Abstract
Nematode parasites of the genus Trichinella are intracellular and distinct life cycle stages invade intestinal epithelial and skeletal muscle cells. Within the genus, Trichinella spiralis and Trichinella pseudospiralis exhibit species-specific differences with respect to host-parasite complex formation and host immune modulation. Parasite excretory-secretory (ES) proteins play important roles at the host-parasite interface and are thought to underpin these differences in biology. Serine proteases are among the most abundant group of T. spiralis ES proteins and multiple isoforms of the muscle larvae-specific TspSP-1 serine protease have been identified. Recently, a similar protein (TppSP-1) in T. pseudospiralis muscle larvae was identified. Here we report the cloning and characterisation of the full-length transcript of TppSP-1 and present comparative data between TspSP-1 and TppSP-1.
Keywords: Trichinella spiralis, Trichinella pseudospiralis, excretory/secretory proteins, Western blot analysis, serine protease
1. Introduction
Trichinella spp. are unusual nematode parasites that occupy two distinct intracellular niches within a single animal host. The infective muscle larvae L1 (ML) invade intestinal epithelial cells and the immature L1, the newborn larvae, invade skeletal muscle cells (Despommier, 1983). A host-parasite complex is formed following invasion of muscle cells, and the presence or absence of a collagen layer around this complex divides the genus into encapsulated species, such as Trichinella spiralis and non-encapsulated species, such as Trichinella pseudospiralis (Pozio and Murrell, 2006). The resulting pathology of these two groups is species-specific. Although both modulate the host muscle cell phenotype, there are differences (Wu et al., 2001; Boonmars et al., 2004; Boonmars et al., 2005) where T. pseudospiralis also modulates the host immune response to suppress inflammation and cellular infiltrations normally associated with a healthy immune response (Wakelin et al., 1994; Li and Ko, 2001; Furze and Selkirk, 2005). It is not known how these species-specific differences are initiated or maintained, but they may be mediated by the excretory-secretory (ES) proteins (Ko et al., 1994; Wu et al., 2001; Boonmars et al., 2004). Among the most abundant group of proteins within the ES of T. spiralis are the serine proteases. Serine proteases are important in a wide variety of biological processes, including digestion, blood coagulation and fibrinolysis, and have been implicated also in tumour growth. In parasites, serine proteases are known to be involved in host tissue and cell invasion (reviewed in Dzik, 2006), and in nematodes are likely to be important in moulting. Several secreted serine proteases have been identified among T. spiralis ES proteins, including the trypsin-like 45 kDa antigen and the serine protease TspSP-1 (Romaris et al., 2002; Robinson and Connolly, 2005). A recent comparison of T. pseudospiralis and T. spiralis ES proteomes revealed that serine proteases, including a putative homologue of TspSP-1, TppSP-1, were also abundant in the T. pseudospiralis ES protein fraction (Robinson et al., 2007). The T. pseudospiralis protein, TppSP-1 is tyvelosylated and cross-reacts with the anti-tyvelose monoclonal antibody (mAb)18H (Ellis et al., 1994; Reason et al., 1994) but may be less abundant where only nine isoforms have thus far been identified compared to the twelve isoforms of TspSP-1. In this study, de novo-derived peptide sequence was used to generate primers to identify the full-length cDNA of the TppSP-1 gene. Preliminary data regarding immunolocalisation of both TspSP-1 and TppSP-1 is also presented.
2. Materials and Methods
2.1 Collection, preparation and analysis of ES proteins
T. pseudospiralis (ISS13) ES proteins were collected and analysed by one-dimensional electrophoresis (1-DE) as described (Robinson and Connolly, 2005). Western analysis was performed using the primary mAb 18H (Appleton et al., 1988), mAb 9H8 and mAb1H7 (Romaris et al., 2002) at dilutions of 1:1500, 1:100 and 1:100, respectively, followed by an anti-mouse-IgG secondary antibody conjugated to alkaline phosphatase (Sigma) at a dilution of 1:10 000.
2.2 PCR
Forward (tspsp1_106F: 5′GAACCATATTTGACAAATCC) and reverse (tspsp1_349R: 5′CTGCTGTCACAGTTATTGC) primers were used in RT-PCR to amplify oligo dT-synthesized cDNA from T. pseudospiralis total RNA. Terminal sequences were identified using 5′-RACE and 3′-RACE and TppSP-1 gene specific primers according to the GeneRacer kit (Invitrogen). PCR products were cloned into pGEM-T Easy vector (Promega) and sequenced by the University of Dundee DNA sequencing facility.
