SUMMARY
Pancreatic cancer is a deadly disease characterized by late diagnosis and resistance to therapy. Much progress has been made in defining gene defects in pancreatic cancer, but a full accounting of its molecular pathogenesis awaits. Here, we show that expression of Ataxia-Telangiectasia Group D Complementing gene (ATDC), also called TRIM29, is elevated in most invasive pancreatic cancers and pancreatic cancer precursor lesions. ATDC promoted cancer cell proliferation in vitro and enhanced tumor growth and metastasis in vivo. ATDC expression correlated with elevated β-catenin levels in pancreatic cancer, and β-catenin function was required for ATDC’s oncogenic effects. ATDC was found to stabilize β-catenin via ATDC-induced effects on the disheveled-2 protein, a negative regulator of GSK3β in the Wnt/β-catenin signaling pathway.
SIGNIFICANCE
Pancreatic cancer is an aggressive malignancy, and an improved understanding of the molecular mechanisms governing its highly aggressive behavior is needed for more effective treatment, early detection and prevention. Defects in Wnt/β-catenin signaling are common in certain cancers, such as colorectal carcinoma, and recent evidence suggests Wnt/β-catenin signaling may contribute to pancreatic cancer. In this report, we show that ATDC is over-expressed in the majority of invasive pancreatic cancers and pancreatic cancer precursor lesions. ATDC contributes to pancreatic cancer via its ability to interact with and stabilize expression of disheveled-2, with resultant stabilization of β-catenin. Besides highlighting ATDC as a potential therapeutic target in pancreatic cancer, our studies have defined a novel mechanism for activating Wnt/β-catenin signaling in cancer.
Keywords: pancreatic cancer, Wnt signaling, β-catenin, cell proliferation, disheveled
INTRODUCTION
Pancreatic cancer is a highly lethal disease that is often diagnosed in an advanced state. In fact, though it is the fourth most common cause of cancer death per year in the United States, affecting 37,000 people (Jemal et al., 2007), it has the worst prognosis of any major malignancy (<5% 5-year survival rate). Recent advances in surgical and medical therapy have had only a modest impact on pancreatic cancer mortality. A major hallmark of pancreatic cancer is extensive local tumor invasion and early systemic dissemination. Pancreatic cancer is also notoriously resistant to chemotherapy and ionizing radiation.
To further understand the molecular pathogenesis of pancreatic cancer, genomic and proteomic profiling has been performed to identify differentially expressed genes and proteins that might represent novel therapeutic targets (Cao et al., 2004; Chen et al., 2005; Crnogorac-Jurcevic et al., 2003; Logsdon et al., 2003; Lowe et al., 2007; Mutter et al., 2001). Using Affymetrix gene expression profiling, we recently found that pancreatic cancer cells over-express the Ataxia-Telangiectasia Group D Complementing gene (ATDC) at an average level 20-fold higher than epithelial cells from normal pancreas or chronic pancreatitis-derived tissues (Logsdon et al., 2003). ATDC was initially described in the hunt for the gene responsible for the genetic disorder ataxia telangiectasia (AT) (Kapp et al., 1992). Both alleles of the ATDC gene in an AT patient-derived cell line were found to contain early stop codon mutations, leading to a truncated and nonfunctional ATDC protein (Tauchi et al., 2000). No germline ATDC mutations were found in AT patients, and subsequently the gene responsible for AT was identified as ATM (ataxia telangiectasia mutated) (Savitsky et al., 1995).
The ATDC gene, located at chromosome 11q23, encodes a 588 amino acid protein with multiple zinc finger motifs and an adjacent leucine zipper motif, that may allow the ATDC protein to form homo- or hetero-dimers. (Savitsky et al., 1995). Northern blot analysis revealed that ATDC is normally expressed in placenta, lung, thymus, prostate, testis, and colon, while no expression is observed in heart, brain, skeletal muscle, pancreas, spleen, ovary, or small intestine (Hosoi and Kapp, 1994). In addition to our observation of high ATDC levels in primary pancreatic cancers and pancreatic cancer cell lines, a search of the Oncomine database (www.oncomine.org) revealed that ATDC has been reported to be overexpressed in lung (Hawthorn et al., 2006), bladder (Dyrskjot et al., 2004), colorectal (Glebov et al., 2006; Ohmachi et al., 2006), ovarian (Santin et al., 2004), endometrial cancers (Mutter et al., 2001) and multiple myeloma (Zhan et al., 2002), with apparent reduced expressionin melanoma (Smith et al., 2005), breast (Nacht et al., 1999), head and neck (Zhang et al., 2006) and prostate cancer (Dyrskjot et al., 2004; LaTulippe et al., 2002; Luo et al., 2001; Mutter et al., 2001; Yu et al., 2004; Zhan et al., 2002). A recent report identified a correlation between ATDC expression in gastric cancer and poor histological grade, large tumor size, extent of tumor invasion, and lymph node metastasis (Kosaka et al., 2007).
The ATDC protein, also known as TRIM29, is a member of the tripartite motif (TRIM) family. TRIM proteins have a series of conserved domains, which include a RING (R), a B-box type 1 (B1) and B-box type 2 (B2), followed by a coiled-coil (CC) region. While some of the domains may be absent or present in the different TRIM proteins (ATDC contains the B1-B2-CC domains but lacks the R domain), their order is always maintained (R-B1-B2-CC) (Reymond et al., 2001). Proteins belonging to the TRIM family have been implicated in a variety of cellular processes, such as development and growth, and in several human diseases, including HIV infection (Stremlau et al., 2004) and leukemia (Goddard et al., 1991). The function of ATDC has not been studied previously in any physiologic or pathologic process, though the ATDC protein appears to be localized primarily to the cytoplasm (Reymond et al., 2001). In this report, we describe our data examining the expression and functional role of ATDC in pancreatic cancer.
RESULTS
ATDC is overexpressed in human pancreatic adenocarcinoma
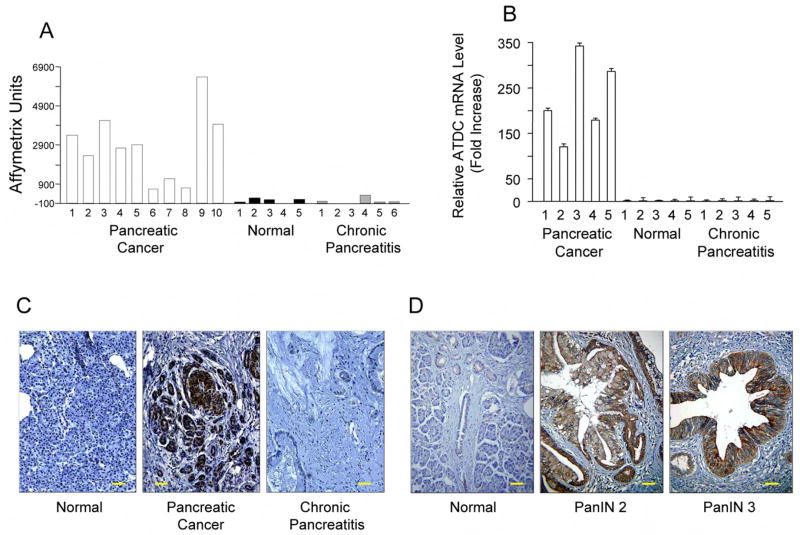
To identify genes with potential roles in the development and progression of pancreatic adenocarcinoma, we assessed gene expression in microdissected samples of human pancreatic carcinoma using Affymetrix arrays. We compared the expression patterns in cancer tissues to those seen in normal pancreas and chronic pancreatitis, the latter which served as an important control for the extensive fibrosis typically observed with pancreatic cancer (Logsdon et al., 2003). One of the most highly up-regulated genes was the ATDC gene, which showed elevated expression in 10 out of 10 samples of pancreatic cancers relative to normal pancreas or chronic pancreatitis samples (Figure 1A). On average, ATDC expression was roughly 20-fold higher in pancreatic adenocarcinomas. The gene expression data were further confirmed in an analysis of ATDC mRNA levels of pancreatic cancer using quantitative real time PCR (qRT-PCR) (Figure 1B). Immunohistochemical staining confirmed that ATDC protein expression was present in the neoplastic epithelium of pancreatic cancer (Figure 1C).
Figure 1. ATDC is highly expressed in human pancreatic cancer.
(A) cDNA microarray analysis (Logsdon et al., 2003) was done using HuGeneFL Arrays containing 7129 probe sets (Affymetrix, Santa Clara, CA). Microdissected samples of human pancreatic cancer (n=10, open bars), normal pancreas (n=5, black bars) and chronic pancreatitis (n=5, gray bars) were analyzed. mRNA expression levels of ATDC were expressed as Affymetrix units. (B) Validation of microarray results of ATDC mRNA levels using quantitative real time RT-PCR analysis. (C) ATDC immunostaining of representative samples of normal pancreas, pancreatic cancer, and chronic pancreatitis. The scale bar indicates 100 μM. (D) Immunohistochemical examination of ATDC expression in normal pancreas or pancreatic intraepithelial neoplasias (PanIN) lesions. The scale bar indicates 100 μM.
A progression model of pancreatic cancer is now widely accepted in which normal pancreatic ductal epithelium progresses to infiltrating cancer through a series of morphologically defined pancreatic precursors called PanINs (Hruban et al., 2000). This progression is associated with accumulation of specific genetic changes, such as K-ras mutations and inactivation of p16, that are observed in invasive pancreatic cancer. We found that ATDC was not expressed in PanIN 1 (0/4) lesions, but was occasionally expressed in PanIN 2 (1/7) lesions and was more often expressed in PanIN 3 lesions (3/6) (Figure 1D). These data suggest that up-regulation of ATDC occurs prior to the development of invasive pancreatic cancer.
