Abstract
The development of skin carcinomas presently is believed to be correlated with mutations in the p53 tumor suppressor and ras gene as well as with the loss of chromosome 9. We now demonstrate that, in addition, loss of chromosome 15 may be a relevant genetic defect. Reintroduction of an extra copy of chromosome 15, but not chromosome 4, into the human skin carcinoma SCL-I cells, lacking one copy of each chromosome, resulted in tumor suppression after s.c. injection in mice. Transfection with thrombospondin-1 (TSP-1), mapped to 15q15, induced the same tumor suppression without affecting cell proliferation in vitro or in vivo. Halted tumors remained as small cysts encapsulated by surrounding stroma and blood vessels. These cysts were characterized by increased TSP-1 matrix deposition at the tumor/stroma border and a complete lack of tumor vascularization. Coinjection of TSP-1 antisense oligonucleotides drastically reduced TSP-1 expression and almost completely abolished matrix deposition at the tumor/stroma border. As a consequence, the tumor phenotype reverted to a well vascularized, progressively expanding, solid carcinoma indistinguishable from that induced by the untransfected SCL-I cells. Thus, these data strongly suggest TSP-1 as a potential tumor suppressor on chromosome 15. The data further propose an unexpected mechanism of TSP-1-mediated tumor suppression. Instead of interfering with angiogenesis in general, in this system TSP-1 acts as a matrix barrier at the tumor/stroma border, which, by halting tumor vascularization, prevents tumor cell invasion and, thus, tumor expansion.
Keywords: keratinocytes, nonmelanoma skin carcinogenesis, angiogenesis
In recent years, skin cancer has become a major threat for Caucasian populations. The increasing number of basal cell carcinomas (BCCs) and squamous cell carcinomas (SCCs), two different types of nonmelanoma skin tumors, is thought to be linked to the depletion of the ozone layer and increasing outdoor activities. The causal role of UV radiation has been substantiated by the finding that a high percentage of skin carcinomas carried specific mutations in the p53 tumor suppressor gene, namely C-to-T transitions in CC sites or CC-to-TT double-base changes (reviewed in ref. 1).
Two other genetic aberrations are considered to be relevant presently. First, oncogenic activation of the ras gene was found in about 15% of the skin carcinomas (reviewed in ref. 2), and, second, loss of heterozygosity (LOH) of 9q frequently is seen in BCCs. The latter is compatible with a loss of the nevoid basal cell carcinoma syndrome locus and its candidate tumor suppressor gene, the human homolog of the Drosophila “patched” gene (reviewed in ref. 3). SCCs, on the other hand, more often show LOH of 9p (4), suggesting that different genes are involved in the development of the two skin carcinoma types.
Data from our experimental in vitro skin carcinogenesis model indicated that loss of material from chromosome 15 may play a role in malignant conversion. The spontaneously immortalized nontumorigenic human skin keratinocyte line HaCaT, which carries UV-type specific mutations in both alleles of the p53 gene (5) and has lost one copy of chromosome 9p, in addition to other chromosomes (6), could be converted to tumorigenicity after the introduction of the c-rasH oncogene. As a result, benign and malignant tumorigenic clones developed (7, 8). However, the malignant phenotype was observed only in clones that had lost copies of chromosome 15 (8).
One possible candidate gene on chromosome 15, already proposed to be connected with carcinogenesis, is thrombospondin-1 (TSP-1), a gene encoding a matrix glycoprotein. So far, the role of TSP-1 in carcinogenesis is somewhat controversial. Whereas in some cell systems TSP-1 overexpression contributed to tumor progression (reviewed in ref. 9), TSP-1 expression was inversely correlated with malignant progression in breast cancer cells, melanomas, and lung carcinomas (10, 11). Furthermore, transfection of TSP-1 resulted in a reduced size of the heterotransplanted primary tumors, a decrease in spontaneous pulmonary metastases of human breast carcinoma cells, and suppressed tumorigenicity of transformed murine endothelial and NIH 3T3 cells (10, 12–14). Inhibition of angiogenesis was suggested as a potential mechanism for tumor suppression because TSP-1 inhibited de novo vascularization in the cornea assay (15). Moreover, blood vessel density was reduced in tumors of breast carcinoma cells overexpressing TSP-1 (10). We now provide evidence for a tumor-suppressive function of TSP-1 also in skin carcinoma cells by demonstrating that transfection-induced overexpression of TSP-1 caused tumor suppression comparable to that seen after transfer of an extra copy of chromosome 15. We demonstrate an indirect mechanism of tumor suppression in that overexpression of TSP-1 did not affect angiogenesis in general but exclusively halted tumor vascularization. This could be abolished completely by coinjecting TSP-1 antisense oligonucleotides, demonstrating a causal role for TSP-1 in tumor suppression by counteracting tumor vascularization.
MATERIALS AND METHODS
Cells and Culture Conditions.
The human carcinoma cell lines SCC-12 and SCC-13 (16), SCC-I (17), and SCL-II (18) and the mouse A9 cell line, which carries a single human chromosome 15 (used for the chromosome transfer studies), were cultivated as described (5). Cells were disaggregated routinely with 0.1% trypsin/EDTA solution and replated at a split ratio of 1:10 and 1:100, respectively.
Microcell-Mediated Chromosome Transfer (MMCT).
MMCT was performed as described (7) using a human chromosome 15 tagged with a neomycin-resistance gene allowing clonal selection and expansion in medium containing 200 μg/ml of G418 (GIBCO/BRL). Clones free of mouse chromosomes (determined by fluorescence in situ hybridization analysis using mouse DNA as probe) from at least two independent MMCT experiments were analyzed further.
Transfection.