2.3 Immunohistochemistry
Samples of skeletal muscle from CD1 mice infected with T. spiralis (ISS003) or T. pseudospiralis (ISS013) were fixed in formalin and embedded in paraffin. Sections were deparaffinized and processed for staining with normal rat immunoglobulins (2 μg ml−1 of immunoglobulins preciptated from serum with 40% ammonium sulfate) or with rat mAbs 18H, 9H8 or 1H7 (culture supernatants diluted 1:2) (Beiting et al., 2004; Duffy et al., 2006). Antibody binding was detected with peroxidase-conjugated goat anti-rat IgG diluted in 10% normal mouse serum, using the substrate 3-amino-9-ethyl-carbazole (AEC) (Sigma). Sections were counterstained with hematoxylin, mounted with Glycergel (Dako Corporation, Carpinteria, CA), and images examined and captured using an Olympus BX51 microscope fitted with a DP12 digital camera. Resolution was adjusted to 300 dpi (Adobe Photoshop).
3. Results
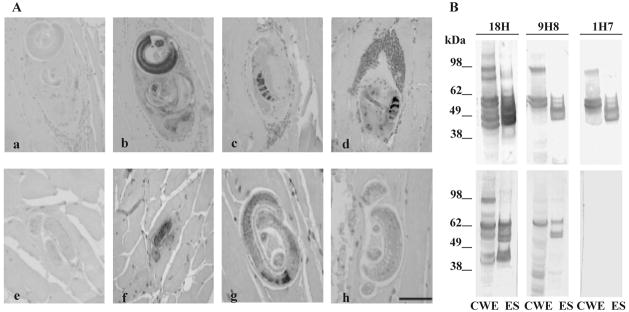
Several T. pseudospiralis peptides (Robinson et al., 2007) with similarity to the T. spiralis TspSP-1 protein, were used to generate PCR primers to amplify a 243 bp cDNA fragment from T. pseudospiralis ML cDNA by RT-PCR. Primers based on this sequence were then used in 5′- and 3′-RACE PCR and ultimately to identify a 1.5 kb sequence representing the full-length cDNA. The cDNA included a 1.3 kb open reading frame encoding a putative ~50.8 kDa protein that showed overall ~63% amino acid sequence identity to TspSP-1 (Fig. 1). The domain structure of the putative protein, TppSP-1, was similar to TspSP-1 with an N-terminal signal peptide and several N-linked glycosylation sites and a C-terminal proline-rich domain conserved between the two proteins. Within the protease domain, the eight characteristic cysteine residues were conserved with respect to the position of the putative catalytic triad; however, the histidine residue of the catalytic triad was replaced with an arginine residue (R98), while the aspartic acid (D150) and serine (S238) were conserved. To investigate the localisation of the TspSP-1 and TppSP-1 proteins, immunohistochemistry was performed on sections of skeletal muscle from mice infected with either T. spiralis or T. pseudospiralis using the anti-TspSP-1 mAb 9H8, mAb1H7 and the control anti-tyvelose mAb18H (Fig. 2A). The mAb 18H reacted strongly with the T. spiralis TSL-1 antigens in the stichosome and on the larval surface as expected and cross-reacted with the T. pseudospiralis antigens present within both α and β stichocytes. Both mAB 9H8 and mAb 1H7 detected TspSP-1 protein within the stichocytes of the T. spiralis larva in the sections. The anti-TspSP-1 mAb 9H8 recognised T. pseudospiralis larval antigens localised in stichocytes, showing a similar pattern to that seen in T. spiralis-infected tissue. However, the anti-TspSP-1 mAb 1H7 antibody failed to bind to the T. pseudospiralis sections. To confirm these results, Western analysis of crude worm extracts (CWE) and ES proteins of T. spiralis and T. pseudospiralis was performed (Fig. 2B). As expected, the mAb 18H bound protein bands in the CWE and ES of both T. spiralis and T. pseudospiralis, and the mAb 9H8 and mAb 1H7 also bound protein bands in T. spiralis CWE and ES as described elsewhere (Romaris et al., 2002). The mAb 9H8 bound to a 64 kDa protein in the CWE of T. pseudospiralis and to three proteins ranging from 49–70 kDa in the ES. In contrast the mAb1H7 did not bind either the CWE or ES consistent with the results from immunohistochemistry.
Fig. 1.
