SUMMARY
Interactions between tumor and immune cells either enhance or inhibit cancer progression. We show here that Stat3 signaling within the tumor microenvironment induces a pro-carcinogenic cytokine, IL-23, while inhibiting a central anti-carcinogenic cytokine, IL-12, thereby shifting the balance of tumor immunity towards carcinogenesis. Stat3 induces expression of IL-23, which is mainly produced by tumor-associated macrophages, via direct transcriptional activation of the IL-23/p19 gene. Furthermore, Stat3 inhibits NF-κB/c-Rel-dependent IL-12/p35 gene expression in tumor-associated dendritic cells. Tumor-associated regulatory T cells (Treg) express IL-23 receptor (IL-23R) which activates Stat3 in this cell type, leading to upregulation of the Treg-specific transcription factor, Foxp3, and the immunosuppressive cytokine, IL-10. These results demonstrate that Stat3 promotes IL-23-mediated pro-carcinogenic immune responses while inhibiting IL-12-dependent anti-tumor immunity.
SIGNIFICANCE
Recent studies suggest that two related cytokines, IL- 23 and IL- 12, play opposite roles in carcinogenesis. However, the underlying mechanisms regulating the balance between these cytokines in the tumor microenvironment have not been elucidated. Mechanisms by which IL-23 promotes tumor immune evasion also remain to be explored. Our results reveal that Stat3 signaling in the tumor microenvironment regulates the IL-12/IL-23 balance and further, that IL- 23 enhances the immunosuppressive activity of regulatory T cells within the tumor microenvironment, in part via IL-23 receptor dependent Stat3 activation. Because Stat3 is a point of convergence for signaling pathways commonly activated in cancer, our data reveal a mechanism by which oncogenic pathways regulate the immune microenvironment to promote tumor development.
INTRODUCTION
Transcription factor Stat3 is constitutively activated in diverse cancers (Yu and Jove, 2004), and its activation in tumors enhances transcription of genes associated with cell cycle progression, anti-apoptosis, angiogenesis and immune evasion (Yu and Jove, 2004; Yu et al., 2007). In addition to tumor cells, Stat3 is constitutively activated within many immune cell types in the tumor microenvironment, including DCs and macrophages (Kortylewski et al., 2005). Ablating Stat3 in myeloid cells allows efficient CD8+ T cell infiltration into tumors while inhibiting accumulation of regulatory T cells (Tregs) (Kortylewski et al., 2005). Activated Stat3 suppresses antitumor immunity by inhibiting the expression of many cytokines and chemokines important for stimulating antitumor immunity, and by upregulating production of several immunosuppressive factors, including IL-10 and VEGF (Takeda et al., 1999; Wang et al., 2004). These immunosuppressive factors are not only Stat3 target genes but also Stat3 activators (Yu et al., 2007). In order to further explore the mechanisms by which Stat3 mediates tumor immunosuppression, we evaluated its role in the regulation of two closely related cytokines, IL-23 and IL-12, which play critical but opposing roles in tumor immunity.
IL-12, a heterodimer composed of α and β subunits (termed p35 and p40 respectively) promotes anti-tumor immunity via activation of NK cells and Th1 T cells, and is characterized by production of interferon-γ, another important cytokine in anti-tumor immunity (Gerosa et al., 2002; Kaplan et al., 1998; Shankaran et al., 2001; Trinchieri, 2003). IL-12 further promotes the expansion and activity of cytotoxic T lymphocytes (CTL) both directly and indirectly by Th1 cells (Colombo and Trinchieri, 2002). IL-23, a more recently discovered IL-12 family member, is composed of the p40 subunit in common with IL-12, paired with a unique p19 subunit (Oppmann et al., 2000). Similarly, IL-12R and IL-23R share a common β subunit, which is paired with a unique α subunit for each receptor (Parham et al., 2002). While IL-23 was originally thought to possess proinflammatory properties similar to IL-12, analysis of mice with selective knockout of the IL-12/p35 gene versus the IL-23/p40 gene revealed very distinct functions for these two cytokines. In particular, a number of experimental autoimmune diseases were shown to be dependent on IL-23 and not on IL-12 (Cua et al., 2003; Ghilardi et al., 2004; Langrish et al., 2005; Murphy et al., 2003). IL-23 has also been shown to promote the expansion of a distinct lineage of helper T cell, termed Th17 (Langrish et al., 2005). Th17 cells are characterized by production of a number of specific cytokines not produced by Th1 or Th2 cells, including IL-17A, IL-17F, IL-21 and IL-22. Interestingly, Stat3 has also been documented to be an essential transcriptional regulator of IL-17, IL-21 and IL-22 production by Th17 cells (Chen et al., 2006; Harris et al., 2007; Laurence et al., 2007; O’Shea and Murray, 2008; Wei et al., 2007; Zheng et al., 2007; Zhou et al., 2007).
An opposite role in carcinogenesis has recently been reported for IL-23, compared to IL-12 (Langowski et al., 2007; Langowski et al., 2006). Carcinogen-induced tumor formation was greater in IL-12/p35 KO mice, confirming the physiologic anti-tumor role of IL-12, however the opposite effect was observed in IL-23/p19 KO mice (Langowski et al., 2006). Whereas IL-12 facilitated tumor infiltration of CD8+ T cells, IL-23 reduces CD8+ T cells in tumor and promoted tumor angiogenesis (Langowski et al., 2006). While these results suggest a pro-carcinogenic role IL-23 production within tumors, its mechanisms of action in tumor regulation has not been elucidated. We therefore investigated how IL-23 and IL-12 expression are differentially regulated in the tumor microenvironment, and how IL-23 further propagates tumor immunosuppressive effects.
RESULTS
Role of Stat3 in the differential expression of IL-23 and IL-12 in tumors
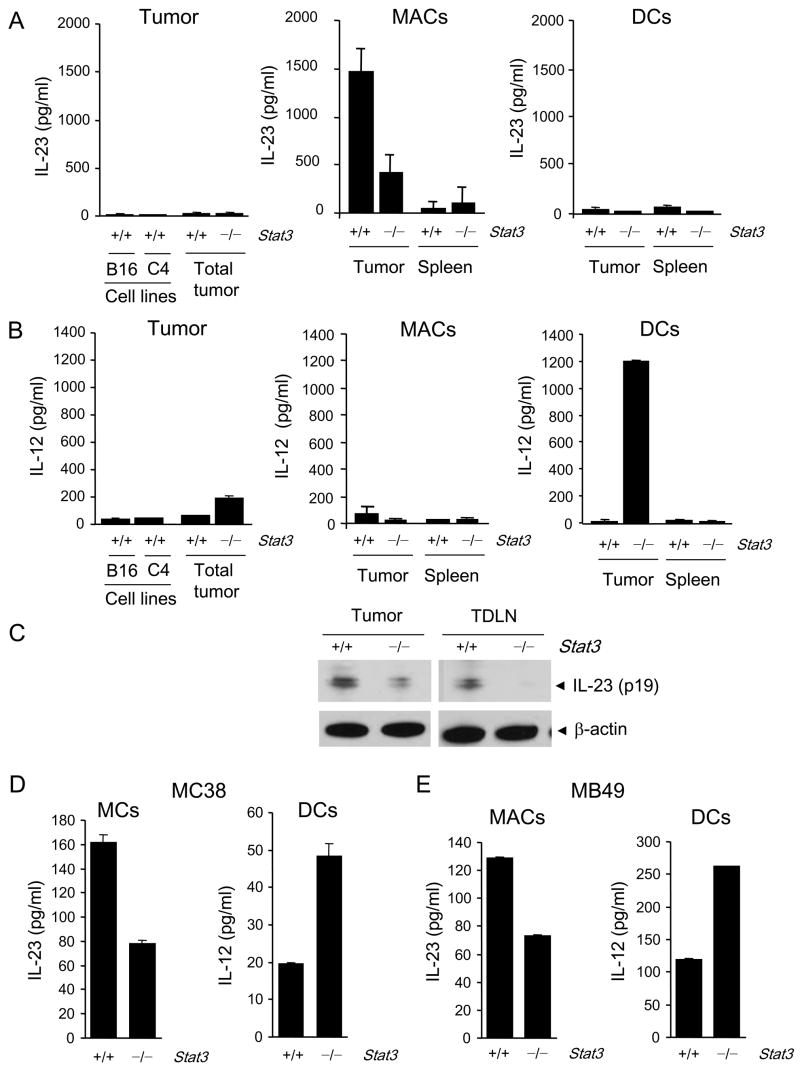
We assessed IL-23 and IL-12 protein secretion by B16 tumor cells in vitro and B16 tumors growing in vivo after implantation into C57BL/6 mice. B16 tumor cells and a second melanoma tumor line, C4, cultured in vitro secreted very little of either cytokine as detected by ELISA (Fig. 1A, left panels). B16 tumors growing in vivo were dissociated into single cell suspensions that consisted of tumor cells as well as hematopoietically-derived cells comprising the tumor microenvironment. Similar to in vitro results, only a small amount of IL-23 or IL-12 secretion was detected by ELISA when the unfractionated cell suspensions were cultured (Fig. 1A, B left panels). When the tumor-associated myeloid component was FACS sorted into macrophages (CD11b+c−) and DCs (CD11b+c+), high levels of IL-23 were found to be produced by the tumor-associated macrophages (Fig. 1A, middle panel) with much lower levels produced by tumor infiltrating DCs (Fig. 1A, right panel). Western blot data demonstrated IL-23 was also detectable in CD11b+ cells isolated from tumors and tumor draining lymph nodes (Fig. 1C). Virtually no IL-12 was secreted by either unfractionated tumor or by purified tumor-associated macrophages or splenic DCs, as determined by ELISA (Fig. 1B).