The expression vector pCEP-4, containing the 3.5-kb fragment of the mouse TSP-1 cDNA under transcriptional control of the cytomegalovirus promoter and a hygromycin-resistance gene, was kindly provided by V. Dixit (University of Michigan, Ann Arbor, MI). TSP-1 cDNA was eliminated by HindIII/NotI digest, and the remaining vector (10.4 kb) was used as control plasmid. Transfection of SCL-I cells and selection of individual clones (200 μg/ml of hygromycin; Boehringer Mannheim) were performed as described (7). Integration of TSP-1 DNA was verified by Southern blot analysis.
In Situ Hybridization on Tumor Sections.
TSP-1 cDNA was inserted into the EcoRI site of pGEM-2 (American Type Culture Collection, Manassas, VA), linearized with SacI, and transcribed with SP6 polymerase to generate antisense or with NheI and T7 polymerase (all Boehringer Mannheim) to generate sense probes in the presence of [35S]CTP (Amersham). In situ hybridization was performed as described by Moorman et al. (19) with an exposure time for autoradiography of ≈4 weeks.
Cytogenetic Analysis.
For fluorescence in situ hybridization, metaphase spreads obtained according to standard protocols were treated with RNase and pepsin (20). The plasmid library from sorted human chromosomes 15, kindly provided by J. Gray (University of California at San Francisco Cancer Center) (21), was amplified according to standard protocols and nick-translated with tetramethylrhodamine (TRITS)-6-dUTP (Boehringer Mannheim). Chromosome-specific library DNA probes for chromosomes 1 and 13 directly conjugated to fluorescein isothiocyanate (FITC) were a generous gift from Vysis (Naperville, IL). Hybridization was carried out as described (22). Comparative genomic hybridization (CGH) was performed as described previously (23). Average FITC/TRITC ratio profiles were calculated from at least 10 metaphase spreads.
In Vitro Growth Curve.
Cells were plated in duplicates in 6-well plates (1 × 105 cells per well), trypsinized, and counted after 24 h to assess plating efficiency and at daily intervals for 7 days.
Tumorigenicity Test.
Cells (5 × 106) in 100 μl culture medium were injected s.c. on each side of the back of ≈7-week-old nude mice (Swiss/c nu/nu backcrosses). Tumor growth was measured at weekly intervals, and tumor size (in mm3) was calculated (24) and plotted against time. Mice received tail-vein injections of BrdUrd and 2-deoxycytidine (65 mM each) 2 h before being sacrificed. Tumors were removed for ethical reasons when they were >150 mm3 or after 1, 3, and 5 weeks, respectively, and were frozen in liquid nitrogen vapor for cryostat sections or snap-frozen in liquid nitrogen for protein extraction.
Indirect Immunofluorescence.
Tumor sections were fixed and stained as described (7, 25) with a rat mAb against TSP-1 (0.2 μg/μl; Immunotech, Marseille, France), a rabbit polyclonal antibody against mouse collagen type IV (dilution 1/100; Institut Pasteur, Lyon, France), a rat mAb against platelet endothelial cell adhesion molecule (MEC 13.3, 26), kindly provided by E. Dejana (Institute of Pharmacological Research, Grenoble, France), and a mouse mAb against BrdUrd (dilution 1/100; Partec, Arlesheim, Switzerland). The respective second antibodies were labeled with either FITC or Texas Red (all purchased from Jackson ImmunoResearch).
Western Blot Analysis.
Twenty-four hours after plating, cultures were washed and incubated in 15 ml of serum-free culture medium for 72 h. TSP-1-conditioned medium was incubated overnight at 4°C with heparin-agarose beads (Sigma). Beads were collected by centrifugation and washed in 0.2 M NaCl/20 mM Tris⋅HCl, pH 7.2, proteins were eluted from the beads in 2× Laemmli buffer, denatured (5 min) at 95°C, and normalized for cell number, and aliquots were stored at −20°C. Tumor nodules were excised, snap-frozen, and crushed in liquid nitrogen, proteins were extracted in Laemmli buffer, and protein concentrations were quantified by using the Bio-Rad protein assay. Protein aliquots were analyzed by SDS/PAGE and blotted onto nitrocellulose membranes. Filters were blocked for 2 h in 10% low-fat milk (in PBS with 0.1% Tween 20), incubated with a rabbit anti-TSP-1 polyclonal antibody (1 μg/ml) for 3 h at room temperature (RT), washed, incubated with a horseradish-peroxidase-conjugated goat anti-rabbit antibody (1:10,000; Jackson ImmunoResearch) for 1 h at RT, washed, and developed by using the Amersham ECL chemiluminescence reagents.
Antisense Experiments.
SCL-ITSP-1 cells (5 × 106 cells in 100 μl medium) were incubated for 2 h at 37°C with 20 nM of either antisense (AAG AAG ATG AAG CCT TTG, nucleotides 466–481) or control phosphorothioate oligonucleotides (ACT ACT ACA CTA GAC TAC), both designed, synthesized, and HPLC-purified by Biognostik GmbH (Göttingen, Germany). Thereafter, cells were injected s.c. into each side of the back of ≈7-week-old nude mice (Swiss/c-nu/nu/backcrosses). At days 3, 5, 7, and 9, 50 nM of oligonucleotides (diluted in NaCl) were injected s.c. at a site remote from the cells, and tumor nodules were excised at weeks 2–6.
RESULTS
Chromosome 15 Aberrations Are Frequent in Skin Carcinoma Lines.
Data from our HaCaT model had indicated that malignant progression correlated with loss of chromosome 15 (8). To determine whether this aberration also was relevant for skin carcinoma-derived cells, we analyzed the status of chromosome 15 in four different SCC lines by CGH and fluorescence in situ hybridization on metaphase chromosomes. Only one line contained a normal set of chromosome 15 in a nearly diploid karyotype (SCC-13; Fig. 1A). Two hypotetraploid cell lines contained one or two chromosome 15 translocation chromosomes in addition to one or two normal copies of chromosome 15 (SCC-12: t(13q;15q), Fig. 1B; SCL-II: t(1q;15q), Fig. 1C). In the fourth hypotetraploid cell line, SCL-I, chromosome 15q was underrepresented altogether (see Fig. 1A) because of a loss of one entire copy (16).
