Abstract
This study investigated the feasibility of automatic image registration of MR high-spatial resolution proximal femur trabecular bone images as well as the effects of gray-level interpolation and volume of interest (VOI) misalignment on MR-derived trabecular bone structure parameters. For six subjects in a short-term study, a baseline scan and a follow-up scan of the proximal femur were acquired on the same day. For ten subjects in a long-term study, a follow-up scan of the proximal femur was acquired 1 year after the baseline. An automatic image registration technique, based on mutual information, utilized a baseline and a follow-up scan to compute transform parameters that aligned the two images. In the short-term study, these parameters were subsequently used to transform the follow-up image with three different gray-level interpolators. Nearest-neighbor interpolation and B-spline approximation did not significantly alter bone parameters, while linear interpolation significantly modified bone parameters (p<0.01). Improvement in image alignment due to the automatic registration for the long-term and short-term study was determined by inspecting difference images and 3D renderings. This work demonstrates the first application of automatic registration, without prior segmentation, of high-spatial resolution trabecular bone MR images of the proximal femur. Additionally, inherent heterogeneity in trabecular bone structure and imprecise positioning of the VOI along the slice (anterior–posterior) direction resulted in significant changes in bone parameters (p<0.01). Results suggest that automatic mutual information registration using B-spline approximation or nearest neighbor gray-level interpolation to transform the final image ensures VOI alignment between baseline and follow-up images and does not compromise the integrity of MR-derived trabecular bone parameters used in this study.
Keywords: registration, trabecular bone, magnetic resonance imaging (MRI), mutual information, osteoporosis, proximal femur
INTRODUCTION
Osteoporosis is a disorder that results in bone with decreased mechanical strength and increased fracture risk. Approximately one in two white women and one in five men are affected by osteoporosis in their lifetime.1, 2 Osteoporosis is responsible for millions of fractures annually involving mostly vertebral bodies, the proximal femur, and the distal radius. The direct financial cost attributable to osteoporotic fracture in the United States is $14 billion and is expected to increase three- to eightfold over the next 50 years.3 Treatments, such as bisphosphonates, are available that have shown significant reduction of fracture incidence.4, 5 Early diagnosis is paramount for intervention, yet signs of the disease cannot be routinely and reliably detected until a fragility fracture occurs. For this reason there exists intense interest within the medical community for developing accurate early diagnostic techniques.
Osteoporosis targets both cortical and trabecular bone. Since trabecular bone has a higher metabolic activity and is highly responsive to hormonal changes, substantial efforts have focused on diagnosing early trabecular bone loss.6 Trabecular bone is spongy bone found in skeletal sites such as the vertebrae and the proximal and distal parts of the appendicular skeleton. Changes in the trabecular bone are characterized by both thinning and loss of structure.7 Trabecular bone microarchitecture is also of particular importance to bone strength.8 Techniques to evaluate trabecular bone microarchitecture using MRI have been established.9, 10 Common MR-derived trabecular bone parameters, analogous to histomorphometric measures,11 are apparent bone volume fraction (App.BV∕TV), apparent trabecular separation (App.Tb.Sp), apparent trabecular thickness (App.Tb.Th), and apparent trabecular number (App.Tb.N). High-spatial resolution MR images of trabecular bone have voxel sizes on the order of trabecular thickness, thus partial volume effects MR-based trabecular structure assessment. For this reason, MR-derived trabecular bone parameters are commonly termed “apparent” measures. MRI studies have confirmed that MR-derived trabecular bone parameters can be used to detect differences in trabecular bone due to age, bone mineral density, and osteoporotic status at peripheral sites.9, 10
The proximal femur is the most important site for osteoporotic fractures. After six months, 85% of hip fracture patients cannot walk across a room without help and 20% of patients die within one year.12 Assessment of osteoporosis at the proximal femur is typically performed using areal bone mineral density derived from dual energy x-ray absorptiometry (DXA); this technique measures the combined mineral density of both cortical and trabecular bone.13 To better characterize early bone loss and increased fracture risk a noninvasive technique to assess trabecular architecture would be desirable. However, until recently there existed no method without ionizing radiation to access in vivo, noninvasively, three-dimensional trabecular bone structure of the proximal femur. Previously, high-spatial resolution MR imaging of trabecular bone was restricted to peripheral sites, and deep-seated regions of the skeleton such as the proximal femur were limited by signal-to-noise ratio. Due to advances in MR pulse sequence development and higher magnetic field strength (3 T), recent studies have been conducted which investigated the feasibility of using high-spatial resolution MRI to evaluate trabecular bone structure of the proximal femur.14, 15 If bone loss or response to therapy is monitored in a longitudinal study, consistent positioning between baseline and follow-up proximal femur scans is required, but it is very difficult due to the complex femoral shape. Primary sources of error for MR-derived trabecular bone parameter reproducibility were previously identified in the distal radius and the distal tibia as involuntary patient motion and failure to accurately match the analysis volumes.16 Despite the complex femoral shape, the same region must be consistently scanned and analyzed between baseline and follow-up image acquisitions.
Image registration may be able to ensure correct volume of interest (VOI) selection for analysis. Techniques for image registration of MR musculoskeletal images have been reported in recent scientific literature,17, 18 however they are not fully automatic since they involve image segmentation. The considerable amount of textural information and limited contrast inherent in high-spatial resolution trabecular bone images makes segmentation time consuming and difficult making the registration technique prone to fail. Investigators have implemented a mutual information registration technique19, 20 which requires no segmentation for the registration of brain MR images.21 This same technique was first adapted to make automatic registration possible for trabecular bone images of the distal tibia22 and later successfully modified to incorporate a cross-correlation metric.23
Image registration involves transforming an image which requires interpolation of the voxel intensities to produce a new image. Since gray-value errors are commonly introduced by interpolation, it is also necessary to evaluate the effects of interpolation on the measurement of trabecular bone parameters.
