Abstract
Capacitative Ca2+ entry (CCE) is Ca2+ entering after stimulation of inositol 1,4,5-trisphosphate (IP3) formation and initiation of Ca2+ store depletion. One hallmark of CCE is that it can also be triggered merely by store depletion, as occurs after inhibition of internal Ca2+ pumps with thapsigargin. Evidence has accumulated in support of a role of transient receptor potential (Trp) proteins as structural subunits of a class of Ca2+-permeable cation channels activated by agonists that stimulate IP3 formation—very likely through a direct interaction between the IP3 receptor and a Trp subunit of the Ca2+ entry channel. The role of Trp’s in Ca2+ entry triggered by store depletion alone is less clear. Only a few of the cloned Trp’s appear to enhance this type of Ca2+ entry, and when they do, the effect requires special conditions to be observed, which native CCE does not. Here we report the full-length cDNA of mouse trp2, the homologue of the human trp2 pseudogene. Mouse Trp2 is shown to be readily activated not only after stimulation with an agonist but also by store depletion in the absence of an agonist. In contrast to other Trp proteins, Trp2-mediated Ca2+ entry activated by store depletion is seen under the same conditions that reveal endogenous store depletion-activated Ca2+ entry, i.e., classical CCE. The findings support the general hypothesis that Trp proteins are subunits of store- and receptor-operated Ca2+ channels.
Keywords: Ca2+ channel, Gq-coupled responses, Ca2+ influx, store operated channels, trp proteins
Mammalian trp genes are homologues of the Drosophila transient receptor potential (trp) gene and were cloned to test the hypothesis (cf. ref. 1, and refs. therein) that they would encode capacitative Ca2+ entry (CCE) channels (2, 3). As originally proposed by Putney (4, 5), CCE channels are Ca2+-permeable cation channels that are activated after phospholipase Cβ stimulation by a G protein-coupled receptor and, independently, by maneuvers that promote depletion of internal Ca2+ stores. These maneuvers include activation of phospholipase Cγ, inhibition of Ca2+/ATPases responsible for pumping Ca2+ into the stores and maintaining resting cytosolic Ca2+ ([Ca2+]i), or by mere introduction into the cytosol of Ca2+ chelators at sufficiently high concentrations to insure quantitative chelation of Ca2+ leaking from internal stores (reviewed in refs. 6 and 7). Six mammalian trp-like genes have been identified (trp1 through trp6). Of these, the full-length cDNAs of all except that of the type 2 trp have been cloned, and their expression has augmented Ca2+ entry evoked by a G protein-coupled receptor in model cells. Involvement of Trp proteins in this entry pathway (very likely as subunits of the entry channels) was inferred from the findings that not only expression of trp cDNAs resulted in enhanced Ca2+ entry but also coexpression of partial cDNAs of Trp-1 through Trp-6 in the antisense direction resulted in complete inhibition of agonist-provoked Ca2+ entry through endogenous channels (3). Further evidence for Trp proteins being integral members of Ca2+ entry channels has come from two recent and related findings. One was that activation of the inositol 3,4,5-trisphosphate (IP3) receptor (IP3R) in excised membrane patches showed enhanced inward currents only when adhering (IP3R-containing) microsomes were exposed to IP3, or, if after extensive washout to remove adhering microsomes, the patches were “reconstituted” with IP3 plus IP3R-containing brain microsomes or with IP3 plus a recombinant cytosolic domain of the IP3R (8). These experiments were performed with cells that expressed both high levels of Trp3 and high levels of IP3R that had resulted as a consequence of overexpression of Trp3, and although they indicated that IP3R activation is functionally involved in activation of membrane Ca2+-permeable cation channels, they did not directly address their structural identity (i.e., whether CCE channels are made up of Trp proteins). We have now obtained evidence (N.Q., D.B., X.Z., and L.B., unpublished data) for structural involvement of Trp proteins as members of the Ca2+ entry channels by showing that IP3R and Trp can be coimmunoprecipitated from detergent extracts, that type-3 IP3R and Trp3 interact to form stable complexes in vitro, and that sequences of either IP3R or Trp3 that interact in vitro (but not sequences that do not) interfere with activation of CCE channels by IP3 formed in response to stimulation by a phospholipase C-stimulating Gq-coupled receptor.