Figure 1. IL-23 and IL-12 expression in tumor-infiltrating myeloid cells is compartmentalized and oppositely regulated by Stat3.
ELISA measurement of protein levels of IL-23 (A) and IL-12 (B) in supernatants of tumor cell lines (B16 and C4 melanoma cells), B16 whole tumor cell suspensions (Total tumor) from mice with Stat3+/+ or Stat3−/− hematopoietic cells, enriched tumor-infiltrating CD11b+CD11c− macrophages (MACs) and CD11c+ dendritic cells (DCs) cultured in vitro for 24 h. Data shown are mean values ± SD from one of three separate experiments performed on cells pooled from four animals per group analyzed. (C) Western blot detection of IL-23/p19 protein levels in the CD11b+ cells isolated from the whole B16 tumor and tumor-draining lymph nodes of Stat3+/+ or Stat3−/− mice. (D, E) IL-23 and IL-12 protein levels measured by ELISA as in Fig. 1A, in supernatants from the enriched tumor-infiltrating CD11b+CD11c− MACs and CD11c+ DCs isolated from the whole MC38 colon carcinoma (D) and MB49 bladder carcinoma (E) tumors; mean ± SEM (n = 3).
The role of Stat3 signaling in IL-23 and IL-12 regulation within the tumor microenvironment was initially determined by conditional knock out of the Stat3 gene in the hematopoietic compartment. To accomplish this, Mx1-Cre/Stat3flox/flox mice were treated with poly(I:C). We have previously shown that Stat3 was efficiently knocked out of myeloid cells under these conditions (Kortylewski et al., 2005). By 24 h after poly(I:C) treatment, there was no evidence of residual activation of either DC or macrophages as measured by either cell membrane markers or cytokine production (Kortylewski et al., 2005). To avoid any confounding effects of immune stimulation by poly(I:C) treatment, Mx1-Cre/Stat3flox/flox and control Stat3flox/flox littermate mice were not challenged with tumors until 4 days after completion of poly(I:C) treatment. IL-12 and IL-23 production in tumors was analyzed 2–3 weeks later. We purified macrophages and DCs in tumors grown in poly(I:C)-treated Mx1CreStat3flox/flox mice with induced Stat3 ablation, as well as poly(I:C)-treated Stat3flox/flox control mice. Hematopoietic Stat3 ablation significantly reduced IL-23 production by B16 tumor-associated macrophages (Fig. 1A). In contrast, IL-12 production was up-regulated in B16 tumors from mice with hematopoietic Stat3 knockout. The majority of IL-12 expression came from the tumor-associated DCs infiltrating B16 tumors (Fig. 1B). Thus, these data implicated that Stat3 signaling drives IL-23 production from the tumor-associated macrophages while restraining IL-12 production by tumor-associated DCs. This effect was specific for the tumor microenvironment since there was no significant production of either IL-12 or IL-23 by splenic macrophages or DC whether or not the Stat3 gene is ablated.
To test the generality of these findings that Stat3 in tumor stromal myeloid cells contributed to regulating IL-23 and IL-12 expression in the tumor, we performed similar experiments using MC38 colon carcinoma (Fig. 1D) and MB49 bladder carcinoma models (Fig. 1E). In vivo experiments showed that the growth of MC38 tumors was inhibited by Stat3 ablation in myeloid cells (Supplementary Fig. 1), confirming previous data in B16 and MB49 tumor models (Kortylewski et al., 2005). Consistent with our results in the B16 tumor model, ablating Stat3 in myeloid cells decreased production of IL-23 by macrophages within MC38 and MB49 tumors (Fig. 1D, E). While the overall level of IL-23 secretion by MC38 and MB49 tumor-infiltrating macrophages in control Stat3flox/flox mice was lower than that observed B16 tumors, the IL-23 that is produced is clearly Stat3-dependent. Likewise, the DCs isolated from MC38 and MB49 tumors showed increased secretion of IL-12 following induced Stat3 ablation. The level of IL-12 secretion by DCs isolated from MC38 and MB49 tumors did not reach levels observed in the B16 tumor model, but the increases observed with hematopoietic Stat3 knockout were highly reproducible. Multiple factors likely contribute to the differences in magnitude of IL-23 and IL-12 production by tumor associated macrophages and Stat3−/− tumor-associated DCs, respectively, in the different tumors. Previous studies indicated that IL-12 expression can be induced in Stat3−/−splenic DCs by TLR activation (Kortylewski et al., 2005). Dying tumor cells have also been shown to stimulate DC IL-12 production (Apetoh et al., 2007). The presence of large necrosis (up to 89% of tumor area) within B16 (Garcia-Hernandez et al., 2002), which was not observed in MC38 or MB49 tumors (Kortylewski M. and Yu H., unpublished observations), might provide cell debris capable of co-stimulating intratumoral DCs to produce IL-12 after deletion of Stat3.
Stat3 directly regulates IL-23/p19 transcription
In order to determine the mechanism for regulation of these cytokines by Stat3 in the tumor microenvironment, we first analyzed IL-23/p19 gene regulation. Stat3 ablation indeed reduced IL-23/p19 mRNA in tumor-associated macrophages by roughly 10 fold with virtually no IL-23/p19 mRNA detectable in Stat3+/+ or Stat3−/− splenic macrophages (Fig. 2A). Despite low levels (26 and 79 times lower in B16 and C4 cells, respectively, relative to tumor-infiltrating macrophages), IL-23/p19 mRNA in the tumor cells was reduced over 80% by Stat3 siRNA knockdown (Supplementary Fig. 2A). These results suggested that Stat3 positively regulates IL-23 at the transcriptional level in different cell types. In order to determine whether this transcriptional regulation was direct, we performed luciferase reporter and chromatin immunoprecipitation (ChIP) analyses. First, 3T3 cells were transfected with a luciferase expression vector driven by the IL-23/p19 promoter (−1159 to +160). Dual-luciferase activity was enhanced significantly by cotransfection with a constitutively active Stat3 mutant, Stat3C (Fig. 2B). B16 cells transfection with the IL-23/p19 promoter-driven luciferase reporter gene resulted in a progressively decreased luciferase activity with successively increasing amounts of Stat3 siRNA (Fig. 2C, left panel). Cotransfection with NF-κB/p65 siRNA also resulted in decreased luciferase activity with the greatest suppression occurring upon cotransfection with both Stat3 and NF-κB siRNAs (Fig. 2C, right panel). These results indicate that NF-κB/p65, which is also constitutively activated in B16 as in many cancers, participates in IL-23/p19 transcription.
Figure 2. Stat3 and NF-κB/p65 synergistically enhance IL-23 expression by directly binding to the IL-23/p19 promoter.