Thus the purpose of this investigation was fourfold: (i) to evaluate the feasibility of proximal femur automatic image registration in short-term and long-term studies, (ii) to compare the effects of three methods of gray-level interpolation on MR-derived trabecular bone parameters, (iii) to determine the effect of misalignment of VOIs between repeat scans on MR-derived trabecular bone parameters, and (iv) to assess the difference in the coefficient of variation (CV) between MR-derived trabecular bone parameters determined from follow-up images with and without automatic image registration in a short-term study.
METHODS
Imaging
Coronal MR scans of the right proximal femur were obtained with a 3 T Signa system (General Electric, Milwaukee, WI) using a four-element phased array coil and a multiacquisition balanced steady state free precession sequence (b-SSFP). Image acquisition and reconstruction were performed using a modified version of generalized autocalibrating partially parallel acquisition (GRAPPA), with an acceleration factor of 2.24 Scans were acquired with a 512×384 matrix, 12 cm FOV, 60° flip angle, TR∕TE 10.3∕3.6 ms, 1 mm slice thickness, a total of 74 slices, and a scan time of approximately 10 min. Image voxel size was 0.234×0.234×1 mm3. Figure 1 shows a representative high-spatial resolution MR image of the proximal femur. A color-coded coil holder and foam foot wedge were used to provide consistent coil and limb positioning during scanning. Six healthy volunteers (age 26±4 years) participated in a short-term study in which the baseline and follow-up images were acquired on the same day after repositioning. After the baseline image was acquired the volunteer was removed from the scanner, allowed to rest for 15 min, and then repositioned for the follow-up scan. One of the volunteers was scanned twice without repositioning in order to assess the robustness of the registration algorithm. Additionally, ten patients (age 55±4 years) were scanned for a long-term study in which the baseline and follow-up images were acquired 1 year apart in order to assess the feasibility of registration for a longitudinal time series.
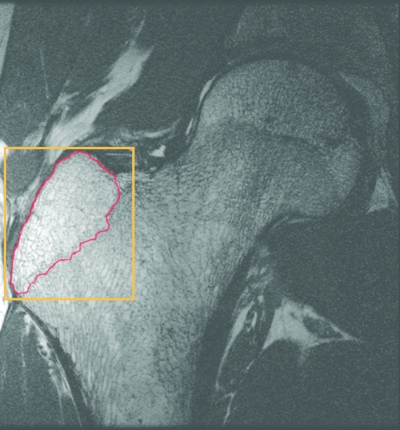
Figure 1.
Representative high-spatial resolution (0.234×0.234×1 mm3) image of the proximal femur. A rectangular parallelepiped masked the baseline image for the calculation of entropy (shown in orange). It was created by selecting a point on the first slice in which the greater trochanter appeared and last slice before the greater trochanter was no longer in view. The analysis region is outlined in red.
Registration
Image registration is the process of aligning two images. The goal is to find a transformation that aligns or matches the anatomical regions of the two images. A rigid transform with a rotation matrix, characterized by three Euler angles, and a translation vector composed of three translation parameters was used to align the proximal femur images. The rotation matrix and the translation vector defined the movement of a point from one image to the other, and image registration was performed by adjusting their parameters. The parameters were adjusted until a mutual information measure function was optimized and the transformation therefore aligned the two images.
The mutual information is computed from the joint probability distribution of the images’ intensity. When two images are aligned, they should provide maximal information about each other and the joint probability distribution results in a high mutual information value. The information contributed by each of the two images, denoted image A and image B, is entropy which measures the dispersion of a probability distribution. The entropy measure of an image is defined as
| (1) |
Here, pA is the marginal probability distribution, the likelihood of finding pixels of a given intensity throughout the imaging volume. The joint entropy of the two imaging volumes A and B is defined as
| (2) |
The mutual information of two images can be defined as the degree of dependence between image A and image B given by the distance between the joint distribution, p(a,b), and the distribution associated with the case of complete independence, pA(a)⋅pB(b),
| (3) |
Before registration, we applied a mask to the baseline image using the shape of a rectangular parallelepiped so that the entropy would only be calculated in this region (Fig. 1). The smallest rectangular parallelepiped which still covered the whole area of the epiphysis of the greater trochanter was selected. Mutual information registration with a conjugate gradient descent optimizer and a linear interpolator was performed using open-source image registration software (INSIGHT TOOLKIT). Registration was applied off-line retrospectively to the high-spatial resolution follow-up images to align them to their corresponding baseline images. Registration was performed on the images from the six volunteers with a short term follow-up and on the ten patients whose follow-up exam was a year later.
We assessed the robustness of the registration with the same technique presented by Studholme et al.25 using the image set with the follow-up image acquired without repositioning. We defined a series of 90 misalignments. Thirty of these corresponded to a misalignment by translation of 5 mm and a rotation of 5°, thirty by 10 mm and 10°, and thirty by 10 mm and 20°. Each set of thirty misalignments was determined by randomly selecting a point on the surface of spheres in translational and rotational parameter space. Each misalignment was selected as the initial starting guess for the registration. The deviation from the expected output was recorded.
