Abstract
Pathogen-specific complement activation requires direct recognition of pathogens and/or the absence of complement control mechanisms on their surfaces. Antibodies direct complement activation to potential pathogens recognized by the cellular innate and adaptive immune systems. Similarly, the plasma proteins MBL and ficolins direct activation to microorganisms expressing common carbohydrate structures. The absence of complement control proteins permits amplification of complement by the alternative pathway on any unprotected surface. The importance of complement recognition molecules (MBL, ficolins, factor H, C3, C1q, properdin, and others) to human disease are becoming clear as analysis of genetic data and knock out animals reveals links between complement proteins and specific diseases.
Keywords: Complement, Antigens, Polysaccharide
Evolution of pathogen/host discrimination
Multicellular organisms developed interiors where cells could trade nutrients and develop specialized functions in a relatively protected space. Invasion of that space threatened survival of the species. Discrimination between the invaders and cells contributing to the collective became critical as did a means of killing or expelling the invaders. There is evidence for complement-like components in primitive coral and sponges [1,2], early multicellular organisms, suggesting that the discriminatory mechanisms of this system have had approximately a billion years to develop. Two of the three pathways of complement activation appear to have been functional long before adaptive immunity developed [3,4].
Complement activation pathways
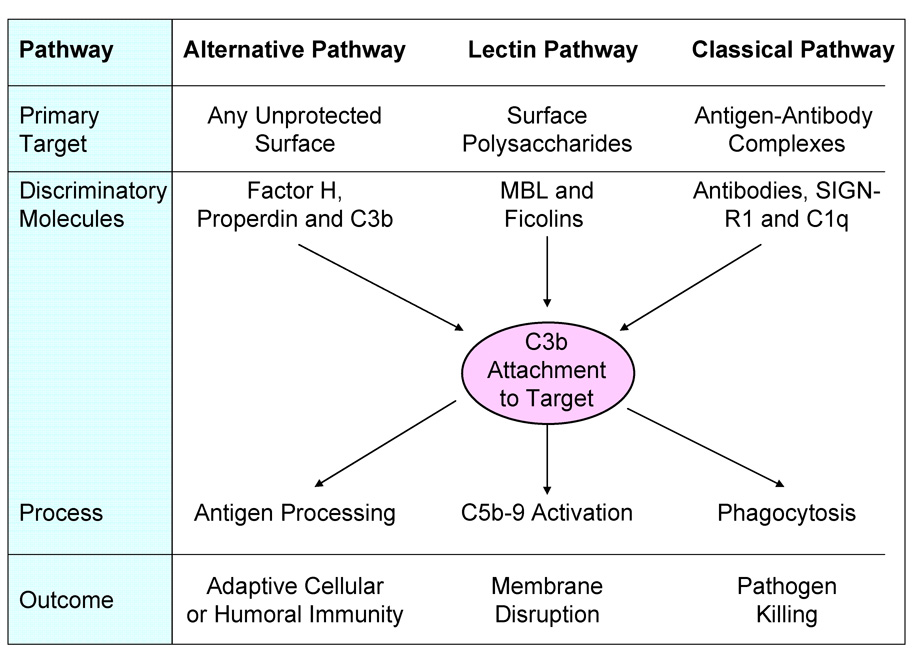
Three pathways of complement activation are known. Each uses its own unique mechanism for target versus host discrimination. All pathways result in covalent attachment of C3b to the target and each is capable of assembling pores in the bilipid layer of the cell being attacked. C3b tags the cell for killing by phagocytes and the pores allow water influx and metabolite efflux from the cell under attack. C3b also covalently tags antigens (opsonization) for processing by antigen presenting cells of the adaptive immune system. Figure 1 summarizes the major targets of each pathway, the discriminatory molecules of each and the different potential outcomes. These outcomes are beneficial when pathogens have been correctly identified by complement, but the same outcomes can all produce severe pathology if misdirected [5].
Figure 1.
Complement activation occurs through one or more of three distinct pathways. The primary targets of each pathway (i.e., the molecular structures that activate that pathway) are different. Each pathway employs a unique set of discriminatory molecules. Some, such as factor H of the alternative pathway, recognize markers on host cells to prevent activation on the host, but allow activation on most other surfaces. Other discriminatory molecules recognize target molecules using pattern recognition mechanisms (properdin, MBL, ficolins, C1q and SIGN-R1) capable of interacting with target surface molecules. Discriminatory specificity developed in the adaptive immune system and expressed in the form of antibodies is utilized by the classical pathway to identify targets for activation of this pathway. All activation pathways lead to C3b attachment to the target and the specificity of the thioester reactive site influences the rate and extent of the activation process. Finally, targets of complement activation undergo numerous processes all directed either at killing or at developing lasting immunity.
The alternative pathway primarily uses reverse recognition. It detects markers on host cells and activates on anything that lacks similar markers [6–8]. Factor H appears to be the primary host pattern recognition molecule and it recognizes host-associated molecular patterns (HAMPs), thought to be primarily surface polyanionic structures (Table 1). Recognition occurs through a number of binding sites on factor H. Mutations in and variants of factor H have been strongly linked to the apparently complement-mediated diseases atypical hemolytic uremic syndrome [9–11] and age-related macular degeneration [12–15]. The connection between altered amino acids in factor H and these diseases has been widely discussed, but the underlying molecular mechanisms remain poorly understood. Another pathogen recognition function, in this case direct recognition, was found just two years ago in another protein of the alternative pathway. Properdin was found to bind to and activate the alternative pathway on Neisseria as well as apoptotic and necrotic human cells [16–18].
Table 1.
Discriminatory molecules of complement and their targets
| Pathways | Discriminatory Molecules | Targets |
|---|---|---|
| Alternative Pathway | Factor H | Host cell surface polyanions (Specific structures undefined) |
| Properdin | Pattern recognition markers in pathogens GAGs?, DND?, Sugars? Sulfated glycoconjugates | |
| C3b | Cell surface carbohydrates (specificity well characterized) | |
| Lectin Pathway | MBL | Mannose-rich polysaccharides, hexoses, glucose, GlcNAc, fucose, etc. |
| L-ficolin | Pattern of acetyl groups, C-reactive protein, lipoteicoic acid | |
| H-ficolin | Pattern of acetyl groups | |
| M-ficolin | Pattern of acetyl groups, CRP, sialic acid | |
| Classical pathway | C1q | Antigen-antibody complexes, (Specificity of the Ab defines target), PAMPS, CRP, |
| Clq & SIGN-R1 | Largely undefined – pattern of mannose/fucose | |
| Clq & Pentraxins (CRP, etc.) | Phosphocholine, chromatin histones, pattern of anionic surface | |
Another unique feature of the alternative pathway is the C3b amplification process [19]. Attachment of small numbers of C3b to all surfaces in contact with blood occurs continuously and spontaneously. Once C3b is attached it forms an enzyme (C3 convertase) capable of activating many C3 molecules and depositing many additional C3b protein molecules on that surface. Each C3b may repeat this process and this amplification can result in rapid, covalent attachment of millions of C3b molecules to the surface in minutes. C3b attaches to proteins and carbohydrates and this opsonization process is important for trafficking and presentation of these molecules to the cells of the adaptive immune system. The specificity of the attachment site, as will be discussed below, may be a significant component in complement-mediated antigen selection and presentation.