Taken together, the data provide strong evidence for a role of Trp proteins as structural elements of Ca2+ entry channels activated by G protein-coupled receptors. In contrast, whether Trp proteins form part of the mechanism that senses and responds to the store-depletion signal that arises as a result of Ca2+ depletion in the absence of phospholipase C stimulation or IP3 formation is less clear. Thus, up to the time of this writing, the expression of only some of the Trp proteins—e.g., Trp4 and Trp5 (9, 10)—have been shown to be stimulated after store depletion elicited without phospholipase C activation, i.e., without stimulation of IP3 formation, as is thought to happen after treatment with thapsigargin (cf. refs. 11–13). Perhaps more importantly, even though activation of endogenous, nonengineered Ca2+ entry channels by thapsigargin is readily seen in experiments where Ca2+ entry is assessed with ratiometric fluorescence methods with fura2 as a [Ca2+]i indicator (3, 12–14), stimulation of Trp-mediated CCE by thapsigargin has been measured for the most part by electrophysiological means and, when measured using fura2, has required special conditions, such as low extracellular Ca2+ (10). Indeed, the same Trp, Trp5 (also CCE2), shown by one group to be stimulated after thapsigargin treatment (10), was reported by another group monitoring Ca2+ entry with fura2 to be insensitive to thapsigargin (15). This raised the question as to whether the two experimental approaches measure the same channels. Alternatively, lack of observable thapsigargin-sensitive Trp-mediated CCE in experiments using fura2 might be caused by limitations imposed by other as-yet-unknown factors. Testing for properties of a newly cloned Trp, mouse Trp2, we now show, however, that in contrast to other Trp proteins, i.e., Trp1 and Trp3-6, transient expression of Trp2 leads to formation of a store depletion-activated CCE channel.
MATERIALS AND METHODS
Cloning of trp2 cDNA.
Reverse transcription (RT) of mouse liver polyA-enriched RNA followed by application of PCR with degenerate primers led to amplification of a 442-bp fragment that coded for a novel sequence that was named trp2. Oligonucleotides DO4 and DO7 were based on regions in Dro-trp, Dro-trp-like, Ce-trp, and hum-trp1 that coded for conserved amino acid sequences (DO4: 5′-GGCNGA[G/A]GGNCTCTT[[T/C]GC-3′ encodes E[G/A/V]L[F/Y]A; DO7: 5′-CGNGC[G/A]AA[C/T]TTCCA[C/T]TC-3′encodes the anitsense strand of EWKFAR). A partial cDNA with 899 codons and lacking an initiator ATG was assembled by Rapid Amplification of cDNA Ends (RACE)–PCR using commercial mouse liver and brain RACE cDNA libraries (CLONTECH). In parallel, a ≈10-kb fragment of mouse genomic DNA containing at its 3′end the 5′end of the incomplete trp2 cDNA was sequenced. Northern analysis for tissues expressing trp2 revealed high expression in testis (Fig. 1A). New rounds of RT-PCR and RACE-PCR using a testis RACE cDNA library (CLONTECH) as template allowed for the cloning of 5′ ends of two N-terminal splice variants of mouse Trp2: clone 14, with an ORF of 1,172 codons, and clone 17, with an ORF of 1,072 codons, each with in-frame stop codons upstream of their respective initiator ATGs and differing from each other in their first 11 and 111 codons, i.e., trp2-17[12–1072] = trp2-14[112–1172]. The sequences were confirmed by isolating multiple partial cDNA sequences from a testis cDNA library. The full-length cDNAs were subcloned as EcoRI/XbaI fragments into pcDNA3 (Invitrogen) downstream of the cytomegalovirus promoter. RT-PCR and RACE-PCR reactions were performed as described (3, 16) using pCRII as the intermediate cloning vector (Invitrogen). DNA was sequenced by the dideoxy-chain termination method of Sanger et al. (17). The nucleotide sequences have been deposited in GenBank under the accession numbers AF111107 (clone 17) and AF 111108 (clone 14).
