Summary
Global energy and environmental problems have stimulated increasing efforts towards synthesizing liquid biofuels as transportation energy. Compared to the traditional biofuel, ethanol, advanced biofuels should offer advantages such as higher energy density, lower hygroscopicity, lower vapor pressure, and compatibility with existing transportation infrastructure. However, these fuels are not synthesized economically using native organisms. Metabolic engineering offers an alternative approach in which synthetic pathways are engineered into user friendly hosts for the production of these fuel molecules. These hosts could be readily manipulated to improve the production efficiency. This review summarizes recent progress in the engineering of Escherichia coli to produce advanced biofuels.
Introduction
The production of fuel substitutes such as bioalcohols and biodiesel from renewable resources has gained significant attention because of the rising energy price and environmental concerns. Currently, ethanol is the major form of biofuel. According to the Renewable Fuels Association, 6.5 billion gallons of bioethanol were produced in the United States in 2007, and both the production capacity and the demand for bioethanol are increasing rapidly. On the other hand, advanced biofuels are being developed rapidly over the past few years (Fig. 1) [1–5]. These biofuels aim to circumvent problems of ethanol, namely, the low energy density (30% lower than gasoline) and incompatibility with existing fuel infrastructure. Because of its tendency to absorb water, ethanol cannot be distributed using existing pipeline and high percentage blends with gasoline (e.g. E85) require vehicle retrofitting in the fuel system. The high vapor pressure of ethanol is also a threat to air quality. These problems present additional barriers for large scale replacement of gasoline. Advanced biofuels typically involve higher carbon chains and are believed to circumvent these problems.
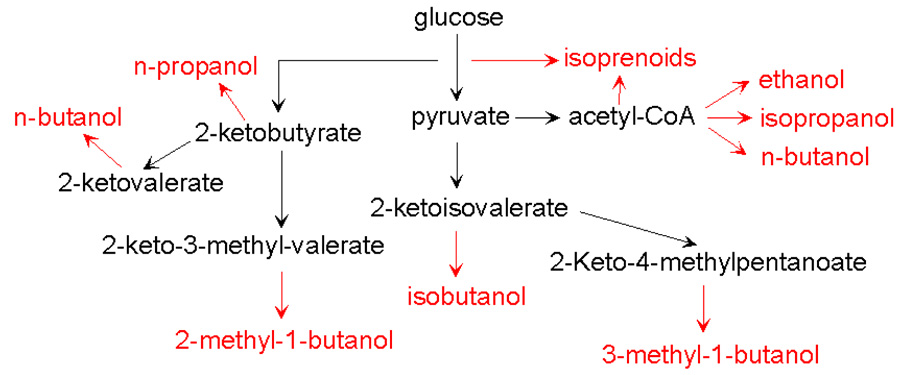
Fig.1. Schematic representation of engineered metabolic pathways to produce candidate biofuels.
Biofuels described in this review are shown in red.
However, higher chain fuel-quality compounds are not commonly produced biologically in large enough quantities for fuel applications, except n-butanol, which is produced by Clostridium species [6,7]. Thanks to the rapidly expanding genomic information, molecular biology techniques, and high throughput tools, metabolic engineers have made significant progress in constructing non-native organisms for production of fuel-grade compounds beyond the scope of what native organisms can produce [1–5]. These compounds include C3 to C5 alcohols[2,4,5], fatty acid esters [1]and isoprenoids [3](Fig. 1). It is believed that user-friendly hosts, such as E. coli or Saccharomyces cerevisiae, can be engineered to produce these fuels efficiently, both to explore the biosynthetic capability and to serve a production platform. This article briefly summarizes the recent progress in these non-traditional biofuels.
Isopropanol production from E. coli using the traditional fermentative pathway
Isopropanol is one of the secondary alcohols which can be produced by microbes [8]. Isopropanol is also used in place of methanol to esterify various fats and oils, which reduces the tendency of biodiesel to crystallize at low temperatures. Finally, isopropanol can be dehydrated to yield propylene that is widely used for plastics and is usually made from petroleum. Isopropanol is produced in Clostridium from the acetone pathway [9]. Since the genetic tools are more limited in Clostridium relative to other hosts such as E. coli, it is of interest to move the pathway to a user-friendly host. Acetone production in E. coli has been demonstrated by introducing four genes from C. acetobutylicum ATCC824, thl, ctfAB, adc coding acetyl-CoA acetyltransferase, acetoacetyl-CoA-transferase, and acetoacetate decarboxylase, respectively [10]. This strain produced 5.4 g/L acetone with 0.5 g/L/hr production rate. This titer is as high as C. acetobutylicum ATCC 824 [10]. This result indicates that E. coli could be a suitable host for acetone and isopropanol production.