The lectin pathway (Figure 1) uses members of the collectin family of proteins (MBL and L-, H-, and M-ficolins) to recognize pathogen-associated molecular patterns (PAMPs) such as polysaccharides on microbial surfaces [20,21]. Binding to their preferred polysaccharides on the surface of pathogens triggers activation of associated MASP proteases and these activate complement through the same protein activation cascade as the classical pathway [22,23]. The specificities of the lectin sites of these proteins have only begun to be characterized [21].
The classical pathway is initiated by the binding of C1q to antigen-antibody complexes (Figure 1), certain pentraxins, as well as some PAMPs. This complement system primarily depends upon the much more sophisticated discriminatory processes of the adaptive immune system to mount responses to pathogens and eliminate responses to autoantigens. Mistakes of this system (i.e. autoantibodies) elicit the same classical pathway response as antibodies to pathogens. The result is increased inflammation and tissue damage in antibody-mediated autoimmunity due to complement activation. Two other target recognition molecules in this pathway are C1q itself, through direct recognition of PAMPs, and the recently identified SIGN-R1 lectin [24] that mediates polysaccharide uptake in spleen (Table 1).
Molecular mechanisms of pathogen/host discrimination
Clearly any errors in target identification that result in complement activation on host cells or tissues can have devastating consequences to the host. Activation attaches the opsonins C3b/C3d on the surface, releases the inflammatory mediators C3a and C5a, and causes cell damage or death due to C5b-9 pores in the cell membrane. Understanding the molecular specificity of the discriminatory molecules is critical to understanding the mechanisms behind mistaken identification of the host and the pathology that follows.
Alternative pathway recognition molecules and their targets
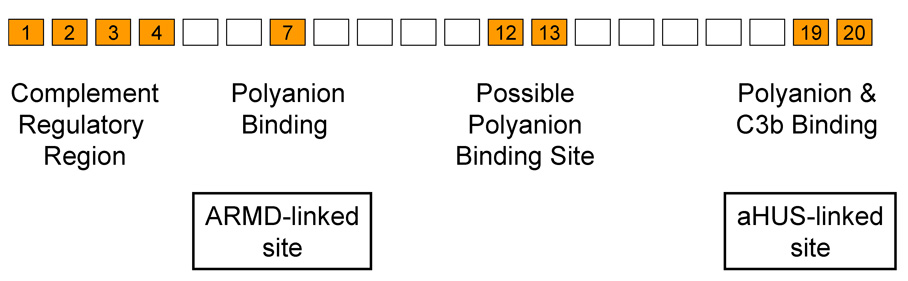
As shown in Table 1 the alternative pathway uses three different recognition molecules to identify the host or targets. Factor H is a protein composed of twenty small domains arranged like beads on a flexible string (Figure 2). The N-terminal four domains regulate the complement amplification system [25–27]. The other sixteen domains contain a variety of binding sites which control the functional effectiveness of domains 1–4 at the N-terminus. At least two sites interact with host cell surface markers, thought to be polyanions, and multiple C3b-binding sites interact with cell-surface deposited C3b/C3d [26–33], allowing factor H to effectively control the spontaneous activation of the alternative pathway on host surfaces. Complement-mediated pathology in the host can be triggered by the lack of proper recognition of markers on host cells due to mutations in the binding sites on factor H, as evidenced by inherited atypical hemolytic uremic syndrome (aHUS) [34,35] and age-related macular degeneration (ARMD) [35]. The specific chemical nature of host markers has yet to be reported, but both regions (domain 7 and 19–20, Figure 2) are thought to interact with host polyanionic molecules, such as sialic acids and glycosoaminoglycans. Complement activating organisms, e.g. yeast, coated with heparin cease to activate human complement [36]. On the other hand, sheep cells, which do not activate human complement, activate the alternative pathway after surface sialic acid is removed [37,38]. Thus, although the human counterparts of these polyanions have never been identified they are assumed to be polyanionic in nature. The primary difficulty in identification appears to be the low affinity between individual markers and factor H. In vivo factor H may utilize both surface-bound C3b and multiple host specific contacts to identify host [39,40]. On surfaces lacking markers recognized by factor H (bacteria, fungi, agarose, glomerular basement membrane, etc.) alternative pathway-mediated C3b amplification proceeds unabated due to the ineffectiveness of factor H on such surfaces.
Figure 2.
Host/target discriminatory sites on factor H. While the N-terminal four domains express all of the complement regulatory functions of factor H, this activity is controlled by other polyanion and C3b binding sites localized to domains 7, 12–13 and 19–20, as indicated. Interactions between these sites and host cell surface polyanion markers increases the effectiveness of the complement regulating domains 1–4 thus preventing inappropriate complement activation on host cells and tissues. Microorganisms lacking molecules recognized by factor H cannot control activation of the alternative complement pathway on their surface.
Properdin is one of the six proteins involved in activation of the alternative pathway. It stabilizes the central enzyme in the amplification process and thus accelerates activation [41]. Although first identified in 1959 [42], it was not until 2007 [17] that it was shown to be a pattern recognition molecule capable of binding to Neisseria gonorreae, necrotic human cells, and fungi and initiating activation of the alternative pathway [16–18]. People with properdin deficiencies have recurrent episodes of meningococcal [43–45] infections suggesting that recognition and elimination of these organisms are uniquely dependent on properdin. Certain molecular structures recognized by properdin have been proposed, such as cell surface GAGs, DNA, as well as non-sulfated glycoconjugates [16,18,46].
