SUMMARY
The mechanisms that allow the body to sense iron levels in order to maintain iron homeostasis are unknown. Patients with the most common form of hereditary iron overload have mutations in the hereditary hemochromatosis protein, HFE. They have lower levels of hepcidin, than unaffected individuals. Hepcidin, a hepatic peptide hormone, negatively regulates iron efflux from the intestines into the blood. We report two hepatic cell lines, WIF-B cells and HepG2 cells transfected with HFE, where hepcidin expression responded to iron-loaded transferrin. The response was abolished when endogenous transferrin receptor 2 (TfR2) was suppressed or in primary hepatocytes lacking either functional TfR2 or HFE. Furthermore, transferrin-treated HepG2 cells transfected with HFE chimeras containing only the α3 and cytoplasmic domains could upregulate hepcidin expression. Since the HFE α3 domain interacts with TfR2, these results supported our finding that TfR2/HFE complex is required for transcriptional regulation of hepcidin by holo-Tf.
Keywords: HFE, TfR1, TfR2, hepcidin, hereditary hemochromatosis
INTRODUCTION
Iron is essential for fundamental metabolic processes in cells and organisms. It is also toxic when in excess. Hereditary hemochromatosis (HH) is a heterogeneous group of iron overload disorders caused by mutations in a variety of proteins involved in iron homeostasis including HFE (Feder et al., 1996), hemojuvelin (Papanikolaou et al., 2004), hepcidin (Roetto et al., 2003), transferrin receptor 2 (TfR2) (Camaschella et al., 2000), or ferroportin (Fpn) (Montosi et al., 2001). HH manifests as increased intestinal iron absorption and liver iron overload (Vujic Spasic et al., 2008). If untreated, HH patients develop hepatic cirrhosis and hepatocellular carcinoma, cardiomyopathy and arrhythmias, diabetes, arthritis and hypogonadotropic hypogonadism (Ajioka and Kushner, 2002; Bothwell and MacPhail, 1998).
The most common form of HH is caused by single base pair mutation in HFE resulting in a C260Y1 substitution (Feder et al., 1996). HFE encodes an atypical major histocompatibility complex class I protein (MHC1). Like the MHC1 proteins, HFE is a membrane protein that consists of a signal sequence, α1–α3 domains followed by a transmembrane domain, and a short cytoplasmic domain. It also forms a heterodimeric complex with β2-microglobulin (Lebron et al., 1998). The mutation disrupts a disulfide bond in the α3 domain leading to misfolding of HFE, lack of association with β2-microglobulin, and failure to traffic to the cell surface (Feder et al., 1997). The generation of a knockout mouse (Hfe−/−), coupled to the finding that the C260Y substitution and the Hfe−/− phenotypes are similar, confirms that C260Y has a loss-of-function phenotype (Muckenthaler et al., 2004). Although the importance of HFE in iron regulation is apparent in HH patients and mouse models (Levy et al., 1999; Zhou et al., 1998), the underlying mechanism by which HFE regulates iron metabolism remains unanswered.
Disease-causing mutations in HFE result in decreased hepcidin production both in HH patients and in Hfe−/− mice when compared to similarly iron-loaded individuals with unmutated HFE (Ahmad et al., 2002; Bridle et al., 2003; Muckenthaler et al., 2003; Nicolas et al., 2003). Hepcidin is a peptide hormone produced predominantly by the liver. It plays a major role in the regulation of iron homeostasis within the body by modulating iron levels through binding to and triggering the internalization and degradation of the iron exporter, Fpn (Nemeth et al., 2004). Thus, hepcidin controls iron loading of Tf by negatively regulating iron efflux from enterocytes, liver macrophages, and hepatocytes into the blood.
Hepcidin production is modulated by many factors including iron levels within the body. In response to iron loading in animal studies, hepcidin expression increases to prevent the further uptake of iron. Conversely, during iron deficiency, hepcidin expression decreases (Pigeon et al., 2001; Weinstein et al., 2002). In the liver, both hepcidin and HFE are predominantly expressed in hepatocytes (Holmstrom et al., 2003; Zhang et al., 2004). Hepatocyte-specific expression of HFE in Hfe−/− mice restores normal iron homeostasis indicating that hepatocyte-specific expression of HFE is sufficient to control iron homeostasis (Vujic Spasic et al., 2008). Thus, HFE appears to function upstream of hepcidin expression to regulate iron homeostasis.
HFE has several binding partners that could participate in iron homeostasis. HFE associates with transferrin receptor 1 (TfR1) (Feder et al., 1998; Waheed et al., 1999) through its α1 and α2 domains (Bennett et al., 2000) and with TfR2 through its α3 domain (Chen et al., 2007). The binding sites on TfR1 for HFE and iron loaded transferrin (holo-Tf) overlap (Giannetti et al., 2003; Lebron and Bjorkman, 1999; West et al., 2001), confirming the competition between HFE and holo-Tf, for binding to TfR1. More recent co-immunoprecipitation studies demonstrate that HFE also interacts with TfR2 (Chen et al., 2007; Goswami and Andrews, 2006). TfR2 is expressed predominantly in hepatocytes and is closely related to TfR1 in sequence and in its ability to bind holo-Tf but not iron-depleted Tf (apo-Tf) (Kawabata et al., 1999). Unlike TfR1, holo-Tf does not compete with HFE for binding to TfR2 (Chen et al., 2007).
Similar to HFE, disease-causing mutations in TfR2 also result in decreased hepcidin levels (Wallace et al., 2007). TfR2 is hypothesized to act as a sensor for iron levels in the body because of its largely hepatocyte-specific expression and its ability to bind holo-Tf (Kawabata et al., 1999). The main limitation in determining how HFE and TfR2 regulate hepcidin expression to date has been the lack of a cell line in which the hepcidin expression is responsive to holo-Tf.
In the present study, we found that WIF-B cells, a rat hepatoma/human fibroblast hybrid, increased the expression of hepcidin in response to holo-Tf. HFE and TfR2 mRNA levels were higher in WIF-B cells compared to HepG2 cells, a human hepatoma cell line whose expression of hepcidin is not sensitive to holo-Tf. We used the HepG2/tTA cells that express HFE under the tight control of tetracycline-inducible promoter and showed that hepcidin levels increase when cells expressing HFE are treated with holo-Tf. The involvement of HFE and TfR2 in this process was investigated using TfR2 siRNA, primary hepatocytes, and HFE chimeras. Our results show that Tf-induced hepcidin expression was dependent on the interaction of TfR2 with HFE.
RESULTS
Holo-Tf induces hepcidin expression in WIF-B cells
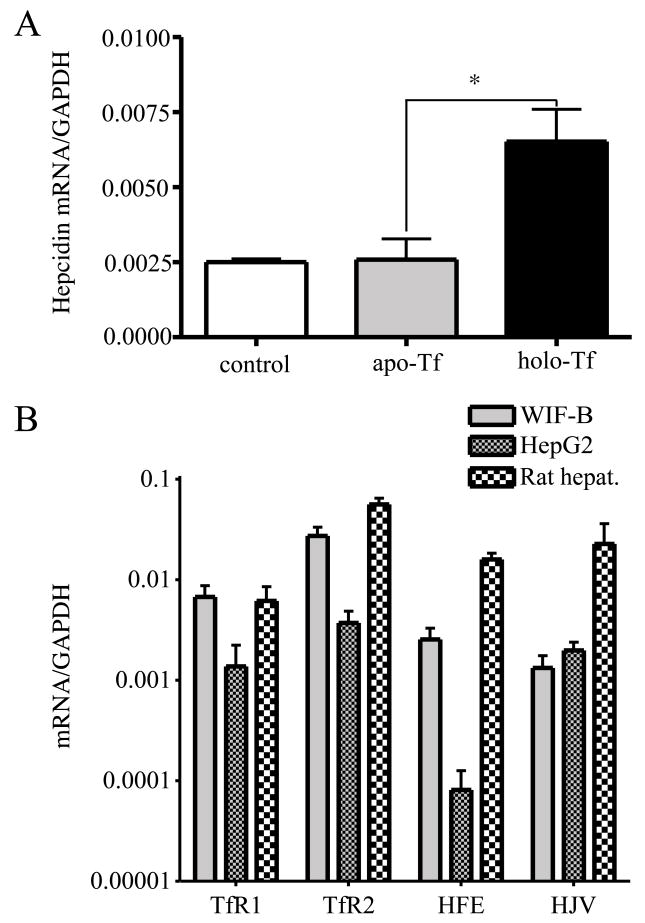
The positive correlation between hepcidin levels and holo-Tf in the blood leads to the hypothesis that the liver senses the level of iron in the body by sensing the amount of holo-Tf. We examined a number of hepatic cell lines for their ability to upregulate hepcidin in response to holo-Tf and found that WIF-B cells met the criterion. WIF-B cells are a rat hepatoma/human fibroblast hybrid with many functional and morphological similarities to hepatocytes (Ihrke et al., 1993). Even though they are rat/human hybrids, others have found that they express predominantly rat genes (Braiterman et al., 2008; Konieczko et al., 1998; Nies et al., 1998) and we could not detect human TfR1, TfR2 or HFE by immunoblot (unpublished results). Primers to rat hepcidin were used to determine levels of the hepcidin transcript. When treated with 25 μM holo-Tf for 24 hours, WIF-B cells expressed ~2.5-fold higher hepcidin mRNA as determined by qRT-PCR compared to untreated cells or cells treated with apo-Tf (Figure 1A). In contrast, HepG2 cells, a human hepatoma cell line, do not upregulate hepcidin in response to Tf (Gehrke et al., 2003). Thus WIF-B cells mimic the regulation of hepcidin levels by holo-Tf that is seen in hepatocytes in vivo.