Amino acid sequences of T. spiralis TspSP-1 and T. pseudospiralis TppSP-1 (GenBank accession numbers AAK31787 and EU245046, respectively). The positions of the signal peptide (italics), catalytic triad (highlighted in black), conserved cysteine residues (grey), predicted N-glycosylation sites (underlined) and proline-rich regions (bold) are shown.
Fig. 2.
(A) Immunohistochemical staining of T. spiralis (a–d) and T. pseudospiralis (e–h) infected mouse muscle tissue: (a, e) negative control rat IgG; (b, f) mAb 18H; (c, g) mAb 9H8; (d & h) mAb 1H7. Bar = 50 μm. (B) 1-DE Western analysis of T. spiralis (upper panel) and T. pseudospiralis (lower panel) CWE and ES proteins with mAb 18H, mAb 9H8 and mAb 1H7 as indicated. Protein molecular weight markers (SeeBlue® Plus2 Prestained Standard, Invitrogen) are shown.
4. Discussion
The TppSP-1 cDNA encodes a homologue of the serine protease TspSP-1 and shares 74% nucleotide sequence identity with the TspSP-1 gene. The putative TppSP-1 protein shows the same domain organisation as TspSP-1 but with some differences. The N-terminal signal peptide and protease domains are highly conserved between the two proteins (74% amino acid identity), whereas the C-terminal domain is less conserved (43% amino acid identity). Primary amino acid sequence differences between TspSP-1 and TppSP-1 may account for the lack of reactivity of mAb 1H7 to T. pseudospiralis antigens. The anti-TspSP-1 monoclonal antibodies were made against deglycosylated T. spiralis ES proteins and in ELISA mAb 9H8 binds only deglycosylated ES proteins, whereas mAb 1H7 binds both native and deglycosylated ES proteins (Romaris et al., 2002). It could be inferred from this that the epitope recognised by mAb 1H7 is present within the C-terminal domain of TspSP-1 and not associated with any of the putative glycosylation sites. Serine proteases are normally secreted as inactive zymogens that are later processed to the mature enzyme by cleavage at a characteristic amino acid motif, IVGG. A similar motif (IVGE) found after amino acid Q47 in TspSP-1 is highly divergent in TppSP-1 (MDGE), questioning whether the T. pseudospiralis protein can be activated by proprotein cleavage. Analysis of the draft T. spiralis genome sequence (http://genome.wustl.edu) showed that TspSP-1 is encoded by a multi-copy gene family (to be published elsewhere). It is likely that TppSP-1 is also multicopy as northern analysis indicates that there are at least two transcripts with sequence similarity to TppSP-1 in T. pseudospiralis ML RNA and preliminary data suggests that the second transcript encodes a protein containing a canonical proprotein cleavage site. In terms of function, the histidine residue of the catalytic triad in TspSP-1 is replaced with an arginine residue in TppSP-1, which may affect its function as a serine protease. A few serine proteases have lost their proteolytic activity following substitution of catalytic triad residues; the protein azurocidin has lost two of the conserved triad residues and no longer functions as a protease but has gained anti-microbial properties and is a strong mediator of inflammation (Watorek, 2003). In this respect, future studies focusing on the role of TppSP-1 during infection with T. pseudospiralis is of particular interest.
Acknowledgments
This work was supported by grants from the BBSRC UK (C20267) to BC and the NIH USA (AI14490) to JAA.
Footnotes
Note: Nucleotide sequence data reported in this paper are available in the Genbank™, EMBL and DDBJ databases under the accession no. EU245046.
