Human spliceosomal U1 snRNP, consisting of U1 snRNA and ten proteins, recognizes the 5′-splice site within precursor-mRNAs and initiates the assembly of the spliceosome for intron excision. An electron density map of the functional core of U1 snRNP at 5.5 Å resolution enabled us to build the RNA and, in conjunction with site-specific labelling of individual proteins, to place the seven Sm proteins, U1C, and U1-70k into the map. Here we present the first detailed structure of a spliceosomal snRNP which reveals a hierarchical network of intricate interactions between subunits. A striking feature is the N-terminal polypeptide of U1-70k, which extends over a distance of 180 Å from its RNA binding domain, wrapping around the core domain consisting of the seven Sm proteins, to finally contact U1C, crucial for 5′-splice site recognition. The structure of U1 snRNP provides insights into U1 snRNP assembly and suggests a possible mechanism of 5′-splice site recognition.
In eukaryotes the majority of protein coding genes are interrupted by non-coding sequences known as introns. The entire length of a gene including its introns is transcribed into precursor-mRNAs (pre-mRNAs). The introns are excised and the exons spliced together to form mRNA with continuous protein coding sequences by a large RNA-protein assembly called the spliceosome1-3. The major components of the spliceosome are five small nuclear ribonucleoprotein particles (U1, U2, U4, U5 and U6 snRNPs), each containing one of five spliceosomal U-type snRNAs, seven Sm or Lsm proteins, and particle-specific proteins1-6. Assembly of the spliceosome is initiated by the binding of U1 snRNP to the 5′-splice site in pre-mRNA, followed by an ATP-dependent binding of U2 snRNP to the branch-point sequence within the intron. Upon further binding of the tri-snRNP, containing U4, U6 and U5 snRNPs, the spliceosome undergoes extensive rearrangements to become catalytically active1-3,5,6.
Mammalian U1 snRNP consists of U1 snRNA, seven Sm proteins (B/B′, D1, D2, D3, E, F and G) and three U1-specific proteins (U1-70k, U1A and U1C). The base-pairing between the 5′-end of U1 snRNA7,8 and the 5′-splice site in pre-mRNA plays a crucial role in 5′-splice site recognition9-11. In higher eukaryotes splicing factors, such as SR proteins, bound to splice enhancer sequences direct U1 snRNP through protein-protein interactions12,13 to the vicinity of 5′-splice sites that differ significantly from the consensus sequence1.
Human U1 snRNA forms four stem-loops (SL)14 (Fig S1). Seven Sm proteins assemble around the Sm site nucleotides, located between SL3 and SL4, to form the core domain3,4. U1-70k and U1A bind to SL1 and SL2, respectively, through their RNA binding domains (RBDs)15-19. U1C does not bind free U1 snRNA but the addition of the first 97 residues of U1-70k enables U1C to bind to the U1 core domain20. We have reported the crystal structure of the N-terminal RBD of U1A bound to SL2 of U1 snRNA19 and the solution structure of the Zn-finger domain of U1C21. We proposed a heptameric ring model of the core domain based on the crystal structures of two Sm protein hetero-dimers (D3B and D1D2)22. However, the three-dimensional organization of these components in U1 snRNP is not known.
Early electron microscopy (EM) studies of negatively stained U1 snRNP revealed a globular core domain with two protuberances attributable to U1-70k and U1A23,24. More recently, Stark et al.25 carried out a cryo-EM single particle analysis of U1 snRNP but the resolution was not sufficient to resolve secondary structure elements of proteins. Therefore, the authors were unable to place protein components unambiguously and thus the proposed arrangement of proteins relied largely on biochemical data.
We have crystallized and solved the structure of the functional core of human U1 snRNP at 5.5 Å. At this resolution the major and minor grooves of double-stranded RNA are evident. In addition, α-helices of proteins can be seen directly and β-sheets appear as flat density26,27, allowing known protein folds to be identified. We have specifically labelled residues in individual proteins and used anomalous difference peaks from selenium, zinc and mercury to place each protein unambiguously into the map. This also allowed us to trace the N-terminal 100 residues of U1-70k with no prior structural information. The crystal structure of human U1 snRNP presented here reveals the first detailed three-dimensional organization of the RNA and protein components within a spliceosomal snRNP and provides important insight into its function.
Structure determination of U1 snRNP
We reconstituted U1 snRNP from proteins expressed in Escherichia coli and U1 snRNA produced by in vitro transcription19,21,22. U1 snRNP is active in a splicing assay when U1A is depleted28 or nucleotides 50-91 of U1 snRNA, containing the U1A binding site, are deleted29. Crystals of the functional core of U1 snRNP were obtained with B (1-174), U1C (1-77), U1-70k (2-215), the full-length constructs of D1, D2, D3, E, F, and G, and U1 snRNA in which the apical part of SL2 is replaced by a kissing loop sequence30 to promote crystal contacts (Fig S1). The crystals belong to the P1 space group with typical unit cell dimensions of a = 127 Å, b = 128 Å, c = 156 Å, α = 96°, β = 107°, γ = 101°, and four complexes in the asymmetric unit. Initial phases were obtained with a Ta6Br12 derivative. After inclusion of other heavy atom derivatives, an electron density map at 6.5 Å resolution allowed the building of all RNA helices and fitting of known protein domains. A mercury-soaked crystal containing a U1C variant (Q39C) diffracted to 5.5 Å. After multi-domain, multi-crystal averaging31 the map calculated with the 5.5 Å data set was of excellent quality (Table S1).
