Abstract
Usher syndrome (USH) is a hereditary disorder associated with sensorineural hearing impairment, progressive loss of vision attributable to retinitis pigmentosa and variable vestibular function. Three clinical types have been described with type I (USH1) being the most severe. To date six USH1 loci have been reported. We ascertained two large Pakistani consanguineous families segregating profound hearing loss, vestibular dysfunction, and retinitis pigmentosa, the defining features of USH1. In these families we excluded linkage of USH to the 11 known USH loci, and subsequently performed a genome-wide linkage screen. We found a novel USH1 locus designated USH1H that mapped to chromosome 15q22-23 in a 4.92 cM interval. This locus overlaps the non-syndromic deafness locus DFNB48 raising the possibility that the two disorders may be caused by allelic mutations.
Keywords: deafness, DFNB48, retinitis pigmentosa, Usher syndrome, USH1H, vestibular dysfunction, 15q22-23
Usher syndrome (USH) is characterized by inherited deafness associated with night blindness due to retinitis pigmentosa (RP) and variable vestibular function (1). USH is estimated to be responsible for more than 50% of deaf-blindness, 8 to 33% of patients thought to have isolated RP, and 3 to 6% of patients thought to have isolated deafness (2–4). USH is classified into three clinical subtypes and type I USH (USH1) is the most genetically heterogeneous. To date, eleven loci have been mapped for Usher syndrome and genes for nine of them, USH1B, USH1C, USH1D, USH1F, USH1G, USH2A, USH2C, USH2D and USH3, have been identified (5–16). More than 340 pathogenic alleles have been reported in these nine USH genes. Although most of the USH mutations are private, a few mutations have a significant carrier frequency in some ethnic groups (11, 15, 17, 18). For some sporadic and familial cases of Usher syndrome, mutations in these USH genes cannot be found, suggesting the possibility of additional novel USH genes (19).
Consanguineous families are an important resource for the identification of novel genes of recessive disorders. Here we report two consanguineous Pakistani families in which the USH1 phenotype is linked to a novel USH1 locus, USH1H, on chromosome 15q22-23 (Fig. 1).
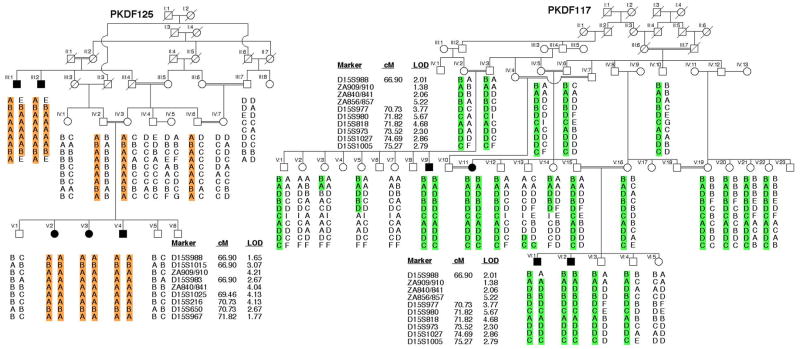
Fig. 1.
Chromosome 15 haplotypes in USH1H families PKDF125 and PKDF117. Filled symbols represent deaf individuals. The USH1H-linked haplotype is color-coded. The STR markers and genetic map positions in centiMorgans (cM) are taken from the Marshfield human genetic map. Haplotype analysis of PKDF125 shows a linkage region of 4.92 cM delimited by markers D15S988 (66.90 cM) and D15S967 (71.82 cM). Affected individuals III:1 and III:2 provided both proximal and distal meiotic breakpoints at marker D15S988 (66.90 cM) and D15S967 (71.82 cM), respectively. In family PKDF117 affected individual VI:1 provided the proximal meiotic breakpoint at marker D15S988 (66.90 cM). The distal breakpoint at marker D15S1005 (75.27 cM) was provided by normal hearing individual V:13. Also given are the maximum two-point lod scores for each STR linked to USH1H.
