SUMMARY
Sonic hedgehog (Shh) and insulin-like growth factor (IGF) signaling are essential for development of many tissues and are implicated in medulloblastoma, the most common solid pediatric malignancy. Cerebellar granule neuron precursors (CGNPs), proposed cells-of-origin for certain classes of medulloblastomas, require Shh and IGF signaling for proliferation and survival during development of the cerebellum. We asked whether Shh regulates IGF pathway components in proliferating CGNPs. We report that Shh-treated CGNPs showed increased levels of insulin receptor substrate 1 (IRS1) protein, which was also present in the germinal layer of the developing mouse cerebellum and in mouse Shh-induced medulloblastomas. Previous roles for IRS1, an oncogenic protein essential for IGF-mediated proliferation in other cell types, have not been described in Shh-mediated CGNP proliferation. We found that IRS1 over-expression can maintain CGNP proliferation in the absence of Shh. Furthermore, lentivirus-mediated knock down experiments showed that IRS1 activity is required for CGNP proliferation in slice explants and dissociated cultures. Contrary to traditional models for Shh signaling which focus on gene transcription, Shh stimulation does not regulate IRS1 transcription but rather stabilizes IRS1 protein by interfering with mTOR-dependent IRS1 turnover and possibly affects IRS1 mRNA translation. Thus, we have identified IRS1 as a novel effector of Shh mitogenic signaling that may serve as a future target for medulloblastoma therapies. Our findings also indicate a previously unreported interaction between the Shh and mTOR pathways and provide an example of a non-classical means for Shh-mediated protein regulation during development.
Keywords: Sonic hedgehog, cerebellum, neural precursor, insulin-like growth factor, insulin receptor substrate 1, proliferation
INTRODUCTION
Medulloblastoma is the most common malignant solid tumor in children. These tumors arise in the cerebellum, a region of the brain with important roles in movement, coordination, and possibly learning. Traditional treatments for medulloblastomas--radiation, surgery and multi-agent chemotherapy--cause devastating side effects in long-term survivors (Packer et al., 1999), including cognitive declines, psychiatric problems, seizures, and movement disorders. The poor understanding of molecular events leading to the formation and maintenance of medulloblastoma has hindered the advancement of treatment options.
CGNPs are proposed cells-of origin for some classes of medulloblastoma (Provias and Becker, 1996). After birth (approximately the first two weeks in mice), CGNPs undergo a rapid expansion phase in the cerebellar external granule layer (EGL). After this expansion CGNPs migrate through the underlying layer of Purkinje neurons with which they will ultimately form synapses. The mature granule neuron cell bodies localize to the internal granule layer (IGL) (Hatten and Heintz, 1995). Normal CGNP proliferation is dependent upon signaling by both Shh and IGF, which are also implicated in medulloblastomas (Altman and Bayer, 1997; Ho and Scott, 2002a; Knoepfler and Kenney, 2006; Marino, 2005; Wetmore, 2003).
Shh is produced by Purkinje neurons and loss of Shh leads to reduced proliferation in the EGL of neonatal mice (Dahmane and Ruiz-i-Altaba, 1999; Wallace, 1999; Wechsler-Reya and Scott, 1999). Treatment of CGNPs in culture with Shh increases BrdU incorporation (Dahmane and Ruiz-i-Altaba, 1999; Wallace, 1999; Wechsler-Reya and Scott, 1999); however the mechanisms underlying Shh mitogenic signaling in CGNPs continue to be subject to ongoing investigation. Classic mitogens such as epidermal growth factor (EGF) or platelet-derived growth factor (PDGF) signal through receptor tyrosine kinases. In contrast, Shh activates a non-receptor tyrosine kinase-associated pathway. In the absence of Shh, the transmembrane protein Patched (Ptch) represses Smoothened (Smo), a G-protein coupled receptor-resembling protein (Alcedo et al., 1996). When Shh binds to Ptch, Smo is released from inhibition and the pathway is activated, resulting in activation of target genes including Ptch itself as well as the transcription factors N-myc, Gli2 and its target Gli1 (Ho and Scott, 2002b). Shh signaling during cerebellar development occurs primarily through the activation of Gli2; mutations in Gli2 result in abnormal CGNP proliferation as well as foliation defects (Corrales et al., 2006; Corrales et al., 2004).