Interpolation
When a transform is applied to an image, resampling is required because the new coordinate points may not line up with the old coordinate points. Image resampling determines pixel values for the new coordinate points by generating a continuous function from a discrete one using an interpolation function and then sampling the continuous signal at a new set of positions.26, 27 Exact resampling is only possible if the interpolation function is an ideal low pass filter (a rect function in the frequency domain). An ideal low pass filter is equivalent to convolving a sinc function which has infinite extent in the spatial domain. However, a sinc function is not physically realizable. Therefore, finite impulse response filters must be used as the interpolation function.28 In this study, all interpolation was performed using open-source image registration software (INSIGHT TOOLKIT).
The simplest interpolation function used in this study was nearest neighbor. Each interpolated output voxel was assigned the intensity of the nearest voxel in the input image. Nearest-neighbor interpolation can be achieved by convolving the input image with a rect function in the spatial domain and is therefore equivalent to multiplying by a sinc function in the frequency domain. Linear interpolation operates in one-dimensional spaces and the output voxel intensity is assigned a value that is weighted by the values of the direct neighbors. In this study, trilinear interpolation,29 the extension of linear interpolation to three-dimensional spaces, was used. Trilinear interpolation computes interslice voxel values as a distance-weighted average of the voxel values assigned to the eight nearest neighbors. B-splines have also been proposed for the interpolation function. B-splines use low-degree polynomials in each of the intervals, and select the polynomial pieces such that they fit smoothly together. A B-spline function of degree n is derived through n convolutions of the box filter. Since, the B-spline function is an approximating function that passes near the points but not necessarily through them, the data will still be modified if the image is resampled to the same grid. The B-spline function is therefore often called B-spline approximation. In this study, a fifth-order B-spline was implemented. The same order of B-spline was used for each dimension and mirror boundary conditions were used.30
The registration algorithm implemented in this study used linear interpolation. The goal of the registration algorithm was to align the shape of the femur in the baseline and follow-up images. The texture information inherent to trabecular bone was not important for alignment and therefore the smoothing due to the linear interpolation is desired. However, for quantifying the trabecular bone structure, preserving the texture information in the image is paramount. Therefore, a key step in aligning proximal femur images for trabecular bone structure analysis was the selection of the interpolation for the final transform. To examine the effect of registration on trabecular bone structure parameters the registration outputs from the six sets of images from the short-term study were used. The output of the mutual information registration algorithm, three rotations and three translations, was applied to the follow-up image with different interpolators: nearest neighbor, linear, and B-spline kernels. Registration and interpolation were performed on a Sun workstation (Sun Ultra 40, AMD Opteron Dual Core processor, 2.4 GHz, 8 Gbytes RAM, Sun Microsystems, CA). The three interpolation methods were compared by analyzing their effect on bone parameters and computational time.
Data analysis
Proximal femur trabecular bone structure analysis was performed on the images using software developed at our institution implemented in IDL (Research Systems, Inc., Boulder, CO).31 A VOI which included only trabecular bone and bone marrow contained between the epiphyseal line and the cortex of the greater trochanter (Fig. 1), consisted of ten slices and was manually defined using a graphics cursor on a slice by slice basis. The same VOI was used on the baseline and registered follow-up images. The unregistered follow-up required a separate VOI definition. Due to the use of surface coils, a correction for the spatial variation in the coil detection sensitivity was required for accurate image analysis. The VOIs were intensity corrected using a low-pass-filter based coil sensitivity correction.31 An image intensity histogram based thresholding technique was used to binarize the VOI into trabecular bone and marrow phases, and previously described methods32 were then used to compute the apparent trabecular bone structural parameters: App.BV∕TV, App.Tb.Sp, App.Tb.Th, and App.Tb.N.
Error simulations
Error simulations were performed to demonstrate the influence of VOI location on MR-derived bone parameters and the importance of accurately aligning the VOI in baseline and follow-up images. The bone parameter variation associated with a shift in the analysis volume along the slice direction (anterior–posterior) was estimated. For one proximal femur image from the short-term study, a VOI spanning ten slices was shifted in one-slice increments. Bone parameters were calculated for the VOI at each increment and the percent variation in trabecular bone parameters from the initial VOI position was calculated. To help explain why MR-derived bone parameters are effected by VOI location, regional variations in MR-derived trabecular bone parameters for the proximal femur were determined. MR-bone parameters, App.BV∕TV, App.Tb.Sp, App.Tb.Th, and App.Tb.N, were measured over a region of 1.5 cm thickness for all of the baseline proximal femur images from the short-term study. The average percent variation in bone parameters was calculated.
Statistical analysis
MR-derived apparent trabecular bone structural parameters determined for the short-term study with each of the three different gray-level interpolators were compared and analyzed using repeated measures of analysis of variance and the Bonferroni t-test. The CV33 was calculated between the baseline and follow-up images with and without image registration for the images in the short-term study.
RESULTS
The total time to register a baseline and follow-up proximal femur image and save the transformation, including uploading the images and applying a mask, is approximately 5 min. The parameters of the transforms that resulted from the registration algorithm in the short-term study are shown in Table 1 and the long-term study in Table 2. Rotations and translations for both the short-term and long-term studies were in the same range. Short-term rotations ranged from −7.74° to 16.83° and long-term rotations ranged from −8.32° to 19.65°. Short-term translations ranged from −9.9 to 9.85 mm and long-term translations ranged from −8.3 to 7.88 mm.
Table 1.