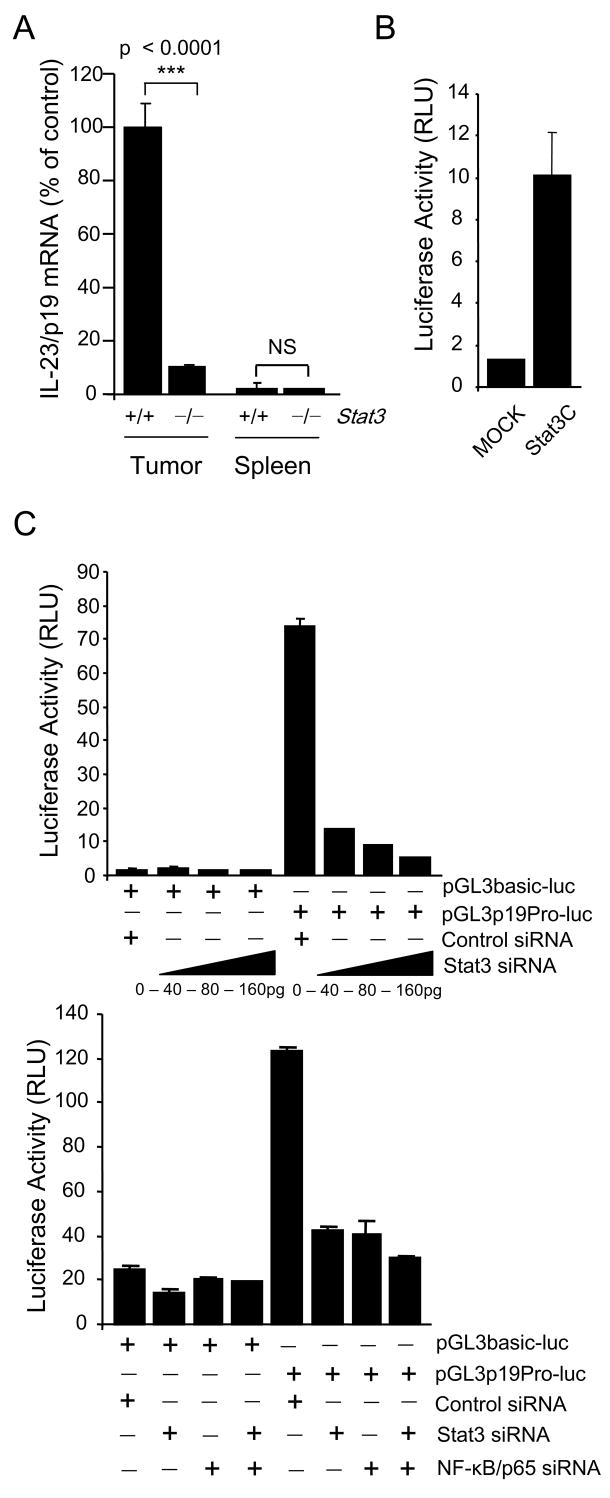
(A) Stat3 upregulates expression of IL-23/p19 mRNA in CD11b+ myeloid cells freshly isolated from B16 tumors. Shown are the results from one of three independent experiments analyzed by real-time PCR, normalized to 18S rRNA. (B) Over expression of constitutively active Stat3 mutant (Stat3C) activates transcription by IL-23/p19 promoter. The fragment from the mouse promoter of IL-23/p19 (sequence from -1159 to +160) was cloned into pGL3 vector with luciferase reporter gene. Dual-luciferase activity was determined 24 h after transfection of various sets of expression vectors into 3T3 fibroblasts. (C) Stat3 and NF-κB/p65 bind directly to the p19 promoter. Top: Stat3 silencing downregulates the activity of IL-23/p19 promoter. Dual-luciferase activity was measured in lysates of B16 cells 24 h after transfection with various concentrations of Stat3 siRNA or with scrambled RNA control. Bottom: Both Stat3 and NF-κB/p65 transcription factors are required for the transcriptional activity of IL-23/p19 promoter. Stat3 siRNA, NF-κB/p65 siRNA or both were transfected into B16 cells together with IL-23/p19 promoter-luciferase construct. Dual-luciferase activity was measured as described above. Data shown are mean values ± SD from experiments performed in triplicates.
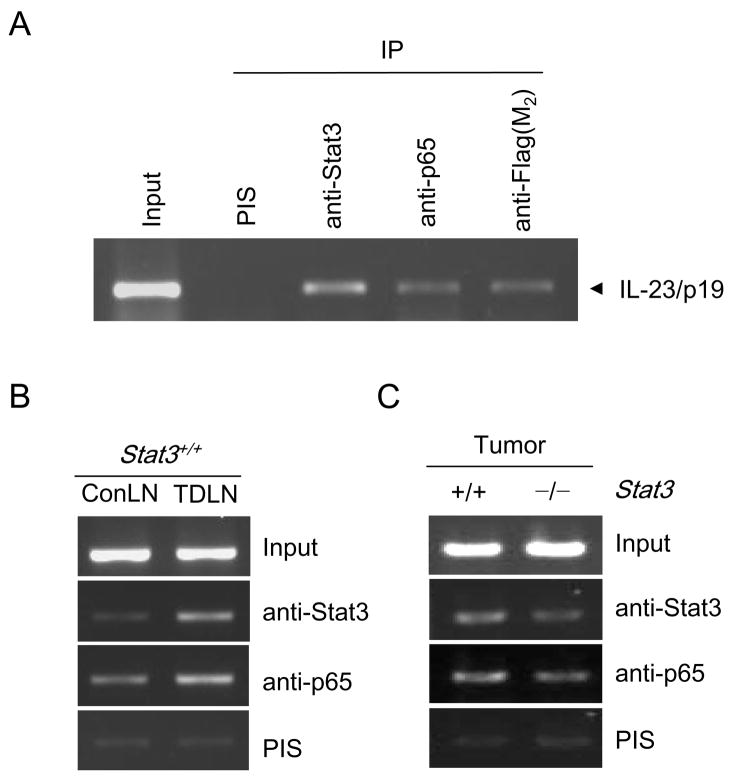
We next performed ChIP analysis on B16 cells using primers for the IL-23/p19 promoter. Both Stat3 and NF-κB/p65 were associated with the IL-23/p19 promoter (Fig. 3A). While it was difficult to obtain enough tumor-associated macrophages to perform ChIP analysis, ChIP of total tumor growing in vivo as well as tumor draining lymph nodes demonstrated Stat3 binding to the IL-23/p19 promoter along with NF-κB/p65 (Fig. 3B, C). NFκB/p65 binding to the IL-23/p19 promoter was also higher in tumor draining lymph nodes relative to non-draining lymph nodes (Fig. 3B). Hematopoietic Stat3 ablation resulted in a decreased ChIP signal for both Stat3 and NF-κB/p65 on the IL-23/p19 promoter in growing B16 tumor (Fig. 3C).
Figure 3. Both Stat3 and NF-κB/p65 bind to the IL-23/p19 promoter.
(A) B16 tumor cells were transfected with pRV-CMV Stat3-Flag or control vector, and antibodies specific for Stat3, Flag-M2, NF-κB/p65 or control IgG as indicated, were used for chromatin immunoprecipitation (ChIP). Chromatin was purified and amplified by PCR using primers specific for Stat3 and NF-κB binding sequence of the mouse IL-23/p19 promoter. (B) The binding of Stat3 and NF-κB/p65 to IL-23/p19 promoter is elevated in tumor-draining but not in distant lymph nodes. Cells isolated from pooled tumor draining or contralateral lymph nodes were fixed with formaldehyde and subjected to ChIP assay performed as described above. (C) Tumors from mice with myeloid cell-specific Stat3 ablation show reduced Stat3 and NF-κB/p65 binding to the IL-23/p19 promoter as assessed by ChIP assay.
Stat3 inhibits NFκBc-Rel mediated IL-12/p35 gene expression
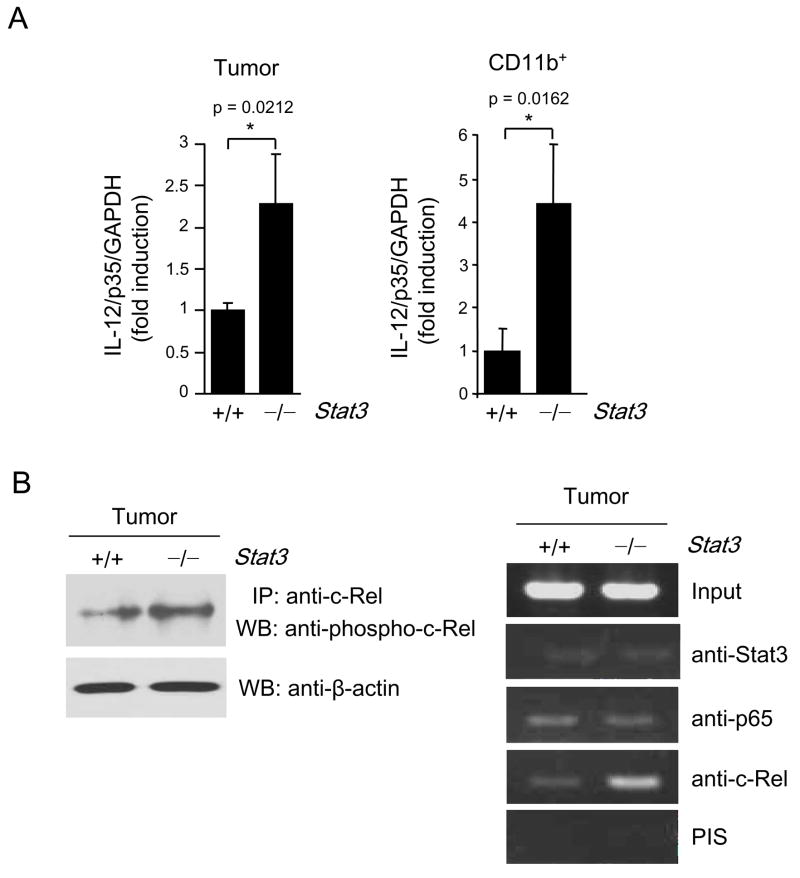
Real-time PCP demonstrated Stat3 reciprocally downregulated IL-12/p35 transcript in whole B16 tumors growing in hematopoietic Stat3KO mice (Fig. 4A, left), in tumor-associated myeloid cells (Fig. 4A, right) and in cultured B16 tumor cells after siRNA transfection (Supplementary Fig. 2B). The NF-κB family transcription factor, c-Rel, has been demonstrated to be essential for transcription of IL-12/p35 (Grumont et al., 2001) and other genes important for DC activation (Wang et al., 2007). However, we found that levels of active c-Rel were low in growing tumor (Fig. 4B). Upon hematopoietic Stat3 ablation, there was increased c-Rel activity (phospho-c-Rel) in tumors (Fig. 4B, left panel), as well as increased binding to the IL-12/p35 promoter of NF-κB/c-Rel (Fig. 4B, right panel). It is of note that Stat3 ablation did not affect p65 binding to the IL-12/p35 promoter.