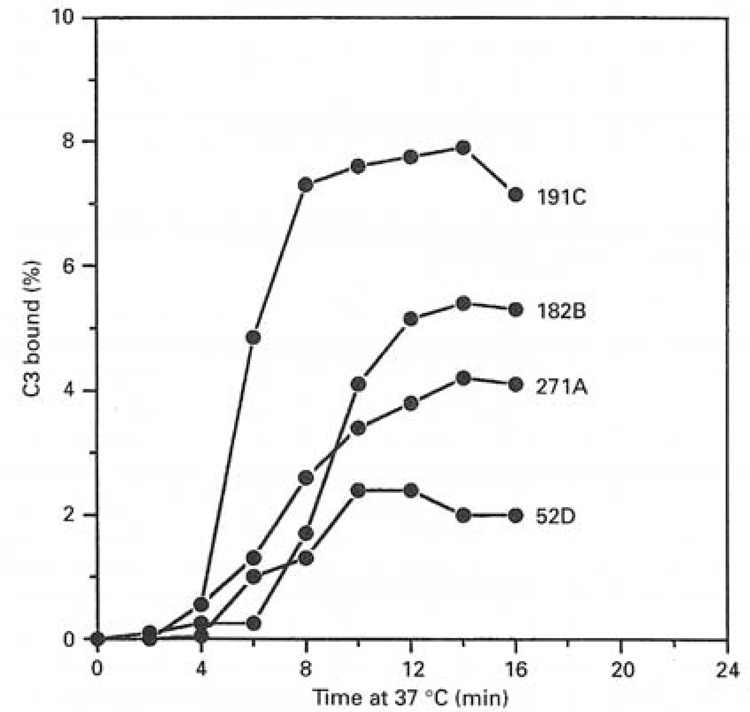
C3b attaches covalently to targets being attacked by all three pathways of complement as illustrated in Figure 1. This critical step in complement activation is not normally thought of as a target recognition event, however, C3b exhibits strong preferences (Table 2) among different sugars (carbohydrates are its preferred attachment site on most targets) and an even stronger preference for amino acid hydroxyl groups (Thr, Ser and Tyr) when they are available [47–51]. Attachment to xylose is preferred 12-fold over attachment to inositol and two fold over attachment to glucose (Table 2). Even preferences for particular hydroxyl positions on sugars have been demonstrated [49]. Attachment to the phenyl hydroxyl group of tyrosine is preferred 50-fold over glucose and the ester link to amino acids is more stable than that to sugars. In one case it was shown (Figure 3) that a series of yeast mutants capable of synthesizing surface polysaccharides terminating with one, two, three or four xylose sugars (the most highly preferred sugar for C3b attachment) exhibited a 4-fold difference in C3b attachment efficiency [49]. The rate and extent of complement activation by these yeast also reflected this quantitative difference suggesting that the specificity measured with sugars is relevant to complement activation [49].
Table 2.
Specificity of C3b Attachment
| Compound | Relative Reactivity* |
|---|---|
| Water | 1 |
| Inositol | 56 |
| Fucose | 179 |
| Glucose | 336 |
| Galactose | 360 |
| Serine | 366 |
| NAc-Glucosamine | 462 |
| Mannose | 491 |
| Xylose | 674 |
| Threonice | 1,575 |
| Tyrosine | 17,273 |
The reactive thioester of activated C3 (nascent C3b) covalently reacts with hydoxyl groups in the compounds listed above. Relative reactivcities were calculated from the IC50 concentrations. Note that amino acids exhibited similar reactivities after incorporation into peptides.
Figure 3.
The specificity of the reactive thioester site of C3 for carbohydrates can determine the rate and extent of complement activation. Different strains of yeast (Cryptococcus neoformans) capable of attaching different numbers of xylose residues per unit of surface polysaccharide exhibit different rates and extents of complement activation [49]. The serotypes attach one (52D), two (271A), three (182B) or four (191C) xylose terminal residues to surface oligosaccharides during growth. The figure shows deposition of radiolabeled C3b on the organisms during activation of the alternative pathway revealing a 2-fold difference in rate of activation and a 4-fold difference in the level of opsonization by C3b. Figure from [49] with permission.
Antigen-processing cells of the adaptive immune system utilize C3b/C3d-tagged antigens as their preferred antigens, through receptors CD21 and the BCR [52,53], especially during the initial low antigen concentration phase of infections. This suggests that the chemical reactivity of C3b for particular sites on an antigen may be an important step governing antigen selection. It may also be important in regulating the pathway of cellular processing (Th1 versus Th2) [54]. The recent discovery of a completely C3 deficient child with severely impaired B-cell, dendritic cell and T-cell responses supports this conclusion [55]. The fact that the ability of activated C3 to attach to invaders occurred early in evolution suggests that the mechanism of invader identification and tagging by C3 might be an important factor in the selection of antigens. It seems reasonable to expect that the selection of specific structures on target surface molecules by C3b, and the covalent tag it attaches, will influence what polysaccharides and protein fragments are utilized for processing by antigen-presenting cells.
All three pathways of complement activation deposit C3b on their respective targets. If that surface lacks host markers or membrane-bound complement regulatory proteins (i.e., DAF, MCP, CR1, CD59) the alternative pathway will amplify the number of C3b on that surface. This system is capable of depositing millions of C3b protein molecules on a strong activator within five to 15 minutes of first contact with host blood. A single cell will elicit such a response suggesting that during early, low antigen dosage responses the molecules identified and tagged by C3b may have a disproportionate influence on what antigens get processed.
Lectin pathway target recognition molecules and their ligands
Recognition of targets by the lectin pathway is more straightforward than target recognition by the alternative pathway. The four known lectin-like proteins that initiate this pathway appear to be traditional pattern recognition molecules specific mainly for different carbohydrates prevalent on microorganisms. Due to the complexity of polysaccharides on microorganisms and the fact that each lectin molecule possesses up to 18 binding sites, it is difficult to define an exact specificity for these proteins. Nevertheless, it has become clear that MBL has a strong preference for polysaccharides with terminal mannose and glucose residues, and more specifically for sugars with 3- and 4-OH groups placed in the equatorial plane of the sugar ring structure [56]. L-ficolin has been well characterized using an array of 279 different glycans [21]. Although the structural preferences are clearly complex, the common feature was the presence of one or more glycans with N-acetylated glucosamine. H-ficolin [57] and M-ficolin [58] are both inhibited by N- acetylated sugars, but their fine specificity has not been well characterized. One unanswered question is why human N-acetylated glycans (primarily terminal sialic acids) are not recognized by the ficolins. Most likely the ficolins rely on complex ligands and the multiplicity of binding sites on each arm of their structure to discriminate host from microorganism. Due to the broad specificity exhibited by the ficolins it seems likely that two or more of them will bind to any particular microorganism. It is not yet clear how much cooperation specific pathogens elicit from the four complement activating lectins.
Classical pathway target recognition molecules and their ligands
The primary target recognition mechanism of the classical pathway is easy to describe. IgM or clusters of IgG bound to their antigens activate this pathway of complement. The specificity expressed by the antibody is determined by the B-cell maturation process. C1q is also known to bind directly to PAMPs on pathogenic targets, such as envelope proteins of certain retroviruses, lipopolysaccharides (LPS), and porins from Gram-negative bacteria [59–62]. In addition, C1 can be activated by binding to pentraxins such as C reactive protein and serum amyloid P [63–65], or by binding to SIGN-R1, a pattern recognition receptor located on spleen marginal zone macrophages. SIGN-R1 has been shown to bind and activate C1 upon binding to its preferred carbohydrate structures on pathogens [24]. C1 activated by SIGN-R1 has been shown to activate complement through the classical pathway.
Complement regulatory proteins
There are many mechanisms by which inadvertent damage to the host occurs following complement activation. All activation in vivo can damage host cells whether that activation occurs by immune complexes, on microorganisms, or due to the spontaneous activation of the alternative pathway. Host cells express a variety of membrane-bound control proteins to limit damage by both the activation process and the membrane attack complex. Damage can occur from C3b attachment on host cells and tissues or due to “reactive lysis” by C5b-9.