Traditional receptor tyrosine kinase signaling mediated by IGF family members has roles in CGNP proliferation and Shh-associated medulloblastomas. IGF1 and IGF2 are expressed in the developing and mature cerebellum. Activation of the IGF pathway is found in medulloblastomas (Reiss, 2002), and IGF2 in particular is required for Shh-mediated medulloblastoma formation (Hahn et al., 2000) in vivo and medulloblastoma cell proliferation in vitro (Hartmann et al., 2005). IGF1 and IGF2 activate the IGF receptor. One way through which IGF-mediated phosphoinositide-3 kinase (PI-3K) signaling cooperates with Shh signaling is by inhibiting GSK-3β (Kenney et al., 2004; Mill et al., 2005), which blocks cell cycle progression in CGNPs by phosphorylating N-myc and targeting it to the proteosome for degradation. The goal of our current study is to identify additional mechanisms through which Shh and IGF pathway members cooperate to promote CGNP proliferation.
IGF binding to its receptor leads to tyrosine phosphorylation of scaffolding proteins that act as downstream effectors, including Gab1 and IRS1-4 (Van Obberghen et al., 2001). Tyrosine phosphorylation of IRS proteins provides multimeric docking sites for src homology-2 (SH2) domain containing proteins. Through this mechanism IRS1-4 and Gab1 can activate PI-3K (White, 1998), which executes many of IGF’s functions. However, in addition to their overlapping ability to activate PI-3K, IGF effectors also have unique effects on cell survival, proliferation and differentiation. In particular functional IRS1 is essential for the proliferative effects of the IGF receptor (Waters et al., 1993). Aberrant IRS1 expression has been associated with several types of human cancer including medulloblastoma (Del Valle et al., 2002; Waters et al., 1993), and its over-expression can drive mammary tumor formation in mice (Dearth et al., 2006b). The role of IRS1 in neural precursor expansion however is poorly understood.
We asked whether Shh treatment alters expression or activity of IGF pathway effectors. Interestingly, we observed no effect of Shh on Akt activity, in contrast to a previous report from a cell line (Riobo et al., 2006). Among IGF receptor substrates we investigated, only IRS1 protein levels were increased in the presence of Shh. In neonatal mouse cerebella we detected IRS1 protein in the germinal layer of the developing cerebellum. Lentivirus-mediated IRS1 knock down reduced Shh proliferative effects on CGNPs without affecting survival. Interestingly, our studies indicate that Shh treatment does not alter IRS1 mRNA expression. Instead, Shh increases IRS1 protein stability by impeding an mTOR-dependent turnover process and may also promote IRS1 mRNA translation. Our results reveal a novel mechanism through which Shh utilizes components of the IGF pathway to drive proliferation, as well as providing evidence that Shh signaling directly or indirectly affects the mTOR pathway.
MATERIALS AND METHODS
Animal studies
Harvest of neural precursors from neonates and preparation of cerebella from wild-type and mutant mice for histological analysis were carried out in compliance with the Memorial Sloan-Kettering Institutional animal care and use committee guidelines.
Cerebellar granule neuron precursor culture
CGNP cultures were generated as previously described (Kenney and Rowitch, 2000). Briefly, cerebella from postnatal day (PN) 5 Swiss-Webster 129 (SW129) mice were dissected into Hanks buffered saline solutions (HBSS) (Gibco) supplemented with glucose. The meninges were removed and cerebella were pooled and treated with Trypsin-EDTA for dissociation by trituration into HBSS. Triturated cells were centrifuged and resuspended in Dulbecco’s modified Eagle’s medium-F-12 (DMEM/F12) (Gibco) supplemented with 25mM KCl, N2 supplement (Gibco), antibiotic, and 10% fetal calf serum (Sigma). Cells were plated in individual poly-DL-ornithine (Sigma) precoated wells of a 6-well plate and for treated samples 3μg/mL Shh (Biogen Inc) was added to the media. After 6-12 hours the media was changed to DMEM/F12/N2/KCl minus serum, with or without Shh as indicated. Unless otherwise stated, cells were left undisturbed for 24 hours prior to further treatment or analysis. For purified cultures, resuspended CGNPs were passed through a Percoll gradient as previously described (Wechsler-Reya and Scott, 1999).