Structure of the U1 snRNP core domain
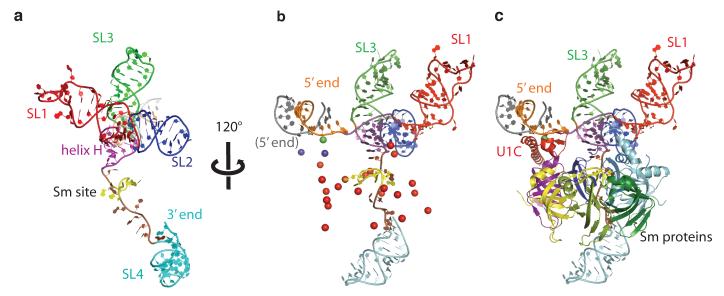
RNA on the 5′ side of the Sm site forms a four-helix junction and SL4 is located on the opposite side of the Sm ring (Fig 1). SL1 and SL2 co-axially stack on each other. Similarly, SL3 and helix H stack co-axially (Fig 1a). These co-axially stacked helices cross at an angle of ~90°. This stacking scheme was proposed for free U1 snRNA based on biochemical studies14,32.
Figure 1. Overall fold of U1 snRNA and the U1 snRNP core domain containing the seven Sm proteins.
a, Human U1 snRNA forms four stem-loop (SL) structures: SL1 (red), SL2 (blue), SL3 (green) and SL4 (cyan). A four-helix junction with two co-axially stacked helices (SL1-SL2; SL3-helix H (magenta)) is separated from SL4 by the single-stranded Sm site (lime green). b, Anomalous difference peaks used to place the seven Sm proteins and U1C into the map. Se atoms of Se-methionine-labelled Sm proteins (red); a Zn atom of the Zn-finger of U1C (green); Hg atoms attached to residues C24 and mutant Q39C of U1C (purple). The 5′-end of U1 snRNA (orange) base-pairs with its counterpart from a neighboring complex (grey). c, Seven Sm proteins and U1C fitted into the map.
The Sm ring consisting of seven Sm proteins is readily recognized in the electron density where selenium anomalous peaks from all Sm proteins are clustered (Fig 1b). The identity of the seven Sm proteins was established unambiguously from the positions of these anomalous peaks. The Sm proteins are arranged in the order E-G-D3-B-D1-D2-F (Fig 1c and 2a), in agreement with our heptameric ring model22. The 5.5 Å resolution averaged map is of excellent quality such that a rod-like density of the N-terminal helix and a curved slab of density for the β-sheet of the Sm-fold are almost perfectly formed in each Sm protein (Fig 2a).
Figure 2. Structure of the U1 snRNP core domain.
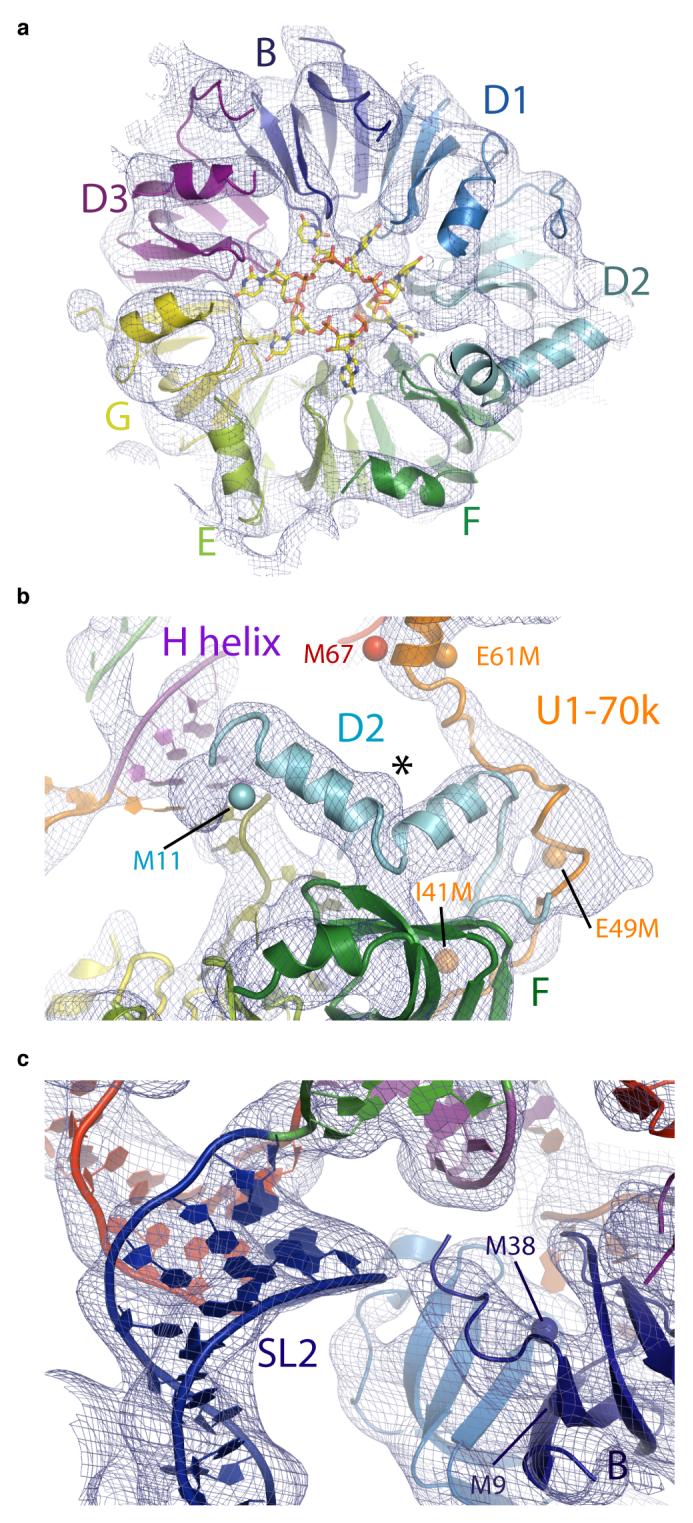
a, The Sm proteins and the experimental electron density map (contoured at 1σ). Cartwheel-shaped density in the central hole of the U1 snRNP core domain is attributed to the seven Sm site nucleotides. b, The N-terminus of D2 supporting helix H. The extended polypeptide of U1-70k interacts with D2. Seleno-methionine anomalous peaks from natural (M67; red) and engineered Met residues (I41M, E49M and E61M; orange) of U1-70k and of Sm protein D2 (Met11; cyan). The asterisk indicates a “bump” due to Phe-24 of D2. c, The N-terminus of Sm B protein interacting with the phosphate backbone of SL2. Navy blue spheres, anomalous peaks from seleno-methionine-labelled B protein.