To synthesize isopropanol, a secondary alcohol dehydrogenase [11] is required to convert acetone to isopropanol in an NADPH-dependent reaction. This secondary alcohol dehydrogenase was overexpressed in E.coli along with the acetone biosynthesis pathway from C. acetobutylicum to produce isopropanol [2]. In addition, the native E. coli acetyl-CoA acetyltransferase (encoded by atoB) [12] and acetoacetyl-CoA transferase (encoded by atoAD) [13] were evaluated to optimize the pathway. Furthermore, the activity of the secondary alcohol dehydrogenase (encoded by adh) from C. beijerinckii NRRL B593 [14] was compared to C. brockii HTD4 [15]. The best combination of these genes produced 4.9 g/L with 0.4 g/L/hr production rate which is higher than the native Clostridium strains.
n-Butanol production from E. coli using the traditional fermentative pathway
n-Butanol is hydrophobic and its energy content (27MJ/L) is similar to that of gasoline (32MJ/L). It can completely replace gasoline or mix with gasoline at any ratio. It can also be stored and transported using existing infrastructure. Furthermore, the vapor pressure of n-butanol (4mmHg at 20 °C) is approximately 11 times less than that of ethanol (45mmHg at 20 °C). As such, n-butanol has been proposed as a substitute and supplement of gasoline as a transportation fuel. n-Butanol is produced by C. acetobutylicum in a pathway branched from the acetone and butyrate pathways [7]. C. acetobutylicum also produces byproducts such as butyrate, acetone, and ethanol [7]. The production of n-butanol from glucose is redox neutral with the release of CO2 (Fig. 2). Therefore, it is possible to reconstruct the n-butanol pathway in a non-native host and achieve homofermentative production of n-butanol.
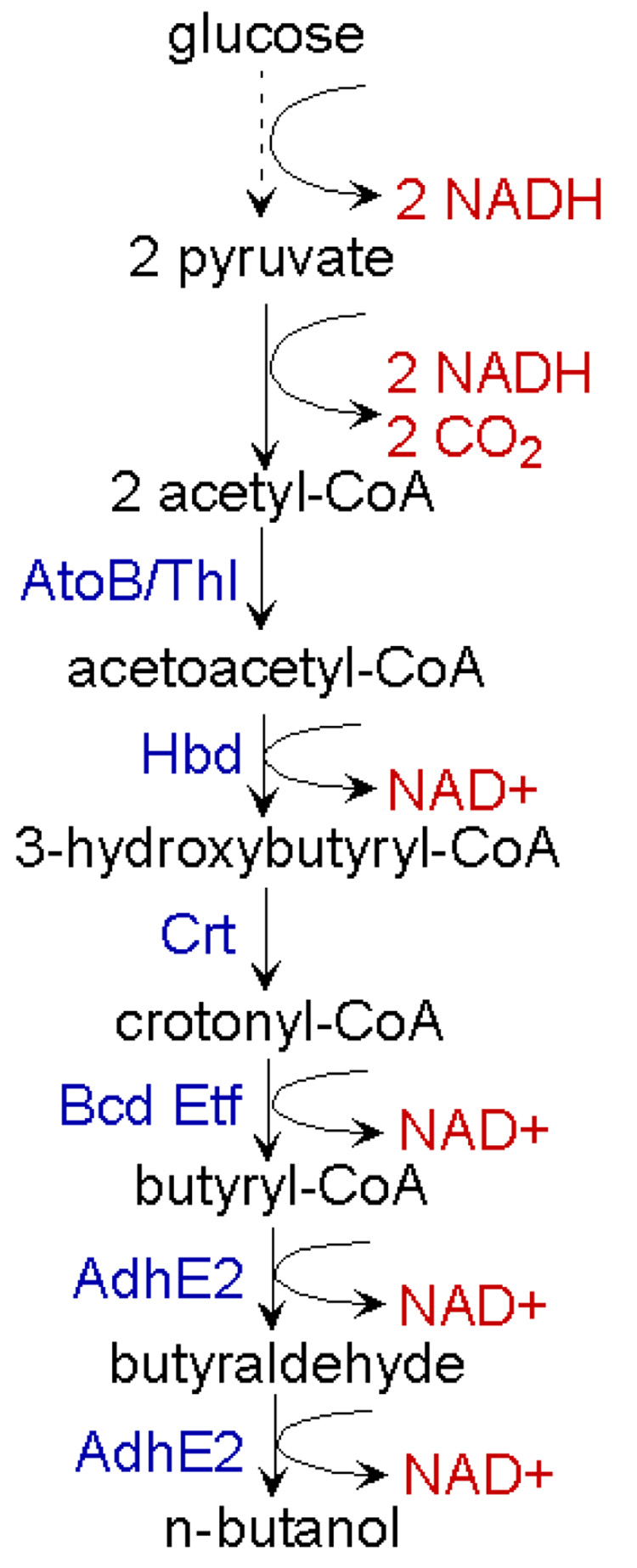
Fig.2. Schematic representation of n-butanol production in engineered E. coli.