Cerebellar slice cultures
Cerebella from PN 5 pups were aseptically removed and embedded in 3% low-melting agarose (Bio-Rad) made with HBSS. Cerebella were cut using a Leica VT1000S vibratome into 300 μm sections and cultured on Whatman nuclopeore track-etched membrane (Fisher Scientific) in serum-free media for 24 hours supplemented with Shh. After 24 hours indicated sections were infected with shRNA lentiviruses targeting IRS1 for 6 hours. The slices were maintained in serum free media for 48 hours and pulsed with BrdU for an additional 4 hours. Slices were flash frozen, sectioned and stained for BrdU and DAPI.
Protein preparation and immunobloting
For immunoblot analysis cells were scraped cells loose into their medium. Cells were washed once in PBS and protein extracts were prepared as previously described (Kenney and Rowitch, 2000). Protein content was determined by using the Bio-Rad protein assay. Assays were performed in duplicate for each sample. 50 μg of each sample was separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) on 8% polyacrylamide gels and then transferred in 20% methanol buffer at 4°C to Immobilon polyvinylidene difluoride (Millipore) membranes. Standard western blot procedures (see Kenney and Rowitch 2000) were used to determine protein levels. Antibodies used for western blotting were: IGFRβ, N-myc, and Cyclin D2 (Santa Cruz), IRS1, IRS2, phosphorylated IRS1 (S636/639), phosphorylated Akt (S473), Akt, Gab1, phosphorylated Erk, phosphorylated p70S6 kinase (T389), phosphorylated ribosomal protein S6 (S235/236) (Cell Signaling) and β-tubulin (Sigma). Peroxidase activity was detected using Amershams’s ECL reagents and exposing membranes to Kodak Biomax film. Multiple exposures were taken to avoid saturating film. The film was scanned and the digitalized images were quantified by densitometry using Adobe Photoshop 9.0 software.
Immunofluorescence
Frozen sections (10 μm) from SW129 pups were dried and then boiled in 0.01M Citric Acid for 15 minutes for antigen retrieval. For paraffin embedded sections tissues were first de-waxed and re-hydrated prior to antigen retrieval. After cooling, slides were washed twice with PBS for 10 minutes. Sections were blocked with 10% normal goat serum (Sigma) in 0.25% Trition X-100/PBS for one hour at room temperature. Primary antibodies for IRS1 (Cell Signaling), GFAP (Cell Signaling), BrdU (Becton Dickinson) and Ki67 (Vector labs) were added to the blocking solution at a 1:100 dilution and incubated overnight at 4°C. After washing in PBS, slides were incubated with either goat anti-rabbit or goat anti-mouse fluorescently tagged secondary antibody (Invitrogen) at a 1:5000 dilution for 1 hour at room temperature. Sections were mounted using Vectashield mounting media with DAPI (Vector Laboratories).
For detecting BrdU incorporation dissociated CGNPs were grown on poly-DL-ornithine coated glass coverslips. Cells were pulsed with 20 μg/mL BrdU for 2 hours. The cells were fixed with 4% paraformaldehyde for 20 minutes followed by two 10-minute washes with PBS. The coverslips were treated with 2N HCl for 2 minutes followed by two 10-minute washes with PBS. Cells were blocked for 30 minutes then exposed to primary and secondary antibodies according to standard methods (see Supplementary methods). All other antibodies, IRS-1 (Cell Signaling or Upstate), Ki67 (Vector Labs), p27 (BD Pharmingen), PCNA (Calbiochem), Zic1 (gift from Rosalind Segal, Harvard) and cleaved caspsase-3 (Cell Signaling) were used at a 1:100 dilution in blocking solution.
Reverse transcriptase and Quantitative PCR
RNA from CGNPs was collected using TRIZOL® reagent according to the manufacturer’s instructions. RNA samples were resuspended in 87.5 μL DEPC-treated water. In order to fully remove any residual DNA from the samples, RNA was further purified using the RNeasy Mini Kit (Qiagen) according to manufacturer’s instructions. DNase (Qiagen) digestion was performed in solution prior to further RNA purification over the RNeasy column.