Figure 1. Hepcidin expression in WIF-B cells responds to holo-Tf.
A. Holo-Tf induces hepcidin expression in WIF-B cells. WIF-B cells were left untreated or incubated with 25 μM human apo-Tf or holo-Tf for 24 hours, prior to RNA isolation. The hepcidin mRNA level was measured by qRT-PCR and normalized to GAPDH. B. Comparison of HFE, TfR1, TfR2 and HJV mRNA levels in WIF-B cells, HepG2 cells, and rat hepatocytes. Levels of TfR1, TfR2, HFE and HJV mRNA, iron related genes upstream of hepcidin, were measured by qRT-PCR. All samples were run in triplicate in three independent experiments. Data are shown as average ± S.D. P-values < 0.05 are indicated by *.
We compared the mRNA levels of the key iron-related genes in WIF-B, HepG2 cells, and isolated rat hepatocytes in order to determine the reason for the differences in hepcidin response to holo-Tf between the two cell lines. While the TfR1 and TfR2 levels in WIF-B cells were comparable to those of freshly isolated rat hepatocytes, HFE and HJV levels were 6~10 fold less in WIF-B cells. Interestingly, the mRNA level of HFE in WIF-B cells was about 32-fold higher than in HepG2 cells. Amongst other genes involved in iron homeostasis in the body, TfR2 mRNA level was eight-fold higher in WIF-B than HepG2 implicating TfR2 in the response. The HJV mRNA level was comparable in the two cell lines and lower than in isolated rat hepatocytes (Figure 1B). TfR1 levels were 5-fold higher in WIF-B cells than in HepG2 cells and isolated hepatocytes also implicating TfR1 in signaling. Since the largest difference in the measured mRNA levels of the two cell lines was the level of HFE mRNA, we created a cell line HepG2/tTA HFE stably transfected with HFE under the control of the tetracycline-activated promoter.
HFE contributes to Tf-sensitive induction of the hepcidin promoter activity independent of TfR1
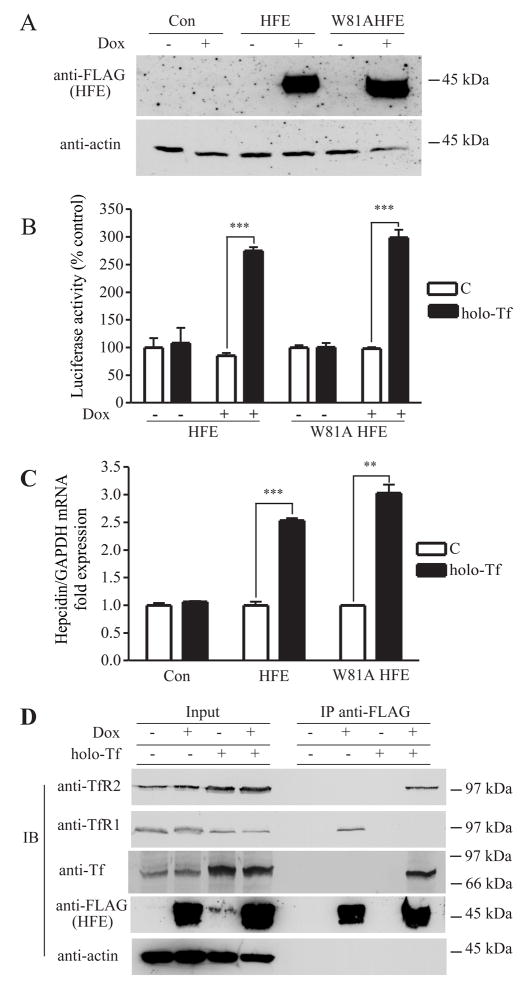
HepG2/tTA HFE cells were induced to express HFE with doxycycline (dox), a tetracycline analog, to test the possible role of HFE in the regulation of hepcidin expression (Figure 2A). To observe the effect of holo-Tf and HFE on hepcidin transcription, HepG2/tTA HFE cells were transiently transfected with a luciferase reporter vector, pLuc-link-HAMP, containing ~2-kb-length human hepcidin promoter. Hepcidin promoter activity was measured using luciferase. Holo-Tf treatment of cells had no effect on the hepcidin promoter activity in HepG2 cells as measured by luciferase, in the absence of HFE expression (Figure 2B). The lack of response is consistent with a previous study in HepG2 cells (Gehrke et al., 2003) which we confirmed. HFE expression alone did not increase hepcidin expression (Figure 2B). However, after the induction of HFE expression with dox, holo-Tf treatment of cells increased hepcidin promoter activity by about 2.7-fold. Similar results were obtained when hepcidin mRNA levels were measured by qRT-PCR (Figure 2C). These results indicated that the changes in mRNA levels were the result of transcriptional activation.
Figure 2. Tf-induced dissociation of HFE from TfR1 results in the association of HFE with TfR2 and stimulates hepcidin promoter activity.
A. Inducible expression of wild-type or W81A mutant HFE in HepG2/tTA HFE and HepG2/tTA W81AHFE cells. Lysates (25 μg) of HepG2/tTA HFE or HepG2/tTA W8AHFE cells, uninduced (−dox) or induced (+dox) to express HFE or W81AHFE, were detected with anti-FLAG antibody. B. HFE is required for Tf to induce hepcidin promoter activity independently of interaction with TfR1. Cells were cotransfected with pLuc-link-HAMP and pCMV-β-gal, treated without (C) or with holo-Tf and analyzed as described in Experimental Procedures. Results are expressed as average ± S.D. of three independent experiments performed in triplicate. C. Hepcidin mRNA expression in HepG2/tTA (Con) HepG2/tTA HFE (HFE) and HepG2/tTA W81AHFE (W81AHFE) cells were treated without (C) or with holo-Tf as described in Experimental Procedures. The expression of hepcidin was measured by qRT-PCR and normalized to GAPDH. The fold of increase in hepcidin mRNA in the dox-induced group was obtained by normalization to uninduced controls. All samples were run in triplicate in three independent experiments. Data are shown as average ± S.D. D. Tf releases HFE from TfR1 to bind to TfR2 in HepG2 cells. Cell lysates (200 μg) from HepG2/tTA HFE treated with or without 25 μM holo-Tf were immunoprecipitated with rabbit anti-FLAG antibodies, and Sepharose-4B/Protein A. Proteins were detected on immunoblots using mouse anti- TfR2, TfR1, FLAG, and actin and goat anti-Tf. The input lanes correspond to 1/5 of the material used for immunoprecipitation. These results were repeated once with similar results. P-values < 0.001 are indicated by *** and < 0.01 by **.
The finding that HepG2/tTA HFE cells increased hepcidin transcription in response to holo-Tf only in the presence of HFE made this an ideal cell line to examine the binding partners of HFE necessary for the holo-Tf-induced hepcidin response. We stably transfected HepG2/tTA cells with W81AHFE to examine the role of the TfR1/HFE complex in the regulation of hepcidin expression. W81AHFE has 5,000 fold lower affinity for TfR1 (Lebron and Bjorkman, 1999) and does not detectably co-immunoprecipitate with TfR1 (Zhang et al., 2003). This mutant form of HFE does bind to TfR2 (Chen et al., 2007). Thus if the TfR1/HFE complex was necessary for signaling to increase hepcidin levels, this cell line would not increase hepcidin mRNA in response to Tf treatment. The HepG2/tTA W81AHFE cells expressed comparable amounts of W81AHFE to cells expressing HFE (Figure 2A), showed similar increased hepcidin transcription (Figure 2B) and increased hepcidin mRNA levels (Figure 2C) compared to cells expressing wild-type HFE. These results suggested that the interaction between HFE and TfR1 is not required for Tf-dependent induction of hepcidin expression in HepG2 cells.