The engineered n-butanol production pathway consists of six enzymatic steps from acetyl-CoA. AtoB, acetyl-CoA acetyltransferase; Thl, acetoacetyl-CoA thiolase; Hbd, 3-hydroxybutyryl-CoA dehydrogenase; Crt, crotonase; Bcd, butyryl-CoA dehydrogenase; Etf, electron transfer flavoprotein; AdhE2, aldehyde/alcohol dehydrogenase.
To do so, the genes responsible in n-butanol production [16,17] were cloned and expressed in E. coli using expression plasmids. The activity of these gene products, except for butyryl-CoA dehydrogenase (Bcd) and an electron transfer flavoprotein (Etf), were detected by enzyme assays. The activity of Bcd was not conclusively demonstrated using crude extract from E. coli that expressed bcd and etfAB, possibly due to the instability of the enzyme. Despite the inconclusive demonstration of Bcd activity, the expression of this synthetic pathway produced 14 mg/L of 1-butanol under anaerobic conditions.
To optimize the pathway, alternative enzymes from other organisms were evaluated. In addition to the C. acetobutylicum thiolase (coded by thl) [18], acetyl-CoA acetyltranserase from E. coli (coded by atoB) [12] was overexpressed. To determine whether homologues and isoenzymes of Bcd from other organisms would be more effective in E. coli, bcd [19] and etfAB [20] from Megasphaera elsdenii and ccr from Streptomyces coelicolor, which encodes a crotonyl-CoA reductase (Ccr) [21], were expressed in place of their counterparts from C. acetobutylicum. However, the M. elsdenii and S. coelicolor genes led to much lower production of 1-butanol in E. coli. Nevertheless, possibilities still exist that alternative genes from other organisms might improve 1-butanol production in E. coli.
The strategy to delete host pathways competing for carbon and reducing power is useful to increase the target products. For n-butanol production, the host pathways that compete with the n-butanol pathway for acetyl-CoA and NADH were deleted. Deletion of ldhA, adhE, frdBC, pta and fnr from wild type E.coli improved n-butanol production nearly three-fold over wild type levels by significantly reducing the amount of acetate, lactate, ethanol, and succinate produced. The use of a user-friendly host facilitates such exploration. The best combination of genes and host produced 0.5 g/L n-butanol. Although the level of production was not as high as expected, this result demonstrates the feasibility of the metabolic engineering approach.
Production of higher-chain alcohols using the keto acid pathways
Expression of heterologous genes may lead to metabolic imbalance, while the accumulation of the heterologous metabolites may cause cytotoxicity. To achieve a high productivity of biofuels, the synthetic pathway should be compatible with the host. Amino acid biosynthesis pathways are universal in almost all organisms. Atsumi et al. took advantage of this capacity in the host E.coli and diverted the metabolic intermediates to higher alcohols by inserting the last two steps in the Ehrlich pathway [22] for 2-ketoacid degradation from other organisms [4].