A 50 μL reaction volume was used for 50 ng RNA of each sample using SuperScript™ One-Step RT-PCR with Platinim® Taq (Invitrogen). Samples were run per manufacturer’s instructions. Absence of genomic DNA was verified by omitting the RT step and using Taq alone. Primer sequences were as follows β-actin sense 5’-CACAGCTACAAAGAGCGGCTCCACC-3’ β-actin antisense 5’–CACTGCATTCTAGTTGTGGTTTGTCC-3’ cyclin D2 sense 5’-CACTTCCTCTCCAAAATGCCA-3’ cyclin D2 antisense 5’-CCTGGCGCAGGCTTGACTC-3’ IRS1 sense 5’-CCCGCGTTCAAGGAGGTCTG-3’ IRS1 antisense 5’-TGGCTGGGTGGAGGGTTGTT-3’ Gli1 sense 5’-CCACGGGGAGCGGAAGGAA-3’ Gli1 antisense 5’-AGGCGGCGAAGCGTGGAGAGT-3’ Gli2 sense 5’-AGCCCCTGCACTGGAGAAGAAAGA-3’ Gli2 antisense 5’-CTGGGGCTGCGAGGCTAAAGAG-3’ N-myc sense 5’-GCCTTCTCGTTCTTCACCAG-3’ N-myc antisense 5’-GCGGTAACCACTTTCACGAT-3’. PCR products were resolved on a 2.5% agarose-ethidium bromide gel.
For quantitative PCR, total mRNA was extracted from untreated and Shh-treated CGNP cultures as described above. cDNA was generated with SuperScript First-Strand Synthesis System for RT-PCR (Invitrogen) per manufacturer’s instructions. TaqMan Gene Expression Assays (Applied Biosystems) using TaqMan custom designed MGB probes for IRS1 (Mm01278327_m1) and β–actin (Mm01191484_m1) were performed in triplicate according to manufacturer’s protocol on an ABI 7000 Sequence Detection System. Data was analyzed with ABI GeneAmp SDS software (Applied Biosystems). The average threshold cycle (CT) was determined to quantify initial transcript levels and results reported as fold changes.
Retroviruses
IRS1-expressing retroviruses were constructed by ligating a mouse IRS1 cDNA (gift of Morris White, Harvard) into the retroviral vector pIG. The construct was verified by sequencing. Gli1-expressing retrovirus was provided by Rob Wechlser-Reya (Duke). For producing viruses, 293e packaging cells (Invitrogen) were co-transfected with 10 μg each of retrovirus construct, vsv-g, and gagpol plasmids using Fugene in DMEM containing 10% fetal calf serum. Twenty-four hours later the medium was aspirated and replaced with fresh medium. Viral supernatent was collected for 3 days and stored at 4 C, then pooled and filtered through a .45 μ filter. CGNPs were infected by removing their medium, exposing them to filtered viral supernatants for 2 hours, then replacing with fresh or conditioned medium as appropriate.
Short hairpin RNA lentiviruses
293e packaging cells were co-transfected with lentiviral constructs expressing short hairpin RNAs targeting IRS1 (The RNAi Consortium) or GFP, delta 8.9, and vesicular stomatitis virus G glycoprotein plasmids, using Fugene 6 transfection reagent (Roche). The media was changed 12 hours after transfection and supernatants (10 mL) were harvested every 24 hours for 72 hours and kept at 4°C until they were pooled, filtered through 0.45 μm syringe filters, aliquoted and stored at -80°C until use. CGNPs were infected as described above.
Image capturing
Staining of cultured primary cells and tissue sections was visualized with a Leica DM5000B microscope and images were taken using Leica FW400 software. For quantification of BrdU uptake into newly synthesized DNA, TIFF images of 4 random fields were taken for each experimental group using the 10X objective. The percentage of cells staining positive for BrdU over the total number of cells was determined using Image Pro Plus software (MediaCybernetics). Con-focal images were visualized with Leica TCS AOBS SP2 (Inverted Stand) and images captured with Leica LCS Lite software.
Statistics
Statistical analysis was performed using one-way ANOVA followed by a Bonferroni-Dunn test for multiple comparisons within a group, or a two tailed t test for comparisons between groups, as indicated by the figure legends; p < 0.05 was considered significant and is marked by an asterisk (*). All results are represented as average with error bars indicating the standard error of the mean. Experiments in vitro were performed at least 3-4 times, with separate litters of mice, to confirm reproducibility and consistency.
