 1997;88:483–492. doi: 10.1016/s0092-8674(00)81888-7. [DOI] [PubMed] [Google Scholar]
- 28.Chen H, Engelman A. J Virol. 2000;74:8188–8193. doi: 10.1128/jvi.74.17.8188-8193.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press; Plainview, New York: 1989. pp. 9.34–9.36. [Google Scholar]
- 30.Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. Current Topics in Molecular Biology. John Wiley & Sons, Inc.; Hoboken, New Jersey: 1998. pp. 2.9.1–2.9.6. [Google Scholar]
- 31.Brussel A, Sonigo P. J Virol. 2003;77:10119–10124. doi: 10.1128/JVI.77.18.10119-10124.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Butler SL, Hansen MST, Bushman FD. Nat Med. 2001;7:631–634. doi: 10.1038/87979. [DOI] [PubMed] [Google Scholar]
- 33.Hazuda DJ, Felock P, Witmer M, Wolfe A, Stillmock K, Grobler JA, Espeseth A, Gabryelski L, Schleif W, Blau C, Miller MD. Science. 2000;287:646–650. doi: 10.1126/science.287.5453.646. [DOI] [PubMed] [Google Scholar]
- 34.Evering TH, Markowitz M. Drugs Today (Barc) 2007;43:865–877. doi: 10.1358/dot.2007.43.12.1146063. [DOI] [PubMed] [Google Scholar]
- 35.Van Maele B, Busschots K, Vandekerckhove L, Christ F, Debyser Z. Trends Biochem Sci. 2006;31:98–105. doi: 10.1016/j.tibs.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 36.Engelman A. Future HIV Ther. 2007;1:415–426. [Google Scholar]
- 37.Brass AL, Dykxhoorn DM, Benita Y, Yan N, Engelman A, Xavier RJ, Lieberman J, Elledge SJ. Science. 2008;319:921–926. doi: 10.1126/science.1152725. [DOI] [PubMed] [Google Scholar]
- 38.Konig R, Zhou Y, Elleder D, Diamond TL, Bonamy GMC, Irelan JT, Chiang C-y, Tu BP, De Jesus PD, Lilley CE, Seidel S, Opaluch AM, Caldwell JS, Weitzman MD, Kuhen KL, Bandyopadhyay S, Ideker T, Orth AP, Miraglia LJ, Bushman FD, Young JA, Chanda SK. Cell. 2008;135:49–60. doi: 10.1016/j.cell.2008.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Engelman A. Adv Virus Res. 1999;52:411–426. doi: 10.1016/s0065-3527(08)60309-7. [DOI] [PubMed] [Google Scholar]
- 40.Katzman M, Katz RA. Adv Virus Res. 1999;52:371–395. doi: 10.1016/s0065-3527(08)60307-3. [DOI] [PubMed] [Google Scholar]
- 41.Vincent KA, York-Higgins D, Quiroga M, Brown PO. Nucleic Acids Res. 1990;18:6045–6047. doi: 10.1093/nar/18.20.6045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vink C, Groenink M, Elgersma Y, Fouchier RA, Tersmette M, Plasterk RH. J Virol. 1990;64:5626–5627. doi: 10.1128/jvi.64.11.5626-5627.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]