Figure 4. Stat3 inhibits IL-12/p35 gene expression.
(A) Stat3 knockdown or ablation augments IL-12/p35 expression in the whole tumor (left panel) and in the tumor infiltrating myeloid cells (right panel); mean ± SD (n = 3). (B) Stat3 inhibits c-Rel activity and binding of c-Rel to IL-12/p35 promoter in tumors. Left panel: Western blot analysis of c-Rel phosphorylation after immunoprecipitation of total c-Rel from spleens and tumors grown in mice with Stat3+/+ and Stat3−/− hematopoietic compartment. Right panel: Ablating Stat3 (in hematopoietic cells) increases binding of NF-κB/c-Rel, but not NF-κB/p65, to IL-12/p35 promoter in whole tumor preparation, as measured by ChIP assay.
IL-23 receptor and Stat3 signaling in tumor-infiltrating Treg
An immune suppressive role for Stat3 in myeloid cells in the tumor microenvironment has been demonstrated in Mx1-Cre/Stat3flox/flox mice (Kortylewski et al., 2005). However, whether intrinsic Stat3 signaling in T cells mediates tumor immunosuppression has not been shown. To address this question, we next determined the role of Stat3 and IL-23 in T cell physiology within the tumor microenvironment. Tumor growth is enhanced by the CD4+FoxP3+ Treg subset, which produces cytokines such as IL-10 and TGF-β and antagonizes Th1 and CD8 mediated anti-tumor responses (Zou, 2006). As has been found in several other tumors, the major subset of CD4 T cells infiltrating growing B16 tumors (>50%) are FoxP3+ Treg cells (Kortylewski et al., 2005). Analysis of B16 tumor Tregs by intracellular staining of phospho-Stat3 (pStat3) demonstrated that Stat3 was constitutively activated in tumor infiltrating Tregs, relative to their splenic counterparts from the same mice (Fig. 5A, top panel). Since the IL-23R is known to signal through Stat3, we tested whether tumor CD4+FoxP3+ Treg express detectable IL-23R in B16 tumors. Whereas splenic Treg did not express detectable IL-23R, Treg from both tumor and tumor-draining lymph nodes were positive for IL-23R (Fig. 5A, bottom). Elevated expression of IL-23R in Treg from tumor and tumor-draining lymph nodes was further confirmed in both the MB49 and MC38 tumor models (Fig. 5A, bottom). Furthermore, although recombinant IL-23 did not induce Stat3 activation in splenic Treg, the already evident endogenous pStat3 of the tumor Treg could be further up-regulated by addition of recombinant IL-23 in vitro (Fig. 5B). These results demonstrated that tumor-associated Tregs are distinct from peripheral Tregs in that they express functional IL-23R and that the IL-23R signals in part through Stat3.
Figure 5. IL-23 receptor and Stat3 signaling in tumor Tregs.
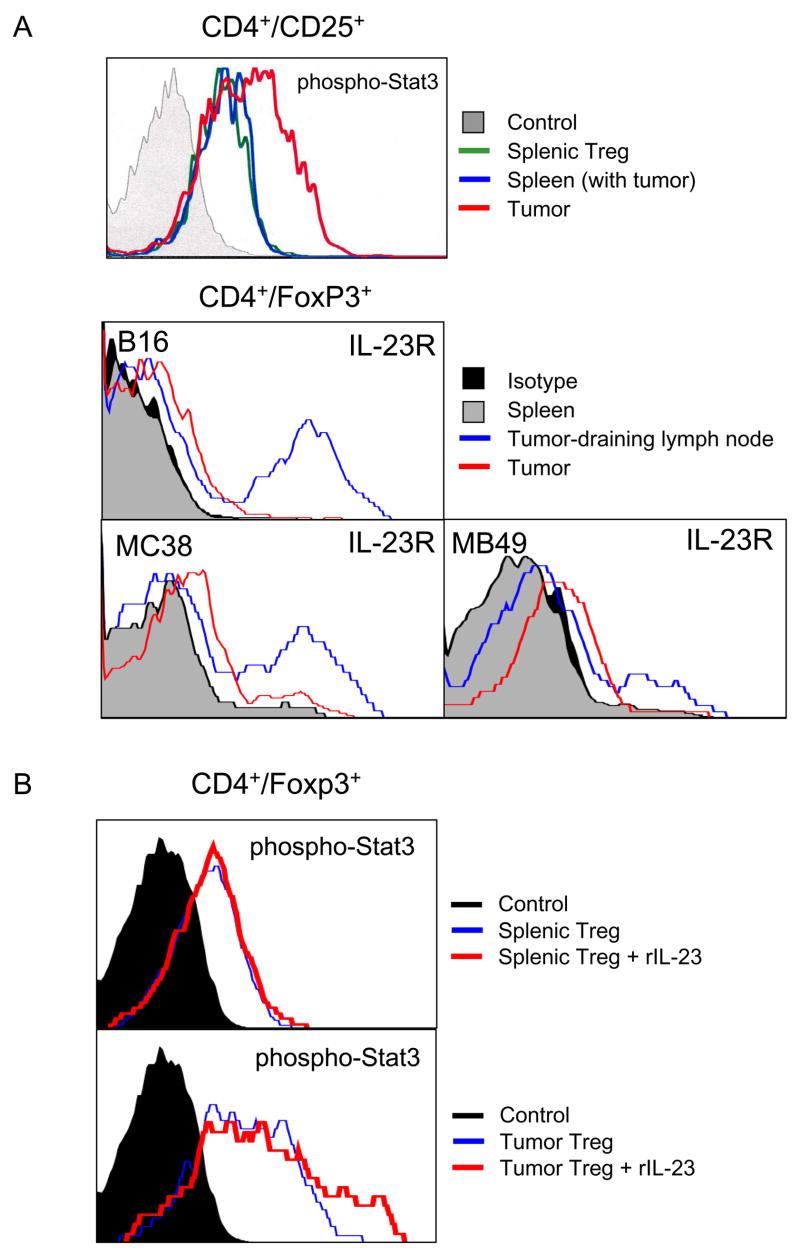
(A) Stat3 is activated in tumor Tregs, which are IL-23R positive. Cell suspensions prepared from spleens and from tumors as well as tumor draining lymph nodes (TDLN) were subjected to flow analyses for phospho-Stat3 (top panel) and IL-23R (three bottom panels). (B) Recombinant IL-23 can further activate Stat3 in CD4+Foxp3+ lymphocytes derived from tumors but not spleens. Shown are the representative results from 2–3 independent experiments.
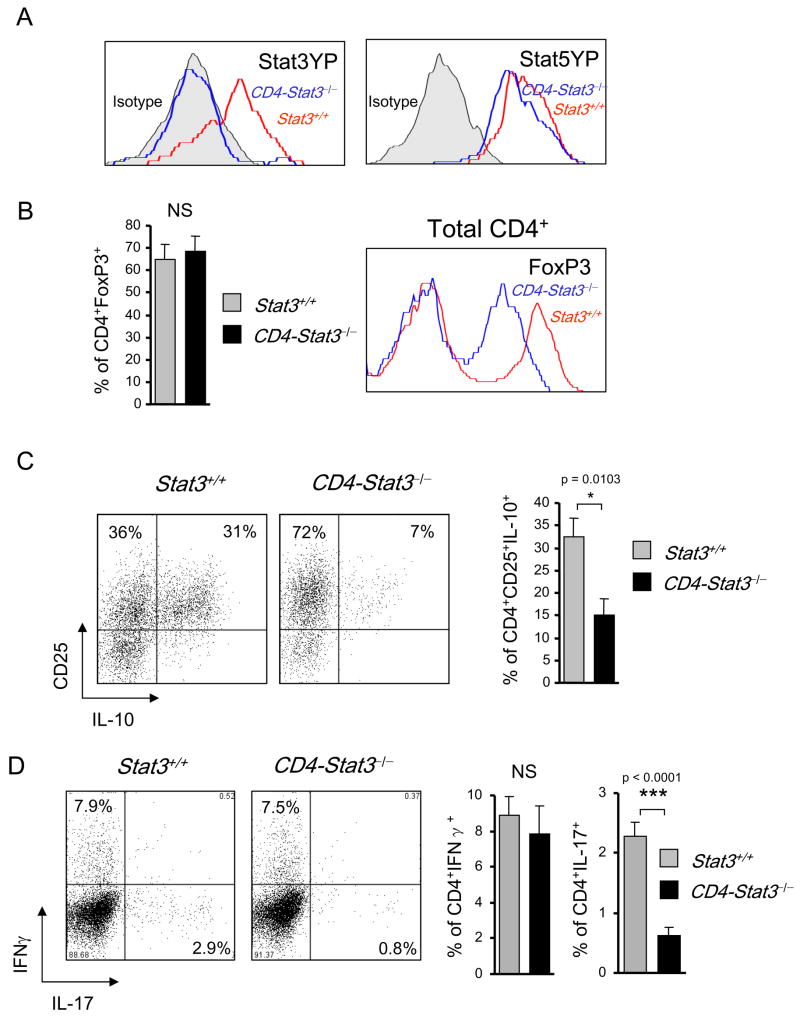
In order to assess the role of Stat3 activity in tumor-infiltrating T cells in vivo, we introduced B16 tumors into CD4-Cre/Stat3flox/flox mice, which selectively ablate Stat3 in the T cell compartment. Figure 6A shows that tumor infiltrating Treg cells from wild-type mice contained significant amounts of pStat3, while no pStat3 was detected in tumor infiltrating Treg from CD4-Cre/Stat3flox/flox mice. Consistent with an important role for Stat5 in maintaining Treg (Yao et al., 2007), we found that B16 tumor-infiltrating Treg also displayed high Stat5 activity (Fig. 6A). Since IL-23 is known to stimulate both Stat3 and Stat5 (Parham et al., 2002), the finding that tumor Treg express functional IL-23R suggests that Stat3-dependent expression of IL-23 in the myeloid compartment can promote tumor growth through an auto-amplification mechanism involving activation of both Stat3 and Stat5 in IL-23R+ tumor-infiltrating Treg. In the mice with Stat3−/− T cells, FoxP3+ Treg were still the dominant tumor infiltrating CD4 subset; however FoxP3 levels were reduced relative to the tumor-bearing wild-type mice (Fig. 6B). Furthermore, tumor-infiltrating Treg from CD4-Cre/Stat3flox/flox mice produced significantly less IL-10 than tumor infiltrating Treg from their Stat3+/+ counterparts (Fig. 6C). Consistent with previous reports on the absolute requirement for Stat3 in Th17 development in vivo (Harris et al., 2007; Wei et al., 2007; Zhou et al., 2007), Th17 cells (as defined by intracellular cytokine staining for IL-17) were completely absent in CD4-Cre/Stat3flox/flox mice while Th1 cells (as defined by intracellular cytokine staining for IFN-γ) were unaffected (Fig. 6D).