To further investigate the binding partners of HFE in the presence of holo-Tf in HepG2 cells, HFE was immunoprecipitated and blots were probed for proteins that coprecipitated with HFE. TfR1 co-immunoprecipitated with HFE in the absence of added holo-Tf but not in the presence of high physiological levels of holo-Tf consistent with the competition between Tf and HFE for binding to TfR1 (Figure 2D). Note that these cells synthesize Tf but not enough holo-Tf to completely abrogate the HFE/TfR1 interaction. Prolonged treatment of cells with holo-Tf increased intracellular iron loading, which decreased TfR1 presumably through the destabilization of TfR1 mRNA by the IRE/IRP pathway (reviewed in (De Domenico et al., 2008)). TfR2 co-immunoprecipitated with HFE in presence of high physiological levels of holo-Tf (25 μM). Holo-Tf treatment of cells resulted in increased TfR2 levels (Figure 2D), consistent with previous reports that holo-Tf stabilizes TfR2 at the protein level (Johnson and Enns, 2004; Robb and Wessling-Resnick, 2004). The lack of detectable TfR2 that coprecipitated with HFE in the absence of added holo-Tf could reflect the lower endogenous levels of TfR2 in this cell line than seen in cell lines transfected with TfR2 (Chen et al., 2007; Johnson and Enns, 2004). The coprecipitation of Tf and TfR2 with HFE in the presence of higher holo-Tf concentrations indicated the possibility that they form a signaling complex to increase hepcidin expression.
TfR2 is necessary for the HFE-mediated Tf induction of hepcidin expression in HepG2 cells
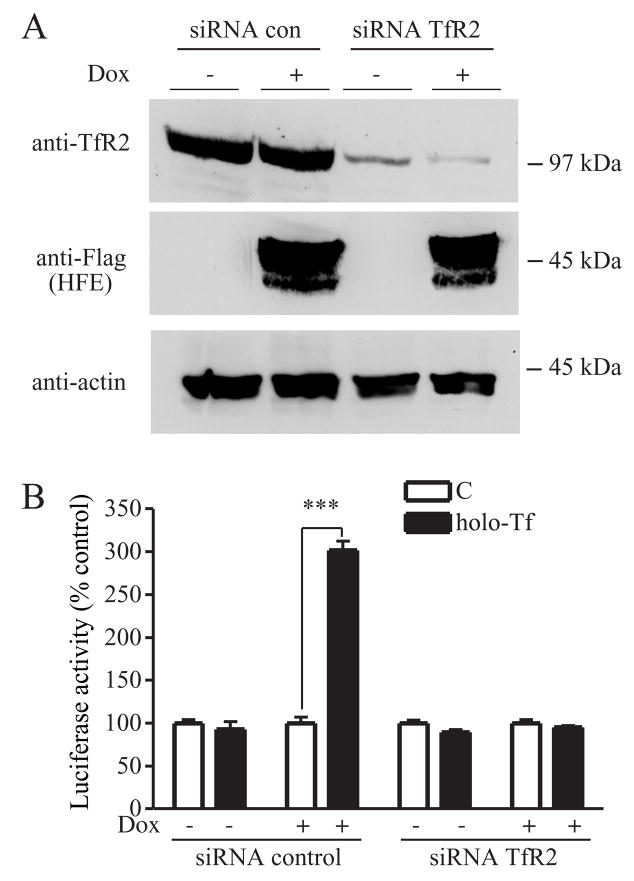
We investigated the role of TfR2 in the Tf-sensitive induction of hepcidin. HepG2/tTA HFE cells were cotransfected with TfR2 siRNA to decrease TfR2 levels and with pLuc-link-HAMP to measure hepcidin promoter activity. Immunoblot analysis of TfR2 showed that TfR2 was significantly reduced after transfection of cells with TfR2 siRNA (Figure 3A). The sensitivity of hepcidin transcription to the addition of holo-Tf as measured by the luciferase reporter activity was abolished by TfR2 siRNA but not by control siRNA (Figure 3B). These results along with our other results imply that both TfR2 and HFE are necessary for Tf-induced expression of hepcidin.
Figure 3. Knockdown of TfR2 blocks Tf-mediated induction of hepcidin expression in HepG2/tTA HFE cells.
A. Knockdown of endogenous TfR2 in HepG2/tTA HFE cells using specific siRNA to TfR2. Cells were transfected with control siRNA or TfR2 siRNA and co-transfected with pLuc-link-HAMP and pCMV-β-gal as described in Experimental Procedures. To observe the efficiency of knockdown of TfR2, cell lysates (50 μg) from TfR2 siRNA or control siRNA transfected HepG2/tTA HFE cells were analyzed by immunoblot for TfR2, HFE and actin expression. These results were representative of one out of three experiments. Human TfR2 siRNA significantly decreased endogenous TfR2 protein level in both HepG2/tTA control cells and HepG2/tTA HFE cells. B. Hepcidin promoter activity assay in HepG2/tTA HFE cells after knockdown of TfR2 and treatment without (C) or with holo-Tf. Luciferase and β-galactosidase activity from cell lysates were measured as described in Experimental Procedures. Relative luciferase activity was obtained by normalization to β-galactosidase activity and the fold increase in luciferase activity of each group was obtained by normalization to the control group without holo-Tf. Results are expressed as average ± S.D. of three independent experiments performed in triplicate. P-values < 0.001 are indicated by ***.
HFE and TfR2 are both necessary for the Tf-mediated induction of hepcidin expression in mouse primary hepatocytes
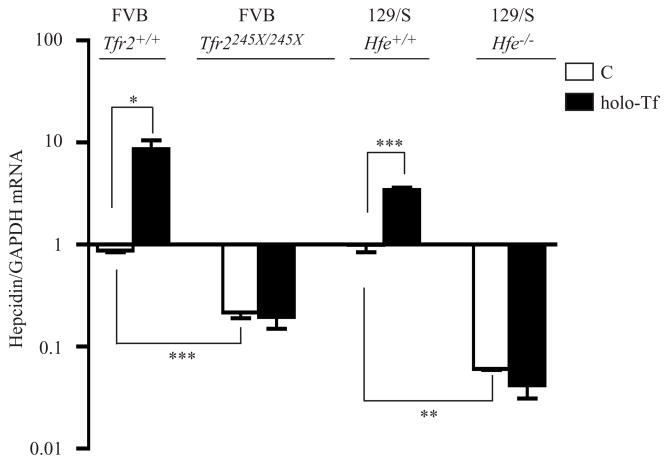
In vitro cultures of primary hepatocytes from wild type, Hfe−/−, and Tfr2245X/245X mice were isolated to measure the hepcidin response of hepatocytes to holo-Tf treatment. The TfR2 (245X) mutation causes iron overload in mice and the orthologous mutation, TfR2 (250X), causes HH in humans. Hfe−/− mice were on the 129/SvEvTac (129/S) background and Tfr2245X/245X mice were on a FVB/NJ (FVB) background. Previous studies indicated that TfR2 (245X) binds HFE but not Tf in vitro (Chen et al., 2007; Goswami and Andrews, 2006). In the primary hepatocytes isolated from 129/S mice and FVB mice, we found that hepcidin mRNA levels increased by about 3.4 and 9.9-fold (Figure 4), respectively, in response to treatment with 25 μM holo-Tf. These differences between the ability to stimulate hepcidin expression in hepatocytes from the different strains of mice could be either strain dependent or due to the expression of the truncated TfR2. However, no significant changes of hepcidin mRNA levels were detected in the primary hepatocytes isolated from Hfe−/− or Tfr2245X/245X mice compared with their corresponding controls (Figure 4). These results strongly support the idea that both HFE and TfR2 are required for Tf-induced hepcidin expression and agree with the finding that HFE is necessary for Tf-induced hepcidin expression in HepG2 cells. Consistent with the reports that hepcidin levels are lower in Hfe−/− and in Tfr2 mutant mice (Ahmad et al., 2002; Bridle et al., 2003; Muckenthaler et al., 2003; Nicolas et al., 2003; Wallace et al., 2007), the hepcidin mRNA levels in the isolated primary hepatocytes from Hfe−/− and Tfr2245X/245X mice were 16.5 and 4-fold lower than those in their corresponding wild type counterparts (Figure 4). Thus, both TfR2 and HFE are necessary for the induction of hepcidin by holo-Tf.
Figure 4. TfR2 and HFE are required for Tf-mediated induction of hepcidin expression in mouse primary hepatocytes.
Mouse primary hepatocytes were treated without (C) or with holo-Tf and qRT-PCR was performed on isolated RNA as described in Experimental Procedures. Average relative level of hepcidin was normalized to GAPDH. Results are expressed as average ± S.D. of two or four independent experiments performed in triplicate. P-values < 0.05, < 0.01 and < 0.001 are indicated by *, ** and ***, respectively.