In order for the host to survive a robust and hopefully lethal complement attack on pathogens, host cells express membrane-bound complement regulators on their surfaces (DAF, MCP, CR1, CD59, and CRIg) or express markers that attract the soluble regulators, factor H and C4 binding protein. DAF (CD55), MCP (CD46), CR1 (CD35) and CRIg all act by inactivating complement enzymes or by inactivating C3b, the essential component of those enzymes. CRIg molecules are membrane-attached immunoglobulin-like IgV domains that inhibit the alternative pathway C3/C5 convertase [66]. CD59 acts differently in protecting host cells. If all the other regulators fail to stop complement enzyme formation on host cells and C5 is activated then the membrane attack complex is formed. CD59 limits host cell damage by binding to C5b-8 or C5b-9 and preventing leakage or lysis of the host cell membrane [67,68].
Host cell defenses can be overwhelmed by aggressive activation such as occurs in antibody-mediated autoimmune diseases. In myasthenia gravis nerve damage is limited or nonexistent if the animal is complement deficient or if the complement system is inactivated [69]. Even if antibodies are passively transferred, the new host suffers little or no pathology so long as the complement system is inactive. Under normal conditions, however, constant immune complex formation results in constant complement activation which eventually overwhelms host defenses. Another situation where host defenses can be overwhelmed is when pathogens undergoing aggressive complement attack are tightly bound to host cells by Toll-like receptors, SIGN-R1, or by receptors (CR1, CR2, CR3, and CRIg) that bind to C4b or C3b attached to the target during the complement activation process. Inadvertent transfer of C3b (which can initiate alternative pathway activation) or C5b,6 to the host cell surface from the surface of the activating pathogen must be controlled by the host cell’s defenses or the host cell will be killed.
The in vivo lifespan of normal human red cells is between 60 to 90 days, however abnormal cells that lack complement regulatory molecules DAF and CD59 (PNH type II and III cells) are lysed in 6-60 days [70,71] due to low level complement activation on their surface [70,72,73]. These cells are not lysed in a shorter period of time due to cell surface protection provided by factor H [74]. The only approved complement regulatory drug, the humanized antibody eculizumab from Alexion, was approved for this disease and it acts by binding to C5 and preventing formation of the membrane attack complex. This disease illustrates the point that pathology can result from either a lack of membrane control factors or from complement activation that overwhelms those factors on host cells as a result of continuous and indiscriminate attack. The aggressive nature of this attack is fundamental to how the alternative pathway defends the host from potential pathogens.
Failures leading to complement attack on host cells and tissues
The three main causes of complement-mediated pathology are 1) decreased host defense against complement activation (e.g., PNH, aHUS, and age related macular degeneration), 2) exposure of cells or tissues that activate the host’s own complement system (e.g., ischemia/reperfusion, burns, apoptotic/necrotic cells), and 3) massive or continuous complement activation that overwhelms normally adequate defenses against complement-mediated damage (e.g., Ab-mediated autoimmunity, immune complex disease, sepsis).
As described in the preceding section, the loss of complement regulators DAF and CD59 on host red cells leads to complement-mediated pathology in the disease PNH. A somatic mutation in bone marrow stem cells prevents the attachment of membrane anchors on DAF and CD59 [72,73] and yields red cells and platelets lacking these protective proteins. The alternative pathway attacks every surface in contact with blood, including red cells. Cell markers recognized by factor H prevent uncontrolled activation [74], but membrane-bound regulators must be present or the complement C3/C5 convertases that escape control by factor H slowly inflict enough damage to lyse the PNH cells.
The inherited form of aHUS is associated with mutations in the C-terminus of factor H (Figure 2) that affect the ability of factor H to recognize host markers [75]. Reduced control of the alternative pathway leads to damage to many tissues, but damage to the kidney is the most rapid and destructive probably due to exposure of the glomerular basement membrane and subsequent aggressive complement attack on this exposed surface, which lacks membrane-bound complement regulatory proteins and relies mainly on factor H for its protection [76]. It has become clear that mutations in other alternative pathway components (factor I, factor B, C3 and CD46/MCP) can also lead to aHUS or can exacerbate or moderate effects of mutations in factor H [77].
Age-related macular degeneration appears to be another example of a disease caused by decreased host defense, although the exact mechanism has yet to be described in molecular detail. ARMD is strongly linked to a single amino acid variant in the seventh domain of factor H (Figure 2) [12,14,15,78]. The effects of mutations in factor H are exacerbated or moderated by mutations in other complement proteins that either intensify complement attacks, slow complement activation, or alter other control proteins. Thus, the genetic complexity of ARMD is much greater than initially thought, but the loss of function and gain of function variants found in multiple complement proteins [79,80] correlating with this disease seem to have straightforward consequences for the progression of this disease. Variants that have been shown to accelerate alternative pathway activation exacerbate pathology while those that moderate activation suppress pathology [81,82].
Another mechanism of failed discrimination is activation of host complement on host tissues exposed due to damage or other pathological events. Altered cell surfaces appear during ischemia and necrosis of cellular tissues. These abnormal surfaces activate one or more of the complement pathways [83–85]. This activation may play a necessary role in clearing damaged tissue after ischemia, following burns or after normal apoptosis. This suggests a reason for evolution to have preserved this function, but damage to bystander normal tissues also occurs [41,86]. In the case of ischemia/reperfusion a significant amount of tissue loss can be prevented by inactivating complement prior to reperfusion [83–85]. This implies that while there is a role for complement in clearing damaged tissue, complement may sometimes be overly aggressive in this process causing loss of tissue that could otherwise recover from the event. Damage to the epithelial layer in the kidney exposes the glomerular basement membrane which has limited ability to protect itself from complement activation. Membranoproliferative glomerulonephritis (MPGN) Type II is accompanied by massive complement activation via the alternative pathway and this can lead to complete organ failure [76]. It is not clear that any positive role is being played by complement activation, suggesting that this is truly a lapse in host discrimination by complement.
The destructive capacity of complement can overwhelm any cellular defenses if activation is rapid or persistent. Most antibody-mediated autoimmune diseases lead to activation of the classical pathway on targets identified by the autoantibodies. Cells with normal levels of the regulatory proteins DAF, MCP, and CD59 defend themselves against immediate lysis, but the attack persists as long as more antigen is expressed, more antibody is available, and complement levels remain sufficient to maintain the attack. Examples of antibody-mediated diseases include myasthenia gravis and rheumatoid arthritis (RA) [87,88]. In RA there appears to be a strong dependence on the alternative pathway. Mice deficient in factor B are resistant to induction of pathology either by immunization with collagen II or by passive transfer of antibodies [89]. Similarly, specific inhibition of the alternative pathway by soluble CRIg not only prevented disease induction, but blocked progression of established rheumatoid arthritis in a mouse model [90].
Bystander lysis (reactive lysis) occurs if a host cell is bound to or is even close to a pathogen or immune complex where the surface is releasing activated complement proteins [86,91–93]. Damage occurs if the C5b,6 complex produced by C5 convertases on the activator diffuse to the host cell surface and insert into the cell membrane after combining with C7. The inserted C5b-7 binds C8 and C9 spontaneously causing damage to the host cell. Host CD59 binds to these complexes on a 1-to-1 basis preventing membrane damage, but if the number of C5b-9 exceeds the amount of CD59 in the host cell then apoptosis and cell death may result.