The output of the registration algorithm, a transform with three translations and three rotations for the six subjects in the short-term study. The three planes are defined as Right∕Left (R∕L), Anterior∕Posterior (A∕P), and Inferior∕Superior (I∕S).
| Subject | R∕L translation (mm) | A∕P translation (mm) | I∕S translation (mm) | R∕L rotation (°) | A∕P rotation (°) | I∕S rotation (°) |
|---|---|---|---|---|---|---|
| 1 | 1.60 | −3.33 | −9.90 | 5.88 | −2.69 | 14.19 |
| 2 | −3.26 | 2.58 | −0.09 | −3.79 | 1.66 | 2.45 |
| 3 | 3.45 | −1.62 | 1.77 | 2.58 | −2.45 | −5.70 |
| 4 | 9.85 | 4.34 | −1.95 | 4.65 | −1.78 | 16.83 |
| 5 | 3.59 | −0.89 | −4.55 | −0.85 | 2.31 | −7.74 |
| 6 | 5.67 | −5.39 | 2.58 | −1.49 | −0.59 | 8.98 |
| Average | 3.48 | −0.72 | −2.02 | 1.16 | −0.59 | 4.84 |
| s.d. | 4.34 | 3.63 | 4.64 | 3.80 | 2.13 | 10.23 |
Table 2.
The output of the registration algorithm for the ten subjects in the long-term study.
| Subject | R∕L translation (mm) | A∕P translation (mm) | I∕S translation (mm) | R∕L rotation (°) | A∕P rotation (°) | I∕S rotation (°) |
|---|---|---|---|---|---|---|
| 1 | 2.59 | −1.35 | 1.23 | −0.82 | −2.13 | 6.19 |
| 2 | −1.76 | −0.86 | −1.82 | −3.51 | −4.87 | −2.96 |
| 3 | −8.3 | −1.53 | 0.92 | 5.1 | 2.13 | −4.6 |
| 4 | 0.25 | −1.64 | 5.4 | 19.65 | 1.85 | −1.43 |
| 5 | −1.92 | −1.06 | 0.68 | −0.09 | 0.41 | 1.15 |
| 6 | −5.58 | −4.78 | −2.95 | 13.21 | 0.2 | −2.24 |
| 7 | −3.13 | 0.66 | 5.97 | −0.6 | 5.14 | −0.7 |
| 8 | 0.72 | 0.06 | 7.88 | −6.35 | −1.29 | −8.32 |
| 9 | 0.36 | −3.29 | −0.8 | −7.6 | 1.04 | 1.33 |
| 10 | 5.6 | −2.3 | 5.85 | 0.1 | −2.8 | −2.8 |
| Average | −1.12 | −1.61 | 2.16 | 1.91 | −0.03 | −1.44 |
| s.d. | 3.98 | 1.57 | 3.86 | 8.57 | 2.86 | 3.88 |
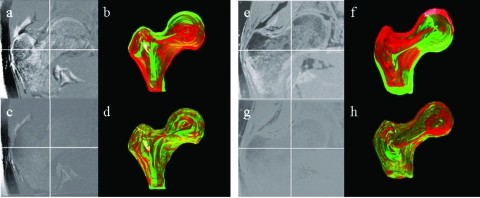
Figure 2 shows the results of the registration with subtraction images and surface renderings from two femurs. Figures 2a, 2b, 2c, 2d show results for a femur from the short-term study and Figs. 2e, 2f, 2g, 2h show results for a femur in the long-term study. As can be assessed by simple image subtraction in Figs. 2a, 2c, and by surface rendering of the proximal femurs in Figs. 2b, 2d, the follow-up image is better aligned to the baseline scan after registration. In Figs. 2a, 2e the edges of the cortical bone are misaligned with higher intensities in the difference image, and clear separation of the red and green femoral renderings in Figs. 2b, 2f is visible. In Figs. 2c, 2g, the high intensity differences within the femoral edges are reduced, and there is considerably more overlap in the red and green femoral renderings in Figs. 2d, 2h. The results from the assessment of the robustness of the registration are shown in Table 3. For the 30 misregistrations of 5 mm and 5° the root mean square error (RMSE) for translations and rotations in Right∕Left (R∕L), Anterior∕Posterior (A∕P), and Inferior∕Superior (I∕S) were within half a pixel and half a degree. For the misregistrations of 10 mm and 10° and 10 mm and 20° the RMSE for translations in R∕L, A∕P, and I∕S were within 1 pixel, while the RMSE for rotations in R∕L and A∕P were within 1° and the rotations in I∕S were within 1.75°. Outside of a translation of 10 mm and a rotation of 20° the registration algorithm fails and is not able to align the images.
Figure 2.
Comparison of follow-up with registration vs follow-up without registration for the short-term study (a)–(d) and the long-term study (e)–(h). (a),(e) Subtraction of baseline and follow-up without registration. (b),(f) 3D rendering of nonregistered proximal femur surfaces. (c),(g) Subtraction of baseline and registered follow-up. (d),(h) 3D rendering of registered proximal femur surfaces (green=baseline, red=follow-up).
Table 3.