Figure 1.
Aberrant chromosomes 15 in SCC cells. (A) Average CGH ratio profiles of chromosomes 15 from SCC-13 and SCL-I and four from SCL-I cells. 4′,6-Diamidino-2-phenylindole-stained normal reference chromosomes, fluorescence ratio images (blue, normal balanced state of chromosome material; red, underrepresentation; and green, overrepresentation in the tumor genome), and idiogram with ratio profiles (left, center, and right vertical lanes represent lower, average, and upper thresholds of the normal range) are shown for each chromosome. Whereas SCC-13 cells show a normal ratio profile of chromosome 15, it is underrepresented in SCL-I cells, as is chromosome 4p. Gray represents chromosomal regions of highly repetitive DNA sequences that are suppressed by Cot I DNA and excluded from evaluation. (B and C) Fluorescence in situ hybridization of metaphase chromosomes. (B) SCC-12 cells (tetraploid) show a translocation chromosome t(13q;15q) (13 in green and 15 in red; upper left corner), two normal copies of chromosomes 15 and 13, an isochromosome [i(13q)], and a translocation chromosome containing material from chromosome 13. (C) SCL-II cells (hypodiploid) contain a translocation chromosome t(1q;15q) (1 in green and 15 in red), one normal copy of chromosomes 15 and 1, and two additional translocation chromosomes containing material from chromosome 1.
Transfer of Chromosome 15 Induces Tumor Suppression.
To determine the functional consequence of the loss of one copy of chromosome 15, we introduced extra copies into the SCL-I cells via microcell-mediated chromosome transfer and selected three clones (SCL-Ichr15) that now contained an additional copy of chromosome 15. Because in vitro proliferation remained unchanged with a population-doubling time of ≈24 h and our previous data had suggested a role of chromosome 15 in malignant conversion (8), all three clones were injected s.c. into nude mice. Whereas the nodules of the parental SCL-I cells steadily increased in size and had to be excised 40–60 days after injection (Fig. 2A), the nodules of all SCL-Ichr15 clones remained stationary and increased only slightly in size toward the end of the observation period as shown for SCL-Ichr15 clone 4 (Fig. 2B).
Figure 2.
Tumor growth curves after s.c. injection of the cells into athymic nude mice. (A) SCL-I cells show steady tumor growth. (B) SCL-I cells with an extra chromosome 15 (SCL-Ichr15 clone 4) were suppressed in their growth during this observation period. (C) An additional copy of chromosome 4 (SCL-Ichr4 clone 1) did not affect tumor growth. Each curve represents an individual animal.
To investigate whether complementation of any “lost” chromosome would cause tumor suppression, we also introduced chromosome 4, which similarly is underrepresented in the SCL-I cells (see Fig. 1A). Four different microcell hybrid clones with an extra copy of chromosome 4 (SCL-Ichr4) were injected s.c. into nude mice. Tumors occurred in all mice with a pattern identical to that of the parental cells (Fig. 2C).
TSP-1, a Putative Suppressor Gene, on Chromosome 15.
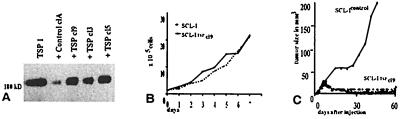
We transfected the SCL-I cells with TSP-1 cDNA, a candidate gene that already was described for its tumor-suppressive potential (reviewed in ref. 27). Three TSP-1 transfectants (SCL-ITSP-1) overexpressing TSP-1 (Fig. 3A) and two control clones, transfected with the plasmid without TSP-1 insert (SCL-Icontrol), were analyzed further. As with the SCL-Ichr15 cells, TSP-1 overexpression did not affect in vitro proliferation. The population-doubling times remained largely unchanged under standard conditions (Fig. 3B). Also under serum-free conditions, the population-doubling time was similar for all cells with ≈72 h. In vivo, however, all three SCL-ITSP-1 clones exhibited tumor-suppressive activity. Whereas the tumors of two control clones expanded to a size of >150 mm3 within less than 2 months, tumor growth of the three SCL-ITSP-1 clones was suppressed completely during this 60-day period as demonstrated for SCL-Icontrol clone A and SCL-ITSP-1 clone 9 cells (Fig. 3C).
Figure 3.
Characterization of the TSP-1-transfected SCL-I cells (SCL-ITSP-1 clone 9). (A) Western blot analysis of control cells (vector clone A cells) and three SCL-ITSP-1 clones (clones 3, 5, and 9). TSP-1 monomers from activated platelets (TSP-1) with a molecular mass of 180-kDa were used as standard. Expression of the 180-kDa TSP-1 protein is up-regulated in all three SCL-ITSP-1 clones. (B) In vitro growth curves are similar for the parental SCL-I and SCL-ITSP-1 clone 9 cells. (C) Tumor growth curve of SCL-ITSP-1 clone 9 cells after s.c. injection of the cells in nude mice. In comparison with the control clone A cells (mean value of 10 animals), which developed large tumors in less than 60 days, SCL-ITSP-1 cells are suppressed in their growth during this time span (10 individual animals).
TSP-1 Overexpression Prevents Tumor Vascularization.