ATDC promotes cellular proliferation in vitro and pancreatic tumorigenesis in vivo
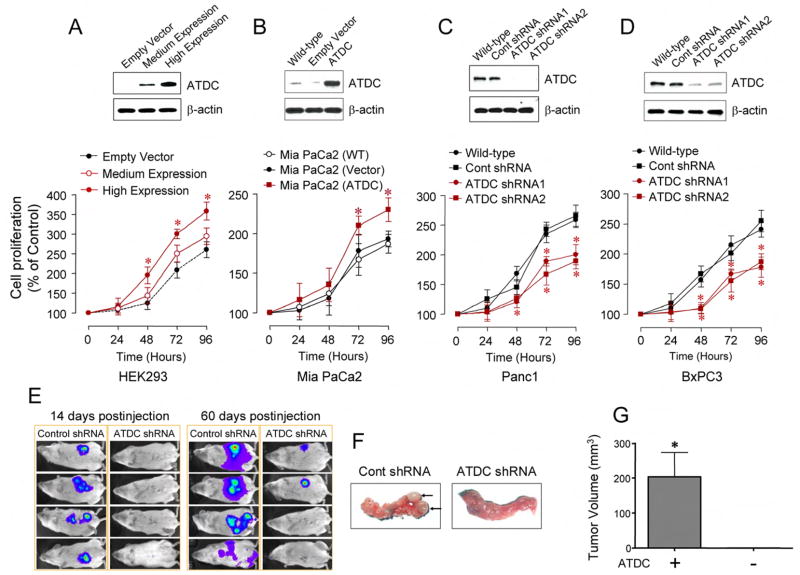
To understand the function of ATDC in pancreatic cancer, we first explored the effect of ATDC ectopic expression on cellular growth in vitro in multiple cell lines with differing levels of endogenous ATDC expression. Following transfection with an ATDC cDNA expression construct, HEK 293 cells, which normally do not express ATDC, and MiaPaCa2 pancreatic cancer cells, which express low endogenous levels of ATDC, demonstrated a significant increase in cellular proliferation (Figure 2A and 2B). Similar changes were observed in monoclonal and polyclonal HEK 293 cells lines stably overexpressing ATDC (Supplemental Figure S1). Conversely, cellular proliferation was attenuated when endogenous ATDC expression was silenced by stable transfection with 2 different shRNA vectors targeting distinct regions of ATDC in Panc1 and BxPC3 pancreatic cancer cell lines, both of which have high endogenous levels of ATDC (Figure 2C and 2D). Expression of ATDC shRNA1 and 2 did not alter basal cell proliferation rates in HEK 293 cells (Supplemental Figure S2), verifying the specificity of the inhibitory function of the ATDC shRNAs on ATDC’s function.
Figure 2. ATDC promotes cell proliferation and pancreatic tumorigenesis.
(A) Upper panel, medium or high expression of ATDC in stably transfected HEK 293 cells. Lower panel, MTS proliferation assay in ATDC-transfected HEK 293 cells (mean±SE, n=4, *p<0.05 vs empty vector-transfected cells). (B) Upper panel, ATDC expression in wild type (WT), empty vector or ATDC transfected Mia PaCa2 cells. Lower panel, MTS proliferation assay in ATDC-transfected Mia PaCa2 cells (mean±SE, n=3, *p<0.05 vs empty vector-transfected cells). (C), (D) Upper panels, ATDC expression in wild type, control shRNA, ATDC shRNA1 or 2 tranfected Panc1 (C) or BxPC3 cells (D). Lower panels, MTS proliferation assays in ATDC shRNA-transfected Panc1 and BxPC3 cells (mean±SE, n=4, *p<0.05 vs wild type cells). (E) Representative bioluminescent images of half of the animals in control or ATDC shRNA group are shown at 14 (left panels) and 60 (right panels) days after injection, depicting the extent of tumor burden. (F) Representative pictures of mouse pancreata injected with control shRNA or ATDC shRNA-transfected Panc1 cells 60 days after injection. Only 25% (2/8) of the mice injected with Panc1 cells expressing ATDC shRNA had evidence of tumor formation. (G) Average tumor volume measured in animals injected with control shRNA or ATDC shRNA-transfected Panc1 cells at 60 days post-injection (mean ± SE, n=3, *p<0.05).
To examine the effects of ATDC silencing on pancreatic tumor growth in vivo, we infected Panc1 cells (expressing a control shRNA or ATDC shRNA1) with a luciferase-expressing lentivirus. Following injection of 5 × 105 cells into the tail of the pancreas, tumor growth was assessed using bioluminescent imaging (n= 8 animals per group). All of the animals injected with Panc1 cells expressing control shRNA demonstrated tumor formation 14 days post-injection, while tumors were not detected in the animals injected with Panc1 ATDCshRNA cells (Figure 2E, left panels). At 60 days post-injection, the control shRNA animals tumors grew significantly larger, with evidence of metastatic spread, while only 25% (2/8) of the ATDCshRNA animals demonstrated evidence of macroscopic tumors (Figure 2E, right panels and 2F). The mean tumor volume was significantly larger in the tumors grown in mice injected with Panc1 cells expressing control shRNA compared to mice injected with Panc1 cells expressing ATDC shRNA (203.2 ± 68.8 vs. 2.01 ± 1.0mm3, *p<0.001) (Figure 2G). These data support the role of ATDC in promoting growth of pancreatic cancer cells.
ATDC activates the Wnt/β-Catenin/TCF signaling cascade
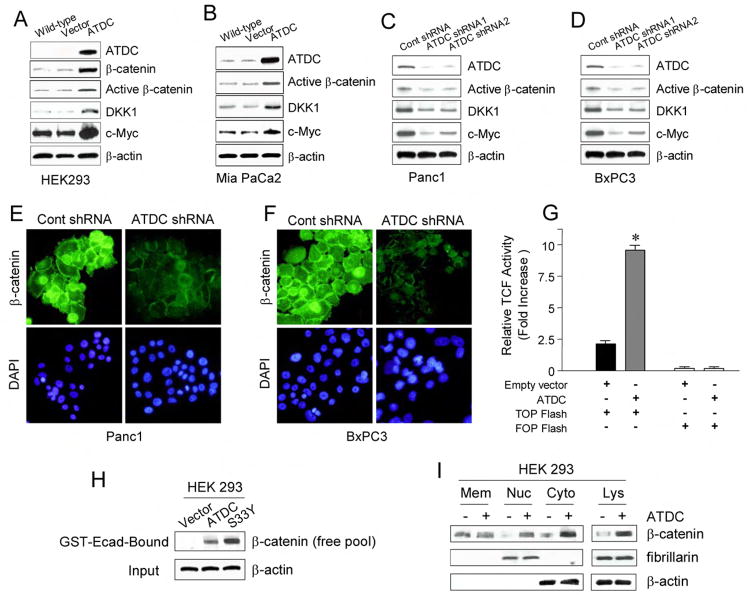
In exploring possible downstream mediators of ATDC’s growth-promoting effects, we noted that over-expression of ATDC resulted in a significant increase in β-catenin levels in HEK 293 cells (Figure 3A). In addition, the over-expression of ATDC increased expression of the under- or non-phosphorylated forms of β-catenin (“active” β-catenin) presumed to be the forms of β-catenin responsible for mediating Wnt signaling in cells (van Noort et al., 2002) (Fig. 3A). Similar results were obtained in Mia PaCa2 pancreatic cancer cells ectopically expressing ATDC (Figure 3B). Conversely, ATDC shRNA1- or 2-mediated silencing of endogenous ATDC expression significantly decreased total β-catenin and active β-catenin levels in Panc1 cells (Figure 3C, 3E) and BxPC3 cells (Figure 3D, 3F).
Figure 3. ATDC upregulates β-catenin levels and TCF transcriptional activity.
(A, B) Representative Western blots of wild type, empty vector, and ATDC-transfected HEK 293 (A) and MiaPaCa2 cells (B). Overexpression of ATDC results in upregulation of β-catenin, active β-catenin, and the TCF target genes DKK1 and c-Myc. β-actin was used as a loading control. (C, D) Representative Western blots of Panc1 (C) and BxPC3 (D) cells expressing control shRNA, ATDC shRNA1 or 2. Silencing of ATDC in Panc1 and BxPC3 cells decreases levels of active β-catenin, DKK1 and c-Myc. β-Actin serves as a loading control. (E, F) Photomicrographs of control shRNA and ATDC shRNA1-expressing Panc1 cells (E) and BxPC3 cells (F) immunostained with an anti-β-catenin antibody (green). Cell nuclei were counterstained with DAPI (blue). (G) TCF reporter activity was assessed by using the β-catenin responsive TOPFLASH reporter and the mutant control FOPFLASH reporter in HEK 293 cells stably transfected with empty vector or an ATDC expression vector (mean±SE, *p<0.05 vs empty vector-transfected cells). (H) The GST-E-cadherin (GST-Ecad) fusion protein detects increases in the free pool of β-catenin. HEK 293 cells expressing ATDC or S33Y β-catenin (S33Y) were harvested. Free β-catenin levels were assessed by western blotting of GST-Ecad-bound fractions of cell lysate using a specific anti-β-catenin antibody. β-actin (input) was used as a loading control. (I) Representative blots of β-catenin levels in membrane (Mem), nuclear (Nuc) and cytoplasmic (Cyto) fractions and total lysates (Lys). β-actin (cytoplasmic expression) and fibrillarin (nuclear expression) were used as loading controls.