The results from testing the robustness of the registration algorithm. The average, standard deviation (s.d.), and the root mean square error (RMSE) with respect to the expected output are shown. Initial misalignments of 5 mm and 5°, 10 mm and 10°, and 10 mm and 20° were used.
| R∕L translation (mm) | A∕P translation (mm) | I∕S translation (mm) | R∕L rotation (°) | A∕P rotation (°) | I∕S rotation (°) | ||
|---|---|---|---|---|---|---|---|
| 5 mm and 5° | Average | 0.08 | −0.04 | −0.06 | 0.22 | 0.15 | 0.02 |
| s.d. | 0.07 | 0.27 | 0.11 | 0.30 | 0.14 | 0.32 | |
| RMSE | 0.10 | 0.26 | 0.12 | 0.36 | 0.24 | 0.31 | |
| 10 mm and 10° | Average | 0.10 | −0.22 | −0.08 | 0.29 | 0.09 | 0.35 |
| s.d. | 0.14 | 1.02 | 0.18 | 0.43 | 0.22 | 1.40 | |
| RMSE | 0.17 | 1.02 | 0.20 | 0.51 | 0.26 | 1.42 | |
| 10 mm and 20° | Average | 0.04 | 0.29 | −0.17 | 0.63 | 0.07 | −0.30 |
| s.d. | 0.22 | 1.28 | 0.12 | 0.80 | 0.19 | 1.74 | |
| RMSE | 0.22 | 1.30 | 0.20 | 1.00 | 0.22 | 1.74 |
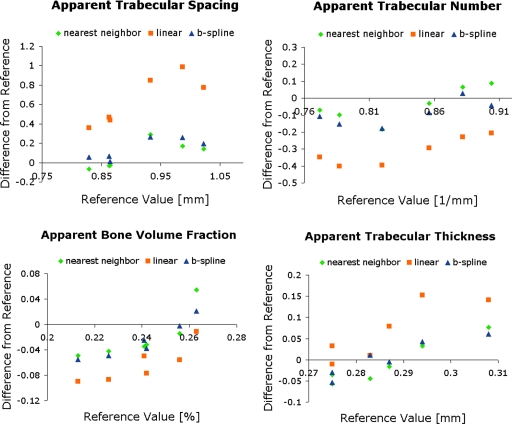
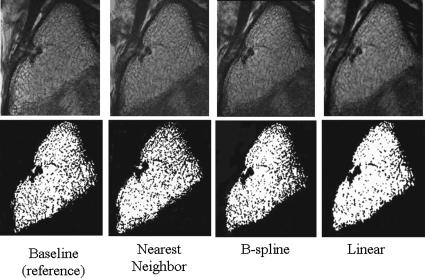
The computation time for nearest neighbor interpolation and linear interpolation was less than 12 s, while for B-spline approximation the computation time was on average 9 min. The effects of different gray-level interpolators on trabecular bone parameters in the short-term study are shown in Figs. 34. Trabecular bone structure parameters determined from follow-up images without and with automatic image registration using linear, nearest neighbor, and B-spline kernels are shown in Fig. 3. Because no interpolation was performed when bone parameters were determined from baseline images, these images served as the reference value. The difference from the reference value for each of the interpolators is shown in Fig. 3. All bone parameters were significantly different from the reference (p<0.01) with linear interpolation. However, nearest neighbor interpolation and B-spline approximation resulted in no significant difference from the reference (p>0.05). Additionally, in Fig. 4 the effects of the gray-level interpolation can be assessed visually. Figure 4 shows that nearest neighbor interpolation blurs out some of the trabeculae in some areas and exaggerates the trabeculae in others, but overall, the same number of pixels are designated to be bone as the in reference image. B-spline approximation maintains the most of the trabecular structure and therefore results in the same bone parameter measurements as the reference image. Linear interpolation clearly blurs out portions of the trabecular structure causing the spacing between the trabeculae to increase and the number of trabeculae to decrease.
Figure 3.
Assessment of different interpolators on the four trabecular bone structure parameters analyzed for the six subjects of the short-term study. Values determined from images with a linear interpolation were significantly different (p<0.01), but B-spline approximation and nearest neighbor interpolation did not change trabecular bone structure parameters significantly (p>0.05).
Figure 4.
The effects of the gray-level interpolation can be assessed visually. One of the steps in the trabecular microarchitecture quantification process is thresholding to create a binary image. The top row displays the gray-scale image and the bottom row displays the same image after thresholding. The baseline image, which served as the reference image, and registered follow-up images with linear interpolation, nearest neighbor interpolation, and B-spline approximation for the final transform are shown.
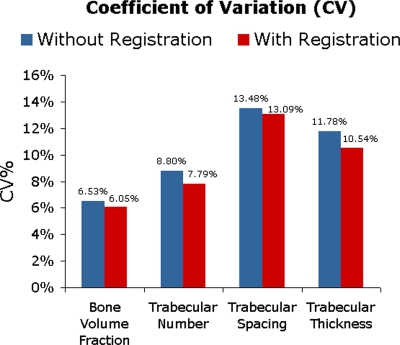
Reproducibility of bone parameters between the repeat scans in the short-term study is shown in Fig. 5. The CV was slightly better for registered follow-up images than for nonregistered follow-up images. The CV improved 0.48%–1.25% when the baseline images were compared to the registered follow-up images versus follow-up images without registration. However, the improvement in CV was not statistically significant (p>0.05).
Figure 5.
The improvement in coefficient of variation (CV) between baseline and follow-up due to registration (between 0.39% and 1.25%) was not statistically significant.
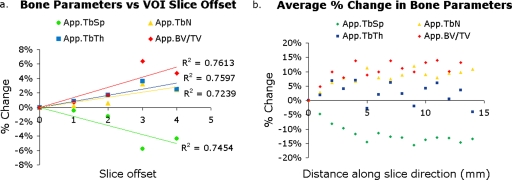
Average errors from a slice offset in VOI placement were significant [Fig. 6a]. The change in App.BV∕TV, App.Tb.N, and App.Tb.Th increased linearly with a shift in the VOI along the slice (A∕P) direction, while the change in App.Tb.Sp decreased linearly. The average percent change in bone parameters associated with a shift in the VOI along the slice direction was up to 6.37%. App. BV∕TV and App.Tb.Sp had a higher percent change (over 5.7%) than App.Tb.N and App.Tb.Th (less than 3.6%).