The Sm protein D2 contains a particularly long N-terminal extension whereas B, D1 and D3 have long C-terminal extensions. The four-helix junction of U1 snRNA lies over a relatively flat face of the Sm protein ring consisting of the N-terminal helices of the Sm proteins. The N-terminus of D2 is not ordered in the D1D2 hetero-dimer22 but in U1 snRNP it forms a kinked helix and interacts with helix H in its minor groove (Fig 2b). The positive end of the α-helix dipole, as well as the conserved Lys-6 and Lys-8, may facilitate the interaction of the N-terminus of D2 with RNA. The N-terminus of B also interacts with the RNA backbone at the base of SL2 (Fig 2c). These two interactions stabilize the four-helix junction of U1 snRNA onto the core domain and stabilize the overall structure of U1 snRNP.
A cartwheel-shaped electron density observed in the central hole of the Sm ring (Fig 2a) is attributed to the Sm site RNA in analogy to the density observed for the penta-uridylate bound to the homo-heptamer of Lsm proteins33. This density exhibits features of bases splayed out towards the Sm proteins. The first and third uracil bases within an Sm site 9-mer oligonucleotide (rAAUUUUUGG) were cross-linked to G and B proteins respectively34. These two positions correspond to U127 and U129 of U1 snRNA and the register of RNA binding within the Sm ring can be inferred based on these two cross-links. Thus the seven nucleotides of the Sm site (AUUUGUG) are likely to interact with E, G, D3, B, D1, D2 and F, respectively.
The Zn-finger domain of U1C
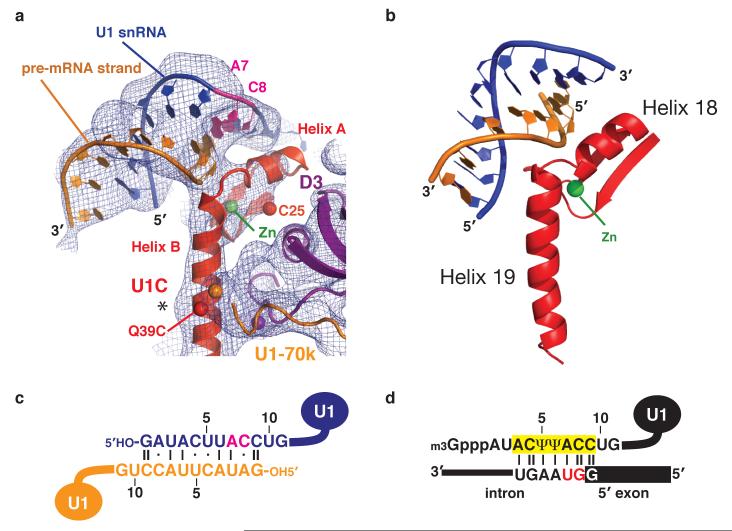
The Zn-finger domain of U1C21 interacts with the Sm protein ring exclusively through D3 (Fig 1c). In solution helix C of U1C folds back onto helix B through clustering of aromatic amino acids21 whereas in U1 snRNP these helices form a long continuous helix (helix B) extending from the Zn-binding site (Fig 3a). In the crystal the 5′-end of U1 snRNA base-pairs with its counterpart from an adjacent complex (Fig 1b). The proposed base-pairing scheme was inferred from the length of the RNA duplex (Fig 3c). This interaction is likely to mimic the binding of the 5′-splice site of pre-mRNA to U1 snRNA (Fig 3d). Helix A of U1C binds across the minor groove of this RNA duplex where C8 and A7 of U1 snRNA base-pair with nucleotides corresponding to the invariant GU dinucleotide of the 5′-splice site (Fig 3a). Importantly, the Zn-finger of U1C is distinct from the TFIIIA-type35 in that there are five intervening residues between the two Zn-coordinating histidines. In order to allow these two histidines to coordinate the Zn ion, the intervening residues in U1C form a loop. This atypical Zn-finger structure21 places the loop and helix A at strategic positions to interact with the duplex between the 5′-end of U1 snRNA and the putative pre-mRNA strand (Fig 3a).