Figure 6. The effects of Stat3 ablation on tumor-infiltrating Tregs.
(A) Stat3 ablation does not significantly affect Stat5 activity in tumor-infiltrating CD4+Foxp3+ Tregs as measured by intracellular staining with Stat3- and Stat5-phosphospecific antibody staining and flow cytometry. (B) The effect of Stat3 signaling in tumor CD4+ T cells on the number of FoxP3+ cells (left) and expression levels of FoxP3 (right), using CD4+ T cells prepared from B16 tumors grown in Stat3flox/flox (Stat3+/+) and CD4cre/Stat3flox/flox mice. Right panel: representative histogram for one of 3 independent experiments with total n = 12; mean ± SEM; NS, not significant. (C) Lower expression of IL-10 by tumor-derived CD4+CD25+ Tregs from Stat3−/− mice. Left two panels – shown are representative results for 3 independent experiments. Right bar graph: combining 3 independent experiments with total n = 12; mean ± SEM; p = 0.0103. (D) Intracellular expression of IFNγ and IL-17 cytokines by tumor-infiltrating Stat3-positive and Stat3-negative CD4+ T cells, as assessed by intracellular staining and flow cytometry using freshly isolated whole tumor cell preparations (left two panels). Right two bar graphs represent 3 independent experiments, with total n = 12, p > 0.05 and p < 0.0001, respectively; NS, not significant. Shown are mean values ± SEM
Effects of IL-23R blockade on tumor Tregs and tumor growth
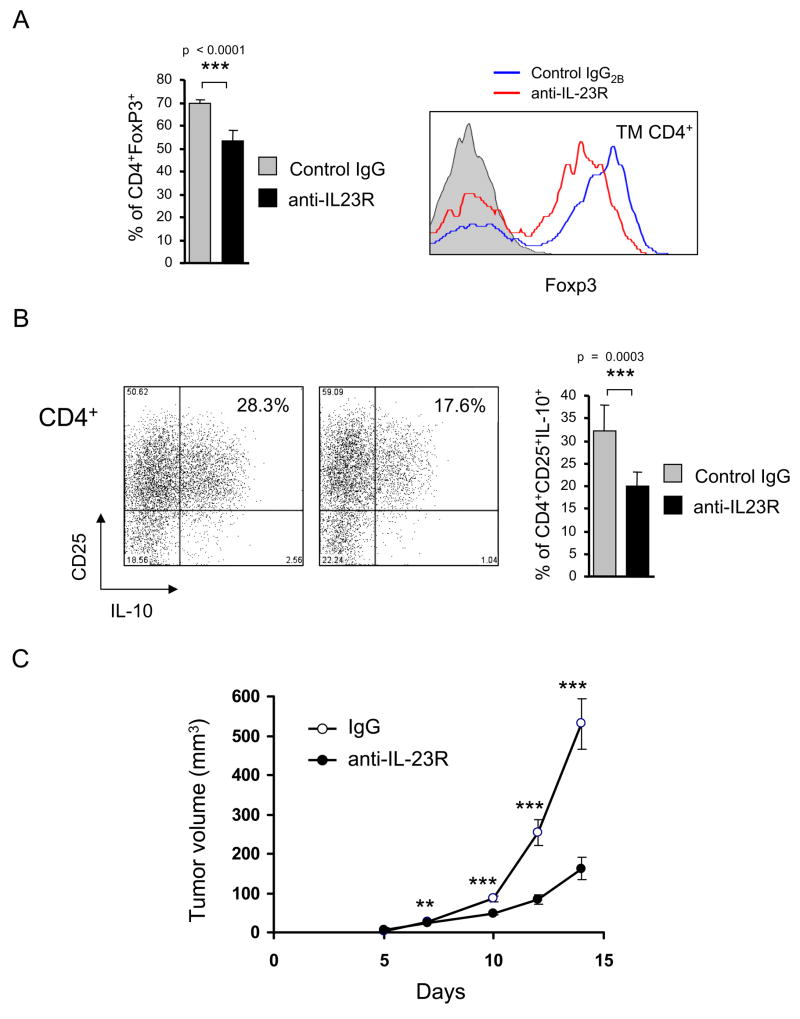
Because IL-23R signals through Stat3 (Parham et al., 2002), and Stat3 can affect expression of FoxP3 and IL-10 in CD4+ T cells (Kinjyo et al., 2006; Pallandre et al., 2007; Yao et al., 2007), we evaluated directly whether IL-23R signaling impacts Treg in the tumor microenvironment and whether this directly enhanced B16 tumor growth in vivo. IL-23R blockade affected tumor infiltrating Tregs in a manner similar but not identical to T cell-specific Stat3 ablation (Fig. 7A, B). Anti-IL-23R treated mice displayed a modest but significant reduction in the number of tumor infiltrating Treg numbers compared with control antibody treated mice. However, FoxP3 levels were diminished (Fig. 7A, right panel), as was IL-10 production (Fig. 7B), after anti-IL-23R treatment. While these results are consistent with a role for IL-23 driven Stat3 activation in tumor-infiltrating Treg although additional IL-23R dependent signaling pathways (such as Stat5) are likely operative in tumor infiltrating Treg. In addition, other cytokines in the tumor microenvironment (such as IL-10) could be additionally contributing to Stat3 activation in tumor infiltrating Treg. The effects of IL-23 within the tumor microenvironment indeed translate to an overall enhancement of tumor growth, as in vivo IL-23R blockade resulted in a significant reduction in tumor growth relative to isotype control antibody (Fig. 7C).
Figure 7. Blocking IL-23R signaling in vivo affects tumor Tregs and tumor growth.
Mice were challenged with B16 tumors injected s.c. and treated with IL-23R-specific neutralizing antibodies or with rat IgG2b control antibodies. (A) Blocking the IL-23 signaling reduces numbers of tumor-infiltrating Tregs (left panel) and, especially expression level of FoxP3 (right panel). (B) IL-23R neutralization reduces IL-10 production by CD4+CD25+ Tregs. The phenotypic analysis of tumor-infiltrating CD4+ T cells were accomplished by cell surface/intracellular staining with specific antibodies and flow cytometry. For both (A) and (B), shown are representative results of FACS analysis from one of two independent experiments. Bar graphs show the means ± SEM with total n = 7, p < 0.0001 (A) and p = 0.0003 (B). (C) Effects of IL-23R blockade on B16 tumor growth. Shown are combined results of two independent experiments as means ± SEM; total n = 10; ***, p < 0.001 and **, p < 0.01.