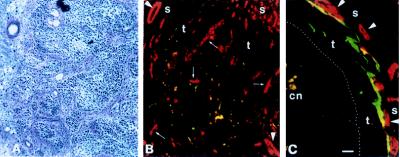
To investigate the mechanism of tumor suppression, we performed a detailed histological and biochemical comparison of nodules from the parental SCL-I and the SCL-ITSP-1 clone 9 cells excised at weekly intervals for a period of 6 weeks. Histological examination revealed the first significant differences in 1-week-old nodules. Whereas the SCL-I cells had grown to solid tumors composed of small tumor-cell islands embedded in stroma (Fig. 4A), SCL-ITSP-1 cells had formed small cyst-like tumors with a rim of vital epithelium surrounding a central necrotic area (Fig. 4B). This difference in tissue morphology was maintained over the entire observation period.
Figure 4.
Comparison of 1-week-old tumor nodules from SCL-I and SCL-ITSP-1 clone 9 cells. (A) Tumor histology of sections from SCL-I cells shows solid tumor nodules with tumor cell islands (t) embedded in stroma ropes (s). (B) SCL-ITSP-1 cells had formed cyst-like nodules consisting of a rim of vital epithelium (t) around massive central necrosis (cn) and surrounded by stroma. (C) In situ hybridization with an antisense TSP-1 probe (bright-field micrograph) showing only scattered cells in SCL-I nodules expressing TSP-I mRNA. (D) In SCL-ITSP-1 nodules, nearly all tumor cells expressed TSP-1. Immunostaining with a TSP-1-specific antibody revealed scattered TSP-1 protein dots throughout the SCL-I nodule (E) whereas SCL-ITSP-1 nodules showed a prominent TSP-1 matrix at the tumor/stroma border (F). (G) The carcinoma-like SCL-I nodules consisted of epithelial islands (high density of Hoechst-stained nuclei) interspersed by stroma (areas of low nuclei content) with blood vessels (yellow) prominent in the stroma around the tumor as well as within the tumor. (H) In SCL-ITSP-1 cysts, vessels double-stained for collagen type IV (in red) and PECAM (in green) (double-labeled vessels appear in yellow) were present in the surrounding stroma but absent in the tumor. s, stroma; t, tumor; cn, central necrosis. (Bar = 100 μm.)
Surprisingly, proliferation profiles established by BrdUrd labeling did not vary significantly in the nodules from SCL-I and SCL-ITSP-1 cells. During the first 2 weeks the mean labeling index was 10% for nodules of the SCL-I cells and 12% for those of the SCL-ITSP-1 cells. Thereafter, the number of BrdUrd-labeled cells increased in both types of tumors up to a mean of 30% and 28%, respectively, and remained at this level.
To study the role of TSP-1 as a causal factor for the different tumor phenotypes, TSP-1 expression and protein deposition were analyzed by in situ hybridization and immunostaining. Whereas in the tumors from the SCL-I cells TSP-1 expression was restricted to single, scattered cells (Fig. 4C), expression was abundant throughout the vital epithelium of SCL-ITSP-1 nodules (Fig. 4D). Similarly, TSP-1 protein was visualized as only small, scattered spots in 1-week-old SCL-I nodules (Fig. 4E) in contrast to a prominent TSP-1 matrix, mostly deposited between the stroma and the SCL-ITSP-1 tumor cells (Fig. 4F). This discriminating pattern of TSP-1 mRNA expression and protein deposition remained unchanged during the 6-week observation period.
To analyze tumor vascularization, blood vessels were labeled with antibodies against collagen type IV (coll IV), a component of basement membranes, or PECAM (CD31), a specific marker for endothelial cells that, as demonstrated previously, reveal identical results (28). In the 1-week-old tumor nodules of the SCL-I cells blood vessels were present throughout the stromal cords surrounding and traversing the solid tumor nodules (Fig. 4G). In marked contrast, nodules of the SCL-ITSP-1 cells, which were characterized by a prominent TSP-1 deposition at the tumor/stroma border (see Fig. 4F), were virtually devoid of blood vessels within the tumor tissue. However, blood vessel density in the stroma surrounding the epithelial tumor nodules was comparable to that of the controls (Fig. 4H). The SCL-ITSP-1 nodules remained completely avascular during the 6-week observation period. In addition, and most likely as a consequence, the nodules were characterized by a large central necrotic area.
Antisense-Mediated TSP-1 Reduction Restores Tumor Vascularization.
The foregoing observations indicated that increased deposition of TSP-1 at the tumor/stroma border in nodules of the SCL-ITSP-1 cells may have been responsible for inhibition of tumor vascularization and that this, in turn, prevented tumor cell invasion into the surrounding stromal tissue. Thus, if the TSP-1 matrix was causal for halting tumor vascularization, reduced TSP-1 deposition during the first 2 weeks should allow tumor vascularization and, as a consequence, tumor expansion. To test this possibility we exposed SCL-ITSP-1 clone 9 cells to antisense or control oligonucleotides (same length and base content and similar hybridization characteristics). Possible toxic effects of the oligonucleotides were excluded by treatment of the cells with 20 nM of control or antisense oligonucleotide in vitro. During the following 3 days the population-doubling time remained unchanged with ≈24 h. To test their efficacy in vivo, the SCL-ITSP-1 cells were incubated with either control nucleotides or oligonucleotides in vitro for 2 h, and after s.c. injection of the cells the mice were injected additionally with antisense and control oligonucleotides at days 3, 5, 7, and 9. After another 5 days (2 weeks after injection of the cells) tumor nodules were excised and analyzed.
In the presence of TSP-1 antisense oligonucleotides the 2-week-old nodules from SCL-ITSP-1 cells were histologically indistinguishable from those of the parental- or control vector-transfected SCL-I cells. They now were solid tumors composed of tumor cell islands dispersed by cords of stroma (Fig. 5A). The same SCL-ITSP-1 cells treated with control oligonucleotides remained suppressed in their tumor growth as untreated SCL-ITSP-1 cells and continued to form encapsulated cystic nodules characterized by only a rim of vital epithelium.