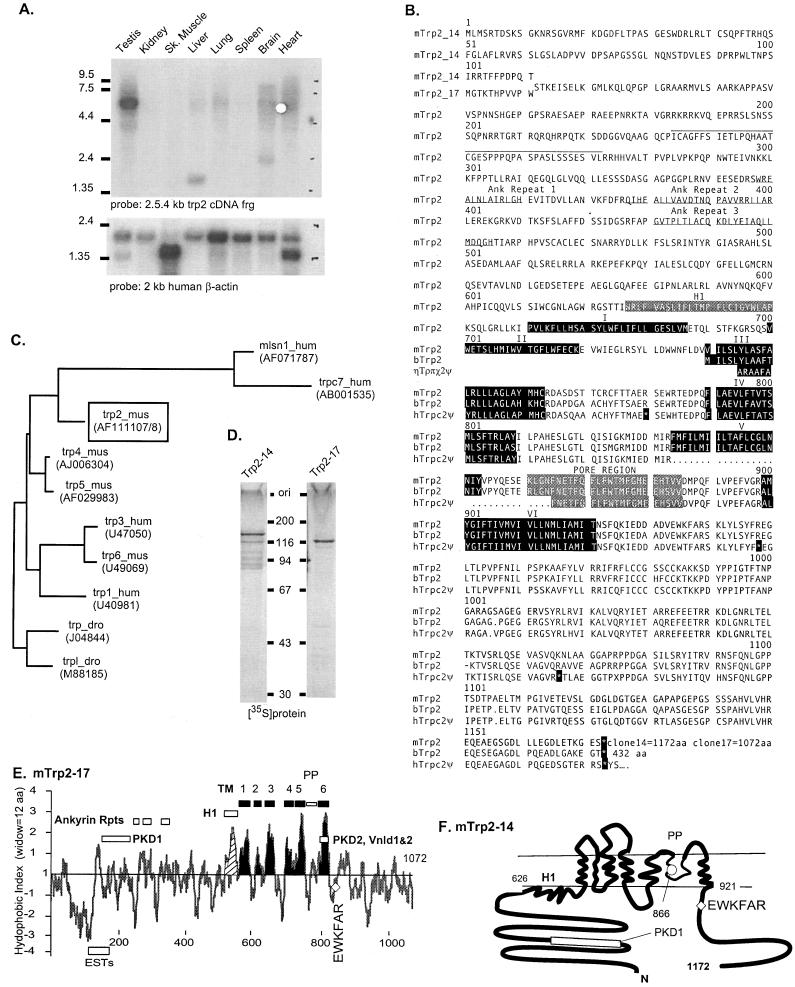
Figure 1.
Structural features of mouse Trp2. (A) Selective expression of trp2 transcripts in testis. (B) Deduced amino acid sequence of clones 14 (1,172 aa) and 17 (1,072 aa) and comparison to the deduced coding sequence of the human trp2 pseudogene (TRPC2Ψ, GenBank accession no. X89067) and a shorter bovine cDNA (GenBank accession no. AJ006304). Sequences predicted to form transmembrane segments and stop codons in the human pseudogene cDNA are shown in black boxes. Segment encoded in ESTs AA145678, AA242713, and AA473022 is overlined. Ankyrin repeats in the N terminus and the postulated pore region (PP) are highlighted in bold and white on gray, respectively. (C) Phylogram based on conserved core sequences encompassing from ≈20 amino acids before the first hydrophobic segment to ≈20 amino acids after the last hydrophobic transmembrane segment shows relationship of Trp2 to other Trp proteins. (D) Coupled transcription and translation (Promega) of clones 14 and 17. (E) Kyte and Doolittle plot of Trp2-17 using a sliding window of 12 amino acids. (F) Model of transmembrane topology of mouse Trp2. Amino acids 626 and 921 delimit the hydrophobic core of the protein. 866, consensus glycolsylation site in the hydrophobic core. Regions with sequence homology to PDK1 and -2 (polycystic kidney disease) and VnldR1 and -2 (vanilloid receptor) and the location of ESTs are highlighted. EWKFAR, highly conserved sequence motif among all Trps.