Material and methods
Subject enrollment
This study was approved by the Institutional Review Board (IRB) at the National Centre of Excellence in Molecular Biology, Lahore, Pakistan (FWA00001758) and the IRB of the NIDCD and NINDS at the National Institutes of Health, USA (OH-93-N-016). Written informed consent was obtained from all adult subjects and parents of minor subjects under the age of 18 years. Subjects were from rural areas of the Punjab province of Pakistan.
Clinical evaluation
We performed medical history interviews to find obvious syndromic and environmental causes of hearing loss. For some of the affected individuals, a physical examination was performed by an internist (P.L.F) to detect signs, symptoms or stigmata of other disorders such as Waardenburg or Pendred syndromes. To rule out obvious chromosomal abnormalities, we performed Giemsa staining and karyotype analyses on chromosomes from peripheral blood leukocytes from one affected member of each family. Affected subjects underwent a general otological examination, including otoscopic examination and audiometry. Hearing was evaluated in some affected and unaffected subjects by pure-tone air- and bone-conduction audiometry with or without tympanometry. No air-bone gaps were observed in any tested individuals. Vestibular function was assessed by tandem gait, Romberg testing and electronystagmography (ENG) with caloric stimulation. Funduscopic and electroretinography (ERG) examinations were performed by an ophthalmologist to confirm the absence or presence of RP. The ages of the affected individuals at the time of examination ranged from 8 to 70 years.
DNA isolation, genotyping and linkage analysis
Genomic DNA was extracted from peripheral blood samples using a standard protocol (20). We first excluded linkage of the USH phenotype to all of the reported USH loci by typing STR (short tandem repeat) markers (http://www.uia.ac.be/dnalab/hhh/) in genomic DNA from affected and unaffected members of the two families. We performed a genome-wide scan for homozygosity among offspring of consanguineous marriages using 388 STR markers (v2.5 ABI Prism Linkage Mapping Set, Applied Biosystems, Foster City, CA) and an ABI Prism 3730 Genetic Analyzer. Alleles were assigned using Genscan and Genotyper software (Applied Biosystems). Fine mapping was performed using additional reported and novel STR markers on chromosome 15q22-23.
LOD score calculations
Marker order and map distances are from the Marshfield genetic map (http://research.marshfieldclinic.org/). Two-point LOD scores were calculated with MLINK (21). We assumed a recessive mode of inheritance, with full penetrance of USH in homozyotes and no phenocopies. The disease allele frequency was set at 0.001 with equal meiotic recombination frequencies for males and females. Short tandem repeat allele frequencies were defined by genotype analyses of 100 unaffected Pakistani individuals.
Candidate genes
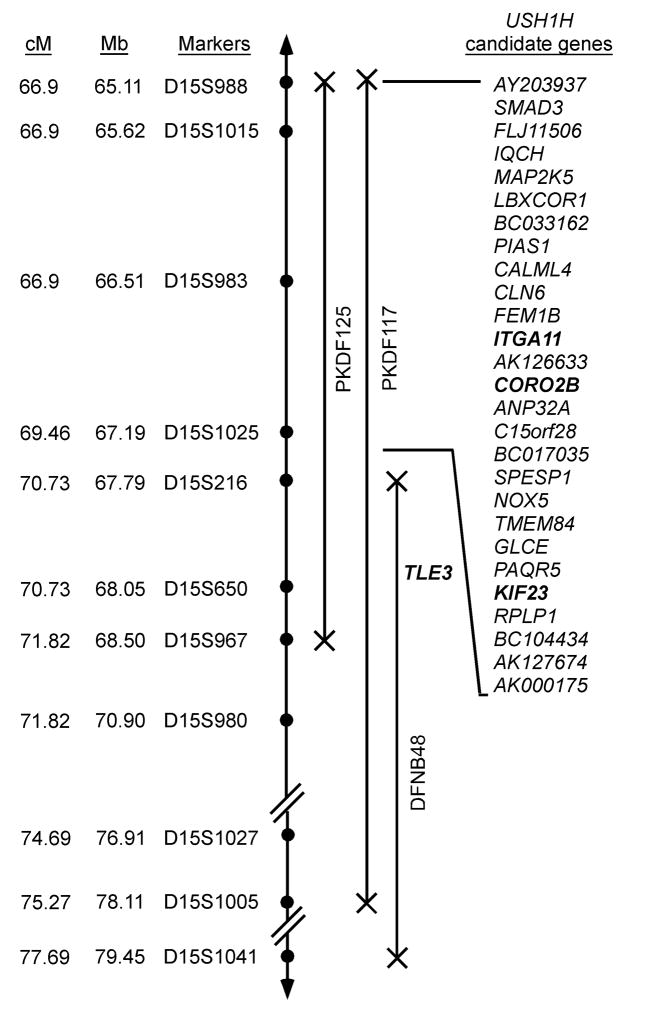