RESULTS
Shh treatment up-regulates IRS1 protein levels in CGNPs
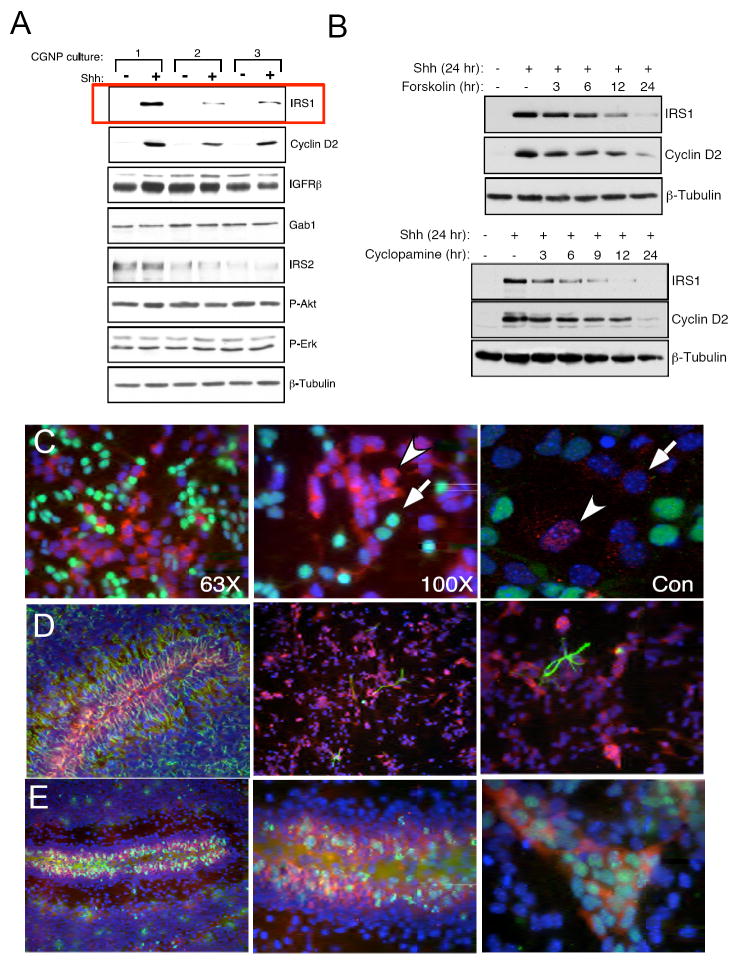
In order to determine whether expression levels or activity of IGF pathway components are altered in CGNPs induced to proliferate by Shh in comparison to levels in differentiating (vehicle-treated) CGNPs, we established CGNP primary cultures from PN 5 SW129 mice and treated them with vehicle (PBS) or added Shh to the media for 24 hours. We then prepared protein lysates and used western blot analysis to assess levels of IGF receptor substrates and Akt phosphorylation, a downstream target of PI-3K (Figure 1A). We also examined levels of cyclin D2, a well-established marker for Shh-induced CGNP proliferation, and Erk phosphorylation, which is known to be unaffected by Shh treatment in CGNPs (Kenney and Rowitch, 2000). We found that Shh-treated CGNPs showed increased levels of IRS1 protein, and that IGF receptor, IRS2 or Gab1 levels did not change in response to Shh (Figure 1A). IRS1 up-regulation correlates with increased cyclin D2 expression. It has been reported that Shh treatment of a fibroblast cell line results in modestly increased phosphorylation of Akt (Riobo et al., 2006). In contrast, we detected no changes in Akt phosphorylation in Shh-treated CGNPs (Figure 1A, 4A, 5A) suggesting functions for IRS1 in CGNPs outside of its Akt-activating role. The up-regulation of IRS1 is also accompanied by activation of IRS1 as seen by its tyrosine phosphorylation, which occurs when the IGF receptor is bound by ligand (Van Obberghen et al., 2001) (Supplementary figure 1).
Figure 1.
IRS1 protein is upregulated in proliferating CGNPs. (A) CGNP cultures were prepared from different litters on different dates. Each preparation of the cells were treated with Shh or left untreated for 24 hours prior to lysis. The autoradiograph depicts a western blot for several downstream components of IGF signaling pathway and the cell cycle progression marker cyclin D2. Only IRS1 levels are upregulated in Shh treated samples, which correlates with increases in cyclin D2. β-Tubulin demonstrates equal protein loading. (B) The autoradiographs show a western blots for CGNPs treated with two Shh signaling pathway inhibitors, forskolin (10 μM) or cyclopamine (1 μg/mL), for increasing time points. In the absence of continuous Shh signaling levels of IRS1 decrease which also correlates with decreased cyclin D2 expression. (C) Shh-treated CGNPs were fixed and immunostained for IRS1 (red) and p27 (green). IRS1 and p27 mark different populations of cells. Confocal imaging (right panel) confirms IRS1 presence in the nucleus (carat) and cytoplasm (arrow). (D) Left: Cerebellar section from post-natal day 7 mouse immunostained for IRS1 (red) and GFAP (green). Middle (low power) and right (high power): Shh-treated primary CGNP cultures immunostained for IRS1 (red) and GFAP (green). IRS1 is excluded from glia both in vivo and in vitro. (E) In the left (low power) and middle (high power) panels, PN 7 mice were pulsed with BrdU and stained for BrdU (green) and IRS1 (red). IRS1 co-localizes to proliferating CGNPs in the EGL. In the far right panel IRS1 (red) is expressed in the cytoplasm of BrdU positive cells (green) in CGNP cultures.