Figure 6.
Results of error simulations. (a) As the VOI was placed in shifted locations along the slice direction (a slice offset), the bone parameters changed up to 6.37%. (b) Within the same VOI, bone parameters have a dependence on the slice position demonstrating the trabecular structural heterogeneity in the proximal femur. The average percent change in bone parameters across a VOI is up to 15.73%.
Effects due to slice location within the proximal femur VOI were also significant [Fig. 6b]. App.BV∕TV and App.Tb.N. increased, App.Tb.Sp. decreased, and App.Tb.Th. remained fairly constant while moving anterior to posterior through the proximal femur image. The average percent change in bone parameters was up to 15.73% between the first and last slices.
DISCUSSION
The study of the progression of metabolic bone diseases or the efficacy of a treatment requires the proper analysis of corresponding regions of interest in repeat MRI scans. This study focused on the accuracy of VOI positioning for the evaluation of MR-derived trabecular bone parameters of the proximal femur. Errors due to VOI placement were assessed and quantified. This work also demonstrated the application of registration, without prior segmentation, of the proximal femur, as well as the effect of gray-level interpolators on trabecular bone parameters. Registration using the proper gray-level interpolator, such as B-spline approximation, to transform the final image may be used to avoid errors due to imprecise VOI placement.
Several investigators have observed significant variations in bone mineral content, morphometric indices, and mechanical properties within the proximal femur.34, 35, 36, 37, 38, 39 Brown et al. were among the first to conclude that trabecular bone of the proximal femur is a heterogeneous, anisotropic material by mapping contours of elastic modulus and yield strength.34 Morgan and Keaveny attributed the observed variation in yield strain across anatomical sites, including the greater trochanter, to intersite variations in trabecular structure.37 In their structural analysis of the greater trochanter, Link et al. noted that the trabecular structure was irregular and anisotropic.39 Similarly, this study found a variation in MR-derived trabecular bone parameters in the greater trochanter of the proximal femur. The variation of trabecular bone parameters in the slice (A∕P) direction suggests that there is a biologically inherent heterogeneity in trabecular bone structure in the proximal femur. This heterogeneity resulted in an average 2.01%–15.73% variation in bone parameters between slices through a VOI 10 mm long. This heterogeneity in trabecular bone structure of the greater trochanter resulted in measurement errors 0.25%–6.37% when there was a slice offset in VOI placement. This source of error demonstrates the importance of accurate VOI placement. These results are in agreement with a study by Gomberg et al.16 which found errors in trabecular bone parameters up to 7.6% in the distal radius due to a VOI mismatch along the z-axis (I∕S) direction. Additionally, Newitt et al.31 determined that analysis region misalignment caused small but significant changes in some structural parameters.
The mutual information image registration algorithm proposed in this study was able to successfully register images from both a short-term study and a long-term study. The resulting transform parameters were within the same range for both studies. When the robustness of the registration was tested, registration was successful for misregistrations less than 10 mm and 20°, which was within the translations and rotations seen in both the long-term and short-term study. The registration algorithm was able to align images in translation within 1 pixel and rotation of 1° in plane (R∕L and A∕P) and 1.75° out of plane (I∕S). For misregistrations of 5 mm and 5°, subpixel accuracy was achieved. Therefore, the registration algorithm will be successful for any likely initial misalignments in short-term and long-term studies.
The interpolation method implemented when transforming the final image can dramatically impact the quantification of trabecular bone structure parameters. Results suggest that when applying a transform to musculoskeletal images acquired for trabecular bone quantification, nearest neighbor interpolation and B-spline approximation will maintain the integrity of the trabecular bone parameters involved in this study. Nearest neighbor interpolation traditionally introduces aliasing and blurring effects to images, so interpolation results may appear surprising. However, as part of the standard trabecular bone structure quantification femoral images are thresholded. Therefore, nearest neighbor interpolation maintains the intensity values in a VOI and maintains the number of pixels thresholded. B-spline approximation performed as expected by maintaining the accuracy of the image information. If computational time becomes an issue, then nearest neighbor interpolation would be preferred for interpolation of registered MR musculoskeletal images for trabecular bone analysis.
Carpenter et al.15 performed a study investigating the short-term reproducibility of trabecular bone structure parameters in the proximal femur. In the study, interoperator variability was small (3.8%) and they suggested that the CVs, ranging from 6.5% to 13.5%, may be partially due to patient repositioning and mismatched analysis volumes between baseline and follow-up images. Krug et al.14 found similar CVs between 2% and 10% for proximal femur trabecular bone structure parameters when volunteers were rescanned four times. Registration is able to ensure accurate VOI placement and therefore eliminate sources of error associated with the imprecision of VOI placement. In this work we have demonstrated the feasibility of using a mutual information based method to automatically register MR images of the proximal femur. Although there was only a 0.48%–1.25% improvement in the CV between parameters determined from follow-up images with and without automatic registration, the registration ensures that the reproducibility error does not originate from inaccurate VOI placement. The improvement in reproducibility and the systematic method of VOI placement with automatic registration is required to establish the adequacy of MRI techniques for longitudinal studies assessing proximal-femur trabecular bone structure.
ACKNOWLEDGMENTS
This work was supported by NIH Grant No. ROI-AG017762 and ARCS and Evnin-Wright Fellowships. The authors would like to acknowledge Dr. Roland Krug for helping to acquire the high-spatial resolution MR images of trabecular bone of the proximal femur and Dr. David Newitt for providing the software for trabecular bone microarchitecture quantification.