We identified candidate USH1H genes on the UCSC Human Genome Browser (http://genome.ucsc.edu/) and used Primer3 (http://frodo.wi.mit.edu/cgi-in/primer3/primer3_www.cgi) to design PCR and sequencing primers flanking all of the exonic and adjacent intronic sequences of TLE3, ITGA11, CORO2B and KIF23 genes (bold font, figure 2). Mutation analysis procedures were performed essentially as described (22).
Fig. 2.
USH1H linkage intervals in families PKDF125 and PKDF117 on human chromosome 15q22-23. STR markers are represented by filled circles. The sex averaged recombination positions in cM and physical map positions in Mb (not drawn to scale) are indicated for STR markers (Center for Medical Genetics, Marshfield Medical Research Foundation, (http://research.marshfieldclinic.org/genetics). Candidate genes in the USH1H interval were identified from the UCSC Human Genome Browser March 2006 assembly (http://genome.ucsc.edu/).
Results
Clinical description
At the time of examination, the ages of the affected individuals in family PKDF117 ranged from 10 to 36 years, and all were reported to be deaf from an early age, perhaps at birth. All affected individuals from families PKDF117 and PKDF125 are segregating bilateral, profound sensorineural hearing loss. Both in family PKDF125 and PKDF117, deaf individuals had delayed onset of independent ambulation, consistent with vestibular dysfunction, which was confirmed by ENG. RP was detected by funduscopy in affected individuals of both families and was further evaluated in four subjects by ERGs. The severity of RP was directly related to the age of the patient and ranged from mild to the complete loss of vision (data not shown).
Linkage mapping
Because the USH1 phenotype segregating in family PKDF125 was not linked to markers for the reported USH loci, we undertook a genome wide linkage analysis. It initially showed suggestive evidence of linkage only to markers on chromosome 15q22-23. Affected individuals were homozygous for markers in this interval while unaffected obligate carriers were heterozygous. Additional markers were genotyped and haplotype analysis revealed a 4.92 cM interval of homozygosity delimited by markers D15S988 (66.90 cM) and D15S967 (71.82 cM; Fig. 1) defining a new locus, which the HUGO nomenclature committee designated USH1H. A maximum two-point lod score (Zmax) of 4.21 at recombination fraction θ=0 was obtained for the marker ZA840/841 (Fig. 1). USH1H-linked STR markers were then used to screen additional families segregating USH or isolated recessive deafness. One additional family, PKDF117, was found to be segregating USH1 linked to markers in this region (Fig. 1). Individual VI:1 of family PKDF117 provided the proximal meiotic breakpoint at marker D15S988 (66.90 cM), whereas individual V:13 provided the distal meiotic breakpoint at marker D15S1005 (75.27 cM) defining a linkage interval of 8.37 cM (Fig. 1). A maximum two-point lod score (Zmax) of 5.67 at recombination fraction θ=0 was obtained for the marker D15S980 (Fig. 1).