The α3 domain of HFE is insufficient for the induction of hepcidin expression in HepG2 cells in response to holo-Tf
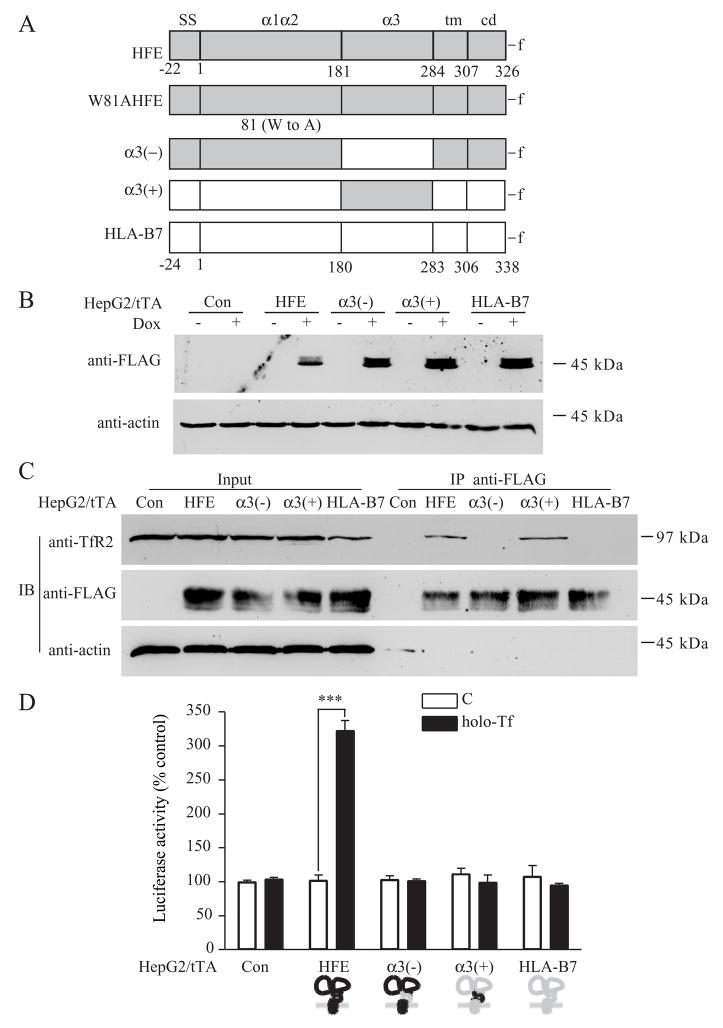
To map the domains of HFE responsible for the Tf-induced upregulation of hepcidin, we took advantage of the findings that HFE binds to TfR2 through its α3 domain (Chen et al., 2007), whereas it binds to TfR1 via its α1α2 domains (Bennett et al., 2000) and that the structures of the HFE and the MHC1 molecules are similar (review in (Wilson and Bjorkman, 1998)). Two HFE-HLA-B7 chimeras were generated; α3(−), in which the HFE α3 domain was replaced with the HLA-B7 α3 domain and α3(+), in which the α3 domain of HFE was substituted for the α3 domain of HLA-B7 (Figure 5A). These two constructs along with HLA-B7 were transfected individually into HepG2/tTA cells under the control of the tetracycline-inducible system (Figure 5B). The abilities of HFE chimeras to co-precipitate TfR2, were detected by immunoblots (Figure 5C). The α3(−) chimera did not show detectable interaction with TfR2 (Figure 5C) but was able to interact with TfR1 indicating that lack of interaction with TfR2 was not due to misfolding of the chimera (Figure S1). TfR2 coprecipitated with both HFE and the chimera containing the HFE α3 domain (α3(+)) but not with the negative control HLA-B7 (Figure 5C). These results confirmed our previous observation that the α3 domain of HFE is critical for HFE binding to TfR2 (Chen et al., 2007). Surprisingly, α3(+) coprecipitated with TfR2, but it did not mediate the Tf-sensitive induction of the hepcidin promoter activity in the luciferase activity assay (Figure 5D). These results imply that α3 domain of HFE is required for binding to TfR2, but it is insufficient for regulation of hepcidin mRNA by Tf.
Figure 5. The α3 domain is required but insufficient for HFE-mediated Tf induction of hepcidin expression in HepG2 cells.
A. Schematic representation of the HFE mutant and chimeras including HFE, W81AHFE, α3(−),α3(+) and HLA-B7. HFE is shown in gray, HLA-B7 is shown in white. Numbers under diagrams for each full-length protein designate the first amino acid in the signal sequence (SS), α1α2 domain (α1α2), α3 domain (α3), transmembrane domain (tm), and cytoplasmic domain (cd). FLAG epitope tag (-f) was added to the C terminus of each protein. HFE-HLA-B7 chimeras were constructed by replacing the appropriate segments between the parent proteins. The first coding methionine in all proteins is labeled as number 1. We refer to individual recombinant proteins used throughout the study by acronyms to left of the construct illustration. B. Induction of HFE chimeras by dox. Lysates (25 μg) of HepG2/tTA HFE, HepG2/tTA α3(−), HepG2/tTA α3(+) and HepG2/tTA HLA-B7 cells, uninduced (−dox) or induced (+dox) to express wtHFE, α3(−),α3(+) or HLA-B7 were detected using mouse anti-FLAG antibody. C. The α3 domain of HFE is critical for interaction with TfR2 in HepG2/tTA cells. Cells were treated without (C) or with 25 μM holo-Tf (holo-Tf) as described in Experimental Procedures. Cells lysates (200 μg) were immunoprecipitated with rabbit anti-FLAG antibody and the blots were probed with mouse antibodies to TfR2, FLAG and actin. The input lanes correspond to 1/5 of the material used for immunoprecipitation. These results were repeated once with similar results. D. The α3 domain of HFE is required but insufficient for HFE-mediated Tf induction of hepcidin expression in HepG2 cells. Cells were cotransfected with pLuc-link-HAMP and pCMV-β-gal plasmids, treated without (C) or with 25 μM holo-Tf (holo-Tf), and analyzed as described in Experimental Procedures. Results are expressed as average ± S.D. of three independent experiments performed in triplicate. P-values < 0.001 are indicated by ***.
A chimera containing both the HFE α3 domain and cytoplasmic domains is sufficient to mediate Tf induction of hepcidin expression in HepG2 cells
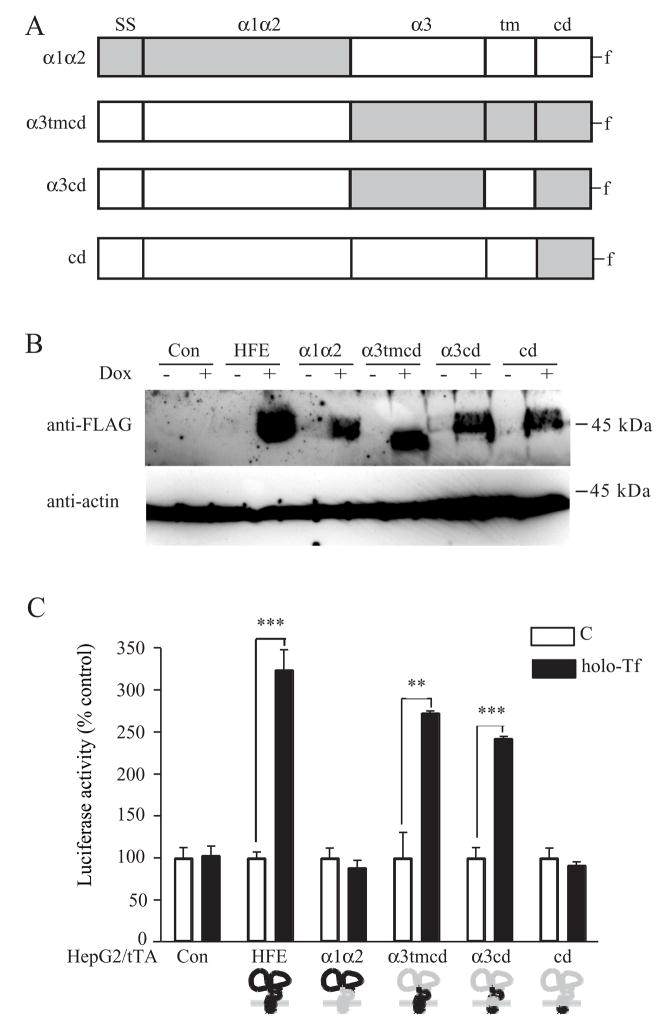
Since the chimera containing only the α3 domain of HFE was not sufficient to mediate the induction of hepcidin by Tf, we generated plasmids encoding chimeras where the domains of HFE and HLA-B7 were exchanged (Figure 6A) and stably transfected them into HepG2/tTA cell lines (Figure 6B). Only the chimeras containing both the α3 and cytoplasmic domains of HFE (α3tmcd and α3cd) were able to induce luciferase activity (Figure 6C). The α1α2 chimera interacted with TfR1 indicating that the lack of stimulation of luciferase activity is not due to misfolding of the chimera (Figure S1). Thus, the α3 and cd domains of HFE on the HLA-B7 backbone are necessary and sufficient to stimulate hepcidin transcription in response to treating cells with holo-Tf.