2-Ketoacids are intermediates in amino acid biosynthesis pathways and can be converted to aldehydes by 2-ketoacid decarboxylases (KDCs) [23] and then to alcohols by alcohol dehydrogenases (ADHs). Only two non-native steps are required to produce biofuels by shunting intermediates from amino acid biosynthesis pathways. Amino acid biosynthesis pathways produce various 2-ketoacids. The E.coli strain with overexpression of KDC and ADH produced longer chain alcohols including n-propanol, isobutanol, n-butanol, 2-methyl-1-butanol, 3-methyl-1-butanol and 2-phenylethanol. Furthermore, addition of specific 2-keto acids to the E. coli culture expressing KDC and ADH confirmed the specific production of the corresponding alcohols. These results indicate that increasing the flux to the 2-keto acids could improve both the productivity and specificity of production of the alcohols.
Isobutanol production from E. coli using the keto acid pathway
Previously, isobutanol was identified only in trace amounts as microbial byproducts. Thus it has not been considered as a practical fuel substitute until recently. Isobutanol has similar physicochemical properties as n-butanol, but has a higher octane number than n-butanol. This compound can be converted from 2-ketoisovalerte, an intermediate in valine biosynthesis (Fig. 3). To produce isobutanol, the carbon source was converted to 2-ketoisovalerate, through overexpressed AlsS (Bacillus subtilis), IlvC (E.coli) and IlvD (E.coli) [4] (Fig. 3). The resulting keto acid is then converted to isobutanol using KDC and ADH discussed above. In addition, genes that are involved in by-product formation were deleted. These deletions were intended to increase the level of pyruvate available for the synthetic isobutanol pathway. This engineered strain produced ~20 g/L isobutanol at a yield of 86% of the theoretical maximum [4] (Fig. 3). This high level production of isobutanol demonstrates the potential of this pathway and opens the possibility for industrial production of this compound as an advanced biofuel. Interestingly, although E. coli is sensitive to 20g/L of isobutanol, it accumulates this high level of alcohol after the growth phase.
Fig.3. Production of isobutanol through the synthetic nonfermentative pathways.
A. Schematic representation of isobutanol production in engineered E. coli. AlsS; acetolactate synthase, IlvC; acetohydroxy acid isomeroreductase, IlvD; dihydroxy-acid dehydratase, Kdc; 2-keto acid decarboxylase, Adh; alcohol dehydrogenase. B–D. Isobutanol production with the engineered E.coli in M9 medium with 3.6% glucose and 0.5 % yeast extract at 30°C. At 40 hr, 3.0% glucose was added to the culture. B: Time profiles of isobutanol production; C: Time profiles of cell growth. D: Time profiles of glucose concentration in medium. This figure was modified from ref[4]
The production of isobutanol in large quantities marks the success of the first non-native biofuel substitute and demonstrates the potential of the keto acid pathways. This strategy opens the door for exploring a variety of alcohols as potential biofuels beyond ethanol. The similar approach for isobutanol production also has been proposed in the patent application (G. K. Donaldson et al. US 2007/0092957) which is currently being examined. The best production titer reported in this application is 0.3 g/L
Production of n-butanol in E. coli using the keto acid pathway
The keto acid strategy can also be used to produce n-butanol, which has been produced by Clostridium using a coenzyme-A-dependent fermentative pathway [5,7]. The ketoacid precursor of n-butanol is 2-ketovalerate, a precursor of norvaline [24], which is produced though a minor side reaction of leucine biosynthesis pathway. To produce n-butanol using this pathway, it is required to increase the upstream precursors of 2-ketovalerate. These include threonine and its deamination product, 2-ketobutyrate.
To increase the L-threonine and 2-ketovalerate level, thrABC, ilvA and leuABCD were overexpressed in the strain with competing pathways deleted. The engineered E.coli produced 0.9 g/L 1-butanol in the initial shake flask experiments without much optimization. Although this productivity is still far below the Clostridium production level, the results demonstrate the feasibility of n-butanol production in E. coli. In particular, since high-efficiency threonine production in E. coli has been achieved in industry, n-butanol production could be improved by modifying a threonine hyperproducing strain [25] with the above strategy.