Northern Blot Analysis.
For Northern blot analysis (Fig. 1A), the probe was a 2,545-kb NcoI/NcoI partial cDNA fragment of mouse trp2 (clone 14 nucleotides 1049–3593, where nucleotide 1 is the A of initiator ATG). The purified fragment was labeled by random priming with [32P]dCTP (DuPont/NEN). The Mouse Multiple Tissue Northern blot (CLONTECH) was prehybridized for 3 hr at 65°C in 500 mM sodium phosphate (pH 7.2), 7% SDS, 1% BSA, and 1 mM EDTA. Hybridization was in the same buffer with 1.1 × 108 cpm/ml 32P-labeled probe for 16 hr at 60°C. The blot was washed two times for 20 min at room temperature with 0.5× standard saline citrate (SSC; 1× SSC = 15 mM sodium citrate, 150 mM NaCl, pH 7.0) and 0.1% SDS and then at 60°C with 0.4× SSC and 0.1% SDS. The blot was exposed to x-ray film for 1 week. After stripping, the membrane was rehybridized to a human β-actin probe furnished by the manufacturer of the multiple tissue blot.
In Vitro Translation of Mouse Trp2-14 and Trp2-17.
The translation reaction was carried out as described by the manufacturer (TNT kit; Promega) using pcDNA-trp2-14 and pcDNA-trp2-17 as templates. The resulting mixtures were stopped by adding 50 μl of Laemmli’s 1× sample buffer (18). Five-microliter samples were analyzed by using SDS/PAGE on 9% polyacrylamide gels. The gel slabs were dried and autoradiographed.
Transfection of COS-M6 cells and Measurement of [Ca2+]i.
COS-M6 cells were transfected by using the DEAE–dextran/chloroquine shock method (19) with changes as described (20). [Ca2+]i transients were measured in individual cells by fluorescence videomicroscopy using the Attofluor Digital Imaging and Photometry attachment of a Carl Zeiss Axiovert inverted microscope (3).
Computer Analysis.
Sequence alignments, restriction maps, and phylogenetic analysis were performed by using the wisconsin Package Software of the Genetics Computer Group (Madison, WI).
RESULTS AND DISCUSSION
Primary Structure of Mouse Trp2.