Figure 6. The α3 and cytoplasmic domains of HFE mediate the Tf induction of hepcidin expression in HepG2 cells.
A. Schematic representation of the HFE chimeras including α1α2, α3tmcd, α3cd and cd constructs. HFE is shown in gray, HLA-B7 is shown in white. HFE chimeras were constructed by replacing the appropriate segments between the parent proteins. Individual recombinant proteins used throughout the study are referred to by the acronyms to left of the construct cartoon. B. Generation of HFE-HLA-B7 chimeras. HFE chimeras including stable cells lines HepG2/tTA α1α2, HepG2/tTA α3tmcd, HepG2/tTA α3cd and HepG2/tTA cd were generated as described in Experimental Procedures. Lysates (25 μg) of cells, uninduced (−dox) or induced (+dox) were used to screen the positive clones by immunoblot using anti-FLAG antibody. C. The α3 and cytoplasmic domain of HFE is required for HFE-mediated Tf induction of hepcidin expression in HepG2 cells. All HepG2/tTA control and HFE chimeras cells were cotransfected with pLuc-link-HAMP and pCMV-β-gal, treated, and analyzed as described in Experimental Procedures. Results are expressed as average ± S.D. of three independent experiments performed in triplicate. P-values < 0.001 are indicated by *** and < 0.01 by **.
DISCUSSION
The liver plays a key role in the sensing of iron levels in the body and in the regulation of iron homeostasis. Hepatocytes within the liver express proteins critical to these processes including HFE, TfR2, HJV, and Tf as well as the peptide hormone, hepcidin. Hepcidin, which negatively regulates iron efflux from cells into the blood, is controlled at the transcriptional level. Thus when the liver senses high iron levels, hepcidin expression increases to inhibit further iron uptake and results in iron sequestration in cells. In this study, we examined the roles of these key proteins in controlling hepcidin expression in cell lines that were engineered to mimic the increase in hepcidin in normal hepatocytes in response to treatment with a physiological concentration of holo-Tf. We found that the Tf/TfR2/HFE complex was necessary to increase hepcidin expression.
The role of Tf in the regulation of hepcidin
The concentration of holo-Tf in blood has been proposed to be a regulator of hepcidin expression (Johnson and Enns, 2004; Robb and Wessling-Resnick, 2004). High holo-Tf levels correlate with increased hepcidin mRNA in the liver (Kawabata et al., 2005). The finding that hepcidin levels increase in freshly isolated mouse hepatocytes (Lin et al., 2007) in response to holo-Tf demonstrates that hepatocytes can respond directly.
Holo-Tf serves two functions in the regulation of hepcidin. First, it alters the interactions between HFE and TfR1. A recent study examined the role of TfR1 in the control of hepcidin mRNA levels by generating mice, which express a mutant TfR1 that does not bind detectably to HFE (Schmidt et al., 2008). A higher level of hepcidin mRNA was detected in the TfR1 mutant mice than in the strain-matched control mice. The authors propose that increased hepcidin mRNA results from unbound HFE or from HFE binding to another partner. They speculate that the binding partner was TfR2 based on previous evidence showing that HFE is capable of binding to TfR2 (Chen et al., 2007; Goswami and Andrews, 2006). They also suggest that the TfR2/HFE complex is responsible for hepcidin regulation and that TfR1 sequesters HFE from binding to TfR2 under low levels of holo-Tf. In our study we showed that disruption of the HFE/TfR1 complex alone did not regulate hepcidin expression using transfected hepatoma cell lines. Upregulation of hepcidin mRNA was achieved only in the presence of holo-Tf and TfR2. In addition, we directly demonstrated that Tf/TfR2/HFE complex is involved in the upregulation of hepcidin. Thus the Tf/TfR2/HFE complex is critical for increased hepcidin expression. How the complex signals to accomplish this remains to be determined.
The second way that Tf can modulate the HFE/TfR2 interactions is through the downregulation of TfR1 and the upregulation of TfR2. Thus under high iron conditions increased saturation of Tf with iron results in an increased amount of Tf/TfR2/HFE complex and together with the results from mice with mutated TfR1, these findings indicate that Tf binding to TfR1 releases HFE to form a complex with Tf and TfR2. This complex is responsible for increased hepcidin expression. This model predicts that HFE is limiting for complex formation. Future experiments will focus on testing this prediction.
The necessity of HFE in the Tf-mediated sensitivity of hepcidin
The changes in hepcidin mRNA in WIF-B and HepG2/tTAHFE cells are within physiological responses. They are similar to the 3–4 fold increase in hepcidin levels in mice fed high iron diets although there is variability in response between strains (Bondi et al., 2005) and our findings in primary hepatocytes. The requirement of HFE for increased hepcidin expression in response to Tf treatment is consistent with the lack of hepcidin response in Hfe−/− mice on a high iron diet (Ahmad et al., 2002). Our results suggest that HFE is required for the Tf-induced hepcidin expression in hepatocytes and are in keeping with the recent finding that hepatocyte-specific expression of HFE increases hepcidin levels in mice (Schmidt et al., 2008; Vujic Spasic et al., 2008). Since HFE affects other aspects of iron homeostasis independent of hepcidin (Gao et al., 2008), lack of HFE could also have other effects on iron metabolism in Hfe−/− mice.
The role of TfR2 in the upregulation of hepcidin in response to Tf
In addition to the role of HFE and Tf in the control of hepcidin mRNA, we also showed that TfR2, which binds to HFE in vitro is required for Tf-dependent stimulation of hepcidin expression. Holo-Tf treatment of cells also increases TfR2 levels by stabilizing TfR2 thus also promoting the formation of the TfR2/HFE complex. Knockdown of TfR2 or lack of functional TfR2 results in loss of hepcidin sensitivity to Tf treatment. These results help to explain why Tfr2−/− mice, have a 3–4 fold lower basal hepcidin levels despite hepatic iron overload and lack a hepcidin response to iron (Wallace et al., 2007) and why Tf failed to increase hepcidin expression in primary hepatocytes isolated from Tfr2245X/245X mice.
Mapping the domains of HFE that are responsible for the Tf-induced upregulation of hepcidin
To further investigate the involvement of TfR2/HFE complex in the Tf induction of hepcidin expression, HFE/HLA-B7 chimeras were used. Intriguingly, the HFE-HLA-B7 chimera containing only the HFE α3 domain bound to TfR2 but did not alter hepcidin expression in presence of holo-Tf. These results suggest that α3 domain of HFE is necessary but not sufficient for Tf-induced stimulation of hepcidin expression. Further domain mapping using HFE/HLA-B7 chimeras showed that both the α3 and cytoplasmic domains of HFE were required for HFE regulating hepcidin. The role of the cytoplasmic domain with respect to hepcidin regulation still remains to be clarified.
Hepcidin mRNA is transcriptionally regulated through at least three pathways. Interleukin 6 stimulates hepcidin expression through the STAT3 signaling pathway. Bone morphogenic proteins, BMPs, increase hepcidin expression through the hemojuvelin (HJV)/BMP receptor/Smad 4 pathway. Tf stimulates hepcidin expression through a TfR2/HFE-mediated pathway. The extent to which these pathways are interconnected is unknown. Mutations in HJV can cause juvenile hemochromatosis, a severe iron overload disease characterized by low hepcidin levels. Similarly Hjv−/− mice have extremely low hepcidin levels, but the levels can be further decreased by phlebotomy (Krijt et al., 2007) implying that iron signaling can be independent of HJV and that HJV participates in establishing a set point for BMP signaling and iron homeostasis. Whether the Tf control of hepcidin signaling is through any of these signaling pathways is still unresolved.
Taken together, our studies suggest that the Tf/TfR2/HFE complex is involved in the sensing of Tf saturation, which leads to the regulation of hepcidin expression. Based on these results, mutations in either HFE or TfR2 should result in a similar iron overload disease with respect to the regulation of hepcidin.
EXPERIMENTAL PROCEDURES
Hepcidin promoter construct
A fragment of the human hepcidin promoter consisting of the first 2043 nucleotides upstream of the translational start site were subcloned into the promoterless luciferase reporter vector pLuc-link as described previously (de Wet et al., 1987; Verga Falzacappa et al., 2008) and detailed in Supplemental Material.
HFE chimera constructs
The plasmids containing HFE-α1α2/HLA-B7-α3/HFE-tmcd (α3(−)), HLA-B7-α1α2/HFE-α3/HLA-B7-tm/HFE-cd (α3cd), and HLA-B7-α1α2α3tm/HFE-cd (cd) were constructed by overlapping PCR using the primers and templates listed in Supplemental Table 1. The wild type HFE, W81AHFE, HFE-α1α2/HLA-B7-α3tmcd (α1α2), HLA-B7-α1α2/HFE-α3tmcd (α3tmcd), HLA-B7-α1α2/HFE-α3/HLA-B7-tmcd (α3(+)), and wild type HLA-B7 constructs were generated as previously described (Chen et al., 2007; Cherry et al., 2008). All constructs contain a C-terminal FLAG tag sequence for immunodetection. In all cases, the gel-purified PCR products were first inserted into the pGEM-T (Promega, Madison, WI), followed by subcloning into the pcDNA4 vector that had been modified by adding tetracycline-inducible promoter (Feng and Longmore, 2005). All of the sequences and orientations were verified.