References
- Siris E. S., Miller P. D., Barrett-Connor E., Faulkner K. G., Wehren L. E., Abbott T. A., Berger M. L., Santora A. C., and Sherwood L. M., “Identification and fracture outcomes of undiagnosed low bone mineral density in postmenopausal women: Results from the National Osteoporosis Risk Assessment,” J. Am. Med. Assoc. 286, 2815–2822 (2001). [DOI] [PubMed] [Google Scholar]
- U.S. Department of Health and Human Services, “Bone health and osteoporosis: A report of the Surgeon General,” U.S. Department of Health and Human Services, Office of the Surgeon General, Washington, DC, 2004.
- Fraser W. D., “The burden of osteoporosis and the case for disease management,” Disease Manage. Health Outcomes 12, 409–418 (2004). [Google Scholar]
- Gourlay M., Richy F., and Reginster J. Y., “Strategies for the prevention of hip fracture,” Am. J. Med. 115, 309–317 (2003). [DOI] [PubMed] [Google Scholar]
- Ettinger M. P., “Aging bone and osteoporosis: Strategies for preventing fractures in the elderly,” Arch. Intern Med. 163, 2237–2246 (2003). [DOI] [PubMed] [Google Scholar]
- Dempster D. W. and Lindsay R., “Pathogenesis of osteoporosis,” Lancet 341, 797–801 (1993). [DOI] [PubMed] [Google Scholar]
- Kleerekoper M., Villanueva A. R., Stanciu J., Rao D. S., and Parfitt A. M., “The role of three-dimensional trabecular microstructure in the pathogenesis of vertebral compression fractures,” Calcif. Tissue Int. 10.1007/BF02554913 37, 594–597 (1985). [DOI] [PubMed] [Google Scholar]
- Ciarelli M. J., Goldstein S. A., Kuhn J. L., Cody D. D., and Brown M. B., “Evaluation of orthogonal mechanical properties and density of human trabecular bone from the major metaphyseal regions with materials testing and computed tomography,” J. Orthop. Res. 10.1002/jor.1100090507 9, 674–682 (1991). [DOI] [PubMed] [Google Scholar]
- Majumdar S., Genant H. K., Grampp S., Newitt D. C., Truong V. H., Lin J. C., and Mathur A., “Correlation of trabecular bone structure with age, bone mineral density, and osteoporotic status: In vivo studies in the distal radius using high resolution magnetic resonance imaging,” J. Bone Miner. Res. 10.1359/jbmr.1997.12.1.111 12, 111–118 (1997). [DOI] [PubMed] [Google Scholar]
- Wehrli F. W., Hwang S. N., Ma J., Song H. K., Ford J. C., and Haddad J. G., “Cancellous bone volume and structure in the forearm: Noninvasive assessment with MR microimaging and image processing,” Radiology 206, 347–357 (1998). [DOI] [PubMed] [Google Scholar]
- Parfitt A. M., Drezner M. K., Glorieux F. H., Kanis J. A., Malluche H., Meunier P. J., Ott S. M., and Recker R. R., “Bone histomorphometry: Standardization of nomenclature, symbols, and units. Report of the ASBMR Histomorphometry Nomenclature Committee,” J. Bone Miner. Res. 2, 595–610 (1987). [DOI] [PubMed] [Google Scholar]
- Cummings S. R. and Melton L. J., “Epidemiology and outcomes of osteoporotic fractures,” Lancet 10.1016/S0140-6736(02)08657-9 359, 1761–1767 (2002). [DOI] [PubMed] [Google Scholar]
- “Assessment of fracture risk and its application to screening for postmenopausal osteoporosis. Report of a WHO Study Group,” W. H. O. Tech. Rep. Ser. 843, 1–129 (1994). [PubMed] [Google Scholar]
- Krug R., Banerjee S., Han E. T., Newitt D. C., Link T. M., and Majumdar S., “Feasibility of in vivo structural analysis of high-resolution magnetic resonance images of the proximal femur,” Osteoporosis Int. 16, 1307–1314 (2005). [DOI] [PubMed] [Google Scholar]
- Carpenter D., Krug R., Banerjee S., and Majumdar S., “Analyzing trabecular bone structure in the proximal femur with high-resolution parallel magnetic resonance imaging,” International Bone Densitometry Workshop, Kyoto, Japan, 2006.
- Gomberg B. R., Wehrli F. W., Vasilic B., Weening R. H., Saha P. K., Song H. K., and Wright A. C., “Reproducibility and error sources of micro-MRI-based trabecular bone structural parameters of the distal radius and tibia,” Bone (N.Y.) 10.1016/j.bone.2004.02.017 35, 266–276 (2004). [DOI] [PubMed] [Google Scholar]
- Takao M., Sugano N., Nishii T., Tanaka H., Masumoto J., Miki H., Sato Y., Tamura S., and Yoshikawa H., “Application of three-dimensional magnetic resonance image registration for monitoring hip joint diseases,” Magn. Reson. Imaging 23, 665–670 (2005). [DOI] [PubMed] [Google Scholar]
- Magland J., Vasilic B., Lin W., and Wehrli F. W., “Automatic 3D registration of trabecular bone images using a collection of regional 2D registrations,” Proceedings of the 14th International Society of Magnetic Resonance Medicine, Seattle, WA, May 6–12th, 2006.