Figure 4.
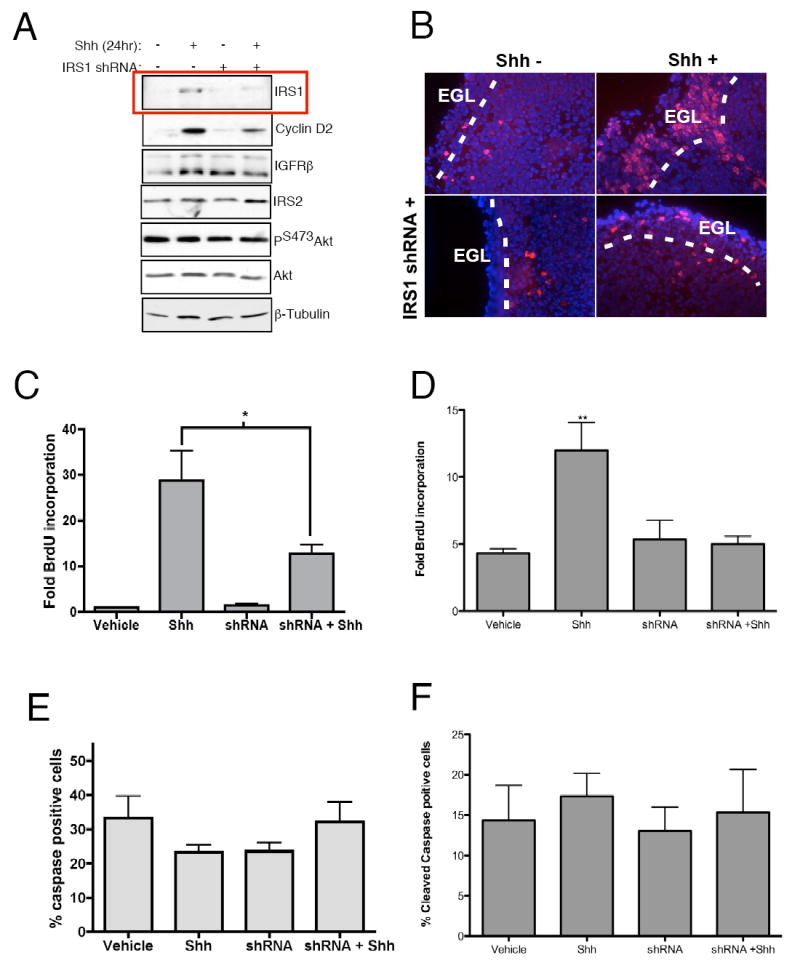
IRS1 is necessary to maintain CGNP proliferation. (A) CGNPs were treated with Shh for 24 hours prior to infection with IRS1 shRNAs for 3 hours at which time the medium was replaced with either Shh treated or untreated medium. Treatment with shRNAs knocked down levels of IRS1 (red box) as well as levels of cyclin D2 without affecting the activation of Akt or levels of IRS2. β-Tubulin confirms equal loading. (B) Infection of cerebellar slices with lentiviruses targeting iRS1 causes redcued thickness of the EGL (white dotted lines) and reduced BrdU incorporation (red) in the EGL. Blue represents Dapi counterstaining. (C) Levels of proliferation in primary CGNP cultures in response to IRS1 shRNA lentivirus infection were determined by measuring BrdU incorporation after a 2-hour BrdU pulse. Reduced BrdU incorporation is evident in shRNA plus Shh compared to Shh-treated CGNPs. BrdU-positive cells were counted and values expressed as percent of total cells per field. Values are represented as fold BrdU positive cells over untreated CGNPs. * statistically significant difference (p<0.05 n=4). (D) Levels of BrdU incorporation were assessed as above in Percoll gradient purified CGNP cultures. Trends of BrdU incorporation remain the same as mixed cultures. (E) Cell survival was assessed by immunostaining for cleaved caspase-3. Based on quantification of cleaved caspase-3 positive cells, there was no change in cell survival regardless of treatment. (F) Cell survival in response to shRNA treatment remained the same in purified cultures. Cleaved caspase-3 positive cells were counted and values are expressed as percent of total cells per field. Error bars represent SEM, n=5.