The active form of β-catenin exerts its growth promoting effects by translocating from the cytoplasm to the nucleus, where it binds to transcription factors such as the TCF/lymphoid enhancer binding factor and thereby stimulates the transcription of Wnt target genes (Clevers, 2006). Over-expression of ATDC in HEK 293 and MiaPaCa2 cells increased expression of the Wnt/β-catenin target genes c-Myc and DKK1 (Figure 3A, 3B), while c-Myc and DKK1 levels were significantly reduced in Panc1 and BxPC3 cells expressing ATDC-targeting shRNA1 or 2 (Figure 3C, 3D). Consistent with the ability of ATDC to increase β-catenin levels in HEK 293 cells, we found that ATDC strongly activated the TCF-dependent TOP-FLASH reporter construct in HEK 293 cells (Figure 3G). Together, these results strongly suggest that ATDC enhances β-catenin levels and activates β-catenin/TCF target gene expression in pancreatic cancer cells.
ATDC increases the free intracellular pool of β-catenin through activation of the canonical Wnt signaling pathway
To address in more detail the means by which ATDC increased β-catenin/TCF-regulated gene expression, we assessed the abundance of the free pool of β-catenin in ATDC-expressing cells compared to control cells. To measure the free pool of β-catenin, we utilized a recombinant GST fusion protein containing the cytoplasmic tail of E-cadherin (GST-Ecad). It has been previously shown that GST-E-cadherin can readily be used to monitor the abundance of the free pool of β-catenin that is stabilized in response to activation of the Wnt signaling pathway (Winer et al., 2006). As show in Figure 3H, following ectopic expression of ATDC or the S33Y mutant form of β-catenin in HEK 293 cells, significant increases in the levels of the free β-catenin pool were seen, as demonstrated by the recovery of β-catenin with GST-E-cadherin. In contrast, no significant β-catenin was recovered from control cell lysates following incubation with the GST-E-cadherin.
To confirm that the increase in the free pool of β-catenin seen with ATDC overexpression was associated with increased cytoplasmic and nuclear levels of β-catenin, extracts from control and ATDC-overexpressing HEK 293 cells were separated into membrane, cytoplasmic, and nuclear fractions and the relative abundance of β-catenin in these fractions analyzed. ATDC overexpression increased both the nuclear and cytoplasmic fractions of β-catenin, while having no effect on the membrane bound fraction of ATDC (Figure 3I). Previous reports have demonstrated an important role for β-catenin in cell adhesion as a part of a protein complex that includes E-cadherin. The E-cadherin expression pattern in pancreatic cancer cells was unaltered with ATDC shRNA (Supplemental Figure S3), suggesting that increased levels of ATDC affect β-catenin levels and β-catenin/TCF-dependent transcription in a fashion similar to that seen following activation of the canonical Wnt signaling pathway.
ATDC stimulates cell proliferation and tumor growth via β-Catenin/TCF activation
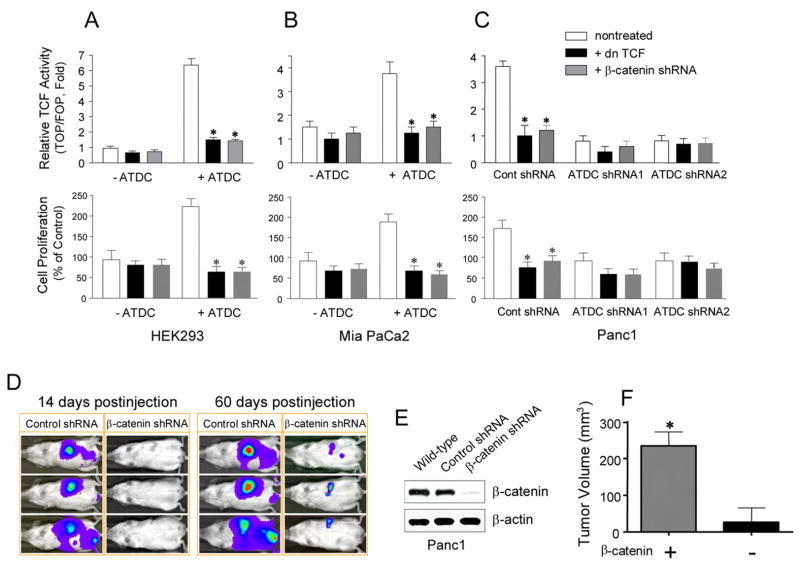
We next sought to determine if the growth-promoting effects of ATDC were mediated by activation of the β-catenin signaling pathway. Constitutive activation of Wnt/β-catenin signaling by mutations in known Wnt pathway components, such as inactivating mutations in the APC (adenomatous polyposis coli) or Axin1 tumor suppressor genes, or activating mutations in β-catenin are commonly seen in certain cancers, such as colorectal or hepatocellular carcinomas, but are rarely seen in pancreatic adenocarcinoma (Gregorieff and Clevers, 2005; Lustig and Behrens, 2003). However, constitutive activation of β-catenin/TCF–dependent transcription, independent of mutations, has been suggested to play an important role in the development of certain human breast and ovarian cancers (Bafico et al., 2004) and in mouse models of basal cell carcinoma (Hoseong Yang et al., 2008) and pancreatic cancer (Pasca di Magliano et al., 2007). To address the contribution of β-catenin/TCF transcription in the growth-promoting effects of ATDC, we ectopically expressed a β-catenin shRNA or a dominant negative TCF (dnTCF) protein in either control vector-transfected or ATDC-expressing HEK 293 and MiaPaCa2 cells. Transfection of either the β-catenin shRNA or dnTCF constructs significantly inhibited ATDC-induced TOPFLASH reporter activity (Figures 4A and 4B, upper panels) and cell proliferation (Figures 4A and 4B, lower panels) in HEK 293 and MiaPaCa2 cells. Similarly, transfection of β-catenin-targeting shRNA or dnTCF in Panc1 cells inhibited TOPFLASH reporter activity (Figure 4C, upper panel) and inhibited the enhanced cellular proliferation seen in Panc1 cells in the setting of elevated endogenous ATDC levels (Figure 4C, lower panel).
Figure 4. ATDC stimulates cell proliferation and tumor growth via β-catenin/TCF activation.
(A, B, C) In the upper panels, TCF reporter activity was measured using the β-catenin responsive TOPFLASH reporter and the mutant control FOPFLASH reporter. The effects of stable transfection of cells with dnTCF (black bars) and β-catenin shRNA (gray bars) on relative TCF activity are shown in HEK 293 (A) and MiaPaCa2 (B) cells (with empty or ATDC expression vector) or Panc1 cells (C) (with ATDC shRNA1 or 2 expression) (mean±SE, n=3, *p<0.05 vs control, non-treated cells). In the lower panels, the effects of stable transfection of dnTCF (black bars) and β-catenin shRNA (gray bars) on cell proliferation are shown in HEK 293 (A) and MiaPaCa2 (B) cells (with empty or ATDC expression vector) or Panc1 cells (C) (with ATDC shRNA1 or 2 expression) (mean ± SE, n=3, *p<0.05 vs control, non-treated cells). (D) Representative bioluminescent images of animals in the control and ATDC shRNA group are shown at 14 (left panels) and 60 (right panels) days after injection, depicting the extent of tumor burden. (E) Western blotting verifies downregulation of β-catenin in tumors derived from β-catenin shRNA transfected Panc1 cells (harvested at 60 days). (F) Average tumor volume measured in animals injected with control shRNA- and β-catenin shRNA-transfected Panc1 cells 60 days post-injection (mean ± SE, n=3, *p<0.05).
To examine the effects of β-catenin silencing on pancreatic tumor growth in vivo, we infected control or β-catenin shRNA-transduced Panc1 cells with a luciferase-expressing lentivirus. Following injection of 5 × 105 cells into the tail of the pancreas, tumor growth was assessed using bioluminescent imaging (n= 8 animals per group). All of the animals injected with Panc1 cells expressing control shRNA demonstrated tumor formation 14 days post-injection, while tumors were not detected in the animals injected with Panc1 cells expressing the β-catenin shRNA (Figure 4D, left panels, with three representative animals shown). At 60 days post-injection, the control tumors grew significantly larger, with evidence of metastatic spread, while the extent of primary tumor size and metastasis was markedly diminished in animals injected with Panc1 cells expressing the β-catenin shRNA (Figure 4D, right panels), similar to the effects observed with silencing ATDC in Panc1 cells (Figure 2E). Western blot analysis of tumors derived from β-catenin shRNA-transfected Panc1 cells harvested at 60 days after injection (Figure 4E) demonstrated effective silencing of β-catenin (Figure 4E). The mean tumor volume was significantly larger in the mice injected with Panc1-Luc cells expressing control shRNA compared to mice injected with Panc1-Luc cells expressing β-catenin shRNA (251.5±79.2 vs. 35.2±7.8 mm3, respectively, *p<0.05) (Figure 4F). Overall, these data indicate that ATDC plays a key role in cell proliferation and tumor growth, and ATDC’s growth promoting effects are dependent, at least in part, on the β-catenin/TCF signaling pathway.