trp2, the second of the mammalian homologues of the Drosophila trp gene to be discovered, was identified in the expressed sequence tag database (EST T67673). As deposited, the sequence presented with a stop codon in the reading frame encoding the homologue, and when aligned with either the Drosophila Trp or the newly cloned human Trp1 required the introduction of a gap. Barring sequencing errors, this suggested that the human trp2 gene might be a pseudogene. That this was the case was confirmed both by us (unpublished data) and Montell and collaborators (21) by cloning and sequencing partial cDNAs containing the EST T67673 sequence. In contrast, sequence analysis of partial mouse cDNAs containing the EST T67673 region showed that mouse trp2 does not have the stop codons found in the human trp2 (3). This observation, in turn, suggested that what is a pseudogene in the human might be a regular gene in the mouse. The mouse gene was found to be highly expressed in testis (Fig. 1A). By a combination of RT-PCR, RACE-PCR, and direct sequencing of a ≈10-kb genomic clone, followed by computer-assisted ORF prediction and another PCR with primers based on the genomic sequence, we have now cloned two full-length cDNAs encoded in mouse trp2. In vitro translation of clones 14 and 17 gives two proteins (Trp2-14 and Trp-17) that when analyzed by SDS/PAGE migrate with apparent Mr of 116 and 131 kDa, very close to the values of 115.521 and 130.468 calculated on the basis of the amino acid sequence predicted by the cDNAs (Fig. 1D). Fig. 1B depicts the deduced amino acid sequence of Trp2-14 and Trp2-17, aligned with that predicted by the human trp2 pseudogene containing three stop codons (GenBank accession no. X89067) and a recently reported bovine sequence, bovine Trp2 (ref. 22, GenBank accession no. AJ006304). Like the mouse sequence, the bovine sequence lacks the stop codons found in the human trpc2 89 fragment, but unlike that of the mouse, it begins well within that of the protein predicted by the two mouse cDNAs. The short length of the clone suggests that the bovine sequence may be a splice variant of a larger form. Mouse Trp2-17 and Trp2-14 are predicted to be 1,072 and 1,172 aa long and differ only in their first 11 and 111 amino acids, respectively. From codon 12 onward to 1,072, trp-14 is identical to trp2-17 from codon 112-1172 (Fig. 1B).
A Kyte–Doolittle hydropathy analysis predicts multiple hydrophobic regions that are candidate transmembrane segments (Fig. 1E). Based on our previous studies with Trp3, we assign transmembrane properties to six of them, the first transmembrane segment being the second hydrophobic region revealed by the Kyte–Doolittle analysis. In contrast to other Trp proteins, each of which has several consensus glycosylation sites within the region that is thought to form the transmembrane domain of the protein, Trp2 has only one: Trp2-14[N866-T868]. All other sites are in the cytosolic N-terminal region (23). The consensus glycosylation site in the transmembrane domain is located in the putative pore region (PP on Fig. 1 E and F). We have not analyzed the glycosylation pattern of Trp2 expressed in COS or HEK cells. Based on published results with the ROMK1 potassium channel (24) as well as on our experience with artificially introduced consensus glycosylation sites (23), we expect to find Trp2 to be glycosylated. By analogy to the deduced topology of human Trp3 (23), we propose the transmembrane topology of Trp2 shown in Fig. 1F.
Sequence homology analysis using the Kimura pairwise near neighbor approach of the GCG wisconsin software package (Fig. 1C), places Trp2 near a branch leading to proteins encoded in the so-called trp-related genes. These include the recently described trpc7 (25) and melastatin1 (mlsn1) genes (26). The proteins predicted by these genes, Trpc7 = 1,576 aa, Mlsn1 = 1,298 aa, are for the most part unrelated to Trp proteins except in a region encompassing the putative pore region and the last transmembrane segment of the hydrophobic channel-forming portion of the protein (transmembrane segments 5–6 with the intervening pore). More distantly related are the two capsaicin-activated vallinoid receptors (Vnld1 and Vnld2) and a small segment of the type 2 polycystic kidney disease gene product (PKD2). The PKD1 gene product, although not showing sequence similarity to the hydrophobic domains of Trp2, is a protein of 4,303 amino acids with 2 leucine-rich repeats, a C-type lectin domain, multiple PKD repeats, and a short segment of 84 amino acids that is 31% identical to a portion of the trp2 N terminus that stretches from amino acid 155 to amino acid 238 of clone 17 (amino acid 4,176–amino acid 4,260 of PKD1; Fig. 1 E and F). This sequence is unique to Trp2 among Trp proteins and is of unknown function. With 1,172 amino acids Trp2-14, is the longest of the six closely related mammalian Trp proteins so far identified, the second longest being Trp6, with 930 amino acids. Trp1, with 793 amino acids, is the shortest. At 626 and 526 amino acids, the N termini of Trp2-14 and Trp2-17 are also the longest of the six mammalian Trps.
Functional Analysis of Trp2.