Failures of pathogen recognition
This volume contains a number of reviews that describe in detail the mechanisms by which pathogens evade or utilize activation of complement (see reviews by A. Blom and S. Ram, J. Ngampasutadol, et al., P.F. Zipfel, et al., M.A. Oliver, et al., P. Avirutnan, et. al., M. Hostetter, J.A. Welsch and S. Ram, E. Kugelberg, et al., S. Meri, et al., M. Pizza, and S. Meri, et al.). The three general strategies for causing complement failure are: defensive, offensive and invisibility. Defensive approaches involve binding host control factors or expressing complement regulating factors on the surface of the pathogen. Offensive strategies involve disabling complement components or products or utilizing them to aid invasion. A cloak of invisibility may be provided by both offensive and defensive strategies or it may be achieved by using complement to gain access to the intracellular space and disappear.
Conclusions
Each pathway of complement activation utilizes specific discriminatory molecules to distinguish host from target. Each pathway utilizes different markers on host or targets making for robust pathogen recognition. Surprisingly, little is known about the specificities of many discriminatory molecules and, similarly, little is known about the target molecules. C3b-tagged antigens feed into the adaptive immune system and it seems, therefore, to be of value to understand how C3b selects antigenic sites for enhanced processing. The identification of markers recognized by complement components on host cells and pathogens, as well as the mechanisms used by the pathogens to protect themselves from complement, will greatly contribute to the development of therapies and vaccines for human diseases.
Acknowledgements
This work was supported by National Institutes of Health Grant DK-35081 and American Heart Association National Scientist Development Grant 0735101N.
Footnotes
Disclosures
Dr. M.K. Pangburn is an officer of and has a financial interest in Complement Technology, Inc. (www.ComplementTech.com) a supplier of complement reagents.
References
- 1.Dishaw LJ, Smith SL, Bigger CH. Characterization of a C3-like cDNA in a coral: phylogenetic implications. Immunogenetics. 2005;57:535–548. doi: 10.1007/s00251-005-0005-1. [DOI] [PubMed] [Google Scholar]
- 2.Vasta GR, Lambris JD. Innate immunity in the Aegean: ancient pathways for today's survival. Dev Comp Immunol. 2002;26:217–225. doi: 10.1016/s0145-305x(01)00072-6. [DOI] [PubMed] [Google Scholar]
- 3.Farries TC, Atkinson JP. Evolution of the complement system. Immunol Today. 1991;12:295–300. doi: 10.1016/0167-5699(91)90002-B. [DOI] [PubMed] [Google Scholar]
- 4.Pinto MR, Melillo D, Giacomelli S, Sfyroera G, Lambris JD. Ancient origin of the complement system: emerging invertebrate models. Adv Exp Med Biol. 2007;598:372–388. doi: 10.1007/978-0-387-71767-8_26. [DOI] [PubMed] [Google Scholar]
- 5.Thurman JM, Holers VM. The central role of the alternative complement pathway in human disease. J Immunol. 2006;176:1305–1310. doi: 10.4049/jimmunol.176.3.1305. [DOI] [PubMed] [Google Scholar]
- 6.Joiner KA. Complement evasion by bacteria and parasites. Ann Rev Microbiol. 1988;42:201–230. doi: 10.1146/annurev.mi.42.100188.001221. [DOI] [PubMed] [Google Scholar]
- 7.Pangburn MK, Müller-Eberhard HJ. The alternative pathway of complement. Springer Seminars in Immunopathology. 1984;7:163–192. doi: 10.1007/BF01893019. [DOI] [PubMed] [Google Scholar]
- 8.Pangburn MK. A fluorimetric assay for native C3. The hemolytically active form of the third component of human complement. J Immunol Methods. 1987;102:7–14. doi: 10.1016/s0022-1759(87)80003-0. [DOI] [PubMed] [Google Scholar]
- 9.Dragon-Durey MA, Fremeaux-Bacchi V, Loirat C, Blouin J, Niaudet P, Deschenes G, et al. Heterozygous and homozygous factor h deficiencies associated with hemolytic uremic syndrome or membranoproliferative glomerulonephritis: report and genetic analysis of 16 cases. J Am Soc Nephrol. 2004;15:787–795. doi: 10.1097/01.asn.0000115702.28859.a7. [DOI] [PubMed] [Google Scholar]
- 10.Perez-Caballero D, Gonzalez-Rubio C, Gallardo ME, Vera M, Lopez-Trascasa M, Rodriguez de Cordoba S, et al. Clustering of missense mutations in the C-terminal region of factor H in atypical hemolytic uremic syndrome. Am J Hum Genet. 2001;68:478–484. doi: 10.1086/318201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Warwicker P, Goodship TH, Donne RL, Pirson Y, Nicholls A, Ward RM, et al. Genetic studies into inherited and sporadic hemolytic uremic syndrome. Kidney Int. 1998;53:836–844. doi: 10.1111/j.1523-1755.1998.00824.x. [DOI] [PubMed] [Google Scholar]
- 12.Edwards AO, Ritter RI, Abel KJ, Manning A, Panhuysen C, Farrer LA. Complement factor H polymorphism and age-related macular degeneration. Science. 2005;308:421–424. doi: 10.1126/science.1110189. [DOI] [PubMed] [Google Scholar]
- 13.Hageman GS, Anderson DH, Johnson LV, Hancox LS, Taiber AJ, Hardisty LI, et al. A common haplotype in the complement regulatory gene factor H (HF1/CFH) predisposes individuals to age-related macular degeneration. Proc Natl Acad Sci USA. 2005;102:7227–7232. doi: 10.1073/pnas.0501536102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haines JL, Hauser MA, Schmidt S, Scott WK, Olson LM, Gallins P, et al. Complement factor H variant increases the risk of age-related macular degeneration. Science. 2005;308:419–421. doi: 10.1126/science.1110359. [DOI] [PubMed] [Google Scholar]
- 15.Klein RJ, Zeiss C, Chew EY, Tsai J-Y, Sackler RS, Haynes C, et al. Complement factor H polymorphism in age-related macular degeneration. Science. 2005;308:385–389. doi: 10.1126/science.1109557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kemper C, Mitchell LM, Zhang L, Hourcade DE. The complement protein properdin binds apoptotic T cells and promotes complement activation and phagocytosis. Proc Natl Acad Sci U S A. 2008;105:9023–9028. doi: 10.1073/pnas.0801015105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Spitzer D, Mitchell LM, Atkinson JP, Hourcade DE. Properdin can initiate complement activation by binding specific target surfaces and providing a platform for de novo convertase assembly. J Immunol. 2007;179:2600–2608. doi: 10.4049/jimmunol.179.4.2600. [DOI] [PubMed] [Google Scholar]