Antibodies
Mouse anti-hTfR2 (9F8-1C11) monoclonal antibodies were described previously (Vogt et al., 2003). M2 anti-FLAG, H68.4 anti-TfR1 and anti-actin were purchased from Sigma-Aldrich (St. Louis, MO), Zymed Laboratories Inc. (San Francisco, CA), and Sigma-Aldrich, respectively. Secondary antibodies against rabbit and mouse IgG conjugated to horseradish peroxidase (HRP) were purchased from Chemicon (Temecula, CA).
Cell culture and transfection
WIF-B cells, obtained from Dr. Ann Hubbard (Johns Hopkins University), were grown in F-12 Coon’s Modification with amphotericin, glutamax and penicillin/streptomycin supplemented with HAT, and 5% FCS and cultured in a humidified 7% CO2 incubator at 37°C (Ihrke et al., 1993). Tetracycline-inducible HepG2 cells (HepG2/tTA) were obtained from Dr Gregory D. Longmore (Washington University in St. Louis) and grown in DMEM medium containing 10% FBS, 2 mM L-glutamate, and 5 μg/ml blasticidin (Feng and Longmore, 2005). To generate HepG2/tTA cells stably expressing HFE chimeras, individual pcDNA4 constructs encoding the specified chimeric HFEs or HLA-B7 were introduced using Nucleofector kit V (Amaxa Biosystems, Gaithersburg, MD). Stable cell clones were obtained under the selection with 1 mg/ml G418. Positive clones in the presence of doxycyline (dox) were screened using anti-FLAG antibody. We generated the following stable cell lines including HepG2/tTA HFE, HepG2/tTA W81AHFE, HepG2/tTA α3(−), HepG2/tTA α3(+), HepG2/tTA α1α2, HepG2/tTA α3tmcd, HepG2/tTA α3cd, HepG2/tTA cd and HepG2/tTA HLA-B7 cells.
Isolation of mouse primary hepatocyte cells
Mouse primary hepatocytes were isolated as described in Supplemental Material.
Tf treatment of cells
WIF-B cells were seeded in 6-well plates and treated with 25 μM holo-Tf or apo-Tf (Athens Research and Technology, Athens, GA) for 24 hours prior to harvesting cells. HepG2/tTA cell lines were seeded in 6-well plates, and after 24 hours the medium was replaced with FBS-free medium in the absence or presence of 25 μM human holo-Tf for 24 hours prior to harvest. To induce expression of HFE, HLA-B7 or the chimeras in HepG2/tTA cell lines, 200 ng/ml dox was added to the medium 24 hours before Tf treatment.
Immunoblot
Preparation of cell lysates are described in Supplemental Materials. Immunoblot analysis was carried out using M2 anti-FLAG (1:10,000), mouse monoclonal anti-TfR2 (1:10,000, 9F8-1C11), H68.4 anti-TfR1 (1:5,000) and mouse anti-actin (1:10,000), followed by anti-mouse secondary antibodies conjugated to horseradish peroxidase (1:10,000). Bands were detected by enhanced chemiluminesense (SuperSignal WestPico; Pierce, Rockford, IL).
Immunoprecipitation
Cell lysate (100 μg protein) was first pre-cleared with Sepharose-4B/Protein A beads (Zymed, San Francisco, CA) for 1 hour at 4°C. The pre-cleared lysates were rotated for 2 hours at 4°C with rabbit anti-FLAG antibody, and Sepharose-4B/Protein A. Immunoprecipitated complexes were washed by centrifugation through 1% NET-Triton buffer containing 15% sucrose and eluted with 2 × Laemmli buffer (Laemmli, 1970). Immunodetection was performed as described above.
Knockdown of TfR2
Lipofectamine 2000 (Invitrogen, Carlsbad, CA) was used to cotransfect siRNA specific for human TfR2 (Dharmacon, Chicago, IL) or negative control siRNA and pLuc-link-HAMP as well as pCMV-β-gal plasmid following the manufacturer’s instructions. Briefly, HepG2/tTA HFE cells in 28-cm2 dishes were transfected with mix of 10 μl of 20 μM TfR2 or control siRNA, 2 μg pLuc-link-HAMP, 0.1 μg pCMV-β-gal plasmids using 10 μl Lipofectamine 2000 (Gao et al., 2008; Zhang et al., 2007). The concentration of siRNA (100 nM) had been optimized to maximally decrease TfR2 levels. One day after the transfection, cells were split into 12-well plates and on the following day, cells were switched to the FBS-free medium and incubated with or without 25 μM holo-Tf for additional 24 hours prior to harvesting.
Quantitative real time reverse transcriptase PCR (qRT-PCR)
Total RNA from HepG2 cells, WIF-B cells, rat hepatocytes, or mouse primary hepatocytes was isolated from cells using the RNAeasy RNA isolation kit (Qiagen, Valencia, CA) and treated with DNase (Roche Applied Science) to remove any contaminating genomic DNA. Rat hepatocytes (n = 3) were from normal male Wistar rats. Hepatocytes were isolated by the Cell Culture Core of the NIDDK-funded USC Research Center for Liver Diseases as previously described (Zhang et al., 2004). Oligo (dT) primers and Superscript II reverse transcriptase were used to synthesize cDNA according manufacturer’s instructions. Hepcidin, TfR1, TfR2, HFE, HJV and GAPDH mRNA were measured using the rat or human primers listed in Supplemental Table 2 and as previously described (Gao et al., 2008; Zhang et al., 2007). Results were expressed as the level relative to the corresponding GAPDH. All primers were verified for linearity of amplification.
Transient transfections and luciferase assays
Control or transfected HepG2/tTA cells were seeded in 78-cm2 dishes and co-transfected with pLuc-link-HAMP (5 μg) and pCMV-β-gal plasmids (0.25 μg) using Lipofectamine (50 μl). On the following day, the cells were split into 12 wells of a 12-well plate. They were treated with FBS-free medium ± 25 βM holo-Tf for 24 hours. Cells were lysed in NET-Triton buffer, followed by centrifugation at 16,000g for 5 min at 4°C to remove cell debris. The luciferase activities of supernatants were measured and normalized to β-galactosidase activity as previously described (Kim et al., 1994; Martin et al., 1996) and normalized with the control (-Tf).
Statistical analysis
Standard deviation (SD) and the paired and two-tailed Student’s T-Test were used to evaluate the statistical significance of qRT-PCR and luciferase assay data.
Supplementary Material
Acknowledgments
We would like to thank Dr. Gregory D. Longmore for the gift of HepG2/tTA cells, Dr. Paul W. Howard for the pLuc-link vector, Dr. Nancy Andrews for the Hfe−/− mice and Dr. Robert Fleming for the Tfr2245X/245X mice. We thank Dr. Maria Chloupkova for assistance in the perfusion of livers for primary hepatocytes culture. We also thank Gary Reiness, Julia Maxson, Kristina Nicholson, and Maria Chloupkova for helpful suggestions on the manuscript. This work was supported by National Institute of Health RO1-DK072166 (to C.A.E) and P50 AA011999 Research Center for Alcoholic Liver and Pancreatic Diseases (to H.T.).