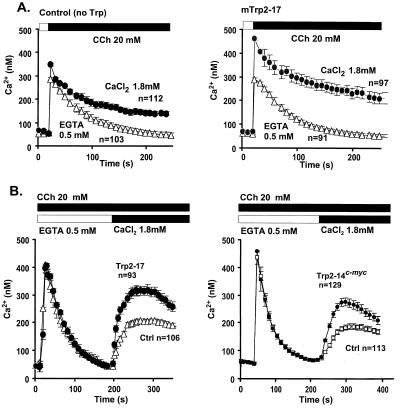
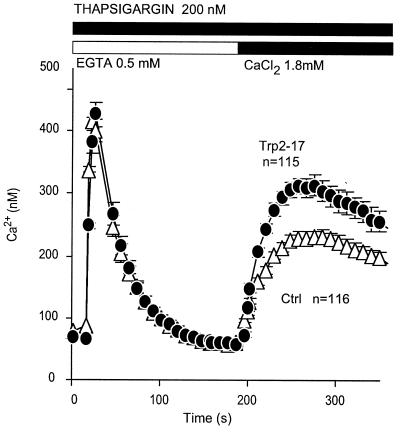
Tests for involvement in CCE showed Trp2 to enhance both CCE elicited by a Gq-coupled phospholipase Cβ-stimulating pathway (Fig. 2) and by a receptor-independent store depletion-induced pathway (Fig. 3). These results show that Trp2 is not only a structural but also a functional Trp homologue. In our hands, the rank order with which Trp-1, -2, -3, and -6, coexpressed in COS-M6 cells with the M5 muscarinic receptor, increase CCE in response to receptor activation is Trp3 = Trp6 > Trp2 > Trp1. In contrast, using the same assay system in which [Ca2+]i changes are measured by using fura2 as the indicator dye, Trp2 is the first that has given an unequivocal response to store depletion. We did not see store depletion-induced Ca2+ influx that could be attributed to Trp6 (15), and increased CCE in Trp3-expressing cells was transient, requiring uncovering by blockade of endogenous channels with gadolinium (27). Because, in Trp3-expressing cells, this expression was accompanied by a marked loss of gadolinium-inhibitable endogenous CCE, we proposed that Trp3 was likely to be acting as a dominant modifier of CCE channels, probably by contributing to the formation of oligomeric CCE channel with an abnormal subunit composition (27). It was thus difficult to ascribe a physiologic role to Trp3 as a participant of natural store depletion-activated CCE channels, and by extension to any Trp. The present data are clear in this respect, as the presence of Trp2 leads to an unequivocal enhancement of store depletion-activated CCE. The data support the hypothesis that the channels mediating store depletion-activated CCE (store-operated channels) and those mediating receptor-activated CCE (receptor activated-channels) may have a common subunit composition and indeed may be one and the same.
Figure 2.
Development of CCE in control and trp2-transfected cells. [Ca2+]i transients were determined by fura2 radiometric fluorescence spectroscopy. COS-M6 cells were cotransfected with M5 muscarinic acetylcholine receptor and either pcDNA3 carrying trp2-17, trp2-14, or murine luteinizing hormone LH receptor (no Trp control), plated onto circular coverslips, exposed to fura2-acetoxymethyl ester, washed, and analyzed by videomicroscopy for agonist-induced Ca2+ transients as described (3). Results are means from 2–3 independent transfections. Data are mean ± SEM of pooled results from 2–3 independent transfections measured in the number of cells shown on the figures. (A) Ca2+ transients induced by carbachol (CCh) in the absence of extracellular Ca2+ are unaltered by trp2-17 but are stimulated in its presence. (B) Postaddition of Ca2+ to the extracellular medium uncovers CCE that is significantly enhanced in cells expressing either trp2-17 (Left) or trp2-14 tagged at its C terminus with the epitope EQKLISEEDL that is recognized by the 9E10 anti-myc monoclonal antibody (Right).
Figure 3.