Figure 5.
The tumor phenotype is reverted by antisense oligonucleotide treatment in vivo. (A) Histology of a 2-week-old tumor nodule of SCL-TSP-1 clone 9 cells treated with antisense oligonucleotides. The originally cyst-like tumor has reverted to a carcinoma. (B) Double-labeling of a corresponding section with antibodies against TSP-1 (green; yellow when colocalized with blood vessels) and collagen type IV (red) showing few TSP-1 dots and extended tumor vascularization (arrow) in addition to blood vessels in the surrounding stroma (arrowhead). (C) SCL-ITSP-1 cells treated with control oligonucleotides remain as cyst-like tumors with a prominent TSP-1 matrix (green) at the tumor/stroma border and blood vessels (red, arrowhead) restricted to the surrounding stroma. (Bar = 100 mm.)
Reversion in tumor phenotype by TSP-1 antisense oligonucleotides also was accompanied by a corresponding TSP-1 protein deposition with only scattered spots of TSP-1 matrix within the tumor tissue and a well developed tumor vascularization (Fig. 5B). The nodules of the control oligonucleotide-treated SCL-ITSP-1 cells, on the other hand, continued to show the characteristic, massive TSP-1 matrix deposited along the tumor/stroma border, with blood vessels restricted to the stroma around the tumor nodules, leaving the tumor epithelium entirely avascular (Fig. 5C).
High TSP-1 Protein Levels Correlate with Halted Tumor Vascularization.
To determine whether quantitative differences exist in TSP-1 protein levels in vivo, Western blot analysis was performed with extracts from different tumor nodules. The 180-kDa TSP-1 band was present only at low levels in the expanding tumors of the parental SCL-I cells (Fig. 6A). In contrast, SCL-ITSP-1 cells expressed high levels of TSP-1 protein already in 1-week-old nodules (see Fig. 6A). Comparably, the nodules of control oligonucleotide-treated cells showed high levels of TSP-1 protein, whereas in nodules of the antisense oligonucleotide-treated SCL-ITSP-1 cells the 180-kDa band was reduced significantly (Fig. 6B), thereby demonstrating the efficacy of the antisense oligonucleotide treatment in vivo.
Figure 6.
Tumor suppression correlates with high levels of TSP-1 protein. (A) Western blot analysis shows high levels of the 180-kDa TSP-1 protein band in 1-week-old nodules from the SCL-ITSP-1 clone 9 cells and low levels in 3- and 5-week-old nodules from the SCL-I cells. The lower-molecular-mass bands likely represent degradation products. (B) The level of TSP-1 protein also is high in 2- and 3-week-old tumor nodules from SCL-ITSP-1 clone 9 cells injected with control oligonucleotides but significantly reduced in the nodules from the TSP-1 antisense oligonucleotide-treated SCL-ITSP-1 cells. Numbers in parentheses represent weeks after injection. Monomeric TSP-1 protein with a molecular mass of 180 kDa was included as standard.
DISCUSSION
Previous studies with our in vitro skin carcinogenesis model had suggested that loss of chromosome 15 was involved in tumor progression from a benign to a malignant phenotype (8), and now we show aberrations in chromosome 15 for three of four skin carcinoma-derived cell lines. This is in good agreement with recent studies demonstrating LOH of chromosome 15 in metastases from lung, breast, and colorectal carcinomas (29) and deletions in chromosome 15 in head and neck carcinomas and pancreatic cancer (30, 31). Furthermore, preliminary CGH studies on primary skin carcinomas indicate loss of material from chromosome 15 in two of seven SCCs, whereas no such loss was seen in the two keratoacanthomas, spontaneously regressing SCC-like skin tumors, analyzed so far (S.P. and P.B., unpublished results). Collectively, these observations may suggest a potential role of chromosome 15-encoded gene(s) in carcinogenesis.
To study the functional consequences of loss of chromosome 15, one cell line, SCL-I, seemed particularly promising because of the lack of an entire copy of chromosome 15. In addition, these cells had lost one copy of chromosome 4, a chromosome that was shown earlier to carry tumor suppressor gene(s) (32). The introduction of chromosome 15 or 4 had no obvious consequence on cell proliferation or long-term propagation in vitro. However, under in vivo conditions chromosome 15 had a significant tumor-suppressive effect, whereas the SCL-Ichr4 exhibited the same progressive tumor growth pattern as the parental SCL-I cells.
We concentrated on TSP-1, a first potential candidate tumor suppressor gene located on chromosome 15 that maps to 15q15. Indeed, overexpression of TSP-1 resulted in a similar tumor suppression as seen with an extra copy of chromosome 15. Thus, our findings correlated well with those reports demonstrating a negative effect on tumor progression (10, 12–14). TSP-1 is a negative regulator of angiogenesis probably through the inhibition of endothelial growth and cell motility (15, 33–39). The antiangiogenic effect of TSP-1 was demonstrated by its potential to inhibit fibroblast growth factor-induced angiogenesis in the cornea assay (15). However, a detailed in vivo analysis of the interaction of tumor cell-derived TSP-1 with its environment has not been documented so far.
Similar to the SCL-Ichr15 cells, TSP-1 transfectants (SCL-ITSP-1) did not show obvious changes in in vitro growth properties. Although this correlated well with an earlier report on TSP-1-transfected breast carcinoma cells (10), it contradicted studies on melanoma cells that were inhibited in their proliferation by TSP-1 peptides (40) or endothelial cells that exhibited a slower growth rate, lower saturation density, and altered morphology after TSP-1 transfection (12). Thus, the effect induced by TSP-1 overexpression may well be cell-type-specific. Under in vivo conditions, however, TSP-1 overexpression in the skin SCL-1 carcinoma cells had significant effects. The continuous tumor growth seen with the parental SCL-I and control vector-transfected cells was inhibited, and suppression was similar, if not more pronounced, than with SCL-Ichr15 cells. Furthermore, we observed a striking difference in tumor histology. Whereas SCL-I cells grew as solid carcinomas from the first week, the SCL-ITSP-1 had developed cystic tumors largely resembling benign epidermal cysts (8, 28).