Fatty acid ethyl esters production from E.coli
Biodiesel is a substitute for petroleum-based diesel fuel. Easily extracted oils from palm trees and soybeans are mainly converted to biodiesel by transesterification of triacylglycerols with short chain alcohols, primarily methanol and ethanol. This reaction results in fatty acid methyl esters (FAMEs) and fatty acid ethyl esters (FAEEs). However, the production of biodiesel is limited by the insufficient availability of cheap vegetable oil feedstocks. To produce biodiesel with low-cost materials, E.coli has been engineered to produce FAEE [1].
To produce FAEE from E.coli, ethanol production was combined with esterification of the ethanol with the acyl moieties of coenzyme A thioesters of fatty acids by overexpressing the promiscuous acyltransferase from Acinetobacter baylyi[26]. The natural host A. baylyi was not a suitable candidate since it is a strictly aerobic bacterium not able to form ethanol. The engineered E.coli produced FAEE concentrations of 1.3 g/L with 0.018 g/L/hr production rate in the presence of glucose and oleic acid by fed-batch fermentation.
Isoprenoids Production from microorganisms
Isoprenoids are a diverse group of metabolites synthesized from isoprenyl pyrophosphate (IPPP) and dimethylallyl pyrophosphate (DMAP) in plants, animals, and bacteria [27]. These molecules are synthesized either from glyceraldehyde-3-phosphate and pyruvate via the methylerythritol pathway, or from acetyl-CoA via the mevalonate pathway. The isoprenoid biosynthesis pathways could produce branched-chain and cyclic alkanes, alkenes and alcohols [3]. However, only trace amount of isoprenoid compounds are produce from natural organisms. To produce isopentenol from E.coli, the isopentenyl pyrophosphate biosynthetic pathway was combined with expression of nudF from B.subtilis. The engineered E.coli produced isopentenol concentrations of 110 mg/L [3]. Engineered strains capable of converting sugars to isoprenoid derivative alcohols at high yields have not yet been reported in open literature although industrial efforts are believed to be under way (e.g. N. S. Renninger et al. US patent application, US 2008/0092829). This application, which is still under examination, disclosed the best isopentenol production as 1.2 g/L.
Conclusions
Metabolic engineering (or a similar field, Synthetic Biology) provides important tools for engineering non-native organisms to produce a broad class of fuel grade molecules. Although further improvement of production is required, advanced biofuels offer significant advantages over the traditional biofuel, ethanol. To make these fuels economically feasible, yield, titer, and productivity need to be further improved. In this regard, continued development of the experimental and computational methods for pathway and strain optimization is important to achieve high efficiency production.
Another approach to produce fuel grade alcohol is to improve production from natural producers. As an example, n-butanol fermentation in Clostridium has been used since the early 1910s [6]. ABE fermentation in C. acetobutylicum produces byproducts such as butyrate, acetone, and ethanol [7]. However, the relatively slow growth rate and spore-forming life cycle of Clostridium create additional problems for industrial fermentation. Furthermore, the relatively unknown genetic system and complex physiology of the microorganism present difficulties in engineering its metabolism for optimal production of n-butanol. Although clearly feasible, this approach has not enjoyed wide success yet.
As such, the problem of biofuels production provides a fertile field for metabolic engineering, particularly in constructing non-native, but user-friendly producing organisms. Furthermore, metabolic engineering offers advantages over traditional strain selection methods because tools for genetic manipulation and genome-scale data for metabolic and regulatory systems are rapidly expanding.
Acknowledgements
This work was supported in part by UCLA-DOE Institute of Genomics and Proteomics and NIH grant 5R01GM076143.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kalscheuer R, Stolting T, Steinbuchel A. Microdiesel: Escherichia coli engineered for fuel production. Microbiology. 2006;152:2529–2536. doi: 10.1099/mic.0.29028-0.This study demonstrates that ethanol can be esterified to a fatty acid in E.coli, which provides significant possibility for a whole-cell catalyst for the production of bio-diesel.
- 2.Hanai T, Atsumi S, Liao JC. Engineered synthetic pathway for isopropanol production in Escherichia coli. Appl Environ Microbiol. 2007;73:7814–7818. doi: 10.1128/AEM.01140-07.This paper demonstrates isopropanol production from E.coli by expressing various combinations of genes from C. acetobutylicum ATCC 824, E. coli K-12 MG1655, C. beijerinckii NRRL B593, and T. brockii HTD4. The isopropanol production from E.coli is higher than those produced by C. acetobutylicum.