The USH1H critical interval overlaps DFNB48, a locus for non-syndromic recessively inherited hearing loss that we previously mapped between markers D15S216 and D15S1041 (23). We next considered the possibility that mutations of a single gene might underlie both USH1H and DFNB48. If so, the mutated gene is located between markers D15S216 and D15S967, which spans 687.32 kb. On this assumption we examined the overlapping linkage interval of USH1H and DFNB48 and found only one known gene, TLE3 (Fig. 1). Sequence analysis of the 21 exons and ~100 nucleotides flanking the exons of TLE3 in affected individuals of USH1H and DFNB48 families did not reveal any pathogenic alleles.
We next examined the 3.36 Mb region of overlapping homozygosity between the two USH1H families in which there are 28 genes (UCSC human genome browser, Fig. 2). Inspection of our massively parallel signature sequencing libraries of mRNA from inner ear tissues (24) show that 12 of the 28 genes are expressed in these libraries. Based upon their function or sequence similarity to reported deafness genes, we have so far analyzed four candidates (ITGA11, CORO2B, TLE3 and KIF23; Fig. 2) and have found no pathogenic variants.
Discussion
Haplotype analysis of two families revealed a 4.92 cM region of homozygosity for USH1H on chromosome 15q22-23. Families PKDF125 and PKDF117 each have unique haplotypes across this region, and therefore probably segregate different mutant USH1 alleles. The USH1H locus overlaps the DFNB48 locus on chromosome 15 (23) and these two hearing disorders may be due to allelic mutations. Fourteen of 38 known genes for non-syndromic deafness are also responsible for a syndromic form of deafness. For example, mutant alleles of four of the known USH1 genes, MYO7A (13, 19), USH1C (7, 12), CDH23 (8, 9) and PCDH15 (5, 6) are responsible for both non-syndromic hearing loss and USH1 (9, 19, 25, 26).
In the linkage interval common to USH1H and DFNB48 there appears to be only one known gene, TLE3, and no pathogenic variants were identified, suggesting either that the mutation might be located in a conserved region of an intron or in a distant regulatory element. Alternatively, there may be separate linked genes responsible for non-syndromic deafness DFNB48 and the USH1H phenotype. Among the remaining candidate deafness genes in the critical USH1H interval are LBXCOR1, PIAS1 and TMEM84 (Fig. 2). LBXCOR1 has an N-terminal cysteine-rich region, a Corl homology (CH1) domain and a C-terminal coiled-coil region and may be a transcriptional repressor (27). PIAS1 contains a putative zinc-binding motif and a highly acidic region (28) and is a suppressor of STAT1 (28). Mutant alleles of transcriptional factors EYA4, POU3F4, POU4F3, GRHL2 are associated with hearing impairment (29–32). TMEM84 encodes a protein with a predicted transmembrane domain. TMIE and LHFPL5 also encode single-pass transmembrane domain proteins and are necessary for sound transduction, but their precise functions are unknown (33–36). The remaining 24 genes and conserved sequences in the USH1H interval will now need to be screened for mutant alleles. Additional USH1H families may refine the locus and reduce the number of candidates to be screened. Mouse models have been helpful in identifying and studying genes for Usher syndrome in humans (5, 6, 13, 14, 37, 38). However, there are no reported deaf mice on mouse chromosome 9 in a region of conserved linkage with human chromosome 15q22-23.
Mapping a new locus for USH1 to chromosome 15 in two Pakistani families emphasizes the genetic heterogeneity of this disorder and is important for several reasons. USH genes have provided unexpected insights into necessary developmental and biochemical processes shared by the eye and ear (26, 39). All of the USH1 proteins, including myosin VIIa, cadherin 23, protocadherin 15, harmonin, SANS, usherin, VLGR1, whirlin and clarin-1, are thought to interact to form a large macromolecular complex (39, 40), which is essential for auditory and visual functions. Following this precedent, we hypothesize the gene underlying USH1H will encode another member of this USH protein network. Further insight into the functions of the USH1 proteins may uncover targets and strategies for therapeutic interventions to prevent or retard the progressive loss of vision due to the RP component of USH.