- 18.Xu W, Berger SP, Trouw LA, de Boer HC, Schlagwein N, Mutsaers C, et al. Properdin binds to late apoptotic and necrotic cells independently of C3b and regulates alternative pathway complement activation. J Immunol. 2008;180:7613–7621. doi: 10.4049/jimmunol.180.11.7613. [DOI] [PubMed] [Google Scholar]
- 19.Gotze O, Müller-Eberhard HJ. The alternative pathway of complement activation. Advances in Immunology. 1976;24:1–35. doi: 10.1016/s0065-2776(08)60328-4. [DOI] [PubMed] [Google Scholar]
- 20.Thiel S. Complement activating soluble pattern recognition molecules with collagen-like regions, mannan-binding lectin, ficolins and associated proteins. Mol Immunol. 2007;44:3875–3888. doi: 10.1016/j.molimm.2007.06.005. [DOI] [PubMed] [Google Scholar]
- 21.Krarup A, Mitchell DA, Sim RB. Recognition of acetylated oligosaccharides by human L-ficolin. Immunol Lett. 2008;118:152–156. doi: 10.1016/j.imlet.2008.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Endo Y, Takahashi M, Fujita T. Lectin complement system and pattern recognition. Immunobiology. 2006;211:283–293. doi: 10.1016/j.imbio.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 23.Takahashi M, Mori S, Shigeta S, Fujita T. Role of MBL-associated serine protease (MASP) on activation of the lectin complement pathway. Adv Exp Med Biol. 2007;598:93–104. doi: 10.1007/978-0-387-71767-8_8. [DOI] [PubMed] [Google Scholar]
- 24.Kang YS, Do Y, Lee HK, Park SH, Cheong C, Lynch RM, et al. A dominant complement fixation pathway for pneumococcal polysaccharides initiated by SIGN-R1 interacting with C1q. Cell. 2006;125:47–58. doi: 10.1016/j.cell.2006.01.046. [DOI] [PubMed] [Google Scholar]
- 25.Alsenz J, Lambris JD, Schulz TF, Dierich MP. Localization of the complement component C3b binding site and the cofactor activity for factor I in the 38 kDa tryptic fragment of factor H. Biochem J. 1984;224:389–398. doi: 10.1042/bj2240389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kuhn S, Zipfel PF. Mapping of the domains required for decay acceleration activity of the human factor H-like protein 1 and factor H. Eur J Immunol. 1996;26:2383–2387. doi: 10.1002/eji.1830261017. [DOI] [PubMed] [Google Scholar]
- 27.Gordon DL, Kaufman RM, Blackmore TK, Kwong J, Lublin DM. Identification of complement regulatory domains in human factor H. J Immunol. 1995;155:348–356. [PubMed] [Google Scholar]
- 28.Kühn S, Skerka C, Zipfel PF. Mapping of the complement regulatory domains in the human factor H-like protein 1 and in factor H. J Immunol. 1995;155:5663–5670. [PubMed] [Google Scholar]
- 29.Sharma AK, Pangburn MK. Identification of three physically and functionally distinct binding sites for C3b in human complement factor H by deletion mutagenesis. Proc Natl Acad Sci USA. 1996;93:10996–11001. doi: 10.1073/pnas.93.20.10996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ram S, Sharma AK, Simpson SD, Gulati S, McQuillen DP, Pangburn MK, et al. A novel sialic acid binding site on factor H mediates serum resistance of sialylated Neisseria gonorrhoeae. J Exp Med. 1998;187:743–752. doi: 10.1084/jem.187.5.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Blackmore TK, Hellwage J, Sadlon TA, Higgs N, Zipfel PF, Ward HM, et al. Identification of the second heparin-binding domain in human complement factor H. J Immunol. 1998;160:3342–3348. [PubMed] [Google Scholar]
- 32.Prosser BE, Johnson S, Roversi P, Herbert AP, Blaum BS, Tyrrell J, et al. Structural basis for complement factor H linked age-related macular degeneration. J Exp Med. 2007;204:2277–2283. doi: 10.1084/jem.20071069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Herbert AP, Deakin JA, Schmidt CQ, Blaum BS, Egan C, Ferreira VP, et al. Structure shows that a glycosaminoglycan and protein recognition site in factor H is perturbed by age-related macular degeneration-linked single nucleotide polymorphism. J Biol Chem. 2007;282:18960–18968. doi: 10.1074/jbc.M609636200. [DOI] [PubMed] [Google Scholar]
- 34.de Cordoba Sr, de Jorge EG. Translational mini-review series on complement factor H: genetics and disease associations of human complement factor H. Clin Exp Immunol. 2008;151:1–13. doi: 10.1111/j.1365-2249.2007.03552.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Holers VM. The spectrum of complement alternative pathway-mediated diseases. Immunol Rev. 2008;223:300–316. doi: 10.1111/j.1600-065X.2008.00641.x. [DOI] [PubMed] [Google Scholar]
- 36.Kazatchkine MD, Fearon DT, Silbert JE, Austen KF. Surface-associated heparin inhibits zymosan-induced activation of the human alternative pathway by augmenting the regulatory action of the control proteins on particle-bound C3b. J Exp Med. 1979;150:1202–1215. doi: 10.1084/jem.150.5.1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fearon DT. Regulation by membrane sialic acid of βIH-dependent decay-dissociation of amplification C3 convertase of the alternative complement pathway. Proc Natl Acad Sci USA. 1978;75:1971–1975. doi: 10.1073/pnas.75.4.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pangburn MK, Müller-Eberhard HJ. Complement C3 convertase: cell surface restriction of βIH control and generation of restriction on neuraminidase treated cells. Proc Natl Acad Sci USA. 1978;75:2416–2420. doi: 10.1073/pnas.75.5.2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jokiranta TS, Zipfel PF, Hakulinen J, Kuhn S, Pangburn MK, Tamerius JD, et al. Analysis of the recognition mechanism of the alternative pathway of complement by monoclonal anti-factor H antibodies: evidence for multiple interactions between H and surface bound C3b. FEBS Lett. 1996;393:297–302. doi: 10.1016/0014-5793(96)00905-2. [DOI] [PubMed] [Google Scholar]
- 40.Pangburn MK, Pangburn KLW, Koistinen V, Meri S, Sharma AK. Molecular mechanisms of target recognition in an innate immune system: interactions among factor H, C3b and target in the alternative pathway of human complement. J Immunol. 2000;164:4742–4751. doi: 10.4049/jimmunol.164.9.4742. [DOI] [PubMed] [Google Scholar]
- 41.Müller-Eberhard HJ. Molecular organization and function of the complement system. Ann Rev Biochem. 1988;57:321–347. doi: 10.1146/annurev.bi.57.070188.001541. [DOI] [PubMed] [Google Scholar]