Figure 3. A mechanism of 5′-splice site recognition.
a, The 5′-end of U1 snRNA (blue) base-pairs with the same region of a neighbouring complex (orange), which mimics the 5′-splice site of pre-mRNA. C8 and A7 of U1 snRNA are highlighted (magenta). The experimental map is contoured at 1σ. Spheres represent anomalous peaks of Zn (green) and Hg (red). The asterisk indicates a “bump” due to Trp-41. b, The Zn-finger of yeast dimethylallyltransferase and the anti-codon stem of a tRNA substrate41. c, Base-pairing between the 5′-ends of two U1 snRNAs within the U1 snRNP crystal. C8 and A7 are highlighted (magenta). d, Base-pairing between the 5′-end of U1 snRNA and a canonical 5′-splice site1. The invariant first and second nucleotides of the intron (GU) are highlighted (red).
U1-70k has an unusual structure and path
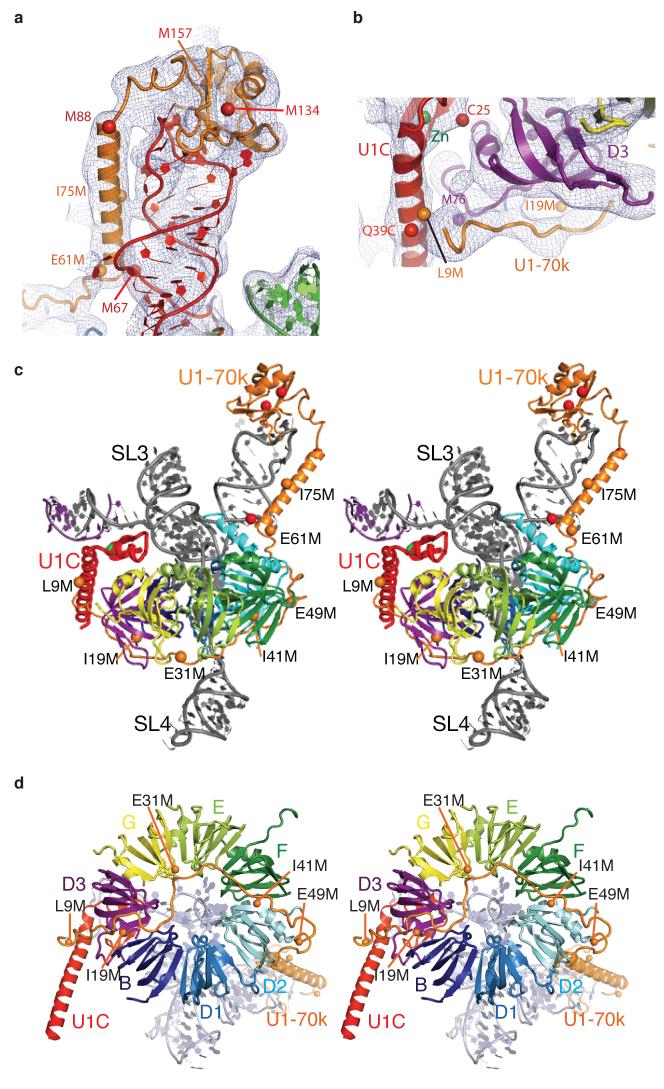
U1-70k contains an RBD between residues 100-180, whereas its N-terminal region (residues 1-100) has no known sequence motif. A homology-modelled RBD of U1-70k was placed into a large density near the SL1 loop of U1 snRNA with the aid of two selenium positions (M134, M157; Fig 4a). Based on the cross-linking data36, the RNA loop of SL1 was built into the density in such a way that G28 and U30 of SL1 are close to Tyr-112 and Leu-175, respectively. The region between residues 61 and 89 of U1-70k, predicted to form an α-helix, was built into the long tubular density extending along the RNA stem from the RBD (Fig. 4a). The register and orientation of the α-helix are determined from the selenium peaks of Met-67 and Met-88. The region around the M67 and I75M landmarks contains nine basic residues. Of these, R63, R66, R70, R71, K74, R68 and R79 are close to the phosphate backbone of SL1. Hence these residues are likely to play an important role in positioning this long α-helix along SL1.
Figure 4. Interaction of U1-70k with SL1 of U1 snRNA, the core domain, and U1C.
a, The RNA binding domain (RBD) of U1-70k and the α-helix extending along SL1 towards the core domain. b, The long α-helix B of U1C (red) held between the N-terminus of U1-70k (orange) and the C-terminal end of D3 (purple). c, Stereo view of U1 snRNP showing the entire path of U1-70k revealed by Se-methionine landmarks (orange spheres). d, Path of the N-terminal region of U1-70k crossing the Sm ring viewed in stereo from SL4 (omitted for clarity). Spheres indicate anomalous peaks from Se-methionines in U1-70k (natural, red; engineered, orange) and D3 (purple). Experimental electron density map is contoured at 1σ.
The N-terminal 97 residues of U1-70k are necessary and sufficient for U1C to bind to the U1 core domain20. The RBD and the long α-helix of U1-70k, distant from U1C, do not provide a plausible explanation for the essential role of U1-70k in U1C binding. To trace the path of the N-terminus, crystals of seven variants of U1 snRNP were grown, each with a Se-Met derivative of single methionine mutations of U1-70k (L9M, I19M, E31M, I41M, E49M, E61M and I75M). These Se-Met anomalous peaks unambiguously established the path of the N-terminal region of U1-70k (Fig 4b-d). Remarkably, the extended polypeptide chain of U1-70k wraps around the core domain and reaches an α-helical density near the Se peak of the L9M mutant. Electron density attributable to U1-70k can be identified along this path from the RBD to U1C (Fig 2b and 4b). The N-terminus of U1-70k and the C-terminus of D3 create a binding groove for the long α-helix of U1C (Fig 4b). This interaction accounts for the essential role of the N-terminus of U1-70k for U1C binding20.