Acknowledgments
The authors are grateful to the families who made this research possible. We thank Julie Schultz, Andrew Griffith, Anne Madeo, Karen Friderici and Dennis Drayna for suggestions regarding this manuscript. This study was supported by the Higher Education Commission, Islamabad, Pakistan; Ministry of Science and Technology, Islamabad, Pakistan and by intramural funds to TBF from the National Institute on Deafness and Other Communication Disorders, NIH (1 ZO1 DC000039-11).
References
- 1.Smith RJ, Berlin CI, Hejtmancik JF, et al. Clinical diagnosis of the Usher syndromes. Usher Syndrome Consortium. Am J Med Genet. 1994;50:32–38. doi: 10.1002/ajmg.1320500107. [DOI] [PubMed] [Google Scholar]
- 2.Boughman JA, Vernon M, Shaver KA. Usher syndrome: definition and estimate of prevalence from two high-risk populations. J Chronic Dis. 1983;36:595–603. doi: 10.1016/0021-9681(83)90147-9. [DOI] [PubMed] [Google Scholar]
- 3.Brownstein Z, Ben-Yosef T, Dagan O, et al. The R245X mutation of PCDH15 in Ashkenazi Jewish children diagnosed with nonsyndromic hearing loss foreshadows retinitis pigmentosa. Pediatr Res. 2004;55:995–1000. doi: 10.1203/01.PDR.0000125258.58267.56. [DOI] [PubMed] [Google Scholar]
- 4.Vernon M. Usher’s syndrome--deafness and progressive blindness. Clinical cases, prevention, theory and literature survey. J Chronic Dis. 1969;22:133–151. doi: 10.1016/0021-9681(69)90055-1. [DOI] [PubMed] [Google Scholar]
- 5.Ahmed ZM, Riazuddin S, Bernstein SL, et al. Mutations of the protocadherin gene PCDH15 cause Usher syndrome type 1F. Am J Hum Genet. 2001;69:25–34. doi: 10.1086/321277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alagramam KN, Yuan H, Kuehn MH, et al. Mutations in the novel protocadherin PCDH15 cause Usher syndrome type 1F. Hum Mol Genet. 2001;10:1709–1718. doi: 10.1093/hmg/10.16.1709. [DOI] [PubMed] [Google Scholar]
- 7.Bitner-Glindzicz M, Lindley KJ, Rutland P, et al. A recessive contiguous gene deletion causing infantile hyperinsulinism, enteropathy and deafness identifies the Usher type 1C gene. Nat Genet. 2000;26:56–60. doi: 10.1038/79178. [DOI] [PubMed] [Google Scholar]
- 8.Bolz H, von Brederlow B, Ramirez A, et al. Mutation of CDH23, encoding a new member of the cadherin gene family, causes Usher syndrome type 1D. Nat Genet. 2001;27:108–112. doi: 10.1038/83667. [DOI] [PubMed] [Google Scholar]
- 9.Bork JM, Peters LM, Riazuddin S, et al. Usher syndrome 1D and nonsyndromic autosomal recessive deafness DFNB12 are caused by allelic mutations of the novel cadherin-like gene CDH23. Am J Hum Genet. 2001;68:26–37. doi: 10.1086/316954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ebermann I, Scholl HP, Charbel Issa P, et al. A novel gene for Usher syndrome type 2: mutations in the long isoform of whirlin are associated with retinitis pigmentosa and sensorineural hearing loss. Hum Genet. 2007;121:203–211. doi: 10.1007/s00439-006-0304-0. [DOI] [PubMed] [Google Scholar]
- 11.Joensuu T, Hamalainen R, Yuan B, et al. Mutations in a novel gene with transmembrane domains underlie Usher syndrome type 3. Am J Hum Genet. 2001;69:673–684. doi: 10.1086/323610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Verpy E, Leibovici M, Zwaenepoel I, et al. A defect in harmonin, a PDZ domain-containing protein expressed in the inner ear sensory hair cells, underlies Usher syndrome type 1C. Nat Genet. 2000;26:51–55. doi: 10.1038/79171. [DOI] [PubMed] [Google Scholar]