- 42.Lepow IH, Pillemer L, Schoenberg MD, Todd EW, Wedgwood RJ. The properdin system and immunity. X. Characterization of partially purified human properdin. J Immunol. 1959;83:428–436. [PubMed] [Google Scholar]
- 43.Sjoholm AG, Braconier JH, Soderstrom C. Properdin deficiency in a family with fulminant meningococcal infections. Clin Exp Immunol. 1982;50:291–297. [PMC free article] [PubMed] [Google Scholar]
- 44.Cunliffe NA, Snowden N, Dunbar EM, Haeney MR. Recurrent meningococcal septicaemia and properdin deficiency. J Infect. 1995;31:67–68. doi: 10.1016/s0163-4453(95)91550-8. [DOI] [PubMed] [Google Scholar]
- 45.Braconier JH, Sjoholm AG, Soderstrom C. Fulminant meningococcal infections in a family with inherited deficiency of properdin. Scand J Infect Dis. 1983;15:339–345. doi: 10.3109/inf.1983.15.issue-4.04. [DOI] [PubMed] [Google Scholar]
- 46.Holt GD, Pangburn MK, Ginsgurg V. Properdin binds to sulfatide [Gal(3-SO4)beta1-1Cer] and has a sequence homology with other proteins that bind sulfated glycoconjugates. J Biol Chem. 1990;265:2852–2855. [PubMed] [Google Scholar]
- 47.Tack BF, Harrison RA, Janatova J, Thomas ML, Prahl JW. Evidence for presence of an internal thiolester bond in third component of human complement. Proc Natl Acad Sci USA. 1980;77:5764–5768. doi: 10.1073/pnas.77.10.5764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Levine RP, Dodds AW. The thioester bond in C3. Curr Top Microbiol Immunol. 1989;153:73–82. doi: 10.1007/978-3-642-74977-3_4. [DOI] [PubMed] [Google Scholar]
- 49.Sahu A, Kozel TR, Pangburn MK. Specificity of the thioester-containing site of human C3 and its significance to complement activation. Biochem J. 1994;302:429–436. doi: 10.1042/bj3020429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sahu A, Pangburn MK. Covalent attachment of human complement C3 to IgG: identification of the amino acid residue involved in ester linkage formation. J Biol Chem. 1994;269:28997–29002. [PubMed] [Google Scholar]
- 51.Sahu A, Pangburn MK. Tyrosine is a potential site for covalent attachment of activated complement component C3. Molec Immunol. 1995;32:711–716. doi: 10.1016/0161-5890(95)98933-f. [DOI] [PubMed] [Google Scholar]
- 52.Dempsey RW, Allison ME, Akkaraju S, Goodnow CC, Fearon DT. C3d of complement as a molecular adjuvant: bridging innate and acquired immunity. Science. 1996;271:348–350. doi: 10.1126/science.271.5247.348. [DOI] [PubMed] [Google Scholar]
- 53.Bergmann-Leitner ES, Leitner WW, Tsokos GC. Complement 3d: from molecular adjuvant to target of immune escape mechanisms. Clin Immunol. 2006;121:177–185. doi: 10.1016/j.clim.2006.07.001. [DOI] [PubMed] [Google Scholar]
- 54.Salehen N, Stover C. The role of complement in the success of vaccination with conjugated vs. unconjugated polysaccharide antigen. Vaccine. 2008;26:451–459. doi: 10.1016/j.vaccine.2007.11.049. [DOI] [PubMed] [Google Scholar]
- 55.Ghannam A, Pernollet M, Fauquert JL, Monnier N, Ponard D, Villiers MB, et al. Human C3 deficiency associated with impairments in dendritic cell differentiation, memory B cells, and regulatory T cells. J Immunol. 2008;181:5158–5166. doi: 10.4049/jimmunol.181.7.5158. [DOI] [PubMed] [Google Scholar]
- 56.Weis WI, Drickamer K, Hendrickson WA. Structure of a C-type mannose-binding protein complexed with an oligosaccharide. Nature. 1992;360:127–134. doi: 10.1038/360127a0. [DOI] [PubMed] [Google Scholar]
- 57.Garlatti V, Belloy N, Martin L, Lacroix M, Matsushita M, Endo Y, et al. Structural insights into the innate immune recognition specificities of L- and H-ficolins. EMBO J. 2007;26:623–633. doi: 10.1038/sj.emboj.7601500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Teh C, Le Y, Lee SH, Lu J. M-ficolin is expressed on monocytes and is a lectin binding to N-acetyl-D-glucosamine and mediates monocyte adhesion and phagocytosis of Escherichia coli. Immunology. 2000;101:225–232. doi: 10.1046/j.1365-2567.2000.00099.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kishore U, Gaboriaud C, Waters P, Shrive AK, Greenhough TJ, Reid KB, et al. C1q and tumor necrosis factor superfamily: modularity and versatility. Trends Immunol. 2004;25:551–561. doi: 10.1016/j.it.2004.08.006. [DOI] [PubMed] [Google Scholar]
- 60.Kishore U, Ghai R, Greenhough TJ, Shrive AK, Bonifati DM, Gadjeva MG, et al. Structural and functional anatomy of the globular domain of complement protein C1q. Immunol Lett. 2004;95:113–128. doi: 10.1016/j.imlet.2004.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Eggleton P, Reid KB, Tenner AJ. C1q--how many functions? How many receptors? Trends Cell Biol. 1998;8:428–431. doi: 10.1016/s0962-8924(98)01373-7. [DOI] [PubMed] [Google Scholar]
- 62.Lu JH, Teh BK, Wang L, Wang YN, Tan YS, Lai MC, et al. The classical and regulatory functions of C1q in immunity and autoimmunity. Cell Mol Immunol. 2008;5:9–21. doi: 10.1038/cmi.2008.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gershov D, Kim S, Brot N, Elkon KB. C-Reactive protein binds to apoptotic cells, protects the cells from assembly of the terminal complement components, and sustains an antiinflammatory innate immune response: implications for systemic autoimmunity. J Exp Med. 2000;192:1353–1364. doi: 10.1084/jem.192.9.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Suresh MV, Singh SK, Ferguson DA, Jr., Agrawal A. Role of the property of C-reactive protein to activate the classical pathway of complement in protecting mice from pneumococcal infection. J Immunol. 2006;176:4369–4374. doi: 10.4049/jimmunol.176.7.4369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Garlanda C, Bottazzi B, Bastone A, Mantovani A. Pentraxins at the crossroads between innate immunity, inflammation, matrix deposition, and female fertility. Annu Rev Immunol. 2005;23:337–366. doi: 10.1146/annurev.immunol.23.021704.115756. [DOI] [PubMed] [Google Scholar]
- 66.Wiesmann C, Katschke KJ, Yin J, Helmy KY, Steffek M, Fairbrother WJ, et al. Structure of C3b in complex with CRIg gives insights into regulation of complement activation. Nature. 2006;444:217–220. doi: 10.1038/nature05263. [DOI] [PubMed] [Google Scholar]
- 67.Harada R, Okada N, Fujita T, Okada H. Purification of 1F5 antigen that prevents complement attack on homologous cell membranes. J Immunol. 1990;144:1823–1828. [PubMed] [Google Scholar]