Footnotes
In this manuscript, we have followed the numbering system of HFE, which starts at the first amino acid after signal peptide cleavage (-22-1) (Lebron et al., 1998) and corresponds to previous numbering systems used in MHC1-like molecules.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ahmad KA, Ahmann JR, Migas MC, Waheed A, Britton RS, Bacon BR, Sly WS, Fleming RE. Decreased liver hepcidin expression in the Hfe knockout mouse. Blood Cells Mol Dis. 2002;29:361–366. doi: 10.1006/bcmd.2002.0575. [DOI] [PubMed] [Google Scholar]
- Ajioka RS, Kushner JP. Hereditary hemochromatosis. Semin Hematol. 2002;39:235–241. doi: 10.1053/shem.2002.35634. [DOI] [PubMed] [Google Scholar]
- Bennett MJ, Lebron JA, Bjorkman PJ. Crystal structure of the hereditary haemochromatosis protein HFE complexed with transferrin receptor. Nature. 2000;403:46–53. doi: 10.1038/47417. [DOI] [PubMed] [Google Scholar]
- Bondi A, Valentino P, Daraio F, Porporato P, Gramaglia E, Carturan S, Gottardi E, Camaschella C, Roetto A. Hepatic expression of hemochromatosis genes in two mouse strains after phlebotomy and iron overload. Haematologica. 2005;90:1161–1167. [PubMed] [Google Scholar]
- Bothwell TH, MacPhail AP. Hereditary hemochromatosis: etiologic, pathologic, and clinical aspects. Semin Hematol. 1998;35:55–71. [PubMed] [Google Scholar]
- Braiterman LT, Heffernan S, Nyasae L, Johns D, See AP, Yutzy R, McNickle A, Herman M, Sharma A, Naik UP, Hubbard AL. JAM-A is both essential and inhibitory to development of hepatic polarity in WIF-B cells. Am J Physiol-Gastr L. 2008;294:G576–588. doi: 10.1152/ajpgi.00159.2007. [DOI] [PubMed] [Google Scholar]
- Bridle KR, Frazer DM, Wilkins SJ, Dixon JL, Purdie DM, Crawford DH, Subramaniam VN, Powell LW, Anderson GJ, Ramm GA. Disrupted hepcidin regulation in HFE-associated haemochromatosis and the liver as a regulator of body iron homoeostasis. Lancet. 2003;361:669–673. doi: 10.1016/S0140-6736(03)12602-5. [DOI] [PubMed] [Google Scholar]
- Camaschella C, Roetto A, Cali A, De Gobbi M, Garozzo G, Carella M, Majorano N, Totaro A, Gasparini P. The gene TFR2 is mutated in a new type of haemochromatosis mapping to 7q22. Nat Genet. 2000;25:14–15. doi: 10.1038/75534. [DOI] [PubMed] [Google Scholar]
- Chen J, Chloupkova M, Gao J, Chapman-Arvedson TL, Enns CA. HFE modulates transferrin receptor 2 levels in hepatoma cells via interactions that differ from transferrin receptor 1-HFE interactions. J Biol Chem. 2007;282:36862–36870. doi: 10.1074/jbc.M706720200. [DOI] [PubMed] [Google Scholar]
- Cherry J, Nieuwenhuijsen BW, Kaftan EJ, Kennedy JD, Chanda PK. A modified method for PCR-directed gene synthesis from large number of overlapping oligodeoxyribonucleotides. J Biochem Biophys Methods. 2008;70:820–822. doi: 10.1016/j.jprot.2007.12.009. [DOI] [PubMed] [Google Scholar]
- De Domenico I, McVey Ward D, Kaplan J. Regulation of iron acquisition and storage: consequences for iron-linked disorders. Nat Rev Mol Cell Biol. 2008;9:72–81. doi: 10.1038/nrm2295. [DOI] [PubMed] [Google Scholar]
- de Wet JR, Wood KV, DeLuca M, Helinski DR, Subramani S. Firefly luciferase gene: structure and expression in mammalian cells. Mol Biol Cell. 1987;7:725–737. doi: 10.1128/mcb.7.2.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feder JN, Gnirke A, Thomas W, Tsuchihashi Z, Ruddy DA, Basava A, Dormishian F, Domingo R, Jr, Ellis MC, Fullan A, Hinton LM, Jones NL, Kimmel BE, Kronmal GS, Lauer P, Lee VK, Loeb DB, Mapa FA, McClelland E, Meyer NC, Mintier GA, Moeller N, Moore T, Morikang E, Prass CE, Quintana L, Starnes SM, Schatzman RC, Brunke KJ, Drayna DT, Risch NJ, Bacon BR, Wolff RK. A novel MHC class I-like gene is mutated in patients with hereditary haemochromatosis. Nat Genet. 1996;13:399–408. doi: 10.1038/ng0896-399. [DOI] [PubMed] [Google Scholar]
- Feder JN, Tsuchihashi Z, Irrinki A, Lee VK, Mapa FA, Morikang E, Prass CE, Starnes SM, Wolff RK, Parkkila S, Sly WS, Schatzman RC. The hemochromatosis founder mutation in HLA-H disrupts beta2-microglobulin interaction and cell surface expression. J Biol Chem. 1997;272:14025–14028. doi: 10.1074/jbc.272.22.14025. [DOI] [PubMed] [Google Scholar]
- Feng Y, Longmore GD. The LIM protein Ajuba influences interleukin-1-induced NF-kappaB activation by affecting the assembly and activity of the protein kinase Czeta/p62/TRAF6 signaling complex. Mol Biol Cell. 2005;25:4010–4022. doi: 10.1128/MCB.25.10.4010-4022.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J, Zhao N, Knutson MD, Enns CA. The Hereditary Hemochromatosis Protein, HFE, Inhibits Iron Uptake via Down-regulation of Zip14 in HepG2 Cells. J Biol Chem. 2008;283:21462–21468. doi: 10.1074/jbc.M803150200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehrke SG, Kulaksiz H, Herrmann T, Riedel HD, Bents K, Veltkamp C, Stremmel W. Expression of hepcidin in hereditary hemochromatosis: evidence for a regulation in response to the serum transferrin saturation and to non-transferrin-bound iron. Blood. 2003;102:371–376. doi: 10.1182/blood-2002-11-3610. [DOI] [PubMed] [Google Scholar]
- Giannetti AM, Snow PM, Zak O, Bjorkman PJ. Mechanism for multiple ligand recognition by the human transferrin receptor. PLoS Biol. 2003;1:341–350. doi: 10.1371/journal.pbio.0000051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goswami T, Andrews NC. Hereditary hemochromatosis protein, HFE, interaction with transferrin receptor 2 suggests a molecular mechanism for mammalian iron sensing. J Biol Chem. 2006;281:28494–28498. doi: 10.1074/jbc.C600197200. [DOI] [PubMed] [Google Scholar]
- Holmstrom P, Dzikaite V, Hultcrantz R, Melefors O, Eckes K, Stal P, Kinnman N, Smedsrod B, Gafvels M, Eggertsen G. Structure and liver cell expression pattern of the HFE gene in the rat. J Hepatol. 2003;39:308–314. doi: 10.1016/s0168-8278(03)00293-9. [DOI] [PubMed] [Google Scholar]
- Ihrke G, Neufeld EB, Meads T, Shanks MR, Cassio D, Laurent M, Schroer TA, Pagano RE, Hubbard AL. WIF-B cells: an in vitro model for studies of hepatocyte polarity. J Cell Biol. 1993;123:1761–1775. doi: 10.1083/jcb.123.6.1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson MB, Enns CA. Diferric transferrin regulates transferrin receptor 2 protein stability. Blood. 2004;104:4287–4293. doi: 10.1182/blood-2004-06-2477. [DOI] [PubMed] [Google Scholar]
- Kawabata H, Fleming RE, Gui D, Moon SY, Saitoh T, O’Kelly J, Umehara Y, Wano Y, Said JW, Koeffler HP. Expression of hepcidin is down-regulated in TfR2 mutant mice manifesting a phenotype of hereditary hemochromatosis. Blood. 2005;105:376–381. doi: 10.1182/blood-2004-04-1416. [DOI] [PubMed] [Google Scholar]
- Kawabata H, Yang R, Hirama T, Vuong PT, Kawano S, Gombart AF, Koeffler HP. Molecular cloning of transferrin receptor 2. A new member of the transferrin receptor-like family. J Biol Chem. 1999;274:20826–20832. doi: 10.1074/jbc.274.30.20826. [DOI] [PubMed] [Google Scholar]
- Kim DS, Ahn SK, Yoon JH, Hong SH, Kim KE, Maurer RA, Park SD. Involvement of a cAMP-responsive DNA element in mediating TRH responsiveness of the human thyrotropin alpha-subunit gene. Mol Endocrinol. 1994;8:528–536. doi: 10.1210/mend.8.4.7519724. [DOI] [PubMed] [Google Scholar]
- Konieczko EM, Ralston AK, Crawford AR, Karpen SJ, Crawford JM. Enhanced Na+-dependent bile salt uptake by WIF-B cells, a rat hepatoma hybrid cell line, following growth in the presence of a physiological bile salt. Hepatology. 1998;27:191–199. doi: 10.1002/hep.510270130. [DOI] [PubMed] [Google Scholar]