One explanation for the observed tumor suppression is that TSP-1 might have altered the kinetics of tumor cell proliferation in vivo. However, the proliferation rate, as determined by BrdUrd labeling index, was indistinguishable for SCL-I and SCL-ITSP-1 cells. Thus, tumor suppression was not a simple consequence of reduced tumor cell proliferation. On the other hand, we were able to demonstrate a significant increase in TSP-1 expression in vivo and, most importantly, a different pattern in TSP-1 protein deposition. Whereas SCL-I cells showed only sparse deposition of protein scattered throughout the tumor, SCL-ITSP-1 cells had deposited a prominent TSP-1 matrix at the tumor/stroma border around the epithelium, giving the impression of a barrier between tumor cells and stroma. In agreement with this interpretation, blood vessels were well established in the stroma around the tumor cysts but did not extend into the epithelium, leaving these tumor nodules completely avascular. Therefore, we suggest, as a novel mechanism of TSP-1-induced tumor suppression, that high levels of TSP-1 protein deposition at the tumor/stroma border during the onset of tumor development cause tumor suppression by halting vessel migration and, with that, blocking tumor vascularization. This, in turn, prevents tumor invasion and expansion. The probability for such a mechanism is supported strongly by the observation that suppressing TSP-1 protein deposition in vivo by means of TSP-1 antisense oligonucleotides completely restored the tumor phenotype of the untransfected parental SCL-I cells with the characteristics of well vascularized and progressively growing tumors whereas in the presence of control oligonucleotides the avascular cyst-like tumor was maintained.
A number of functions have been ascribed to TSP-1 which may have been causal for the observed block of tumor vascularization (27, 41, 42). Because of its heparin-binding affinity, TSP-1 is able to prevent diffusion and/or activation of heparin-binding angiogenic factors such as basic fibroblast growth factor, platelet-derived growth factor, and vascular endothelial growth factor (43–46). SCL-ITSP-1 cells express good levels of vascular endothelial growth factor mRNA (K.B. and B.P., unpublished results), and it appears unlikely that this amount may be insufficient to override inactivation by TSP-1, as discussed for a different cell system (46). Along the same line, the vascular endothelial growth factor receptor flk-1 is expressed by endothelial cells in the surrounding stroma, suggesting an active paracrine stimulation. Finally, TSP-1-mediated “inactivation” of angiogenic factors should not be restricted to inhibition of tumor vascularization but should affect angiogenesis in general. Angiogenesis in the surrounding stroma, however, was not altered in nodules of the TSP-1-overexpressing SCL-I cells. Blood vessels similarly were frequent, as in the vicinity of tumors of the parental SCL-I cells. Correspondingly, the proliferative activity of endothelial cells did not seem to be different in the surrounding stroma of both tumor types. The only obvious difference was the blocked blood vessel ingrowth into the SCL-ITSP-1 tumors. Because this correlated not only with an increased amount of TSP-1 protein but also, specifically, with its matrix deposition at the tumor/stroma border, it may indicate that TSP-1 is able to interfere with endothelial cell migration, chemotaxis, and/or attachment to extracellular matrix as suggested recently by Bouck and coworkers from in vitro experiments (38, 39). Thus, increased TSP-1 matrix deposition seems to be able to counteract one or all of these processes either directly, by functioning as a physical barrier, or indirectly via regulation of other growth factors.
One important factor in this context may be the transforming growth factor β 1 (TGF-β1). It has been shown that TGF-β1 is released from its latent form by TSP-1 in a protease-independent manner (47, 48) and that TSP-1 also is a major activator of TGF-β1 in vivo (49). In addition, both, TGF-β1 and TSP-1 can modulate protein expression in several cell types and, for example, increase the secretion of protease inhibitor PAI-1 (50, 51), which counteracts protease activity (27). Preliminary experiments suggest that the protein levels of the two metalloproteases, MMP3 and MMP9, are reduced in the stroma around the SCL-ITSP-1 nodules (C. P. Sommerhoff, D. Ludolph-Hauser, and P.B., unpublished results). This is in good agreement with an in vitro study by Qian et al. (52) demonstrating activation of MMP9 by low doses of TSP-1 and inhibitory high doses. Thus, our data highlight an important mechanism of tumor/stroma interactions, namely regulating tumor expansion via tumor vascularization. The inhibitory activity of TSP-1 seems to be dose-dependent and determined by the localization of the matrix, which acts as a shield by hindering blood vessel penetration into the tumor. Furthermore, complete reversion of the antiangiogenic phenotype by mere down-regulation of TSP-1 expression in vivo, using antisense oligonucleotides, supports the direct role of TSP-1 in this scenario. Whether TSP-1 itself or TSP-1-activated TGF-β may additionally modulate gene expression of the stroma cells to hinder protease expression, as a prerequisite for blood vessel migration and matrix degradation, remains to be determined.
Acknowledgments
We thank E. Tomakidi, H. Steinbauer, N. Forster, and S. Heid for their expert technical assistance. We thank Dr. M. Skobe and F. Bringold for their help in setting up the in situ hybridization and Western blot analysis. We further thank Drs. V. Dixit, B. Frazier, J. Gray, E. Dejana, and Vysis for generously providing us with the expression vector, antibodies, and the chromosome-specific library DNA probes and particularly acknowledge the members of the “Fotoabteilung” for excellent photo work and B. Plagens for critically reading this manuscript. This work was supported by a Deutsche Forschungsgemeinschaft grant (Bo 1246/3–1) to P.B., a predoctoral grant from the Boehringer Ingelheim Fonds (to K.B.), a European Union grant (BIO4 CT96 0464) (to N.E.F.), and Grants CA19401 and CA 69515 from the U.S. National Cancer Institute (to E.J.S.).