- 3.Withers ST, Gottlieb SS, Lieu B, Newman JD, Keasling JD. Identification of isopentenol biosynthetic genes from Bacillus subtilis by a screening method based on isoprenoid precursor toxicity. Appl Environ Microbiol. 2007;73:6277–6283. doi: 10.1128/AEM.00861-07.The authors developed a screening method to isolate isoprenoid biosynthetic genes. With this method, two terpene synthase genes are cloned from the isoprene-producing bacterium Bacillus subtilis strain 6051.
- 4.Atsumi S, Hanai T, Liao JC. Non-fermentative pathways for synthesis of branched-chain higher alcohols as biofuels. Nature. 2008;451:86–89. doi: 10.1038/nature06450.This paper presents a metabolic engineering approach using E. coli to produce higher alcohols including isobutanol, n-butanol, 2-methyl-1-butanol and 3-methyl-1-butanol from glucose, a renewable carbon source. This strategy uses the host’s amino acid biosynthetic pathway and diverts its 2-keto acid intermediates for alcohol synthesis.
- 5.Atsumi S, Cann AF, Connor MR, Shen CR, Smith KM, Brynildsen MP, Chou KJ, Hanai T, Liao JC. Metabolic engineering of Escherichia coli for 1-butanol production. Metab Eng. 2008 doi: 10.1016/j.ymben.2007.08.003. doi:10.1016/j.ymben.2007.08.003.Clostridium is a natural producer of n-butanol. However, the relatively unknown genetic system and complex physiology of Clostridium present difficulties in engineering its metabolism for optimal production of n-butanol. To deal with these problems, the authors engineered a synthetic pathway in E. coli and demonstrated the production of 1-butanol from this non-native user-friendly host.
- 6.Lin YL, Blaschek HP. Butanol Production by a Butanol-Tolerant Strain of Clostridium acetobutylicum in Extruded Corn Broth. Appl Environ Microbiol. 1983;45:966–973. doi: 10.1128/aem.45.3.966-973.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jones DT, Woods DR. Acetone-butanol fermentation revisited. Microbiol Rev. 1986;50:484–524. doi: 10.1128/mr.50.4.484-524.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Osburn OL, Brown RW, Werkman CH. The butyl alcohol-isopropyl alcohol fermentation. J. Biol. Chem. 1937;121:685–695. [Google Scholar]
- 9.Chen JS, Hiu SF. Acetone-butanol-isopropanol production by Clostridium beijerinckii (synonym, Clostridium butylicum) Biotechnol. Lett. 1986;8:371–376. [Google Scholar]
- 10.Bermejo LL, Welker NE, Papoutsakis ET. Expression of Clostridium acetobutylicum ATCC 824 genes in Escherichia coli for acetone production and acetate detoxification. Appl Environ Microbiol. 1998;64:1079–1085. doi: 10.1128/aem.64.3.1079-1085.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Peretz M, Bogin O, Tel-Or S, Cohen A, Li G, Chen JS, Burstein Y. Molecular cloning, nucleotide sequencing, and expression of genes encoding alcohol dehydrogenases from the thermophile Thermoanaerobacter brockii and the mesophile Clostridium beijerinckii. Anaerobe. 1997;3:259–270. doi: 10.1006/anae.1997.0083. [DOI] [PubMed] [Google Scholar]
- 12.Duncombe GR, Frerman FE. Molecular and catalytic properties of the acetoacetyl-coenzyme A thiolase of Escherichia coli. Arch Biochem Biophys. 1976;176:159–170. doi: 10.1016/0003-9861(76)90152-1. [DOI] [PubMed] [Google Scholar]