The 5′-splice site recognition
The fortuitous interaction between the 5′-ends of U1 snRNA from two U1 snRNPs provides important insights into the mechanism of 5′-splice site recognition. U1C is crucial for the formation of E complex28 in which the 5′-splice site of pre-mRNA base-pairs with the 5′-end of U1 snRNA9-11. The structure of U1 snRNP suggests a role for U1C in stabilizing this base-pairing. A double mutant of U1C (R28G, K29S) fails to promote the E complex formation29. These residues are within the loop between helices A and B, ideally located to interact with the putative pre-mRNA strand, consistent with their crucial role in 5′-splice site recognition (Fig 3a). In the crystal structure of dimethylallyltransferase-tRNA complex, a Zn-finger domain interacts with the anti-codon stem of tRNA37 (Fig 3b). This Zn-finger also contains a loop formed by five intervening residues between the two Zn-coordinating histidines, and the loop and the two flanking helices interact with the anti-codon stem. The remarkable similarities in RNA binding between the two Zn-fingers support the notion that the Zn-finger of U1C interacts with and stabilizes the RNA duplex between the 5′-end of U1 snRNA and 5′-splice site observed in our crystal structure. The cross-linking between U1C and the 5′-splice site is consistent with the structure38.
The first and second nucleotides of the intron (the invariant GU dinucleotide) form Watson-Crick base-pairs with C8 and A7 of U1 snRNA, respectively (Fig 3a and 3d). An intriguing possibility is that the conserved residues in the loop region and helix A of U1C probe hydrogen-bonding groups of these two base-pairs in the minor groove and contribute to a discrimination of nucleotides at these two positions. In yeast the function of an otherwise essential DEAD box helicase, Prp28p, can be bypassed by mutations in U1 snRNA or U1C39. It is plausible that the binding of U1C to a correctly paired 5′-splice site and U1 snRNA could be coupled to an activation of a DEAD-box helicase within B complex to allow a transfer of the 5′-splice site from U1 to U6 snRNA40. Such a proofreading mechanism to discriminate incoming nucleotide substrates in the minor groove has been observed for both DNA and RNA polymerases41.
Implications for assembly of snRNPs
The crystal structure reported here represents the functional core of U1 snRNP, lacking the apical region of SL2. U1A consists of two RBDs linked by a proline-rich sequence, the N-terminal RBD binds to the ten-nucleotide loop of SL218,19. In order to complete the model of U1 snRNP, the crystal structure of the U1A-RNA complex19 was added onto an extended SL2 helix (Fig 5a). Interestingly, the internal loop in SL2 consisting of four conserved non-canonical base-pairs (Fig 1S) would be in a position to interact with B and D1 on the rim of the Sm ring (Fig 5a). This interaction could further stabilize the RNA structure onto the core domain. There is no known function for the C-terminal RBD of U1A nor for the highly similar C-terminal RBD of U2B”, a component of U2 snRNP18,42. The C-terminal RBDs of these proteins may bind to a common binding partner, possibly the Sm ring.
Figure 5. Model of the complete human U1 snRNP.
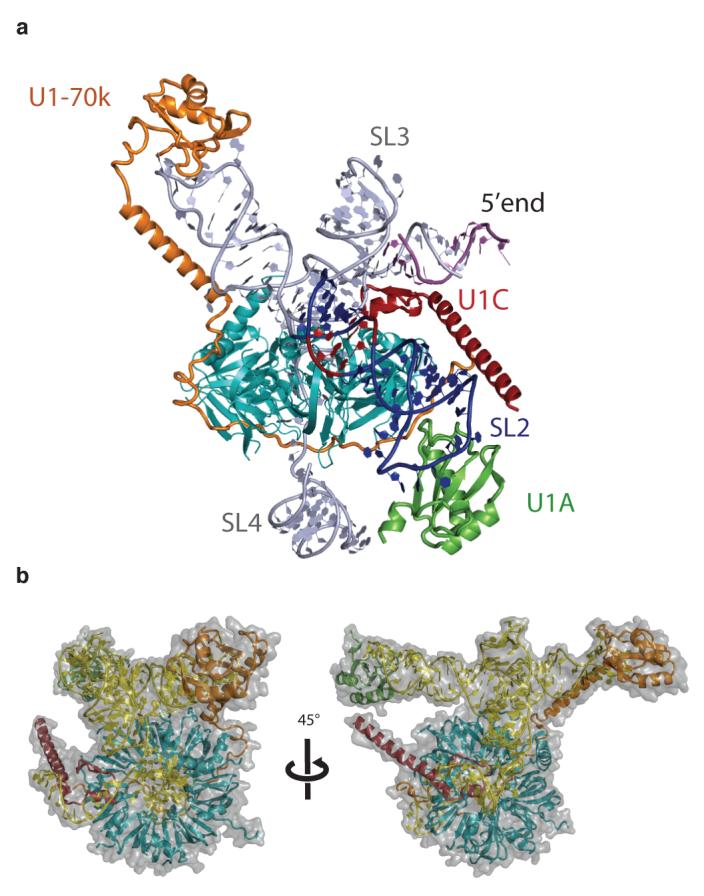
a, Overview of a model of the complete U1 snRNP. Truncated SL2 was extended with A-form RNA and the crystal structure of the U1A-RNA complex19 was appended to the extended helix. The internal loop of SL2, consisting of four consecutive non-Watson-Crick base-pairs (red), is in a position to interact with B and D1. b, Two views of the complete U1 snRNP model approximately 45° apart with surface representation superimposed. Closely matching images are found in the gallery of negative stain images of U1 snRNP reported previously23,24.