ABBREVIATIONS
- SCC
squamous cell carcinoma
- TSP-1
thrombospondin-1
- CGH
comparative genomic hybridization
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
References
- 1.Brash D E. Trends Genet. 1997;13:410–414. doi: 10.1016/s0168-9525(97)01246-8. [DOI] [PubMed] [Google Scholar]
- 2.Ananthaswamy H N, Pierceall W E. Prog Clin Biol Res. 1992;376:61–84. [PubMed] [Google Scholar]
- 3.Gailani M R, Bale S J. J Natl Cancer Inst. 1997;89:1103–1109. doi: 10.1093/jnci/89.15.1103. [DOI] [PubMed] [Google Scholar]
- 4.Quinn A G, Sikkink S, Rees J L. Cancer Res. 1994;54:4756–4759. [PubMed] [Google Scholar]
- 5.Lehman T A, Modali R, Boukamp P, Stanek J, Bennett W P, Welsh J A, Metcalf R A, Stampfer M R, Fusenig N E, Rogan M, et al. Carcinogenesis. 1993;14:833–839. doi: 10.1093/carcin/14.5.833. [DOI] [PubMed] [Google Scholar]
- 6.Boukamp P, Petrussevska R T, Breitkreutz D, Hornung J, Markham A, Fusenig N E. J Cell Biol. 1988;106:761–771. doi: 10.1083/jcb.106.3.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boukamp P, Stanbridge E J, Foo D Y, Cerutti P A, Fusenig N E. Cancer Res. 1990;50:2840–2847. [PubMed] [Google Scholar]
- 8.Boukamp P, Peter W, Pascheberg U, Altmeier S, Fasching C, Stanbridge E J, Fusenig N E. Oncogene. 1995;11:973–978. [PubMed] [Google Scholar]
- 9.Qian X, Tuszynski G P. Proc Soc Exp Biol Med. 1996;212:199–207. doi: 10.3181/00379727-212-44008. [DOI] [PubMed] [Google Scholar]
- 10.Weinstat-Saslow D L, Zabrenetzky V S, VanHoutte K, Frazier W A, Roberts D D, Steeg P S. Cancer Res. 1994;54:6504–6511. [PubMed] [Google Scholar]
- 11.Zabrenetzky V, Harris C C, Steeg P S, Roberts D D. Int J Cancer. 1994;59:191–195. doi: 10.1002/ijc.2910590209. [DOI] [PubMed] [Google Scholar]
- 12.Sheibani N, Frazier W A. Proc Natl Acad Sci USA. 1995;92:6788–6792. doi: 10.1073/pnas.92.15.6788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Castle V, Varani J, Fligiel S, Prochownik E V, Dixit V. J Clin Invest. 1991;87:1883–1888. doi: 10.1172/JCI115212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Castle V P, Dixit V M, Polverini P J. Lab Invest. 1997;77:51–61. [PubMed] [Google Scholar]
- 15.Good D J, Polverini P J, Rastinejad F, Le Beau M M, Lemons R S, Frazier W A, Bouck N P. Proc Natl Acad Sci USA. 1990;87:6624–6628. doi: 10.1073/pnas.87.17.6624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rheinwald J G, Beckett M A. Cancer Res. 1981;41:1657–1663. [PubMed] [Google Scholar]
- 17.Boukamp P, Tilgen W, Dzarlieva R T, Breitkreutz D, Haag D, Riehl R K, Bohnert A, Fusenig N E. J Natl Cancer Inst. 1982;68:415–427. [PubMed] [Google Scholar]
- 18.Tilgen W, Boukamp P, Breitkreutz D, Dzarlieva R T, Engstner M, Haag M D, Fusenig N E. Cancer Res. 1983;43:5995–6011. [PubMed] [Google Scholar]
- 19.Moorman A F M, De Boer P A J, Vermeulen J L M, Lamers W H. Histochem J. 1993;25:251–266. doi: 10.1007/BF00159117. [DOI] [PubMed] [Google Scholar]
- 20.Ried T, Lengauer C, Cremer T, Wiegant J, Raap A K, van der Ploeg M, Groitl P, Lipp M. Genes Chromosomes Cancer. 1992;4:1–6. doi: 10.1002/gcc.2870040109. [DOI] [PubMed] [Google Scholar]
- 21.Collins C C, Kuo W L, Segraves R, Fuscoe J C, Pinkel D, Gray J W. Genomics. 1991;11:997–1006. doi: 10.1016/0888-7543(91)90025-a. [DOI] [PubMed] [Google Scholar]
- 22.Lichter P, Boyle A L, Cremer T, Ward D C. Genet Anal Tech Appl. 1991;8:24–35. doi: 10.1016/1050-3862(91)90005-c. [DOI] [PubMed] [Google Scholar]
- 23.Boukamp P, Popp S, Altmeyer S, Hülsen A, Fasching C, Cremer T, Fusenig N E. Genes Chromosomes Cancer. 1997;19:201–214. doi: 10.1002/(sici)1098-2264(199708)19:4<201::aid-gcc1>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- 24.Parangi S, O’Reilly M, Christophori G, Holmgren L, Grosfeld J, Folkman J, Hanahan D. Proc Natl Acad Sci USA. 1996;93:2002–2007. doi: 10.1073/pnas.93.5.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Skobe M, Rockwell P, Goldstein N, Vosseler S, Fusenig N E. Nat Med. 1997;3:1222–1227. doi: 10.1038/nm1197-1222. [DOI] [PubMed] [Google Scholar]