- 68.Meri S, Morgan BP, Davies A, Daniels RH, Olavesen MG, Waldmann H, et al. Human protectin (CD59), an 18,000-20,000 MW complement lysis restricting factor, inhibits C5b-8 catalysed insertion of C9 into lipid bilayers. Immunology. 1990;71:1–9. [PMC free article] [PubMed] [Google Scholar]
- 69.Zhou Y, Gong B, Lin F, Rother RP, Medof ME, Kaminski HJ. Anti-C5 antibody treatment ameliorates weakness in experimentally acquired myasthenia gravis. J Immunol. 2007;179:8562–8567. doi: 10.4049/jimmunol.179.12.8562. [DOI] [PubMed] [Google Scholar]
- 70.Navenot JM, Muller JY, Blanchard D. Investigation of the survival of paroxysmal nocturnal hemoglobinuria red cells through the immunophenotyping of reticulocytes. Transfusion. 1998;38:337–342. doi: 10.1046/j.1537-2995.1998.38498257371.x. [DOI] [PubMed] [Google Scholar]
- 71.Rosse WF. The life-span of complement-sensitive and -insensitive red cells in paroxysmal nocturnal hemoglobinuria. Blood. 1971;37:556–562. [PubMed] [Google Scholar]
- 72.Rosse WF. New insights into paroxysmal nocturnal hemoglobinuria. Curr Opin Hematol. 2001;8:61–67. doi: 10.1097/00062752-200103000-00001. [DOI] [PubMed] [Google Scholar]
- 73.Luzzatto L, Gianfaldoni G. Recent advances in biological and clinical aspects of paroxysmal nocturnal hemoglobinuria. Int J Hematol. 2006;84:104–112. doi: 10.1532/IJH97.06117. [DOI] [PubMed] [Google Scholar]
- 74.Ferreira VP, Pangburn MK. Factor H mediated cell surface protection from complement is critical for the survival of PNH erythrocytes. Blood. 2007;110:2190–2192. doi: 10.1182/blood-2007-04-083170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kavanagh D, Goodship TH, Richards A. Atypical haemolytic uraemic syndrome. Br Med Bull. 2006:77–78. 5–22. doi: 10.1093/bmb/ldl004. [DOI] [PubMed] [Google Scholar]
- 76.Pickering MC, Cook HT, Warren J, Bygrave AE, Moss J, Walport MJ, et al. Uncontrolled C3 activation causes membranoproliferative glomerulonephritis in mice deficient in complement factor H. Nat Genet. 2002;31:424–428. doi: 10.1038/ng912. [DOI] [PubMed] [Google Scholar]
- 77.Kavanagh D, Richards A, Atkinson J. Complement regulatory genes and hemolytic uremic syndromes. Annu Rev Med. 2008;59:293–309. doi: 10.1146/annurev.med.59.060106.185110. [DOI] [PubMed] [Google Scholar]
- 78.Hageman GS, Anderson DH, Johnson LV, Hancox LS, Taiber AJ, Hardisty LI, et al. A common haplotype in the complement regulatory gene factor H (HF1/CFH) predisposes individuals to age-related macular degeneration. Proc Natl Acad Sci U S A. 2005;102:7227–7232. doi: 10.1073/pnas.0501536102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gold B, Merriam JE, Zernant J, Hancox LS, Taiber AJ, Gehrs KM, et al. Variation in factor B (BF) and complement component 2 (C2) genes is associated with age-related macular degeneration. Nature Genetics. 2006;38:458–462. doi: 10.1038/ng1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Montezuma SR, Sobrin L, Seddon JM. Review of genetics in age related macular degeneration. Semin Ophthalmol. 2007;22:229–240. doi: 10.1080/08820530701745140. [DOI] [PubMed] [Google Scholar]
- 81.Skerka C, Lauer N, Weinberger AA, Keilhauer CN, Suhnel J, Smith R, et al. Defective complement control of factor H (Y402H) and FHL-1 in age-related macular degeneration. Mol Immunol. 2007;44:3398–3406. doi: 10.1016/j.molimm.2007.02.012. [DOI] [PubMed] [Google Scholar]
- 82.Haapasalo K, Jarva H, Siljander T, Tewodros W, Vuopio-Varkila J, Jokiranta TS. Complement Factor H allotype 402H is associated with increased C3b opsonization and phagocytosis of Streptococcus pyogenes. Mol Microbiol. 2008 doi: 10.1111/j.1365-2958.2008.06347.x. [DOI] [PubMed] [Google Scholar]
- 83.Stahl GL, Xu Y, Hao L, Miller M, Buras JA, Fung M, et al. Role for the alternative complement pathway in ischemia/reperfusion injury. Am J Pathol. 2003;162:449–455. doi: 10.1016/S0002-9440(10)63839-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Thurman JM, Ljubanovic D, Edelstein CL, Gilkeson GS, Holers VM. Lack of a functional alternative complement pathway ameliorates ischemic acute renal failure in mice. J Immunol. 2003;170:1517–1523. doi: 10.4049/jimmunol.170.3.1517. [DOI] [PubMed] [Google Scholar]
- 85.Thurman JM, Ljubanovic D, Royer PA, Kraus DM, Molina H, Barry NP, et al. Altered renal tubular expression of the complement inhibitor Crry permits complement activation after ischemia/reperfusion. J Clin Invest. 2006;116:357–368. doi: 10.1172/JCI24521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Greene DL, Khan S. Reactive lysis--a phenomenon of delayed hemolytic transfusion reactions. Immunohematology. 1993;9:74–77. [PubMed] [Google Scholar]
- 87.Majka DS, Holers VM. Can we accurately predict the development of rheumatoid arthritis in the preclinical phase? Arthritis Rheum. 2003;48:2701–2705. doi: 10.1002/art.11224. [DOI] [PubMed] [Google Scholar]
- 88.Conti-Fine BM, Milani M, Kaminski HJ. Myasthenia gravis: past, present, and future. J Clin Invest. 2006;116:2843–2854. doi: 10.1172/JCI29894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hietala MA, Jonsson IM, Tarkowski A, Kleinau S, Pekna M. Complement deficiency ameliorates collagen-induced arthritis in mice. J Immunol. 2002;169:454–459. doi: 10.4049/jimmunol.169.1.454. [DOI] [PubMed] [Google Scholar]
- 90.Katschke KJ, Jr., Helmy KY, Steffek M, Xi H, Yin J, Lee WP, et al. A novel inhibitor of the alternative pathway of complement reverses inflammation and bone destruction in experimental arthritis. J Exp Med. 2007;204:1319–1325. doi: 10.1084/jem.20070432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Rosenfeld SI, Jenkins DE, Jr, Leddy JP. Enhanced reactive lysis of paroxysmal nocturnal hemoglobinuria erythrocytes by C5b-9 does not involve increased C7 binding or cell-bound C3b. J Immunol. 1985;134:506–511. [PubMed] [Google Scholar]
- 92.Jenkins DE, Leddy JP. Enhanced reactive lysis of paroxysmal nocturnal hemoglobinuria erythrocytes by C5b-9 does not involve increased C7 binding or cell-bound C3b. J Immunol. 1985;134:506–511. [PubMed] [Google Scholar]
- 93.Baker PJ, Lint TF, McLeod BC, Behrends CL, Gewurz H. Studies on the inhibition of C56-induced lysis (reactive lysis) VI. Modulation of C56-induced lysis by polyanions and polycations. J Immunol. 1975;114:554–558. [PubMed] [Google Scholar]