A cryo-EM structure of human U1 snRNP25 shows a ring-shaped core domain with two large protuberances that were previously evident in negatively-stained images of U1 snRNP23,24. One striking feature in the cryo-EM structure is the funnel-shaped central hole in the Sm ring. In the crystal structure, a large RNA structure that includes the four-helix junction is located directly above the centre of the Sm ring and no funnel-shaped hole is observed. No class averages are presented in the cryo-EM paper25 but a gallery of negatively-stained images of U1 snRNP23,24 provides different views of U1 snRNP. Closely matching images of these views (Fig 3 in ref. 23) can be generated by rotating our model of the complete U1 snRNP (Fig 5b).
The first crystal structure of a spliceosomal snRNP presented here provides insights into general principles of snRNP assembly. As observed in the U1 snRNP structure the Sm protein ring assembled on U2, U4 and U5 snRNAs is likely to function as a platform for further protein assembly1,3. Functionally important RNA sequences, which make up the catalytic centre of the spliceosome together with pre-mRNA substrates1-3, are located on the 5′-side of the Sm or Lsm protein binding site in all spliceosomal snRNAs1,3. In U1 snRNP the region of U1 snRNA that includes the four-helix junction and the 5′-end are stabilized in a particular orientation relative to the Sm ring by the N-terminal helices of D2 and B (Fig 2b and 2c). The N-terminal helix of D2, not ordered in the D1D2 heterodimer22, becomes ordered through its interaction with helix H of U1 snRNA. In U2, U4 and U5 snRNAs, different RNA structures flank the Sm site on the 5′-side1,3. Hence the N-terminal helices of D2 and B are likely to play a similar role in stabilizing different RNA structures in other spliceosomal snRNPs.
U1C predominantly interacts with D3 but the additional interaction with the N-terminus of U1-70k is crucial in promoting its binding to the core domain. Assembly of U1 snRNP therefore occurs in a hierarchical process, like the ribosome43. U1-70k facilitates the binding of U1C by providing an additional interaction surface for U1C. What is the function of the N-terminal polypeptide of U1-70k, which spans such a long distance? One possibility is that it functions as a harness to restrict the movement of SL1 with respect to the core domain thus stabilizing the overall structure of U1 snRNP. Other snRNP specific proteins may play an analogous role.
The structure presented here reveals the architecture of U1 snRNP, held together by an intricate network of RNA-protein and protein-protein interactions. It has also provided important insights into the mechanisms of 5′-splice site selection by U1 snRNP. During different stages of spliceosomal assembly, U1 snRNP interacts with other snRNPs either directly or through other proteins1-3. Our structure can now be utilized to study and provide an understanding of the interaction of U1 snRNP with constitutive and alternative splicing factors12,13,44, as well as other snRNPs45, during assembly of the spliceosome.
Summary Methods
Crystallisation of U1 snRNP
The preparation of U1 snRNP proteins, U1 snRNA, and the reconstitution and purification of U1 snRNP have been described previously19-21. The eluate of U1 snRNP from a monoQ column was concentrated to 5 mg/ml, mixed with an equal volume of a reservoir solution (MES-KOH pH 6.2-6.6; 300 mM KCl; 38-42% MPD) and equilibrated with the reservoir solution at 4°C by the hanging drop method. Crystals suitable for data collection were grown by streak seeding using a feline whisker at 4°C. Crystals were harvested and cryo-cooled directly from mother liquor.
Data Collection
Data used for structure determination were collected at Swiss Light Source beamlines X06SA and X10SA at 100 K on a mar225 CCD detector. For all crystals, datasets were obtained with the beam focused on the detector (defocused) and using inverse beam mode such that Friedel pairs could be collected with similar radiation damage.
Structural determination
Data processing, phasing and modelling are as described in supplementary methods (Table S1).
Author information
Coordinates of the protein Cα and the RNA phosphorus atoms have been deposited to the PDB database together with structure factors under accession code: 3CW1. Reprints and permissions information is available at www.nature.com/reprints. Correspondence and requests for materials should be addressed to K.N. (kn@mrc-lmb.cam.ac.uk).
Supplementary Material
Acknowledgements
We are grateful to Timm Jessen, Christian Kambach, Jo Avis, Robert Young, Yutaka Muto, Stefan Walke and Tijana Ignjatovic for expressing U1 snRNP proteins and laying the foundation of this project. We thank SLS, Daresbury and ESRF beamline staff, particularly Clemens Schulze-Briese, Takashi Tomizaki, Anuschka Pauluhn at SLS for their essential support; Venki Ramakrishnan, Andy Newman, Antonina Andreeva, Alexey Murzin, and Clemens Vonrhein for helpful discussions; the current members of the Nagai group for help and Soma Sengupta for support, Holger Stark for making the cryoEM structure available. This project has been funded by the MRC and HFSP. DAPK was a recipient of a HFSP long-term fellowship.