DISCUSSION
The immune system acts as an extrinsic tumor suppressor (Dunn et al., 2005; Kaplan et al., 1998; Koebel et al., 2007). However, tumor acquires various mechanisms to facilitate immune escape (Dunn et al., 2002; Zou, 2005). Our results demonstrate that Stat3 signaling in the tumor microenvironment shifts inflammation from an anti-tumor IL-12 program to a tumor-enhancing IL-23 program via transcriptional activation of the IL-23-specific p19 gene and concomitant transcriptional suppression of the IL-12-specific p35 gene. Stat3 appears to co-activate the p19 promoter with NF-κB/p65. In contrast, Stat3 also appears to suppress c-Rel in tumors, thereby reducing binding of c-Rel to the IL-12/p35 promoter. This is in agreement with a recent report indicating that NF-κB subunits have unique roles in regulating gene expression in DCs (Wang et al., 2007). The p50 and c-Rel subunits were found to be critical for the induction of T cell stimulatory gene expression, whereas the NF-κB/p65 subunit regulated expression of a distinct group of inflammatory genes, some of which are procarcinogenic (Wang et al., 2007). While Stat3 transcriptionally activates the IL-23/p19 gene in both tumor cells and myeloid cells within the tumor microenvironment, the major source of Stat3 driven IL-23 is tumor associated macrophages. Stat3 restrains IL-12 transcription in both tumor and infiltrating myeloid cells, though the major IL-12 producing cells in the absence of Stat3 activity are tumor-associated DCs. It is notable that, while these effects of Stat3 are not observed systemically, they appear to be operative in tumor draining lymph nodes. We are currently testing the hypothesis that a preponderance of IL-23 production relative to IL-12 production by APC in tumor draining lymph nodes ultimately skews the differentiation of tumor-specific T cells, possibly leading to dominance of Treg and Th17 responses over anti-tumor Th1 responses.
IL-12 has been validated in numerous studies as a central cytokine in anti-tumor immunity and in anti-viral immunity due to its role in activating NK cells, Th1 and CTL responses (Trinchieri, 2003). The emergence of a second IL-12 family member, IL-23, which shares the p40 subunit with IL-12, has significantly altered our view of autoimmune disease, since a number of murine autoimmune syndromes originally thought to be IL-12 dependent are instead IL-23 dependent. These more recent studies have suggested that IL-12 vs. IL-23 driven immunity is not only qualitatively distinct but may be mutually antagonistic. This notion also pertains to tumor immunity as emphasized by Langowski et al., who showed that tumorigenesis was diminished in IL-23/p19KO mice and increased in IL-12/p35KO mice (Langowski et al., 2006). The studies presented here support the notion that IL-23 has a pro-carcinogenic role in contrast to the well-established anti-carcinogenic role of IL-12. Importantly, they indicate that Stat3 signaling in both the tumor and the hematopoietic/immune microenvironment of the tumor is critical in shifting the balance from IL-12 to IL-23 production.
An additional unexpected finding in our study was the up-regulation of IL-23R on tumor-associated Treg. Although the underlying mechanism responsible for IL-23R upregulation in tumor-associated Tregs remains to be elucidated, our limited data indicated that the increase in IL-23R expression in tumor Treg correlated with Stat3 activity. In addition to its central role in IL-23 up-regulation, Stat3 appears to also play a role in IL-23R signaling within the tumor microenvironment. The role for Stat3 signaling in IL-10 production by Treg has not been previously appreciated though a recent report demonstrates that IL-27 driven IL-10 production by T cells is Stat3 dependent (Stumhofer et al., 2007). While T cell specific Stat3 ablation also diminished the number of tumor-infiltrating Th17 cells, this cell population is relatively minor in the B16 system. Recent studies indicated that Th17 can contribute to tumor progression (Kryczek et al., 2007), however, it appears that, at least with B16 melanoma, the major role of T cell-specific Stat3 signaling (and also IL-23R signaling) lies within the Treg compartment. Indeed, ablation of Treg has been shown to significantly enhance anti-tumor responses in B16 melanoma (Sutmuller et al., 2001; Turk et al., 2004). Whether IL-23R dependent Stat3 signaling has a larger role for Th17 responses in other tumor systems remains to be determined. The IL-23R signals through multiple other Stats, which also undoubtedly play a role in how IL-23 modulates the tumor microenvironment. For example, IL-23R also activates Stat5, which O’Shea and colleagues demonstrated to be very important in Treg expansion and possibly also differentiation (Yao et al., 2007). Indeed, tumor-infiltrating Tregs have high levels of tyrosine-phosphorylated Stat5. In this context, it was somewhat surprising that Stat3 signaling appeared to enhance FoxP3 expression by tumor-infiltrating Tregs. This finding contrasts with studies suggesting Stat3 activation (driven by IL-6 or IL-27) inhibited differentiation to the Treg lineage (Huber et al., 2008; Laurence et al., 2007). However, the role of Stat3 in FoxP3 gene regulation may be quite distinct in tumor-infiltrating Treg (where the cytokine milieu and ambient signals are quite distinct) than in naïve T cells being induced into various differentiation pathways in vitro. Indeed, Pallandre et al. demonstrated that, similar to our findings here, Stat3 ablation decreased FoxP3 expression by Tregs in a graft-vs.-host disease model (Pallandre et al., 2007). Zorn et al. showed that both Stat3 and Stat5 could bind to a Stat consensus site in the FoxP3 promoter (Zorn et al., 2006). Thus, the role of Stat3 in regulating FoxP3 expression by Treg appears to be context dependent. In the context of the signals present within the tumor microenvironment, Stat3 activity in Treg appears to upregulate FoxP3 levels, which Flavell and colleagues have shown to be important in maintaining their inhibitory functions (Wan and Flavell, 2007).
It should also be emphasized that in addition to IL-23, at least two additional cytokines, IL-6 and IL-10, appear to play an important role in enhancing cancer progression (Yu et al., 2007). These cytokines are somewhat interlinked in that they are all Stat3 inducible and also their receptors activate Stat3 (O’Shea and Murray, 2008). While there has been much written about the role of IL-6 and IL-10 in promoting tumorigenesis and inhibiting anti-tumor immunity, our data focuses attention on IL-23 as an important mediator of procarcinogenesis by Stat3 signaling.
EXPERIMENTAL PROCEDURES
Cells
Mouse B16 melanoma cells were purchased from American Type Culture Collection, C4 mouse melanoma, MB49 bladder carcinoma and MC38 colon carcinoma cells were generous gifts from J. Fidler (M. D. Anderson), T. Ratliff (University of Iowa), and M. Shurin (University of Pittsburg), respectively. DC2.4 cells were originally obtained from Dr. Kenneth Rock (University of Massachusetts Medical School, MA). The generation of v-Src-transformed Balb/C 3T3 fibroblasts has been previously reported (Wang et al., 2004).
In vivo experiments
Mouse care and experimental procedures were performed under pathogen-free conditions in accordance with established institutional guidance and approved protocols from Research Animal Care Committees of the City of Hope and John Hopkins University. We obtained Mx1-Cre mice from the Jackson Laboratory, CD4-Cre mice from Taconic and Stat3flox/flox mice from S. Akira and K. Takeda (Osaka University). All transgenic mice were on C57BL/6J background. Generation of mice with Stat3−/− hematopoietic cells by inducible Mx1-Cre recombinase system transgene has been reported (Kortylewski et al., 2005). We generated CD4-Cre/Stat3flox/flox mice by standard interbreeding procedures. We verified specific Stat3 deletion in T cells by PCR, using primer sets that distinguish Stat3, Stat3loxP and Stat3-deleted alleles, and by FACS analysis Stat3 phosphorylation in IL-6-treated T cells. For tumor challenge, we injected 1 ×105 of B16 or 5 × 105 of MB49 and MC38 tumor cells s.c. into 7–8 weeks old wild-type or transgenic mice and tumor growth was monitored three times a week. For IL-23R neutralization experiments in vivo, mice were treated with 250 μg of anti-IL-23Rα antibodies (R&D) injected i.p. every third day during the course of the experiment, starting from the day of tumor inoculation. Mice were sacrificed after two weeks from tumor inoculation. We prepared single cell suspensions of spleen, lymph node or tumor tissues by mechanic dispersion followed by collagenase D/DNase I treatment as described before (Kortylewski et al., 2005).
Cytokine ELISA and Western blotting
We enriched CD11b+c+ or CD11b+c− immune cell subsets from spleens and tumors from Stat3+/+ and Stat3−/− (Mx1-Cre/Stat3flox/flox) mice using specific antibodies in combination with magnetic magnetic nanoparticles from StemCell Technologies as described previously (Kortylewski et al., 2005). For IL-12 and IL-23 measurement by ELISA (eBiosciences), we cultured the enriched cell populations for 24 hrs to collect supernatants. Equal protein amounts of lysates prepared from enzymatically digested tumors and/or tumor-draining lymph nodes, were analyzed by western blotting using antibodies specific to IL-23/p19 (R&D), phospho-S503-c-Rel (Thermo Scientific) and β-actin (Sigma).
Quantitative real-time PCR
We isolated total RNAs from various cell populations using RNeasy System (Qiagen) and then transcribed them into cDNAs with iScript cDNA Synthesis Kit (Bio-Rad). PCR reactions were set up using specific primer pairs for mouse IL-23/p19: 5′-TCCCTACTAGGACTCAGCCAACTC-3′ (forward) and 5′-ACTCAGGCTGGGCACTG-3′ (reverse); or commercially available primers for mouse IL-12/p35, 18S rRNA or GAPDH (SuperArray). Sequence-specific amplification was assessed by measuring fluorescent signal of SYBR Green using Chromo4 Real-Time PCR Detector (Bio-Rad).