Correlation between ATDC and β-catenin expression in pancreatic adenocarcinoma
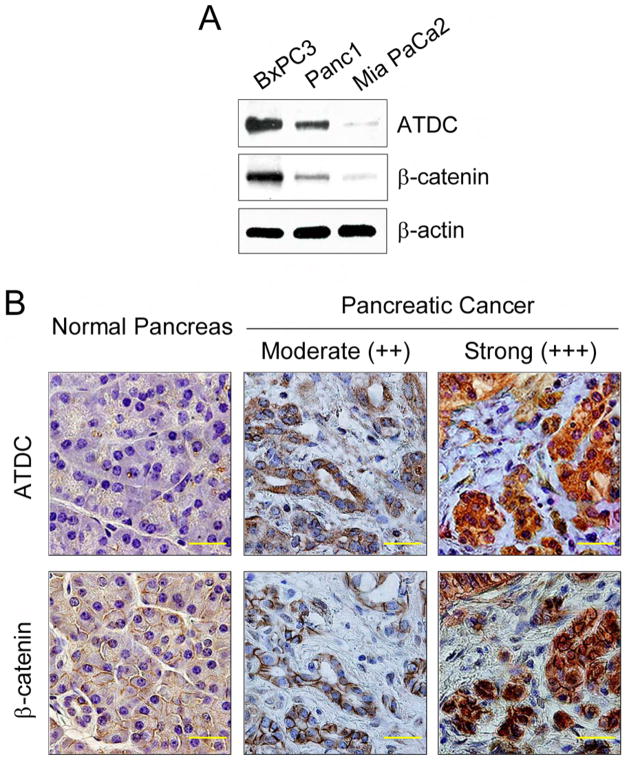
Based on our findings that ATDC is highly expressed in human pancreatic cancers and ATDC-mediated cancer cell growth in pancreatic cancer cells is dependent on β-catenin signaling, we predicted that ATDC and β-catenin expression levels would be well-correlated in pancreatic cancer cell lines and primary pancreatic cancers. As shown in Figure 5A, the levels of ATDC and β-catenin in pancreatic cancer cell lines correlated well, with the highest levels of β-catenin observed in BxPC3 cells, which have high levels of endogenous ATDC. Intermediate levels of β-catenin and ATDC proteins were found in Panc1 cells, and both β-catenin and ATDC were minimally expressed in MiaPaCa2 cells. To study further the relationship of ATDC and β-catenin expression in pancreatic cancer, immunohistochemical analysis of ATDC and β-catenin in human pancreatic normal and adenocarcinoma tissues was performed. We did not observe ATDC staining in normal human pancreas, but did observe expression of β-catenin localized to the cell membrane (Figure 5B, left panels), as has been previously described (Pasca di Magliano et al., 2007). It is commonly thought that both cytoplasmic and nuclear localization of β-catenin is an indicator of active β-catenin signaling in the Wnt pathway (Fodde and Brabletz, 2007). To assess the correlation between ATDC expression and β-catenin, we analyzed a pancreatic cancer tissue microarray (TMA) containing 47 pancreatic carcinoma samples. We observed focal nuclear staining for β-catenin in 5 (11%) tumors and elevated cytoplasmic levels of β-catenin in 24 (51%) tumors. These results correlate well with those previously published on β-catenin expression in pancreatic cancer (Pasca di Magliano et al., 2007). We observed a strong correlation between ATDC and β-catenin expression in the 47 pancreatic carcinoma cases, with moderate to high expression of ATDC in the cancer samples that showed elevated cytoplasmic and/or nuclear β-catenin expression (Supplemental Table 1). Representative samples of pancreatic cancers shown in Figure 5B (middle and right panels) demonstrate the correlation in ATDC and β-catenin expression in cancer samples with moderate and high levels of ATDC. Importantly, we did not find any evidence of pancreatic cancers expressing elevated levels of β–catenin without expressing high levels of ATDC, suggesting that elevated cytoplasmic and nuclear levels of β-catenin expression may in fact be dependent on elevated expression of ATDC.
Figure 5. Correlation between ATDC and β-catenin expression in pancreatic cancer.
(A) Western blot analysis of ATDC and β-catenin expression in BxPC-3, Panc-1, and MiaPaCa-2 cells. β-actin served as a loading control. (B) Immunohistochemical (IHC) staining of samples of normal human pancreas (left panels) and human pancreatic adenocarcinomas (middle and right panels). A correlation between ATDC and β-catenin expression in pancreatic adenocarcinoma samples is evident. The scale bar indicates 50 μm. IHC scores are: moderate (++, intermediate intensity staining) or strong (+++, intense staining).
ATDC interacts with disheveled-2 and components of the β-catenin destruction complex to stablize beta-catenin
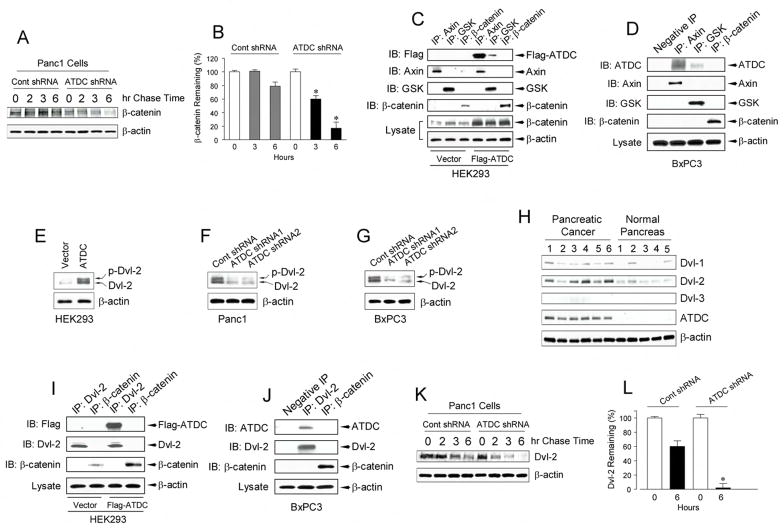
We were interested in defining the mechanism by which elevated levels of ATDC resulted in increased β-catenin expression. In the absence of Wnt ligands, cytoplasmic levels of β-catenin are regulated by a multi-protein complex, termed the destruction complex and which contains the Axin, APC, and glycogen synthase kinase 3-β (GSK3β). Axin and APC are believed to facilitate efficient phosphorylation of β-catenin by GSK3β at multiple serine and threonine residues in its N-terminus. Phosphorylated β-catenin is then ubiquinated, leading to its rapid proteosomal degradation (Gordon et al., 2006). To determine if ATDC increased β-catenin levels by stabilizing β-catenin, Panc1 cells (with or without ATDC silencing) were incubated with cycloheximide (CHX) (10 μg/ml) to prevent new β-catenin synthesis and β-catenin levels were then measured, with levels reflective of the rate of β-catenin protein degradation. Silencing of ATDC expression significantly increased the β-catenin degradation rate, resulting in a marked reduction in the levels of remaining β-catenin (Supplemental Figure S4A, B). This effect of ATDC on β-catenin stability was validated by performing a [35S]-methionine pulse-chase assay. We found that β-catenin was rapidly degraded in Panc1 cells where ATDC expression had been silenced compared to control Panc1 cells (Figure 6A, B).
Figure 6. ATDC stabilizes β-catenin by interacting with disheveled-2 and the β-catenin destruction complex.
(A) Pulse-chase assays in Panc1 cells (with or without ATDC silencing) were performed to determine β-catenin stability. (B) β-catenin remaining in (A) was quantitated by densitometry at hours 0, 3 and 6 and normalized relative to the 0 hour time point. Results are the mean ± SE of three independent experiments (*p<0.05 vs control shRNA cells at 6 hours). (C, D) Cell lysates from HEK 293 cells (C) transfected with empty vector or Flag-ATDC, and BxPC3 cells (D) were subjected to immunoprecipitation (IP) with Axin, GSK3β or β-catenin antibodies. Immunocomplexes were resolved by SDS-PAGE and subjected to western analysis with anti-Flag antibody for HEK293 cells (C) and ATDC antibody for BxPC3 cells (D). Blotting with an anti-β-actin antibody revealed equal loading. (E–G) Lysates of HEK 293 cells (E) transfected with empty vector or ATDC expression vector, and control shRNA- or ATDC shRNA1 or 2- expressing Panc1 (F) and BxPC3 (G) cells were subjected to Western blotting with an anti-Dvl-2 antibody. The upper arrow indicates the phosphorylated form and the lower arrow indicates the non-phosphorylated form of Dvl-2. Experiments were performed twice with similar results. (H) Western blotting of 5 samples each of pancreatic adenocarcinoma and normal pancreas. Dvl-1, Dvl-2, Dvl-3 and ATDC expression in pancreatic tissue samples was measured. The experiments were repeated twice with similar results. (I, J) Cell lysates from HEK 293 cells (I) transfected with empty vector or Flag-ATDC and BxPC3 cells (J) were subjected to immunoprecipitation (IP) with Dvl2 or β-catenin antibodies. Immunocomplexes were resolved by SDS-PAGE and subjected to western analysis with an anti-Flag antibody for HEK293 cells (I) and ATDC antibody for BxPC3 cells (J). Blotting with an anti-β-actin antibody showed equal loading. (K) Pulse-chase assays were performed in Panc1 cells (with or without ATDC silencing) to determine Dvl-2 stability. (L) Dvl-2 remaining in (K) was quantitated by densitometry at hours 0 and 6 and normalized relative to the 0 hour time point. Results are the mean ± SE of three independent experiments (*p<0.05 vs control shRNA cells at 6 hours).
To ascertain the mechanism by which ATDC stabilizes β-catenin, we first examined whether ATDC interacted with the components of the multi-protein complex that regulates β-catenin stability. In co-immunoprecipitation experiments using antibodies directed to Axin, GSK-3β and β-catenin, evidence of complexes containing ATDC and Axin and complexes containing ATDC and GSK-3β were observed in HEK 293 cells transfected with a Flag-tagged ATDC construct (Figure 6C). To verify that this interaction was physiologically relevant, we tested the ability of endogenous ATDC in BxPC3 cells to interact with Axin and GSK3β and similar results were obtained (Figure 6D). ATDC did not interact with β-catenin in either HEK 293 cells over-expressing ATDC or in BxPC3 cells (Figures 6C, 6D). These results are consistent with the possibility that ATDC may interfere with GSK-3β– dependent phosphorylation of β-catenin by the destruction complex.