References
- 1.Burge CB, Tuschl T, Sharp PA. In: The RNA World II. Gesteland RR, Cech TR, Atkins JF, editors. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1999. pp. 525–560. [Google Scholar]
- 2.Will CL, Lührmann R. In: The RNA World. Gesteland RF, Cech TR, Atkins JF, editors. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2006. pp. 369–400. [Google Scholar]
- 3.Yu Y-T, Scharl EC, Smith CM, Steitz JA. In: The RNA World II. Gesteland RR, Cech TR, Atkins JF, editors. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1999. pp. 487–524. [Google Scholar]
- 4.Bringmann P, Lührmann R. Purification of the individual snRNPs U1, U2, U5 and U4/U6 from HeLa cells and characterization of their protein constituents. EMBO J. 1986;5:3509–3516. doi: 10.1002/j.1460-2075.1986.tb04676.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jurica MS, Moore MJ. Pre-mRNA splicing: awash in a sea of proteins. Mol. Cell. 2003;12:5–14. doi: 10.1016/s1097-2765(03)00270-3. [DOI] [PubMed] [Google Scholar]
- 6.Makarov EM, et al. Small nuclear ribonucleoprotein remodeling during catalytic activation of the spliceosome. Science. 2002;298:2205–2208. doi: 10.1126/science.1077783. [DOI] [PubMed] [Google Scholar]
- 7.Guthrie C, Patterson B. Spliceosomal snRNAs. Annu. Rev. Genet. 1988;22:387–419. doi: 10.1146/annurev.ge.22.120188.002131. [DOI] [PubMed] [Google Scholar]
- 8.Griffiths-Jones S, et al. Rfam: annotating non-coding RNAs in complete genomes. Nucleic Acids Res. 2005;33:D121–D124. doi: 10.1093/nar/gki081. (Database issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhuang Y, Weiner AM. A compensatory base change in U1 snRNA suppresses a 5′ splice site mutation. Cell. 1986;46:827–835. doi: 10.1016/0092-8674(86)90064-4. [DOI] [PubMed] [Google Scholar]
- 10.Ruby SW, Abelson J. An early hierarchic role of U1 small nuclear ribonucleoprotein in spliceosome assembly. Science. 1988;242:1028–1035. doi: 10.1126/science.2973660. [DOI] [PubMed] [Google Scholar]
- 11.Séraphin B, Rosbash M. Identification of functional U1 snRNA-pre-mRNA complexes committed to spliceosome assembly and splicing. Cell. 1989;59:349–358. doi: 10.1016/0092-8674(89)90296-1. [DOI] [PubMed] [Google Scholar]
- 12.Kohtz JD, Jamison SF, Will CL, Zuo P, Lührmann R, Garcia-Blanco MA, Manley JL. Protein-protein interactions and 5′-splice-site recognition in mammalian mRNA precursors. Nature. 1994;368:119–124. doi: 10.1038/368119a0. [DOI] [PubMed] [Google Scholar]
- 13.Graveley BR. Sorting out the complexity of SR protein functions. RNA. 2000;6:1197–1211. doi: 10.1017/s1355838200000960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Krol A, et al. Solution structure of human U1 snRNA. Derivation of a possible three-dimensional model. Nucleic Acids Res. 1990;18:3803–3811. doi: 10.1093/nar/18.13.3803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Patton JR, Pederson T. The Mr 70,000 protein of the U1 small nuclear ribonucleoprotein particle binds to the 5′ stem-loop of U1 RNA and interacts with Sm domain proteins. Proc. Natl. Acad. Sci. USA. 1988;85:747–751. doi: 10.1073/pnas.85.3.747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Query CC, Bentley RC, Keene JD. A common RNA recognition motif identified within a defined U1 RNA binding domain of the 70K U1 snRNP protein. Cell. 1989;57:89–101. doi: 10.1016/0092-8674(89)90175-x. [DOI] [PubMed] [Google Scholar]
- 17.Bach M, Krol A, Lührmann R. Structure-probing of U1 snRNPs gradually depleted of the U1-specific proteins A, C and 70k. Evidence that A interacts differentially with developmentally regulated mouse U1 snRNA variants. Nucleic Acids Res. 1990;18:449–457. doi: 10.1093/nar/18.3.449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Scherly D, Boelens W, Dathan NA, van Venrooij WJ, Mattaj IW. Major determinants of the specificity of interaction between small nuclear ribonucleoproteins U1A and U2B” and their cognate RNAs. Nature. 1990;345:502–506. doi: 10.1038/345502a0. [DOI] [PubMed] [Google Scholar]
- 19.Oubridge C, Ito N, Evans PR, Teo CH, Nagai K. Crystal structure at 1.92 Å resolution of the RNA-binding domain of the U1A spliceosomal protein complexed with an RNA hairpin. Nature. 1994;372:432–438. doi: 10.1038/372432a0. [DOI] [PubMed] [Google Scholar]
- 20.Nelissen RL, Will CL, van Venrooij WJ, Lührmann R. The association of the U1-specific 70K and C proteins with U1 snRNPs is mediated in part by common U snRNP proteins. EMBO J. 1994;13:4113–4125. doi: 10.1002/j.1460-2075.1994.tb06729.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Muto Y, et al. The structure and biochemical properties of the human spliceosomal protein U1C. J. Mol. Biol. 2004;341:185–198. doi: 10.1016/j.jmb.2004.04.078. [DOI] [PubMed] [Google Scholar]
- 22.Kambach C, et al. Crystal structures of two Sm protein complexes and their implications for the assembly of the spliceosomal snRNPs. Cell. 1999;96:375–387. doi: 10.1016/s0092-8674(00)80550-4. [DOI] [PubMed] [Google Scholar]