- Krijt J, Niederkofler V, Salie R, Sefc L, Pelichovska T, Vokurka M, Necas E. Effect of phlebotomy on hepcidin expression in hemojuvelin-mutant mice. Blood Cells Mol Dis. 2007;39:92–95. doi: 10.1016/j.bcmd.2007.02.004. [DOI] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lebron JA, Bennett MJ, Vaughn DE, Chirino AJ, Snow PM, Mintier GA, Feder JN, Bjorkman PJ. Crystal structure of the hemochromatosis protein HFE and characterization of its interaction with transferrin receptor. Cell. 1998;93:111–123. doi: 10.1016/s0092-8674(00)81151-4. [DOI] [PubMed] [Google Scholar]
- Lebron JA, Bjorkman PJ. The transferrin receptor binding site on HFE, the class I MHC-related protein mutated in hereditary hemochromatosis. J Mol Biol. 1999;289:1109–1118. doi: 10.1006/jmbi.1999.2842. [DOI] [PubMed] [Google Scholar]
- Levy JE, Montross LK, Cohen DE, Fleming MD, Andrews NC. The C282Y mutation causing hereditary hemochromatosis does not produce a null allele. Blood. 1999;94:9–11. [PubMed] [Google Scholar]
- Lin L, Valore EV, Nemeth E, Goodnough JB, Gabayan V, Ganz T. Iron transferrin regulates hepcidin synthesis in primary hepatocyte culture through hemojuvelin and BMP2/4. Blood. 2007;110:2182–2189. doi: 10.1182/blood-2007-04-087593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin CS, Wight PA, Dobretsova A, Bronstein I. Dual luminescence-based reporter gene assay for luciferase and beta-galactosidase. Biotechniques. 1996;21:520–524. doi: 10.2144/96213pf01. [DOI] [PubMed] [Google Scholar]
- Montosi G, Donovan A, Totaro A, Garuti C, Pignatti E, Cassanelli S, Trenor CC, Gasparini P, Andrews NC, Pietrangelo A. Autosomal-dominant hemochromatosis is associated with a mutation in the ferroportin (SLC11A3) gene. J Clin Invest. 2001;108:619–623. doi: 10.1172/JCI13468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muckenthaler M, Roy CN, Custodio AO, Minana B, deGraaf J, Montross LK, Andrews NC, Hentze MW. Regulatory defects in liver and intestine implicate abnormal hepcidin and Cybrd1 expression in mouse hemochromatosis. Nat Genet. 2003;34:102–107. doi: 10.1038/ng1152. [DOI] [PubMed] [Google Scholar]
- Muckenthaler MU, Rodrigues P, Macedo MG, Minana B, Brennan K, Cardoso EM, Hentze MW, de Sousa M. Molecular analysis of iron overload in beta2-microglobulin-deficient mice. Blood Cells Mol Dis. 2004;33:125–131. doi: 10.1016/j.bcmd.2004.05.003. [DOI] [PubMed] [Google Scholar]
- Nemeth E, Tuttle MS, Powelson J, Vaughn MB, Donovan A, Ward DM, Ganz T, Kaplan J. Hepcidin regulates cellular iron efflux by binding to ferroportin and inducing its internalization. Science. 2004;306:2090–2093. doi: 10.1126/science.1104742. [DOI] [PubMed] [Google Scholar]
- Nicolas G, Viatte L, Lou DQ, Bennoun M, Beaumont C, Kahn A, Andrews NC, Vaulont S. Constitutive hepcidin expression prevents iron overload in a mouse model of hemochromatosis. Nat Genet. 2003;34:97–101. doi: 10.1038/ng1150. [DOI] [PubMed] [Google Scholar]
- Nies AT, Cantz T, Brom M, Leier I, Keppler D. Expression of the apical conjugate export pump, Mrp2, in the polarized hepatoma cell line, WIF-B. Hepatology. 1998;28:1332–1340. doi: 10.1002/hep.510280523. [DOI] [PubMed] [Google Scholar]
- Papanikolaou G, Samuels ME, Ludwig EH, MacDonald ML, Franchini PL, Dube MP, Andres L, MacFarlane J, Sakellaropoulos N, Politou M, Nemeth E, Thompson J, Risler JK, Zaborowska C, Babakaiff R, Radomski CC, Pape TD, Davidas O, Christakis J, Brissot P, Lockitch G, Ganz T, Hayden MR, Goldberg YP. Mutations in HFE2 cause iron overload in chromosome 1q-linked juvenile hemochromatosis. Nat Genet. 2004;36:77–82. doi: 10.1038/ng1274. [DOI] [PubMed] [Google Scholar]
- Pigeon C, Ilyin G, Courselaud B, Leroyer P, Turlin B, Brissot P, Loreal O. A new mouse liver-specific gene, encoding a protein homologous to human antimicrobial peptide hepcidin, is overexpressed during iron overload. J Biol Chem. 2001;276:7811–7819. doi: 10.1074/jbc.M008923200. [DOI] [PubMed] [Google Scholar]
- Robb A, Wessling-Resnick M. Regulation of transferrin receptor 2 protein levels by transferrin. Blood. 2004;104:4294–4299. doi: 10.1182/blood-2004-06-2481. [DOI] [PubMed] [Google Scholar]
- Roetto A, Papanikolaou G, Politou M, Alberti F, Girelli D, Christakis J, Loukopoulos D, Camaschella C. Mutant antimicrobial peptide hepcidin is associated with severe juvenile hemochromatosis. Nat Genet. 2003;33:21–22. doi: 10.1038/ng1053. [DOI] [PubMed] [Google Scholar]
- Schmidt PJ, Toran PT, Giannetti AM, Bjorkman PJ, Andrews NC. The transferrin receptor modulates Hfe-dependent regulation of hepcidin expression. Cell Metab. 2008;7:205–214. doi: 10.1016/j.cmet.2007.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verga Falzacappa MV, Casanovas G, Hentze MW, Muckenthaler MU. A bone morphogenetic protein (BMP)-responsive element in the hepcidin promoter controls HFE2-mediated hepatic hepcidin expression and its response to IL-6 in cultured cells. J Mol Med. 2008;86:531–540. doi: 10.1007/s00109-008-0313-7. [DOI] [PubMed] [Google Scholar]
- Vogt TM, Blackwell AD, Giannetti AM, Bjorkman PJ, Enns CA. Heterotypic interactions between transferrin receptor and transferrin receptor 2. Blood. 2003;101:2008–2014. doi: 10.1182/blood-2002-09-2742. [DOI] [PubMed] [Google Scholar]
- Vujic Spasic M, Kiss J, Herrmann T, Galy B, Martinache S, Stolte J, Grone HJ, Stremmel W, Hentze MW, Muckenthaler MU. Hfe acts in hepatocytes to prevent hemochromatosis. Cell Metab. 2008;7:173–178. doi: 10.1016/j.cmet.2007.11.014. [DOI] [PubMed] [Google Scholar]
- Waheed A, Parkkila S, Saarnio J, Fleming RE, Zhou XY, Tomatsu S, Britton RS, Bacon BR, Sly WS. Association of HFE protein with transferrin receptor in crypt enterocytes of human duodenum. Proc Natl Acad Sci USA. 1999;96:1579–1584. doi: 10.1073/pnas.96.4.1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace DF, Summerville L, Subramaniam VN. Targeted disruption of the hepatic transferrin receptor 2 gene in mice leads to iron overload. Gastroenterology. 2007;132:301–310. doi: 10.1053/j.gastro.2006.11.028. [DOI] [PubMed] [Google Scholar]
- Weinstein DA, Roy CN, Fleming MD, Loda MF, Wolfsdorf JI, Andrews NC. Inappropriate expression of hepcidin is associated with iron refractory anemia: implications for the anemia of chronic disease. Blood. 2002;100:3776–3781. doi: 10.1182/blood-2002-04-1260. [DOI] [PubMed] [Google Scholar]
- West AP, Jr, Giannetti AM, Herr AB, Bennett MJ, Nangiana JS, Pierce JR, Weiner LP, Snow PM, Bjorkman PJ. Mutational analysis of the transferrin receptor reveals overlapping HFE and transferrin binding sites. J Mol Biol. 2001;313:385–397. doi: 10.1006/jmbi.2001.5048. [DOI] [PubMed] [Google Scholar]
- Wilson IA, Bjorkman PJ. Unusual MHC-like molecules: CD1, Fc receptor, the hemochromatosis gene product, and viral homologs. Curr Opin Immunol. 1998;10:67–73. doi: 10.1016/s0952-7915(98)80034-4. [DOI] [PubMed] [Google Scholar]
- Zhang AS, Anderson SA, Meyers KR, Hernandez C, Eisenstein RS, Enns CA. Evidence that inhibition of hemojuvelin shedding in response to iron is mediated through neogenin. J Biol Chem. 2007;282:12547–12556. doi: 10.1074/jbc.M608788200. [DOI] [PubMed] [Google Scholar]
- Zhang AS, Davies PS, Carlson HL, Enns CA. Mechanisms of HFE-induced regulation of iron homeostasis: Insights from the W81A HFE mutation. Proc Natl Acad Sci USA. 2003;100:9500–9505. doi: 10.1073/pnas.1233675100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang AS, Xiong S, Tsukamoto H, Enns CA. Localization of iron metabolism-related mRNAs in rat liver indicate that HFE is expressed predominantly in hepatocytes. Blood. 2004;103:1509–1514. doi: 10.1182/blood-2003-07-2378. [DOI] [PubMed] [Google Scholar]
- Zhou XY, Tomatsu S, Fleming RE, Parkkila S, Waheed A, Jiang J, Fei Y, Brunt EM, Ruddy DA, Prass CE, Schatzman RC, O’Neill R, Britton RS, Bacon BR, Sly WS. HFE gene knockout produces mouse model of hereditary hemochromatosis. Proc Natl Acad Sci USA. 1998;95:2492–2497. doi: 10.1073/pnas.95.5.2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.