- 13.Jenkins LS, Nunn WD. Genetic and molecular characterization of the genes involved in short-chain fatty acid degradation in Escherichia coli: the ato system. J Bacteriol. 1987;169:42–52. doi: 10.1128/jb.169.1.42-52.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ismaiel AA, Zhu CX, Colby GD, Chen JS. Purification and characterization of a primary-secondary alcohol dehydrogenase from two strains of Clostridium beijerinckii. J Bacteriol. 1993;175:5097–5105. doi: 10.1128/jb.175.16.5097-5105.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lamed RJ, Zeikus JG. Novel NADP-linked alcohol--aldehyde/ketone oxidoreductase in thermophilic ethanologenic bacteria. Biochem J. 1981;195:183–190. doi: 10.1042/bj1950183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boynton ZL, Bennett GN, Rudolph FB. Cloning, sequencing, and expression of genes encoding phosphotransacetylase and acetate kinase from Clostridium acetobutylicum ATCC 824. Appl Environ Microbiol. 1996;62:2758–2766. doi: 10.1128/aem.62.8.2758-2766.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fontaine L, Meynial-Salles I, Girbal L, Yang X, Croux C, Soucaille P. Molecular characterization and transcriptional analysis of adhE2, the gene encoding the NADH-dependent aldehyde/alcohol dehydrogenase responsible for butanol production in alcohologenic cultures of Clostridium acetobutylicum ATCC 824. J Bacteriol. 2002;184:821–830. doi: 10.1128/JB.184.3.821-830.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wiesenborn DP, Rudolph FB, Papoutsakis ET. Thiolase from Clostridium acetobutylicum ATCC 824 and Its Role in the Synthesis of Acids and Solvents. Appl Environ Microbiol. 1988;54:2717–2722. doi: 10.1128/aem.54.11.2717-2722.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Becker DF, Fuchs JA, Banfield DK, Funk WD, MacGillivray RT, Stankovich MT. Characterization of wild-type and an active-site mutant in Escherichia coli of short-chain acyl-CoA dehydrogenase from Megasphaera elsdenii. Biochemistry. 1993;32:10736–10742. doi: 10.1021/bi00091a026. [DOI] [PubMed] [Google Scholar]
- 20.O'Neill H, Mayhew SG, Butler G. Cloning and analysis of the genes for a novel electron-transferring flavoprotein from Megasphaera elsdenii. Expression and characterization of the recombinant protein. J Biol Chem. 1998;273:21015–21024. doi: 10.1074/jbc.273.33.21015. [DOI] [PubMed] [Google Scholar]
- 21.Wallace KK, Bao ZY, Dai H, Digate R, Schuler G, Speedie MK, Reynolds KA. Purification of crotonyl-CoA reductase from Streptomyces collinus and cloning, sequencing and expression of the corresponding gene in Escherichia coli. Eur J Biochem. 1995;233:954–962. doi: 10.1111/j.1432-1033.1995.954_3.x. [DOI] [PubMed] [Google Scholar]
- 22.Sentheshanuganathan S. The mechanism of the formation of higher alcohols from amino acids by Saccharomyces cerevisiae. Biochem J. 1960;74:568–576. doi: 10.1042/bj0740568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.de la Plaza M, Fernandez de Palencia P, Pelaez C, Requena T. Biochemical and molecular characterization of alpha-ketoisovalerate decarboxylase, an enzyme involved in the formation of aldehydes from amino acids by Lactococcus lactis. FEMS Microbiol Lett. 2004;238:367–374. doi: 10.1016/j.femsle.2004.07.057. [DOI] [PubMed] [Google Scholar]
- 24.Kisumi M, Sugiura M, Chibata I. Biosynthesis of norvaline, norleucine, and homoisoleucine in Serratia marcescens. J Biochem. 1976;80:333–339. doi: 10.1093/oxfordjournals.jbchem.a131281. [DOI] [PubMed] [Google Scholar]
- 25.Miwa K, Tsuchida T, Kurahashi O, Nakamori S, Sano K, Momose H. Construction of L-Threonine Overproducing Strains of Escherichia coli K-12 Using Recombinant. Agric. Biol. Chem. 1983;47:2329–2334. [Google Scholar]
- 26.Vaneechoutte M, Young DM, Ornston LN, De Baere T, Nemec A, Van Der Reijden T, Carr E, Tjernberg I, Dijkshoorn L. Naturally transformable Acinetobacter sp. strain ADP1 belongs to the newly described species Acinetobacter baylyi. Appl Environ Microbiol. 2006;72:932–936. doi: 10.1128/AEM.72.1.932-936.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sacchettini JC, Poulter CD. Creating isoprenoid diversity. Science. 1997;277:1788–1789. doi: 10.1126/science.277.5333.1788. [DOI] [PubMed] [Google Scholar]