- 23.Kastner B, Lührmann R. Electron microscopy of U1 small nuclear ribonucleoprotein particles: shape of the particle and position of the 5′ RNA terminus. EMBO J. 1989;8:277–286. doi: 10.1002/j.1460-2075.1989.tb03374.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kastner B, Kornstädt U, Bach M, Lührmann R. Structure of the small nuclear RNP particle U1: identification of the two structural protuberances with RNP-antigens A and 70K. J. Cell Biol. 1992;116:839–849. doi: 10.1083/jcb.116.4.839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stark H, Dube P, Lührmann R, Kastner B. Arrangement of RNA and proteins in the spliceosomal U1 small nuclear ribonucleoprotein particle. Nature. 2001;409:539–542. doi: 10.1038/35054102. [DOI] [PubMed] [Google Scholar]
- 26.Clemons WM, et al. Structure of a bacterial 30S ribosomal subunit at 5.5 Å resolution. Nature. 1999;400:833–840. doi: 10.1038/23631. [DOI] [PubMed] [Google Scholar]
- 27.Ban N, et al. Placement of protein and RNA structures into a 5 Å-resolution map of the 50S ribosomal subunit. Nature. 1999;400:841–847. doi: 10.1038/23641. [DOI] [PubMed] [Google Scholar]
- 28.Heinrichs V, Bach M, Winkelmann G, Lührmann R. U1-specific protein C needed for efficient complex formation of U1 snRNP with a 5′ splice site. Science. 1990;247:69–72. doi: 10.1126/science.2136774. [DOI] [PubMed] [Google Scholar]
- 29.Will CL, et al. In vitro reconstitution of mammalian U1 snRNPs active in splicing: the U1-C protein enhances the formation of early (E) spliceosomal complexes. Nucleic Acids Res. 1996;24:4614–4623. doi: 10.1093/nar/24.23.4614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim CH, Tinoco I., Jr. A retroviral RNA kissing complex containing only two G-C base pairs. Proc. Natl. Acad. Sci. USA. 2000;97:9396–9401. doi: 10.1073/pnas.170283697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cowtan KD, Zhang KYJ, Main P. In: International Tables for Crystallography. Rossmann MG, Arnold E, editors. Volume F. Crystallography of Biological Macromolecules. Dordrecht: Kluwer Academic Publishers, The Netherlands; 2001. pp. 705–710. [Google Scholar]
- 32.Duckett DR, Murchie AIH, Lilley DMJ. The global folding of four-way helical junctions in RNA, including that in U1 snRNA. Cell. 1995;63:1027–1036. doi: 10.1016/0092-8674(95)90218-x. [DOI] [PubMed] [Google Scholar]
- 33.Törö I, et al. RNA binding in an Sm core domain: X-ray structure and functional analysis of an archaeal Sm protein complex. EMBO J. 2001;20:2293–2303. doi: 10.1093/emboj/20.9.2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Urlaub H, Raker VA, Kostka S, Lührmann R. Sm protein-Sm site RNA interactions within the inner ring of the spliceosomal snRNP core structure. EMBO J. 2001;20:187–196. doi: 10.1093/emboj/20.1.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lu D, Searles MA, Klug A. Crystal structure of a Zinc-finger-RNA complex reveals two modes of molecular recognition. Nature. 2003;426:96–100. doi: 10.1038/nature02088. [DOI] [PubMed] [Google Scholar]
- 36.Urlaub H, Hartmuth K, Kostka S, Grelle G, Lührmann R. A general approach for identification of RNA-protein crosslinking sites within native human spliceosomal small nuclear ribonucleoproteins (snRNPs) J. Biol. Chem. 2000;275:41458–41468. doi: 10.1074/jbc.M007434200. [DOI] [PubMed] [Google Scholar]
- 37.Zhou C, Huang RH. Crystallographic snapshots of eukaryotic dimethylallyltransferase acting on tRNA: Insight into tRNA recognition and reaction mechanism. Proc. Natl. Acad. Sci. USA. 2008;105:16142–16147. doi: 10.1073/pnas.0805680105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Du H, Rosbash M. The U1 snRNP protein U1C recognizes the 5′ splice site in the absence of base pairing. Nature. 2002;419:86–90. doi: 10.1038/nature00947. [DOI] [PubMed] [Google Scholar]
- 39.Chen JY-F, et al. Specific alterations of U1-C protein or U1 small nuclear RNA can eliminate the requirement of Prp28p, an essential DEAD box splicing factor. Mol. Cell. 2001;7:227–232. doi: 10.1016/s1097-2765(01)00170-8. [DOI] [PubMed] [Google Scholar]
- 40.Ismaïli N, Sha M, Gustafson EH, Konarska MM. The 100-kDa U5 snRNP protein (hPrp28p) contacts the 5′ splice site through its APRase site. RNA. 2001;7:182–193. doi: 10.1017/s1355838201001807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Steitz TA. Visualizing polynucleotide polymerase machines. EMBO J. 2006;25:3458–3468. doi: 10.1038/sj.emboj.7601211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Price SR, Evans PR, Nagai K. Crystal structure of the spliceosomal U2B”-U2A’ protein complex bound to a fragment of U2 small nuclear RNA. Nature. 1998;394:645–650. doi: 10.1038/29234. [DOI] [PubMed] [Google Scholar]
- 43.Mizushima S, Nomura M. Assembly mapping of 30S ribosomal proteins from E. coli. Nature. 1970;226:1214–1218. doi: 10.1038/2261214a0. [DOI] [PubMed] [Google Scholar]
- 44.Bedford MT, Reed R, Leder P. WW domain-mediated interactions reveal a spliceosome-associated protein that binds a third class of proline-rich motif: The proline, glycine and methionine-rich motif. Proc. Natl. Acad. Sci. USA. 1998;95:10602–10607. doi: 10.1073/pnas.95.18.10602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dönmez G, Hartmuth K, Kastner B, Will CL, Lührmann R. The 5′ end of U2 snRNA is in close proximity to U1 and functional sites of the pre-mRNA in early spliceosomal complexes. Mol. Cell. 2007;25:399–411. doi: 10.1016/j.molcel.2006.12.019. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.