Thapsigargin treatment stimulates CCE in trp2-expressing cells. COS-M6 cells were transfected with pcDNA3 carrying either the trp2-17 or the luteinizing hormone (LH) receptor cDNA, plated onto circular coverslips, loaded with fura2, washed, and analyzed by video microscopy for changes in [Ca2+]i. The results are means from three independent transfections. n, total number of cells analyzed.
As visualized by Northern blotting, mouse trp2 appears to be expressed only in testis. The test used, however, is not very sensitive and only detects expression in tissues where accumulation of its mRNA is relatively high. ESTs derived from trp2 mRNA have been cloned from kidney (GenBank accession no. AA473022) and spleen (GenBank accession no. AA145678), indicating that this message is likely to be present at low levels in many other tissues than testis in which it obviously is expressed at much higher levels than in somatic tissues. It may therefore play a significant role on the general makeup of CCE channels.
In conclusion, Trp2 joins five other members of the mammalian family of Trp proteins. Its expression in cells leads to enhanced CCE stimulated by both agonists and store depletion. Based on studies with its family members, it is likely to form Ca2+-permeable cation channels either by itself or by forming multimeric complexes—most likely tetrameres. It remains for future studies to determine the exact expression pattern of Trp2, its contribution to normal CCE channels, and the role these channels play in regulation of cellular metabolism and responsiveness to extracellular signals.
Acknowledgments
This work was supported in part by National Institutes of Health Grants HL45198 to L.B. and GM-54235 to X.Z. and by a National Institutes of Health National Research Service Award and an American Heart Association grant to N.Q.; B.V. was a fellow of the French Association for Research Against Cancer (ARC) and G.B. was a fellow of the Canadian Medical Research Council.
ABBREVIATIONS
- CCE
capacitative Ca2+ entry
- IP3
inositol 3,4,5-triphosphate
- Trp
transient receptor protein
- IP3R
IP3 receptor
- RT
reverse transcription
- [Ca2+]i
cytosolic Ca2+
- EST
expressed sequence tag
- PKD
polycystic kidney disease
Footnotes
References
- 1.Suss E, Barash S, Stavenga D G, Stieve H, Selinger Z, Minke B. J Gen Physiol. 1989;94:465–491. doi: 10.1085/jgp.94.3.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhu X, Chu P B, Peyton M, Birnbaumer L. FEBS Lett. 1995;373:193–198. doi: 10.1016/0014-5793(95)01038-g. [DOI] [PubMed] [Google Scholar]
- 3.Zhu X, Jiang M, Peyton M J, Boulay G, Hurst R, Stefani E, Birnbaumer L. Cell. 1996;85:661–671. doi: 10.1016/s0092-8674(00)81233-7. [DOI] [PubMed] [Google Scholar]
- 4.Putney J W., Jr Cell Calcium. 1986;7:1–12. doi: 10.1016/0143-4160(86)90026-6. [DOI] [PubMed] [Google Scholar]
- 5.Putney J W., Jr Cell Calcium. 1990;11:611–624. doi: 10.1016/0143-4160(90)90016-n. [DOI] [PubMed] [Google Scholar]
- 6.Putney J W, Jr, Bird G, St J. Trends Endocrinol Met. 1994;5:256–260. doi: 10.1016/1043-2760(94)p3085-l. [DOI] [PubMed] [Google Scholar]