Based on the findings, we examined if ATDC might activate disheveled (Dvl), a cytoplasmic protein that is activated by binding of Wnt ligands to the frizzled/LRP co-receptor at the cell surface. Activated Dvl binds to the Axin/GSK3β complex and antagonizes GSK-3β dependent phosphorylation of β-catenin in a manner not dissimilar to what we observed with ATDC. Indeed, we found increased Dvl-2 levels (total and phosphorylated forms) in HEK 293 cells over-expressing ATDC as well as in Panc1 and BXPC3 cells (Figures 6E–G). Furthermore, silencing of endogenous ATDC expression in Panc1 and BxPC3 cells reduced the expression of Dvl-2 (Figures 6F, 6G). Increased levels of Dvl-2 were present in primary pancreatic cancer samples that had elevated expression of ATDC (Figure 6H). Finally, immunohistochemical analysis revealed co-localization of ATDC and Dvl-2 in ATDC-transfected HEK 293 cells and Panc1 cells (Supplemental Figure S5). Co-immunoprecipation experiments in HEK 293 cells and BxPC3 cells demonstrated that ATDC formed a complex with Dvl-2 (Figures 6I, 6J). Complex formation between ATDC and Dvl-1 or Dvl-3 in either ATDC-transfected HEK 293 or BxPC3 cells was not seen (data not shown). To determine which region of the ATDC molecule interacted with Dvl-2, we created a series of Flag-tagged ATDC truncation mutants and performed co-immunoprecipitation experiments. We found that the ATDCΔ260 deletion mutant was able to interact with Dvl-2 while the ATDCΔ348 mutant did not, suggesting that amino acids 260–348 of ATDC, in, a region which contains a coiled-coil domain, interacted with Dvl-2 (Supplemental Figure S6).
We then tested whether modulating the levels of ATDC in cells affected the abundance of Dvl-2 transcripts. Dvl-2 gene expression, as measured by quantitative real time RT-PCR, was not altered in either HEK 293 cells overexpressing ATDC or in Panc1 cells with ATDC knockdown compared to control cells (Supplemental Figure S7). To examine if ATDC might be altering Dvl-2 levels by affecting protein stability. [35S]-methionine pulse-chase assays were performed. Dvl-2 was rapidly degraded in Panc1 cells with ATDC knockdown compared to control Panc1 cells (6K,6L), suggesting that ATDC forms a complex with Dvl-2 and increases Dvl-2 levels by regulating Dvl-2 post-transcriptionally, perhaps via direct effects on Dvl-2 protein stability.
The oncogenic effects of ATDC are mediated by Dvl-2
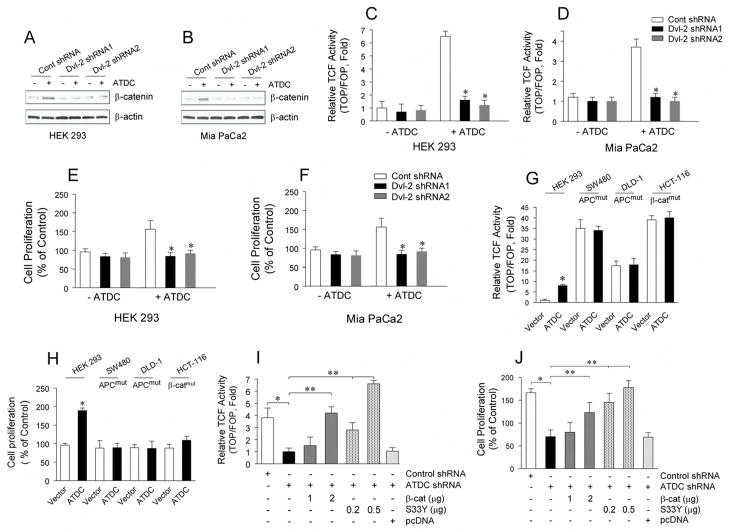
To verify that the ability of ATDC to increase TCF activity and cellular proliferation is mediated through Dvl-2, we transfected two Dvl-2 shRNA constructs targeting different regions of Dvl-2 into HEK 293 and MiaPaCa2 cells and examined the effects of Dvl-2 knockdown on ATDC function. Both Dvl-2 shRNA1 and 2 were effective in knocking down levels of Dvl-2 in HEK 293 and MiaPaCa2 cells (Supplemental Figure S8). Knockdown of Dvl-2 in both HEK 293 and MiaPaCa2 cells overexpressing ATDC inhibited increases in β-catenin levels (Figures 7A,7B), TCF activity (Figures 7C,7D) and cell proliferation (Figure 7E,7F) induced by ATDC.
Figure 7. The oncogenic effects of ATDC are mediated by Dvl-2.
(A. B) The effects of Dvl-2 shRNA 1 or 2 on β-catenin expression is shown in representative western blots of HEK 293 (A) and MiaPaCa2 cells (B) with or without ATDC overexpression. (C.D) TCF reporter activity was assessed in Dvl-2 shRNA 1 or 2-transfected HEK 293 (C) or Mia PaCa2 cells (D) with (+) or without (−) ATDC overexpression. (mean±SE, n=3, *p<0.05 vs empty vector-transfected cells). (E. F) MTS proliferation assays in Dvl-2 shRNA1 or 2-transfected HEK 293 (E) and MiaPaCa2 (F) cells with (+) or without (−)ATDC overexpression (mean±SE, n=3, *p<0.05 vs wild type cells). (G) TCF reporter activity was measured in HEK 293, SW480, DLD-1 and HCT-116 cells with vector (white bars) or ATDC (black bars) transfection using the β-catenin responsive TOPFLASH reporter and the mutant control FOPFLASH reporter (mean±SE, n=3, *p<0.05). (H) Cell proliferation of empty vector- and ATDC expression vector-transfected HEK 293, SW480, DLD-1 and HCT-116 cells are shown (mean±SE, n=3, *p<0.05 vs control, non-treated cells). (I) Varying amounts of wild type β-catenin (β-Cat) or constitutively active mutant β-catenin (S33Y) constructs with TOPFLASH or FOPFLASH reporter constructs were co-transfected into Panc1 cells with control shRNA or ATDC shRNA expression. 48 hours after transfection, TOPFLASH reporter assays were performed. (mean±SE, n=3, *p<0.05, control shRNA vs ATDC shRNA; **p<0.01, Panc1 cells (ATDC shRNA) with β-catenin vs. without β-catenin; and ***p<0.001, Panc1 cells (ATDC shRNA) with S33Y vs without S33Y). (J) Varying amounts of wild type β-catenin (β-Cat) or constitutively active mutant β-catenin (S33Y) constructs were co-transfected into Panc1 cells with control shRNA or ATDC shRNA expression. 48 hours after transfection, cell growth rates were assessed. The experiments were repeated three times and the data is expressed as the mean + SE. (*p<0.05, control shRNA vs ATDC shRNA; **p<0.01, Panc1 cells (ATDC shRNA) with β-catenin vs. without β-catenin; and ***p<0.001, Panc1 cells (ATDC shRNA) with S33Y vs without S33Y).
If ATDC stabilizes β-catenin levels by acting on upstream signaling events mediated by Dvl-2, then modulating ATDC levels should not influence β-catenin levels in cell lines with either APC or β-catenin mutations. Indeed, over-expression of ATDC in the APC mutant SW480 and DLD-1 cell lines or the β-catenin mutant HCT-116 cell line did not alter TCF activity or cell proliferation as compared to control cells (7G and 7H). Our findings also predicted that the decrease in TOPFLASH reporter caused by silencing of ATDC in Panc1 cells would be reversed by increasing the levels of β-catenin in the cells. Silencing of ATDC in Panc1 cells significantly inhibited the increase in TOPFLASH reporter activity seen in control Panc1 cells and transfection with wild type or a mutant oncogenic form of β-catenin (S33Y β-catenin) was able to reverse the inhibitory effect of silencing of ATDC on TOPFLASH reporter activity (Figure 7I) and cell proliferation (Figure 7J) in a dose-dependent fashion.
DISCUSSION
Our studies have identified ATDC as a protein highly expressed in the majority of human pancreatic adenocarcinomas and pancreatic cancer precursor lesions. We also demonstrated that expression of ATDC in pancreatic cancer cells promoted cellular proliferation and enhanced tumor growth and metastasis. Additionally, we provided evidence that elevated levels of ATDC expression correlated with elevated β-catenin levels in pancreatic cancer cell lines and primary pancreatic cancers, and that silencing of ATDC via shRNA approaches antagonized β-catenin/TCF-mediated reporter activation and activation of TCF target genes. β-catenin was implicated in the oncogenic effects of ATDC in vitro and in vivo, and the ability of ATDC to increase β-catenin levels appeared to be attributable to ATDC’s effects on disheveled-2 protein expression. In summary, our findings implicate ATDC as an important positive regulator of β-catenin-dependent signaling in pancreatic cancer.
ATDC has been reported to be up-regulated in a number of different cancer types, including lung, bladder, colorectal, ovarian and endometrial cancers and multiple myeloma (Dyrskjot et al., 2004; Glebov et al., 2006; Hawthorn et al., 2006; Mutter et al., 2001; Ohmachi et al., 2006; Santin et al., 2004; Zhan et al., 2002). A recent report correlated ATDC expression in gastric cancer and poor histological grade, large tumor size, extent of tumor invasion, and lymph node metastasis (Kosaka et al., 2007). ATDC has also been reported to be down-regulated in some cancer types (Smith et al., 2005, Nacht et al., 1999; Zhang et al., 2006; Ernst et al., 2002) suggesting the function of ATDC may be depend on cellular context. In none of these reports was the role of ATDC in tumorigenesis examined in functional studies.