- 26.Vecchi A, Garlanda C, Lampugnani M G, Resnati M, Matteucci C, Stoppacciaro A, Schnurch H, Risau W, Ruco L, Mantovani L, Dejana E. Eur J Cell Biol. 1994;63:247–254. [PubMed] [Google Scholar]
- 27.Roberts D D. FASEB J. 1996;10:1183–1191. [PubMed] [Google Scholar]
- 28.Skobe M, Fusenig N E. Proc Natl Acad Sci USA. 1998;95:1050–1055. doi: 10.1073/pnas.95.3.1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wick W, Petersen I, Schmutzler R K, Wolfahrt B, Lenartz D, Bierhoff E, Hümmerich J, Müller D J, Stangl A P, Schramm J, et al. Oncogene. 1996;12:973–978. [PubMed] [Google Scholar]
- 30.Bockmuhl U, Petersen S, Schmidt S, Wolf G, Jahnke V, Dietel M, Petersen I. Cancer Res. 1997;57:5213–5216. [PubMed] [Google Scholar]
- 31.Mahlamaki E H, Hoglund M, Gorunova L, Karhu R, Dawiskiba S, Andren-Sandberg A, Kallioniemi O P, Johansson B. Genes Chromosomes Cancer. 1997;20:383–391. doi: 10.1002/(sici)1098-2264(199712)20:4<383::aid-gcc10>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- 32.McGowan-Jordan I J, Speevak M D, Blakey D, Chevrette M. Cancer Res. 1994;54:2568–2572. [PubMed] [Google Scholar]
- 33.Rastinejad F, Polverini P J, Bouck N P. Cell. 1989;56:345–355. doi: 10.1016/0092-8674(89)90238-9. [DOI] [PubMed] [Google Scholar]
- 34.Tolsma S S, Volpert O V, Good D J, Frazier W A, Polverini P J, Bouck N P. J Cell Biol. 1993;122:497–511. doi: 10.1083/jcb.122.2.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vogel T, Guo N H, Krutzsch H C, Blake D A, Hartman J, Mendelovitz S, Panet A, Roberts D D. J Cell Biochem. 1993;53:74–84. doi: 10.1002/jcb.240530109. [DOI] [PubMed] [Google Scholar]
- 36.DiPietro L A, Nebgen D R, Polverini P J. J Vasc Res. 1994;31:178–185. doi: 10.1159/000319585. [DOI] [PubMed] [Google Scholar]
- 37.Dawson D W, Pearce S F, Zhong R, Silverstein R L, Frazier W A, Bouck N P. J Cell Biol. 1997;138:707–717. doi: 10.1083/jcb.138.3.707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tolsma S S, Stack M S, Bouck N P. Microvasc Res. 1997;54:13–26. doi: 10.1006/mvre.1997.2015. [DOI] [PubMed] [Google Scholar]
- 39.Campbell S C, Volpert O V, Ivanovich M, Bouck N P. Cancer Res. 1998;58:1298–1304. [PubMed] [Google Scholar]
- 40.Guo N, Zabrenetzky V S, Chandrasekaran L, Sipes J M, Lawler J, Krutzsch H C, Roberts D D. Cancer Res. 1998;58:3154–3162. [PubMed] [Google Scholar]
- 41.Bornstein P. FASEB J. 1992;6:3290–3299. doi: 10.1096/fasebj.6.14.1426766. [DOI] [PubMed] [Google Scholar]
- 42.Bornstein P. J Cell Biol. 1995;130:503–506. doi: 10.1083/jcb.130.3.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Folkman J, Klagsbrun M, Sasse J, Wadzinski M, Ingber D, Vlodavsky I. Am J Pathol. 1988;130:393–400. [PMC free article] [PubMed] [Google Scholar]
- 44.Senger D R, Conolly D T, Van de Water L, Feder J, Orona U. Cancer Res. 1990;50:1774–1778. [PubMed] [Google Scholar]
- 45.Ferrara N, Houck K, Jakema L, Leung D W. Endocrinology. 1992;13:18–32. doi: 10.1210/edrv-13-1-18. [DOI] [PubMed] [Google Scholar]
- 46.Volpert O V, Dameron K M, Bouck N. Oncogene. 1997;14:1495–1502. doi: 10.1038/sj.onc.1200977. [DOI] [PubMed] [Google Scholar]
- 47.Schultz-Cherry S, Lawler J, Murphy-Ullrich J E. J Biol Chem. 1994;269:26783–26788. [PubMed] [Google Scholar]
- 48.Schultz-Cherry S, Chen H, Mosher D F, Misenheimer T M, Krutzsch H C, Roberts D D, Murphy-Ullrich J E. J Biol Chem. 1995;270:7304–7310. doi: 10.1074/jbc.270.13.7304. [DOI] [PubMed] [Google Scholar]
- 49.Crawford S E, Stellmach V, Murphy-Ullrich J E, Ribeiro S M, Lawler J, Hynes R O, Boivin G P, Bouck N. Cell. 1998;93:1159–1170. doi: 10.1016/s0092-8674(00)81460-9. [DOI] [PubMed] [Google Scholar]
- 50.Sandler M A, Zhang J N, Westerhausen D R, Billadello J J. J Biol Chem. 1994;269:21500–21504. [PubMed] [Google Scholar]
- 51.Arnoletti J P, Albo D, Granick M S, Solomon M P, Castiglioni A, Rothman V L, Tuszynski G P. Cancer. 1995;76:998–1005. doi: 10.1002/1097-0142(19950915)76:6<998::aid-cncr2820760613>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- 52.Qian X, Wang T N, Rothman V L, Nicosia R F, Tuszynski G P. Exp Cell Res. 1997;235:403–412. doi: 10.1006/excr.1997.3681. [DOI] [PubMed] [Google Scholar]