- 7.Birnbaumer L, Zhu X, Jiang M, Boulay G, Peyton M, Vannier B, Brown D, Platano D, Sadeghi H, Stefani E, Birnbaumer M. Proc Natl Acad Sci USA. 1996;93:15195–15202. doi: 10.1073/pnas.93.26.15195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kiselyov K, Xu X, Mozhayeva G, Kuo T, Pessah I, Mignery G, Zhu X, Birnbaumer L, Muallem S. Nature (London) 1998;396:478–482. doi: 10.1038/24890. [DOI] [PubMed] [Google Scholar]
- 9.Philipp S, Cavalie A, Freichel M, Wissenbach U, Zimmer S, Trost C, Marquart A, Murakami M, Flockerzi V. EMBO J. 1996;15:6166–6171. [PMC free article] [PubMed] [Google Scholar]
- 10.Philipp S, Hambrecht J, Braslavski L, Schroth G, Freichel., Murakami M, Cavalie A, Flockerzi V. EMBO J. 1998;77:4274–4282. doi: 10.1093/emboj/17.15.4274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thastrup O, Dawson A P, Scharff O, Foder B, Cullen P J, Drobak B K, Bjerrum B J, Christensen S B, Hanley M R. Agents Actions. 1989;27:17–23. doi: 10.1007/BF02222186. [DOI] [PubMed] [Google Scholar]
- 12.Takemura H, Hughes A R, Thastrup O, Putney J W., Jr J Biol Chem. 1989;246:12266–12271. [PubMed] [Google Scholar]
- 13.Kwan C Y, Takemura H, Obie J F, Thastrup O, Putney J W., Jr Am J Physiol. 1990;258:C1006–C1015. doi: 10.1152/ajpcell.1990.258.6.C1006. [DOI] [PubMed] [Google Scholar]
- 14.Boulay G, Zhu X, Peyton M, Jiang M, Hurst R, Stefani E, Birnbaumer L. J Biol Chem. 1997;272:29672–29680. doi: 10.1074/jbc.272.47.29672. [DOI] [PubMed] [Google Scholar]
- 15.Okada T, Shimizu S, Wakamori M, Maeda A, Kurosaki T, Takada N, Imoto K, Mori Y. J Biol Chem. 1998;273:10279–10287. doi: 10.1074/jbc.273.17.10279. [DOI] [PubMed] [Google Scholar]
- 16.Tareilus E, Roux M, Qin N, Olcese R, Zou J, Stefani E, Birnbaumer L. Proc Natl Acad Sci USA. 1997;94:1703–1708. doi: 10.1073/pnas.94.5.1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sanger F, Nicklen S, Coulson A B. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Laemmli U. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 19.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 20.Zhu X, Birnbaumer L. Proc Natl Acad Sci USA. 1996;93:2827–2831. doi: 10.1073/pnas.93.7.2827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wes P D, Chevesich J, Jeromin A, Rosenberg C, Stetten G, Montell C. Proc Natl Acad Sci USA. 1995;92:9652–9656. doi: 10.1073/pnas.92.21.9652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wissenbach U, Schroth G, Philipp S, Flockerzi V. FEBS Lett. 1998;429:61–66. doi: 10.1016/s0014-5793(98)00561-4. [DOI] [PubMed] [Google Scholar]
- 23.Vannier B, Zhu X, Brown D, Birnbaumer L. J Biol Chem. 1998;273:8675–8679. doi: 10.1074/jbc.273.15.8675. [DOI] [PubMed] [Google Scholar]
- 24.Schwalbe R A, Wang Z, Bianchi L, Brown A, M. J Chem Biol. 1996;271:24201–24206. doi: 10.1074/jbc.271.39.24201. [DOI] [PubMed] [Google Scholar]
- 25.Nagamine K, Kudoh J, Minoshima S, Kawasaki K, Asakwa S, Ito F, Shimizu N. Genomics. 1998;54:124–131. doi: 10.1006/geno.1998.5551. [DOI] [PubMed] [Google Scholar]
- 26.Hunter J J, Shao J, Smutko J S, Dussault B J, Nagle D L, Woolf E A, Holmgren L M, Moore K J, Shyjan A W. Genomics. 1998;54:116–123. doi: 10.1006/geno.1998.5549. [DOI] [PubMed] [Google Scholar]
- 27.Zhu X, Jiang M, Birnbaumer L. J Biol Chem. 1998;273:133–142. doi: 10.1074/jbc.273.1.133. [DOI] [PubMed] [Google Scholar]