We found that ATDC upregulated β-catenin levels in pancreatic cancer cell lines and primary pancreatic cancers. A large body of data supports the contribution of activation of the canonical (β-catenin-dependent) Wnt signaling pathway in the development of colorectal cancer. Sustained β-catenin pathway activation independent of APC, Axin1 or β-catenin mutations has been demonstrated in a subset of breast and ovarian cancer (Bafico et al., 2004). Mutations in APC or β-catenin appear to be rare in pancreatic adenocarcinoma (Zeng et al., 2006). While robust activation of the pathway due to signature mutations in components of the β-catenin signaling cascade that are commonly observed in other gastrointestinal cancers are not present in pancreatic adenocarcinoma, immunohistochemical analysis of β-catenin suggests a possible contribution of β-catenin signaling during PanIN progression and the development of invasive pancreatic cancer. Increased levels of both cytoplasmic and nuclear β–catenin, indicative of β-catenin signaling activity, have been reported in a substantial group of pancreatic adenocarcinomas and PanIN s (Zeng et al., 2006; Pasca di Magliano et al., 2007). Pasca di Magliano and colleagues reported that 65% of pancreatic cancers have an increase in either cytoplasmic and/or nuclear β-catenin. Similar results were obtained in Pdx-Cre;KrasG12D and Pdx-cre;KrasG12D;p53f/+ transgenic mice that developed PanIN lesions and subsequent invasive pancreatic cancers that were phenotypically indistinguishable from human pancreatic adenocarcinomas. Moreover, the authors showed that cancer cell survival and proliferation, depended on β-catenin signaling activity in multiple human pancreatic cancer cell lines.
The mechanisms by which ATDC levels are upregulated in pancreatic adenocarcinomas remain unclear. Interestingly, Pasca di Magliano and colleagues demonstrated that increased hedgehog signaling, one of the earliest changes in PanIN lesions, activated β-catenin signaling in transgenic mice and untransformed pancreatic duct cells, suggesting that hedgehog may play a role in upregulating β-catenin activity in some pancreatic adenocarcinomas. A possible connection between hedgehog upregulation and ATDC expression in human pancreatic cancer cell lines or primary tumors remains to be explored.
The levels of the free cytoplasmic pool of β-catenin are known to be regulated by Wnt ligands. In the absence of an activating Wnt signal, mediated via Wnt binding to the frizzled-low density lipoprotein-related (LRP) 5/6 co-receptor complex, cytoplasmic β-catenin is destabilized by a multiprotein complex containing axin, GSK3β, and APC. Axin acts as the scaffold of this complex and interacts with the other components- β catenin, APC, and GSK3β. Interaction of GSK3β with Axin in the complex facilitates efficient phosphorylation of β-catenin by GSK3β. Phosphorylated β-catenin is then ubiquinated, leading to its rapid proteosomal degradation. We found that ATDC bound to Axin and GSK-3β in pancreatic cancer cells, suggesting that ATDC interacted with the destruction complex to prevent phosphorylation and subsequent ubiquination of β-catenin.
When Wnt binds to the frizzled/LRP co-receptors at the cell surface, a cytoplasmic protein, Dvl, antagonizes GSK-3β dependent phosphorylation of β-catenin. Although it is not known if Dvl binds directly to the frizzled/LRP co-receptor or whether intermediary proteins are involved in the signal transduction between frizzled and Dvl, Dvl appears to bind to axin and inhibit GSK-3β-dependent phosphorylation of β-catenin, APC and axin. Once the phosphorylation of β-catenin is reduced, β-catenin dissociates from the axin complex, resulting in its accumulation in the cytoplasm. Once stabilized, a fraction of the β-catenin is translocated to the nucleus, where it binds to transcription factors such as the TCF/lymphoid enhancer binding factor and thereby stimulates the transcription of β-catenin target genes. We noted that ATDC expression in HEK 293 cells induced expression of Dvl-2, and demonstrated that ATDC formed a complex with Dvl-2 in pancreatic cancer cells. We demonstrated that levels of ATDC in primary pancreatic cancers correlated well with Dvl-2 levels, suggesting that ATDC upregulates Dvl-2 levels in primary pancreatic cancers, and increases β-catenin levels by this mechanism. We further show that knockdown of Dvl-2 in ATDC expressing cells abrogates enhanced TCF activity and cell proliferation induced by ATDC, directly implicating Dvl-2 as an intermediary in this process.
The regulation of the disheveled protein is still poorly understood, but recent data suggest that disheveled, like β-catenin, may be controlled by ubiquination and degradation by the proteosome (Hershko et al., 1998; Simons et al., 2005; Miyazaki et al., 2004). Dvl-1 has been reported to interact with the neuronal Homologous to E6AP carboxyl terminus (HECT)-type ubiquitin ligase NEDL1 (Miyazaki et al., 2004). The proteins inversin and the interactions between PP2A phosphatase and the Wnt-induced antagonist naked cuticle have been shown to modulate the stability of Dvl-1 (Simons et al., 2005; Creyghton et al., 2005). And, in a recent manuscript published by Angers and colleagues, KLHL12-Cullin-3 ubiquitin ligase was shown to negatively regulate the Wnt-β-catenin pathway by targeting disheveled for degradation (Angers et al., 2006). We found that ATDC did not increase Dvl-2 levels by changes in Dvl-2 gene expression but rather by enhancing the stability of the Dvl-2 protein, supporting changes in Dvl stability serve as a important mechanism in regulating the Wnt/β-catenin signaling pathway.
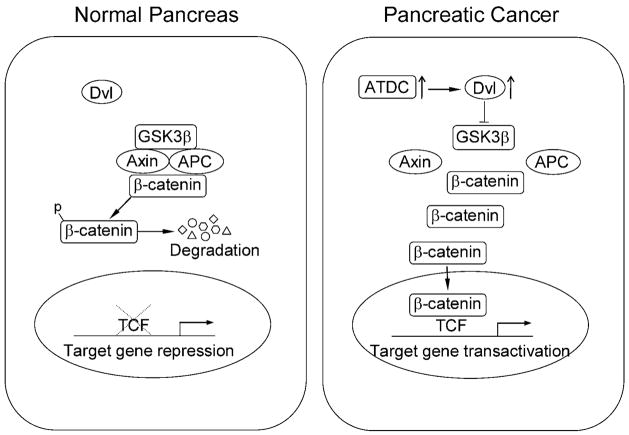
The data in the present study support a model for the mechanism by which ATDC functions to promote the oncogenesis of pancreatic cancer cells (Figure 8). In unstimulated, normal pancreatic cells lacking ATDC, Dvl-2 is in the cytoplasm and is not bound to the Axin/Gsk-3β/APC destruction complex. This allows the destruction complex to phosphorylate β-catenin and target it for ubiquitin-mediated degradation. In pancreatic cancer cells expressing high levels of ATDC, ATDC binds to and stabilizes Dvl-2, resulting in the release of β-catenin from the destruction complex, increased β-catenin levels and subsequent activation of downstream β-catenin/TCF-regulated target genes These studies define for the first time a functional role of ATDC in human tumorigenesis and besides highlighting ATDC as a potential therapeutic target in pancreatic cancer, have defined a novel mechanism for activating Wnt/β-catenin signaling in cancer.
Figure 8. Model of how ATDC mediates activation of β-catenin signaling in pancreatic cancer cells.
(Left panel). In unstimulated normal pancreatic cells lacking ATDC, disheveled-2 (Dvl-2) is in the cytoplasm and is not bound to the Axin/GSK-3β/APC destruction complex. This allows the destruction complex to phosphorylate β-catenin and target it for ubiquitin-mediated degradation. (Right panel). In pancreatic cancer cells expressing high levels of ATDC, ATDC binds to a stabilized Dvl-2, bringing it to the β-catenin destruction complex. Binding of the ATDC and Dvl-2 to the destruction complex inhibits destruction complex function, resulting in the release of β-catenin from the destruction complex, leading to increased β-catenin levels and subsequent activation of downstream target genes.
EXPERIMENTAL PROCEDURES
Cell Lines and Human Samples
The human pancreatic ductal adenocarcinoma cell lines Panc1, Mia PaCa2 and BxPC3 and the human embryonic kidney cell line HEK 293 were purchased from American Type Culture Collection (ATCC, Manassas, VA). Fresh frozen and paraffin-embedded human pancreatic tissues were obtained from patients undergoing surgical resection at the University of Michigan Medical Center. All human samples were obtained following approval by the Institutional Review Board of the University of Michigan Medical Center. A pancreatic cancer tissue microarray constructed by the University of Michigan Medical Center’s Tissue Procurement Facility contained 5 samples of normal pancreas and 47 samples of pancreatic adenocarcinoma and was used for immunohistochemical analysis of ATDC expression. Twenty-five paraffin-embedded human pancreatic tissue samples containing pancreatic intraductal neoplasia (PanIn) lesions of various stages were analyzed for ATDC expression using an anti-ATDC antibody. ATDC expression in PanIn lesions was evaluated by an experienced pancreatic pathologist and graded as absent or present.
Constructs
The complementary DNA (cDNA) of human ATDC (kindly provided by J. Murnane, University of California, San Franscisco) was subcloned into the pcDNA3.1 expression vector (Invitrogen, San Diego, CA). Sequence analysis after cloning showed 100% homology to the published sequence of ATDC. The methods used to create the control shRNA and ATDC shRNA constructs are listed in the Supplemental Experimental Procedures Endogenous Dvl-2 was knocked down by transient transfection of specific Dvl-2 shRNA1 and 2 (OriGene Technologies, Rockville, MD) in HEK 293 and Mia PaCa2 cells with or without ATDC overexpression. A reporter plasmid carrying three TCF binding sites upstream of a minimal c-fos promoter driving the firefly luciferase gene (TOP Flash), the plasmid carrying the mutated TCF binding sites upstream of a minimal c-fos promoter driving luciferase expression (FOP Flash), and the expression constructs of wild type or constitutively active β-catenin containing a missense mutation of tyrosine for serine at codon 33 (S33Y) were used (Caca et al., 1999). The PGEX-2T vector containing C-terminal E-cadgherin/glutathione S-transferase (GST) was generated as previously described (Winer et al., 2006).