Conflict of interest statement
None declared.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Appleton JA, Schain LR, McGregor DD. Rapid expulsion of Trichinella spiralis in suckling rats: Mediation by monoclonal antibodies. Immunol. 1988;65:487–492. [PMC free article] [PubMed] [Google Scholar]
- Beiting DP, Bliss SK, Schlafer DH, Roberts VL, Appleton JA. Interleukin-10 limits local and body cavity inflammation during infection with muscle-stage Trichinella spiralis. Infect Immunol. 2004;72:3129–3137. doi: 10.1128/IAI.72.6.3129-3137.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boonmars T, Wu Z, Nagano I, Nakada T, Takahashi Y. Differences and similarities of nurse cells in cysts of Trichinella spiralis and T. pseudospiralis. J Helminthol. 2004;78:7–16. doi: 10.1079/joh2003203. [DOI] [PubMed] [Google Scholar]
- Boonmars T, Wu Z, Nagano I, Takahashi Y. Trichinella pseudospiralis infection is characterized by more continuous and diffuse myopathy than T. spiralis infection. Parasitol Res. 2005;97:13–20. doi: 10.1007/s00436-005-1359-x. [DOI] [PubMed] [Google Scholar]
- Despommier DD. Biology. In: Campbell WC, editor. Trichinella and trichinosis. Plenum Press; New York and London: 1983. pp. 75–151. [Google Scholar]
- Duffy MS, Cevasco DK, Zarlenga DS, Sukhumavasi W, Appleton JA. A cathepsin B homologue at the interface between a parasitic nematode and its intermediate host. Infect Immunol. 2006;74:1297–1304. doi: 10.1128/IAI.74.2.1297-1304.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dzik JM. Molecules released by helminth parasites involved in host colonization. Acta Biochim Pol. 2006;53:33–64. [PubMed] [Google Scholar]
- Ellis LA, Reason AJ, Morris HR, Dell A, Iglesias R, Ubeira FM, Appleton JA. Glycans as targets for monoclonal antibodies that protect rats against Trichinella spiralis. Glycobiol. 1994;4:585–592. doi: 10.1093/glycob/4.5.585. [DOI] [PubMed] [Google Scholar]
- Furze RC, Selkirk ME. Comparative dynamics and phenotype of the murine immune response to Trichinella spiralis and Trichinella pseudospiralis. Parasite Immunol. 2005;27:181–188. doi: 10.1111/j.1365-3024.2005.00761.x. [DOI] [PubMed] [Google Scholar]
- Ko RC, Fan L, Lee DL, Compton H. Changes in host muscles induced by excretory/secretory products of larval Trichinella spiralis and Trichinella pseudospiralis. Parasitol. 1994;108:195–205. doi: 10.1017/s0031182000068293. [DOI] [PubMed] [Google Scholar]
- Li CK, Ko RC. Inflammatory response during the muscle phase of Trichinella spiralis and T. pseudospiralis infections. Parasitol Res. 2001;87:708–714. doi: 10.1007/s004360100420. [DOI] [PubMed] [Google Scholar]
- Pozio E, Darwin Murrell K. Systematics and epidemiology of Trichinella. Adv Parasitol. 2006;63:367–439. doi: 10.1016/S0065-308X(06)63005-4. [DOI] [PubMed] [Google Scholar]
- Reason AJ, Ellis LA, Appleton JA, Wisnewski N, Grieve RB, McNeil M, Wassom DL, Morris HR, Dell A. Novel tyvelose-containing tri- and tetra-antennary N-glycans in the immunodominant antigens of the intracellular parasite Trichinella spiralis. Glycobiol. 1994;4:593–603. doi: 10.1093/glycob/4.5.593. [DOI] [PubMed] [Google Scholar]
- Robinson MW, Connolly B. Proteomic analysis of the excretory-secretory proteins of the Trichinella spiralis L1 larva, a nematode parasite of skeletal muscle. Proteomics. 2005;5:4525–4532. doi: 10.1002/pmic.200402057. [DOI] [PubMed] [Google Scholar]
- Robinson MW, Greig R, Beattie KA, Lamont DJ, Connolly B. Comparative analysis of the excretory-secretory proteome of the muscle larva of Trichinella pseudospiralis and Trichinella spiralis. Int J Parasitol. 2007;37:139–148. doi: 10.1016/j.ijpara.2006.08.007. [DOI] [PubMed] [Google Scholar]
- Romaris F, North SJ, Gagliardo LF, Butcher BA, Ghosh K, Beiting DP, Panico M, Arasu P, Dell A, Morris HR, Appleton JA. A putative serine protease among the excretory-secretory glycoproteins of L1 Trichinella spiralis. Mol Biochem Parasitol. 2002;122:149–160. doi: 10.1016/s0166-6851(02)00094-4. [DOI] [PubMed] [Google Scholar]
- Wakelin D, Goyal PK, Dehlawi MS, Hermanek J. Immune responses to Trichinella spiralis and T. pseudospiralis in mice. Immunol. 1994;81:475–479. [PMC free article] [PubMed] [Google Scholar]
- Watorek W. Azurocidin - inactive serine proteinase homolog acting as a multifunctional inflammatory mediator. Acta Biochim Pol. 2003;50:743–752. [PubMed] [Google Scholar]
- Wu Z, Matsuo A, Nakada T, Nagano I, Takahashi Y. Different response of satellite cells in the kinetics of myogenic regulatory factors and ultrastructural pathology after Trichinella spiralis and T. pseudospiralis infection. Parasitol. 2001;123:85–94. doi: 10.1017/s0031182001007958. [DOI] [PubMed] [Google Scholar]