- Maes F., Collignon A., Vandermeulen D., Marchal G., and Suetens P., “Multimodality image registration by maximization of mutual information,” IEEE Trans. Med. Imaging 10.1109/42.563664 16, 187–198 (1997). [DOI] [PubMed] [Google Scholar]
- Viola P. and Wells W. M., “Alignment by maximization of mutual information,” Int. J. Comput. Vis. 10.1023/A:1007958904918 24, 137–154 (1997). [DOI] [Google Scholar]
- Hancu I., Blezek D. J., and Dumoulin M. C., “Automatic repositioning of single voxels in longitudinal 1H MRS studies,” NMR Biomed. 18, 352–361 (2005). [DOI] [PubMed] [Google Scholar]
- Blumenfeld J., Carballido-Gamio J., Krug R., Blezek D. J., Hancu I., and Majumdar S., “Automatic prospective registration of high-resolution trabecular bone images of the tibia,” Ann. Biomed. Eng. 35, 1924–1931 (2007). [DOI] [PubMed] [Google Scholar]
- Rajapakse C. S., Magland J. F., and Wehrli F. W., “Fast prospective registration of in vivo MR images of trabecular bone microstructure in longitudinal studies,” Magn. Reson. Med. 10.1002/mrm.21593 59, 1120–1126 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee S., Choudhury S., Han E. T., Brau A. C., Morze C. V., Vigneron D. B., and Majumdar S., “Autocalibrating parallel imaging of in vivo trabecular bone microarchitecture at 3 Tesla,” Magn. Reson. Med. 10.1002/mrm.21059 56, 1075–1084 (2006). [DOI] [PubMed] [Google Scholar]
- Studholme C., Hill D. L., and Hawkes D. J., “Automated three-dimensional registration of magnetic resonance and positron emission tomography brain images by multiresolution optimization of voxel similarity measures,” Med. Phys. 10.1118/1.598130 24, 25–35 (1997). [DOI] [PubMed] [Google Scholar]
- Parker J., Kenyon R., and Troxel D., “Comparison of interpolating methods for image resampling,” IEEE Trans. Med. Imaging 2, 31–39 (1983). [DOI] [PubMed] [Google Scholar]
- Wolberg G., Digital Image Warping (IEEE Computer Society Press, New York, 1990). [Google Scholar]
- Lehmann T. M., Gonner C., and Spitzer K., “Survey: Interpolation methods in medical image processing,” IEEE Trans. Med. Imaging 10.1109/42.816070 18, 1049–1075 (1999). [DOI] [PubMed] [Google Scholar]
- Parrot R. W., Stytz M. R., Amburn P., and Robinson D., “Towards statistically optimal interpolation for 3-D medical imaging,” IEEE Eng. Med. Biol. Mag. 12, 49–59 (1993). [Google Scholar]
- Thévenaz P., Blu T., and Unser M., “Image interpolation and resampling,” in Handbook of Medical Imaging, Processing and Analysis (Academic Press, San Diego, 2000), pp. 393–420. [Google Scholar]
- Newitt D. C., Van Rietbergen B., and Majumdar S., “Processing and analysis of in vivo high-resolution MR images of trabecular bone for longitudinal studies: Reproducibility of structural measures and micro-finite element analysis derived mechanical properties,” Osteoporosis Int. 10.1007/s001980200027 13, 278–287 (2002). [DOI] [PubMed] [Google Scholar]
- Majumdar S. and Genant H. K., “Assessment of trabecular structure using high resolution magnetic resonance imaging,” Stud. Health Technol. Inform. 40, 81–96 (1997). [PubMed] [Google Scholar]
- Gluer C. C., Blake G., Lu Y., Blunt B. A., Jergas M., and Genant H. K., “Accurate assessment of precision errors: How to measure the reproducibility of bone densitometry techniques,” Osteoporosis Int. 10.1007/BF01774016 5, 262–270 (1995). [DOI] [PubMed] [Google Scholar]
- Brown T. D. and A. B.Ferguson, Jr., “Mechanical property distributions in the cancellous bone of the human proximal femur,” Acta Orthop. Scand. 51, 429–437 (1980). [DOI] [PubMed] [Google Scholar]
- Nazarian A., Muller J., Zurakowski D., Muller R., and Snyder B. D., “Densitometric, morphometric and mechanical distributions in the human proximal femur,” J. Biomech. 40, 2573–2579 (2007). [DOI] [PubMed] [Google Scholar]
- Ciarelli M. J., Goldstein S. A., Kuhn J. L., Cody D. D., and Brown M. B., “Evaluation of orthogonal mechanical properties and density of human trabecular bone from the major metaphyseal regions with materials testing and computed tomography,” J. Orthop. Res. 10.1002/jor.1100090507 9, 674–682 (1991). [DOI] [PubMed] [Google Scholar]
- Morgan E. F. and Keaveny T. M., “Dependence of yield strain of human trabecular bone on anatomic site,” J. Biomech. 10.1016/S0021-9290(01)00011-2 34, 569–577 (2001). [DOI] [PubMed] [Google Scholar]
- Issever A. S., Vieth V., Lotter A., Meier N., Laib A., Newitt D., Majumdar S., and Link T. M., “Local differences in the trabecular bone structure of the proximal femur depicted with high-spatial-resolution MR imaging and multisection CT,” Acad. Radiol. 9, 1395–1406 (2002). [DOI] [PubMed] [Google Scholar]
- Link T. M., Vieth V., Langenberg R., Meier N., Lotter A., Newitt D., and Majumdar S., “Structure analysis of high resolution magnetic resonance imaging of the proximal femur: In vitro correlation with biomechanical strength and BMD,” Calcif. Tissue Int. 72, 156–165 (2003). [DOI] [PubMed] [Google Scholar]