Reporter gene assays
We inserted the mouse IL-23/p19 promoter fragment (−1159 - +160) by PCR into the Xho I/HindIII site of the pGL3-basic vector, generating pGL3-p19-Pro-luciferase plasmid. For luciferase assays, we co-transfected 0.4μg/ml pGL3-p19-Pro-luciferase containing 0.02μg/ml pRL-TK normalization construct and 0.4μg/ml of either the indicated pRC plasmid or siRNAs (80pmol/ml) into Balb/C 3T3 fibroblasts, B16 and C4 mouse melanoma cells using Lipofectamine 2000 (Invitrogen). We analyzed luciferase activities in whole cell lysates using the Dual-Luciferase Reporter Assay (Promega) and quantified using Mikrotek Laborsysteme microplate reader. Data were presented as firefly luciferase activity normalized to the Renilla luciferase activity in each triplicate sample.
Chromatin immunoprecipitation (ChIP) assays
We performed chromatin immunoprecipitations using the ChIP Assay Kit (Upstate Biotechnology) according to manufacturer’s protocol. Briefly, cultured cells were fixed with 1% formaldehyde at 37°C for 10 min before lysis. For ChIP assays on freshly isolated whole tumors, tumor draining or distant lymph nodes, prior to cross-linking with formaldehyde, tissues were frozen in liquid nitrogen and homogenized to enable nuclei isolation. We incubated the sonicated chromatin solutions with 2μg of Stat3-, p65- or c-Rel-specific antibodies (Santa Cruz) or with control rabbit IgG. Following immunoprecipitation and reversed crosslinking, DNA was extracted and analyzed by PCR using primer sets for mouse IL-23/p19 promoter: 5′-GGATTCCCGTCCCTCGGTCTC-3′ (forward) and 5′-GGGCCAAGGCGCTTGGCACAG-3′ (reverse); mouse IL-12/p35 promoter: 5′-GACAGT GGAGGCACCAGGCC-3′ (forward) and 5′-CAGACATCGCTGTCCCGGCG-3′ (reverse).
Flow cytometry
For extracellular staining of immune markers, we prepared single-cell suspensions by mechanic dispersion and enzymatic digestion of spleen, lymph node or tumor tissues. We preincubated 1 × 106 of freshly prepared cells suspended in a mixture of PBS, 2% FCS and 0.1% (wt/vol) sodium azide with FcγIII/IIR-specific antibody to block non-specific binding and stained with different combinations of fluorochrome-coupled antibodies to CD3, CD4, CD25 (BD Biosciences) or IL-23R (Imgenex). Staining with the anti-IL-23R antibody required earlier labeling of the unconjugated antibody with a fluorescent dye, using Zenon Labeling Kit (Invitrogen). Prior to intracellular staining with antibodies to phosphotyrosine-Stat3 or -Stat5 (BD Biosciences) and FoxP3 (eBioscience), we fixed cells in paraformaldehyde and permeated them in methanol. For intracellular staining of IFNγ, IL-10 and IL-17 (BD Pharmingen), we followed the manufacturer’s protocol after 4 h incubation in the presence of Leukocyte Activation Coctail (BD Pharmingen). Fluorescence data were collected on FACScalibur (Beckton Dickinson) and analyzed using FlowJo software (Tree Star).
Statistical analysis
Unpaired t test was used to calculate two-tailed p value to estimate statistical significance of differences between two treatment groups. Statistically significant p values are indicated in figures and labeled as follows: ***; p < 0.001; **, p < 0.01 and *, p < 0.05. Data were analyzed using Prism software (GraphPad).
Supplementary Material
Acknowledgments
We are most grateful to Drs. S. Akira and K. Takeda (Osaka University) for providing the Stat3flox mice, and the Pathology Core at the City of Hope for technical assistance. We also acknowledge dedication of staff members at the animal facilities and flow cytometry core of Beckman Research Institute at the City of Hope National Medical Center.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Apetoh L, Ghiringhelli F, Tesniere A, Obeid M, Ortiz C, Criollo A, Mignot G, Maiuri MC, Ullrich E, Saulnier P, et al. Toll-like receptor 4-dependent contribution of the immune system to anticancer chemotherapy and radiotherapy. Nat Med. 2007;13:1050–1059. doi: 10.1038/nm1622. [DOI] [PubMed] [Google Scholar]
- Chen Z, Laurence A, Kanno Y, Pacher-Zavisin M, Zhu BM, Tato C, Yoshimura A, Hennighausen L, O’Shea JJ. Selective regulatory function of Socs3 in the formation of IL-17-secreting T cells. Proc Natl Acad Sci USA. 2006;103:8137–8142. doi: 10.1073/pnas.0600666103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombo MP, Trinchieri G. Interleukin-12 in anti-tumor immunity and immunotherapy. Cytokine Growth Factor Rev. 2002;13:155–168. doi: 10.1016/s1359-6101(01)00032-6. [DOI] [PubMed] [Google Scholar]
- Cua DJ, Sherlock J, Chen Y, Murphy CA, Joyce B, Seymour B, Lucian L, To W, Kwan S, Churakova T, et al. Interleukin-23 rather than interleukin-12 is the critical cytokine for autoimmune inflammation of the brain. Nature. 2003;421:744–748. doi: 10.1038/nature01355. [DOI] [PubMed] [Google Scholar]
- Dunn GP, Bruce AT, Ikeda H, Old LJ, Schreiber RD. Cancer immunoediting: from immunosurveillance to tumor escape. Nat Immunol. 2002;3:991–998. doi: 10.1038/ni1102-991. [DOI] [PubMed] [Google Scholar]
- Dunn GP, Bruce AT, Sheehan KC, Shankaran V, Uppaluri R, Bui JD, Diamond MS, Koebel CM, Arthur C, White JM, Schreiber RD. A critical function for type I interferons in cancer immunoediting. Nat Immunol. 2005;6:722–729. doi: 10.1038/ni1213. [DOI] [PubMed] [Google Scholar]
- Garcia-Hernandez ML, Hernandez-Pando R, Gariglio P, Berumen J. Interleukin-10 promotes B16-melanoma growth by inhibition of macrophage functions and induction of tumour and vascular cell proliferation. Immunology. 2002;105:231–243. doi: 10.1046/j.1365-2567.2002.01363.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerosa F, Baldani-Guerra B, Nisii C, Marchesini V, Carra G, Trinchieri G. Reciprocal activating interaction between natural killer cells and dendritic cells. J Exp Med. 2002;195:327–333. doi: 10.1084/jem.20010938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghilardi N, Kljavin N, Chen Q, Lucas S, Gurney AL, De Sauvage FJ. Compromised humoral and delayed-type hypersensitivity responses in IL-23-deficient mice. J Immunol. 2004;172:2827–2833. doi: 10.4049/jimmunol.172.5.2827. [DOI] [PubMed] [Google Scholar]
- Grumont R, Hochrein H, O’Keeffe M, Gugasyan R, White C, Caminschi I, Cook W, Gerondakis S. c-Rel regulates interleukin 12 p70 expression in CD8(+) dendritic cells by specifically inducing p35 gene transcription. J Exp Med. 2001;194:1021–1032. doi: 10.1084/jem.194.8.1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris TJ, Grosso JF, Yen HR, Xin H, Kortylewski M, Albesiano E, Hipkiss EL, Getnet D, Goldberg MV, Maris CH, et al. Cutting edge: An in vivo requirement for STAT3 signaling in TH17 development and TH17-dependent autoimmunity. J Immunol. 2007;179:4313–4317. doi: 10.4049/jimmunol.179.7.4313. [DOI] [PubMed] [Google Scholar]
- Huber M, Steinwald V, Guralnik A, Brustle A, Kleemann P, Rosenplanter C, Decker T, Lohoff M. IL-27 inhibits the development of regulatory T cells via STAT3. Int Immunol. 2008;20:223–234. doi: 10.1093/intimm/dxm139. [DOI] [PubMed] [Google Scholar]
- Kaplan DH, Shankaran V, Dighe AS, Stockert E, Aguet M, Old LJ, Schreiber RD. Demonstration of an interferon gamma-dependent tumor surveillance system in immunocompetent mice. Proc Natl Acad Sci USA. 1998;95:7556–7561. doi: 10.1073/pnas.95.13.7556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinjyo I, Inoue H, Hamano S, Fukuyama S, Yoshimura T, Koga K, Takaki H, Himeno K, Takaesu G, Kobayashi T, Yoshimura A. Loss of SOCS3 in T helper cells resulted in reduced immune responses and hyperproduction of interleukin 10 and transforming growth factor-beta 1. J Exp Med. 2006;203:1021–1031. doi: 10.1084/jem.20052333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koebel CM, Vermi W, Swann JB, Zerafa N, Rodig SJ, Old LJ, Smyth MJ, Schreiber RD. Adaptive immunity maintains occult cancer in an equilibrium state. Nature. 2007;450:903–907. doi: 10.1038/nature06309. [DOI] [PubMed] [Google Scholar]