- 13.Weil D, Blanchard S, Kaplan J, et al. Defective myosin VIIA gene responsible for Usher syndrome type 1B. Nature. 1995;374:60–61. doi: 10.1038/374060a0. [DOI] [PubMed] [Google Scholar]
- 14.Weil D, El-Amraoui A, Masmoudi S, et al. Usher syndrome type I G (USH1G) is caused by mutations in the gene encoding SANS, a protein that associates with the USH1C protein, harmonin. Hum Mol Genet. 2003;12:463–471. doi: 10.1093/hmg/ddg051. [DOI] [PubMed] [Google Scholar]
- 15.Weston MD, Eudy JD, Fujita S, et al. Genomic structure and identification of novel mutations in usherin, the gene responsible for Usher syndrome type IIa. Am J Hum Genet. 2000;66:1199–1210. doi: 10.1086/302855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weston MD, Luijendijk MW, Humphrey KD, et al. Mutations in the VLGR1 gene implicate G-protein signaling in the pathogenesis of Usher syndrome type II. Am J Hum Genet. 2004;74:357–366. doi: 10.1086/381685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ben-Yosef T, Ness SL, Madeo AC, et al. A mutation of PCDH15 among Ashkenazi Jews with the type 1 Usher syndrome. N Engl J Med. 2003;348:1664–1670. doi: 10.1056/NEJMoa021502. [DOI] [PubMed] [Google Scholar]
- 18.Ness SL, Ben-Yosef T, Bar-Lev A, et al. Genetic homogeneity and phenotypic variability among Ashkenazi Jews with Usher syndrome type III. J Med Genet. 2003;40:767–772. doi: 10.1136/jmg.40.10.767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Riazuddin S, Nazli S, Ahmed ZM, et al. Mutation spectrum of MYO7A and evaluation of a novel nonsyndromic deafness DFNB2 allele with residual function. Hum Mutat. 2008;29:502–511. doi: 10.1002/humu.20677. [DOI] [PubMed] [Google Scholar]
- 20.Grimberg J, Nawoschik S, Belluscio L, et al. A simple and efficient non-organic procedure for the isolation of genomic DNA from blood. Nucleic Acids Res. 1989;17:8390. doi: 10.1093/nar/17.20.8390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schaffer AA. Faster linkage analysis computations for pedigrees with loops or unused alleles. Hum Hered. 1996;46:226–235. doi: 10.1159/000154358. [DOI] [PubMed] [Google Scholar]
- 22.Khan SY, Ahmed ZM, Shabbir MI, et al. Mutations of the RDX gene cause nonsyndromic hearing loss at the DFNB24 locus. Hum Mutat. 2007;28:417–423. doi: 10.1002/humu.20469. [DOI] [PubMed] [Google Scholar]
- 23.Ahmad J, Khan SN, Khan SY, et al. DFNB48, a new nonsyndromic recessive deafness locus, maps to chromosome 15q23-q25.1. Hum Genet. 2005;116:407–412. doi: 10.1007/s00439-004-1247-y. [DOI] [PubMed] [Google Scholar]
- 24.Peters LM, Belyantseva IA, Lagziel A, et al. Signatures from tissue-specific MPSS libraries identify transcripts preferentially expressed in the mouse inner ear. Genomics. 2007;89:197–206. doi: 10.1016/j.ygeno.2006.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ahmed ZM, Riazuddin S, Ahmad J, et al. PCDH15 is expressed in the neurosensory epithelium of the eye and ear and mutant alleles are responsible for both USH1F and DFNB23. Hum Mol Genet. 2003;12:3215–3223. doi: 10.1093/hmg/ddg358. [DOI] [PubMed] [Google Scholar]
- 26.Petit C. Usher syndrome: from genetics to pathogenesis. Annu Rev Genomics Hum Genet. 2001;2:271–297. doi: 10.1146/annurev.genom.2.1.271. [DOI] [PubMed] [Google Scholar]