Creation of Stable Cell Lines
Details regarding the creation of HEK 293 and MiaPaCa2 cell lines with stable expression of ATDC and Panc1 and BxPC-3 cells with silencing of ATDC using shRNA can be found in the Supplemental Experimental Procedures.
Quantitative Real-Time RT-PCR
Total RNA from the human pancreatic ductal adenocarcinoma, normal pancreas and chronic pancreatitis specimens or Panc1 cells (with or without ATDC shRNA) were isolated using TRIzol reagent (Invitrogen, Carlsbad, CA). To avoid amplification of genomic DNA, RNA was pretreated with DNase (DNA-free kit from Applied Biosystems, Foster City, CA). cDNA synthesis was performed using a High Capacity cDNA Archive Kit (Applied Biosystems, Foster City, CA). The cDNA served as a template in quantitative real-time PCR utilizing TaqMan Fast Universal PCR Master Mix and TaqMan Gene Expression assay probes for ATDC (Hs00232590_m1), Dvl-2 (Hs01005253_ml) or ribosomal protein S6 (RPS6)(Hs02339423_g1) (Applied Biosystems Foster City, CA) and an ABI 7500 Fast Sequence Detection System. All reactions were done in triplicate. ATDC mRNA expression of different group specimens were normalized to endogenous ribosomal protein S6. Ct is the threshold cycle value defined as the fractional cycle number at which the target fluorescent signal passes a fixed threshold above baseline. Relative ATDC mRNA levels were presented as unit values of 2−ΔCt =2−(Ct (Ribosomal protein S6) −Ct (ATDC)).
Immunohistochemical Analysis
The paraffin-embedded pancreatic tissue sections (4-μm-thick) were cut, deparaffinized, and subjected to a heat-induced epitope retrieval step. Endogenous peroxidase activity was blocked with 1% (v/v) hydrogen peroxide in distilled water. Details of the immunohistochemical analysis can be found in the Supplemental Experimental Procedures.
Luciferase Reporter Gene Assays
Pancreatic cancer cells (with or without β-catenin silencing) or HEK 293 and Mia PaCa2 cells (with or without ATDC overexpression) were transfected using the LipofectAMINE 2000 Transfection kit (Invitrogen Carlsbad, CA) according to the manufacturer’s instructions with 0.2 μg of the TOPFLASH or FOPFLASH reporter constructs with or without varying amounts (0.2–2 μg) of constructs expressing either wild type β-catenin or the S33Y β-catenin (constitutively activated) mutant and 25 ng of a β-galactosidase construct as an internal control to normal luciferase activity to tranfection efficiency. The total DNA for each transfection was kept constant by adding empty pcDNA vector. Forty eight hours after transfection, luciferase activity was measured in a luminometer and normalized to β-galactosidase expression. Endogenous β-catenin or Dvl-2 was silenced by stably expression β-catenin shRNA or transient expression of Dvl-2 shRNAs (OriGene Technologies, Rockville, MD). TCF transcriptional activity was inhibited by transiently transfection of the dnTCF vector (Upstate, Temecula, CA).
Nuclear and Membrane Fractionation
Nuclear and cytoplasmic proteins were extracted using NE-PER Nuclear and Cytoplasmic Extraction Reagents (Pierce, Rockford, IL). Membrane proteins were extracted using the Mem-PER Eukaryotic Membrane Protein Extraction Kit (Pierce, Rockford, IL) according to the manufacturer’s instructions.
C-terminal E-cadherin/Glutathione S-Transferase (GST) Fusion-mediated precipitation of the free β-catenin pool
The free pool of β-catenin in ATDC transfected and control HEK 293 cells was measured using a C-terminal E-cadherin/glutathione S-transferase fusion protein as previously described (Winer et al., 2006).
Immunoblot analysis
Immunoblot analysis was done as previously described (Zhang et al., 2004). Details of the immunoblot analysis analysis can be found in the Supplemental Experimental Procedures.
Proliferation Assay
Cell proliferation was measured using a CellTiter 96 AQ nonradioactive cell proliferation assay (Promega, Madison, WI) as we have previously described (Zhang et al., 2004).
Co-Immunoprecipitation Experiments
Cells were lysed by sonicating for 5 sec in 1 ml of detergent free lysis buffer (PBS, 5 mM EDTA, 0.02% sodium azide, 10 mM iodoacetamide, 1 mM PMSF and 2 μg leupeptin) at 4°C. The lysates were cleared by microcentrifuging for 15 minutes at 16,000 × g at 4°C. Antibody conjugated beads were prepared by combining 1 μg of polyclonal antibodies with 30 μl of a 50% protein A-Sepharose bead slurry in 0.5 ml of ice-cold PBS for 1 hr at 4°C in a tube rotator and then were washed two times with 1 ml of lysis buffer. The antibodies used for co-immunoprecipitation were: Dvl-2, β-catenin (Cell Signaling Technology, Beverly, MA), axin, and GSK3β (Upstate, Temecula, CA). Cell lysates (500 μg) were incubated with the prepared beads and 10 μl of 10% BSA overnight at 4°C. The beads were washed four times with washing buffer (50 mM Tris-Hcl [pH 7.4], 300 mM NaCl, 5 mM EDTA, 0.02% sodium azide, 0.1% Triton X-100) and one time with ice-cold PBS. Proteins were revealed after SDS-PAGE and Western blotting with antibodies to Flag (Sigma, St Louis, MO), Dvl-2, β-catenin, axin, GSK3β, and ATDC (Santa Cruz Biotechnology, Santa Cruz, CA). Images were visualized using an ECL detection system.
Pulse Chase Assays
Cells were cultured in 6 well plates to 70% confluence and then starved in 2 ml methionine-deficient DMEM (Sigma; Madison, WI) for 30 min at 37°C with 5% CO2 Cells were pulse-labeled with 100 μCi/ml [35S]-methionine (Amersham Pharmacia, Pittsburgh, PA) for 30 minutes at 37°C. Labeled cells were chased in DMEM with a saturating amount of cold methionine (2 mM) for various times and lysed in RIPA buffer (10 mM Tris, pH 7.2, 158 mM NaCl, 1 mM EGTA, 1% Triton X-100, 1% deoxycholate, 0.1% SDS, 1mM PMSF, and 1x concentration of protease inhibitors (Complete Protease Inhibitor Cocktail, Roche Molecular Biochemicals)). Total cell lysates were then prepared and immunoprecipitated with a β-catenin antibody or Dvl-2 antibody and protein G-Sepharose beads (Invitrogen, San Diego, CA) for 3 hours at 4°C. Immunocomplexes were separated on SDS-PAGE (4–20% gradient gel) and transferred to nitrocellulose. The precipitates were subjected to autoradiography, and then the densities of the labeled proteins were analyzed by Kodak Gel Documentation System (model 1D 3.6).
In Vivo Tumorigenicity Studies
Six-week-old male NOD/SCID (non-obese diabetic/severe combined immunodeficient) mice (Taconic, Germantown, NY) were housed under pathogen-free conditions. Animal experiments were approved by the University of Michigan Animal Care and Use Committee and were performed in accordance with their established guidelines. Mice were anesthetized with an i.p injection of xylazine (9 mg/kg) and ketamine (100 mg/kg). A median laparotomy was done and 5 × 105 Panc1 cells infected with a lentivirus encoding luciferase (control shRNA, ATDC-shRNA1 or β-Catenin shRNA stably transfected) in a volume of 30 μl were injected into the pancreatic tail using a 30-gauge needle (n= 8 per group). To prevent leak at the injection site, the needle was slowly withdrawn and a sterile cotton swab was applied to the injection site for 30 sec. Bioluminescent imaging of the mice was performed bi-weekly using a Xenogen IVIS 200 imaging system (Xenogen Biosciences, Cranbury, NJ). Sixty days following cancer cell injection, mice were euthanized with carbon dioxide inhalation, and autopsies were performed to assess the extent of primary tumor growth and metastasis.
Statistical Analysis
Data are represented as mean ± SE from at least three independent experiments. The significance of the difference between groups was evaluated by Student’s t test or ANOVA test. p < 0.05 was considered significant.
Supplementary Material
Acknowledgments
This work was funded by NIH grant R01 CA131045 (DMS) and grant support from the Lustgarten Foundation (DMS). Special thanks to Dr. Marina Pasca di Magliano for her useful comments regarding β-catenin signaling in pancreatic cancer.