- Kortylewski M, Kujawski M, Wang T, Wei S, Zhang S, Pilon-Thomas S, Niu G, Kay H, Mule J, Kerr WG, et al. Inhibiting Stat3 signaling in the hematopoietic system elicits multicomponent antitumor immunity. Nat Med. 2005;11:1314–1321. doi: 10.1038/nm1325. [DOI] [PubMed] [Google Scholar]
- Kryczek I, Wei S, Zou L, Altuwaijri S, Szeliga W, Kolls J, Chang A, Zou W. Cutting edge: Th17 and regulatory T cell dynamics and the regulation by IL-2 in the tumor microenvironment. J Immunol. 2007;178:6730–6733. doi: 10.4049/jimmunol.178.11.6730. [DOI] [PubMed] [Google Scholar]
- Langowski JL, Kastelein RA, Oft M. Swords into plowshares: IL-23 repurposes tumor immune surveillance. Trends Immunol. 2007;28:207–212. doi: 10.1016/j.it.2007.03.006. [DOI] [PubMed] [Google Scholar]
- Langowski JL, Zhang X, Wu L, Mattson JD, Chen T, Smith K, Basham B, McClanahan T, Kastelein RA, Oft M. IL-23 promotes tumour incidence and growth. Nature. 2006;442:461–465. doi: 10.1038/nature04808. [DOI] [PubMed] [Google Scholar]
- Langrish CL, Chen Y, Blumenschein WM, Mattson J, Basham B, Sedgwick JD, McClanahan T, Kastelein RA, Cua DJ. IL-23 drives a pathogenic T cell population that induces autoimmune inflammation. J Exp Med. 2005;201:233–240. doi: 10.1084/jem.20041257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurence A, Tato CM, Davidson TS, Kanno Y, Chen Z, Yao Z, Blank RB, Meylan F, Siegel R, Hennighausen L, et al. Interleukin-2 signaling via STAT5 constrains T helper 17 cell generation. Immunity. 2007;26:371–381. doi: 10.1016/j.immuni.2007.02.009. [DOI] [PubMed] [Google Scholar]
- Murphy CA, Langrish CL, Chen Y, Blumenschein W, McClanahan T, Kastelein RA, Sedgwick JD, Cua DJ. Divergent pro- and antiinflammatory roles for IL-23 and IL-12 in joint autoimmune inflammation. J Exp Med. 2003;198:1951–1957. doi: 10.1084/jem.20030896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Shea JJ, Murray PJ. Cytokine signaling modules in inflammatory responses. Immunity. 2008;28:477–487. doi: 10.1016/j.immuni.2008.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oppmann B, Lesley R, Blom B, Timans JC, Xu Y, Hunte B, Vega F, Yu N, Wang J, Singh K, et al. Novel p19 protein engages IL-12p40 to form a cytokine, IL-23, with biological activities similar as well as distinct from IL-12. Immunity. 2000;13:715–725. doi: 10.1016/s1074-7613(00)00070-4. [DOI] [PubMed] [Google Scholar]
- Pallandre JR, Brillard E, Crehange G, Radlovic A, Remy-Martin JP, Saas P, Rohrlich PS, Pivot X, Ling X, Tiberghien P, Borg C. Role of STAT3 in CD4+CD25+FOXP3+ regulatory lymphocyte generation: implications in graft-versus-host disease and antitumor immunity. J Immunol. 2007;179:7593–7604. doi: 10.4049/jimmunol.179.11.7593. [DOI] [PubMed] [Google Scholar]
- Parham C, Chirica M, Timans J, Vaisberg E, Travis M, Cheung J, Pflanz S, Zhang R, Singh KP, Vega F, et al. A receptor for the heterodimeric cytokine IL-23 is composed of IL-12Rbeta1 and a novel cytokine receptor subunit, IL-23R. J Immunol. 2002;168:5699–5708. doi: 10.4049/jimmunol.168.11.5699. [DOI] [PubMed] [Google Scholar]
- Shankaran V, Ikeda H, Bruce AT, White JM, Swanson PE, Old LJ, Schreiber RD. IFNgamma and lymphocytes prevent primary tumour development and shape tumour immunogenicity. Nature. 2001;410:1107–1111. doi: 10.1038/35074122. [DOI] [PubMed] [Google Scholar]
- Stumhofer JS, Silver JS, Laurence A, Porrett PM, Harris TH, Turka LA, Ernst M, Saris CJ, O’Shea JJ, Hunter CA. Interleukins 27 and 6 induce STAT3-mediated T cell production of interleukin 10. Nat Immunol. 2007;8:1363–1371. doi: 10.1038/ni1537. [DOI] [PubMed] [Google Scholar]
- Sutmuller RP, van Duivenvoorde LM, van Elsas A, Schumacher TN, Wildenberg ME, Allison JP, Toes RE, Offringa R, Melief CJ. Synergism of cytotoxic T lymphocyte-associated antigen 4 blockade and depletion of CD25(+) regulatory T cells in antitumor therapy reveals alternative pathways for suppression of autoreactive cytotoxic T lymphocyte responses. J Exp Med. 2001;194:823–832. doi: 10.1084/jem.194.6.823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda K, Clausen BE, Kaisho T, Tsujimura T, Terada N, Forster I, Akira S. Enhanced Th1 activity and development of chronic enterocolitis in mice devoid of Stat3 in macrophages and neutrophils. Immunity. 1999;10:39–49. doi: 10.1016/s1074-7613(00)80005-9. [DOI] [PubMed] [Google Scholar]
- Trinchieri G. Interleukin-12 and the regulation of innate resistance and adaptive immunity. Nat Rev Immunol. 2003;3:133–146. doi: 10.1038/nri1001. [DOI] [PubMed] [Google Scholar]
- Turk MJ, Guevara-Patino JA, Rizzuto GA, Engelhorn ME, Sakaguchi S, Houghton AN. Concomitant tumor immunity to a poorly immunogenic melanoma is prevented by regulatory T cells. J Exp Med. 2004;200:771–782. doi: 10.1084/jem.20041130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan YY, Flavell RA. Regulatory T-cell functions are subverted and converted owing to attenuated Foxp3 expression. Nature. 2007;445:766–770. doi: 10.1038/nature05479. [DOI] [PubMed] [Google Scholar]
- Wang J, Wang X, Hussain S, Zheng Y, Sanjabi S, Ouaaz F, Beg AA. Distinct roles of different NF-kappa B subunits in regulating inflammatory and T cell stimulatory gene expression in dendritic cells. J Immunol. 2007;178:6777–6788. doi: 10.4049/jimmunol.178.11.6777. [DOI] [PubMed] [Google Scholar]
- Wang T, Niu G, Kortylewski M, Burdelya L, Shain K, Zhang S, Bhattacharya R, Gabrilovich D, Heller R, Coppola D, et al. Regulation of the innate and adaptive immune responses by Stat-3 signaling in tumor cells. Nat Med. 2004;10:48–54. doi: 10.1038/nm976. [DOI] [PubMed] [Google Scholar]
- Wei L, Laurence A, Elias KM, O’Shea JJ. IL-21 is produced by TH17 cells and drives IL-17 production in a STAT3-dependent manner. J Biol Chem. 2007;282:34605–34610. doi: 10.1074/jbc.M705100200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Z, Kanno Y, Kerenyi M, Stephens G, Durant L, Watford WT, Laurence A, Robinson GW, Shevach EM, Moriggl R, et al. Nonredundant roles for Stat5a/b in directly regulating Foxp3. Blood. 2007;109:4368–4375. doi: 10.1182/blood-2006-11-055756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Jove R. The STATs of cancer--new molecular targets come of age. Nat Rev Cancer. 2004;4:97–105. doi: 10.1038/nrc1275. [DOI] [PubMed] [Google Scholar]
- Yu H, Kortylewski M, Pardoll D. Crosstalk between cancer and immune cells: role of STAT3 in the tumour microenvironment. Nat Rev Immunol. 2007;7:41–51. doi: 10.1038/nri1995. [DOI] [PubMed] [Google Scholar]
- Zheng Y, Danilenko DM, Valdez P, Kasman I, Eastham-Anderson J, Wu J, Ouyang W. Interleukin-22, a T(H)17 cytokine, mediates IL-23-induced dermal inflammation and acanthosis. Nature. 2007;445:648–651. doi: 10.1038/nature05505. [DOI] [PubMed] [Google Scholar]
- Zhou L, Ivanov II, Spolski R, Min R, Shenderov K, Egawa T, Levy DE, Leonard WJ, Littman DR. IL-6 programs T(H)-17 cell differentiation by promoting sequential engagement of the IL-21 and IL-23 pathways. Nat Immunol. 2007;8:967–974. doi: 10.1038/ni1488. [DOI] [PubMed] [Google Scholar]
- Zorn E, Nelson EA, Mohseni M, Porcheray F, Kim H, Litsa D, Bellucci R, Raderschall E, Canning C, Soiffer RJ, et al. IL-2 regulates FOXP3 expression in human CD4+CD25+ regulatory T cells through a STAT-dependent mechanism and induces the expansion of these cells in vivo. Blood. 2006;108:1571–1579. doi: 10.1182/blood-2006-02-004747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou W. Immunosuppressive networks in the tumour environment and their therapeutic relevance. Nat Rev Cancer. 2005;5:263–274. doi: 10.1038/nrc1586. [DOI] [PubMed] [Google Scholar]
- Zou W. Regulatory T cells, tumour immunity and immunotherapy. Nat Rev Immunol. 2006;6:295–307. doi: 10.1038/nri1806. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.