- 27.Mizuhara E, Nakatani T, Minaki Y, et al. Corl1, a novel neuronal lineage-specific transcriptional corepressor for the homeodomain transcription factor Lbx1. J Biol Chem. 2005;280:3645–3655. doi: 10.1074/jbc.M411652200. [DOI] [PubMed] [Google Scholar]
- 28.Liu B, Liao J, Rao X, et al. Inhibition of Stat1-mediated gene activation by PIAS1. Proc Natl Acad Sci U S A. 1998;95:10626–10631. doi: 10.1073/pnas.95.18.10626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.de Kok YJ, van der Maarel SM, Bitner-Glindzicz M, et al. Association between X-linked mixed deafness and mutations in the POU domain gene POU3F4. Science. 1995;267:685–688. doi: 10.1126/science.7839145. [DOI] [PubMed] [Google Scholar]
- 30.Peters LM, Anderson DW, Griffith AJ, et al. Mutation of a transcription factor, TFCP2L3, causes progressive autosomal dominant hearing loss, DFNA28. Hum Mol Genet. 2002;11:2877–2885. doi: 10.1093/hmg/11.23.2877. [DOI] [PubMed] [Google Scholar]
- 31.Vahava O, Morell R, Lynch ED, et al. Mutation in transcription factor POU4F3 associated with inherited progressive hearing loss in humans. Science. 1998;279:1950–1954. doi: 10.1126/science.279.5358.1950. [DOI] [PubMed] [Google Scholar]
- 32.Wayne S, Robertson NG, DeClau F, et al. Mutations in the transcriptional activator EYA4 cause late-onset deafness at the DFNA10 locus. Hum Mol Genet. 2001;10:195–200. doi: 10.1093/hmg/10.3.195. [DOI] [PubMed] [Google Scholar]
- 33.Kalay E, Li Y, Uzumcu A, et al. Mutations in the lipoma HMGIC fusion partner-like 5 (LHFPL5) gene cause autosomal recessive nonsyndromic hearing loss. Hum Mutat. 2006;27:633–639. doi: 10.1002/humu.20368. [DOI] [PubMed] [Google Scholar]
- 34.Naz S, Giguere CM, Kohrman DC, et al. Mutations in a novel gene, TMIE, are associated with hearing loss linked to the DFNB6 locus. Am J Hum Genet. 2002;71:632–636. doi: 10.1086/342193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shabbir MI, Ahmed ZM, Khan SY, et al. Mutations of human TMHS cause recessively inherited non-syndromic hearing loss. J Med Genet. 2006;43:634–640. doi: 10.1136/jmg.2005.039834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tlili A, Mannikko M, Charfedine I, et al. A novel autosomal recessive non-syndromic deafness locus, DFNB66, maps to chromosome 6p21.2–22.3 in a large Tunisian consanguineous family. Hum Hered. 2005;60:123–128. doi: 10.1159/000088974. [DOI] [PubMed] [Google Scholar]
- 37.Gibson F, Walsh J, Mburu P, et al. A type VII myosin encoded by the mouse deafness gene shaker-1. Nature. 1995;374:62–64. doi: 10.1038/374062a0. [DOI] [PubMed] [Google Scholar]
- 38.Kikkawa Y, Shitara H, Wakana S, et al. Mutations in a new scaffold protein Sans cause deafness in Jackson shaker mice. Hum Mol Genet. 2003;12:453–461. doi: 10.1093/hmg/ddg042. [DOI] [PubMed] [Google Scholar]
- 39.Kremer H, van Wijk E, Marker T, et al. Usher syndrome: molecular links of pathogenesis, proteins and pathways. Hum Mol Genet. 2006;(15 Spec No 2):R262–270. doi: 10.1093/hmg/ddl205. [DOI] [PubMed] [Google Scholar]
- 40.El-Amraoui A, Petit C. Usher I syndrome: unravelling the mechanisms that underlie the cohesion of the growing hair bundle in inner ear sensory cells. J Cell Sci. 2005;118:4593–4603. doi: 10.1242/jcs.02636. [DOI] [PubMed] [Google Scholar]