Abbreviations
- ATDC
ataxia telangiectasia group D complementing gene
- TCF
T-cell factor
- DKK1
Dickkopf homolog 1
- Dvl
Disheveled
- GSK3β
Glycogen synthase kinase 3β
Footnotes
Supplemental Data
The Supplemental Data can be found with this article online at
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Angers S, Thorpe CJ, Biechele TL, Goldenberg SJ, Zheng N, MacCoss MJ, Moon RT. The KLHL12-Cullin-3 ubiquitin ligase negatively regulates the Wnt-β-catenin pathway by targeting dishevelled for degradation. Nature Cell Biol. 2006;8:348–357. doi: 10.1038/ncb1381. [DOI] [PubMed] [Google Scholar]
- Arumugam T, Simeone DM, Van Golen K, Logsdon CD. S100P promotes pancreatic cancer growth, survival, and invasion. Clin Cancer Res. 2005;11:5356–5364. doi: 10.1158/1078-0432.CCR-05-0092. [DOI] [PubMed] [Google Scholar]
- Bafico A, Liu G, Goldin L, Harris V, Aaronson SA. An autocrine mechanism for constitutive Wnt pathway activation in human cancer cells. Cancer Cell. 2004;6:497–506. doi: 10.1016/j.ccr.2004.09.032. [DOI] [PubMed] [Google Scholar]
- Caca K, Kolligs FT, Ji X, Hayes M, Qian J, Yahanda A, Rimm DL, Costa J, Fearon ER. Beta- and gamma-catenin mutations, but not E-cadherin inactivation, underlie T-cell factor/lymphoid enhancer factor transcriptional deregulation in gastric and pancreatic cancer. Cell Growth Differ. 1999;10:369–376. [PubMed] [Google Scholar]
- Cao D, Hustinx SR, Sui G, Bala P, Sato N, Martin S, Maitra A, Murphy KM, Cameron JL, Yeo CJ, et al. Identification of novel highly expressed genes in pancreatic ductal adenocarcinomas through a bioinformatics analysis of expressed sequence tags. Cancer Biol Ther . 2004;3:1081–1089. doi: 10.4161/cbt.3.11.1175. discussion 1090–1081. [DOI] [PubMed] [Google Scholar]
- Chen R, Yi EC, Donohoe S, Pan S, Eng J, Cooke K, Crispin DA, Lane Z, Goodlett DR, Bronner MP, et al. Pancreatic cancer proteome: the proteins that underlie invasion, metastasis, and immunologic escape. Gastroenterology. 2005;129:1187–1197. doi: 10.1053/j.gastro.2005.08.001. [DOI] [PubMed] [Google Scholar]
- Crnogorac-Jurcevic T, Missiaglia E, Blaveri E, Gangeswaran R, Jones M, Terris B, Costello E, Neoptolemos JP, Lemoine NR. Molecular alterations in pancreatic carcinoma: expression profiling shows that dysregulated expression of S100 genes is highly prevalent. J Pathol. 2003;201:63–74. doi: 10.1002/path.1418. [DOI] [PubMed] [Google Scholar]
- Dyrskjot L, Kruhoffer M, Thykjaer T, Marcussen N, Jensen JL, Moller K, Orntoft TF. Gene expression in the urinary bladder: a common carcinoma in situ gene expression signature exists disregarding histopathological classification. Cancer Res. 2004;64:4040–4048. doi: 10.1158/0008-5472.CAN-03-3620. [DOI] [PubMed] [Google Scholar]
- Fodde R, Brabletz T. Wnt/beta-catenin signaling in cancer stemness and malignant behavior. Curr Opin Cell Biol. 2007;19:150–158. doi: 10.1016/j.ceb.2007.02.007. [DOI] [PubMed] [Google Scholar]
- Goddard AD, Borrow J, Freemont PS, Solomon E. Characterization of a zinc finger gene disrupted by the t(15;17) in acute promyelocytic leukemia. Science. 1991;254:1371–1374. doi: 10.1126/science.1720570. [DOI] [PubMed] [Google Scholar]
- Gregorieff A, Clevers H. Wnt signaling in the intestinal epithelium: from endoderm to cancer. Genes Dev. 2005;19:877–890. doi: 10.1101/gad.1295405. [DOI] [PubMed] [Google Scholar]
- Hoseong Yang S, Andl T, Grachtchouk V, Wang A, Liu J, Syu LJ, Ferris J, Wang TS, Glick AB, Millar SE, Dlugosz AA. Pathological responses to oncogenic Hedgehog signaling in skin are dependent on canonical Wnt/beta-catenin signaling. Nat Genet. 2008 doi: 10.1038/ng.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosoi Y, Kapp LN. Expression of a candidate ataxia-telangiectasia group D gene in cultured fibroblast cell lines and human tissues. Int J Radiat Biol. 1994;66:S71–76. [PubMed] [Google Scholar]
- Hruban RH, Goggins M, Parsons J, Kern SE. Progression model for pancreatic cancer. Clin Cancer Res. 2000;6:2969–2972. [PubMed] [Google Scholar]
- Jemal A, Siegel R, Ward E, Murray T, Xu J, Thun MJ. Cancer statistics, 2007. CA Cancer J Clin. 2007;57:43–66. doi: 10.3322/canjclin.57.1.43. [DOI] [PubMed] [Google Scholar]
- Kosaka Y, Inoue H, Ohmachi T, Yokoe T, Matsumoto T, Mimori K, Tanaka F, Watanabe M, Mori M. Tripartite motif-containing 29 (TRIM29) is a novel marker for lymph node metastasis in gastric cancer. Ann Surg Oncol. 2007;14:2543–2549. doi: 10.1245/s10434-007-9461-1. [DOI] [PubMed] [Google Scholar]
- LaTulippe E, Satagopan J, Smith A, Scher H, Scardino P, Reuter V, Gerald WL. Comprehensive gene expression analysis of prostate cancer reveals distinct transcriptional programs associated with metastatic disease. Cancer Res. 2002;62:4499–4506. [PubMed] [Google Scholar]
- Logsdon CD, Simeone DM, Binkley C, Arumugam T, Greenson JK, Giordano TJ, Misek DE, Kuick R, Hanash S. Molecular profiling of pancreatic adenocarcinoma and chronic pancreatitis identifies multiple genes differentially regulated in pancreatic cancer. Cancer Res. 2003;63:2649–2657. [PubMed] [Google Scholar]
- Lowe AW, Olsen M, Hao Y, Lee SP, Taek Lee K, Chen X, van de Rijn M, Brown PO. Gene expression patterns in pancreatic tumors, cells and tissues. PLoS ONE. 2007;2:e323. doi: 10.1371/journal.pone.0000323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo J, Duggan DJ, Chen Y, Sauvageot J, Ewing CM, Bittner ML, Trent JM, Isaacs WB. Human prostate cancer and benign prostatic hyperplasia: molecular dissection by gene expression profiling. Cancer Res. 2001;61:4683–4688. [PubMed] [Google Scholar]
- Lustig B, Behrens J. The Wnt signaling pathway and its role in tumor development. J Cancer Res Clin Oncol. 2003;129:199–221. doi: 10.1007/s00432-003-0431-0. [DOI] [PubMed] [Google Scholar]
- Mutter GL, Baak JP, Fitzgerald JT, Gray R, Neuberg D, Kust GA, Gentleman R, Gullans SR, Wei LJ, Wilcox M. Global expression changes of constitutive and hormonally regulated genes during endometrial neoplastic transformation. Gynecol Oncol. 2001;83:177–185. doi: 10.1006/gyno.2001.6352. [DOI] [PubMed] [Google Scholar]
- Pasca di Magliano M, Biankin AV, Heiser PW, Cano DA, Gutierrez PJ, Deramaudt T, Segara D, Dawson AC, Kench JG, Henshall SM, et al. Common activation of canonical wnt signaling in pancreatic adenocarcinoma. PLoS ONE. 2007;2:e1155. doi: 10.1371/journal.pone.0001155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reymond A, Meroni G, Fantozzi A, Merla G, Cairo S, Luzi L, Riganelli D, Zanaria E, Messali S, Cainarca S, et al. The tripartite motif family identifies cell compartments. Embo J. 2001;20:2140–2151. doi: 10.1093/emboj/20.9.2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savitsky K, Bar-Shira A, Gilad S, Rotman G, Ziv Y, Vanagaite L, Tagle DA, Smith S, Uziel T, Sfez S, et al. A single ataxia telangiectasia gene with a product similar to PI-3 kinase. Science. 1995;268:1749–1753. doi: 10.1126/science.7792600. [DOI] [PubMed] [Google Scholar]
- Stremlau M, Owens CM, Perron MJ, Kiessling M, Autissier P, Sodroski J. The cytoplasmic body component TRIM5alpha restricts HIV-1 infection in Old World monkeys. Nature. 2004;427:848–853. doi: 10.1038/nature02343. [DOI] [PubMed] [Google Scholar]
- van Noort M, Meeldijk J, van der Zee R, Destree O, Clevers H. Wnt signaling controls the phosphorylation status of beta-catenin. J Biol Chem. 2002;277:17901–17905. doi: 10.1074/jbc.M111635200. [DOI] [PubMed] [Google Scholar]
- Winer IS, Bommer GT, Gonik N, Fearon ER. Lysine residues Lys-19 and Lys-49 of beta-catenin regulate its levels and function in T cell factor transcriptional activation and neoplastic transformation. J Biol Chem. 2006;281:26181–26187. doi: 10.1074/jbc.M604217200. [DOI] [PubMed] [Google Scholar]
- Yu YP, Landsittel D, Jing L, Nelson J, Ren B, Liu L, McDonald C, Thomas R, Dhir R, Finkelstein S, et al. Gene expression alterations in prostate cancer predicting tumor aggression and preceding development of malignancy. J Clin Oncol. 2004;22:2790–2799. doi: 10.1200/JCO.2004.05.158. [DOI] [PubMed] [Google Scholar]
- Zhan F, Hardin J, Kordsmeier B, Bumm K, Zheng M, Tian E, Sanderson R, Yang Y, Wilson C, Zangari M, et al. Global gene expression profiling of multiple myeloma, monoclonal gammopathy of undetermined significance, and normal bone marrow plasma cells. Blood. 2002;99:1745–1757. doi: 10.1182/blood.v99.5.1745. [DOI] [PubMed] [Google Scholar]
- Zhang L, Duan CJ, Binkley C, Li G, Uhler MD, Logsdon CD, Simeone DM. A transforming growth factor beta-induced Smad3/Smad4 complex directly activates protein kinase A. Mol Cell Biol. 2004;24:2169–2180. doi: 10.1128/MCB.24.5.2169-2180.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.