Figure 5.
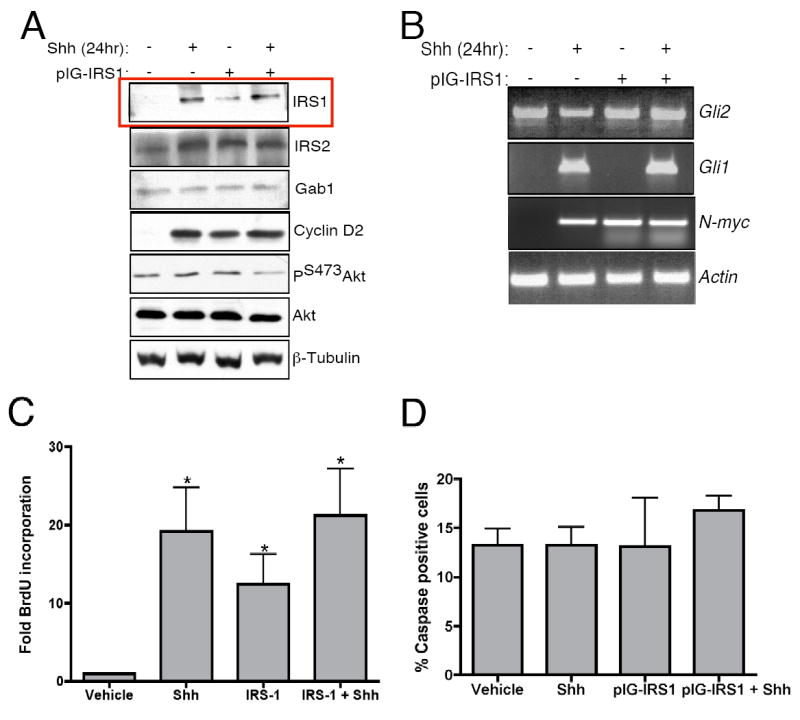
IRS1 over-expression maintains CGNP proliferation in the absence of Shh. CGNPs were treated with Shh for 24 hours prior to infection with retrovirus expressing IRS1. (A) Western blotting shows that IRS1 over-expression maintains increased levels of cyclin D2 after Shh withdrawal. (B) RT-PCR analysis shows no effect of IRS1 overexpression on Gli1 or Gli2 expression but did show an increase in N-Myc expression (C) Levels of CGNP proliferation in response to IRS1-expressing retrovirus infection were assessed with BrdU incorporation. Numbers of BrdU positive cells are increased with treatment of Shh as well as treatment with IRS1 retrovirus. Quantification of BrdU incorporation was performed as described in Figure 4D. (D) To assess effects of virus treatment on cell death CGNPs were stained with antibodies to activated caspase-3. Levels of cleaved caspase-3 remain the same for all treatment groups. Cleaved caspase-3 positive cells were counted and values are expressed as percent of total cells per field. Error bars represent SEM, n=5. * statistically significant difference to untreated CGNPs (p<0.05 n=4).
We next asked whether maintenance of increased IRS1 levels requires ongoing Shh signaling. We treated CGNPs with two well-characterized Shh pathway inhibitors, forskolin and cyclopamine, for increasing lengths of time. Forskolin serves as a potent inhibitor of Shh signaling by increasing cAMP levels which in turn activates PKA leading to CGNP cell cycle exit and differentiation (Cai et al., 1999). Cyclopamine is an antagonist of Smo, the activator of Shh signaling (Chen et al., 2002). Treatment with either inhibitor reduced IRS1 and cyclin D2 protein levels in a time dependent manner (Figure 1B), with IRS1 loss apparent 6-9 hours after addition of inhibitors. This delayed, rather than immediate, response of IRS1 to Shh inhibition may be a result of the long half-life of IRS1, which has been reported to be up to 10 hours (Lee et al., 2000) or it may suggest that IRS1 is sensitive to cell cycle exit, as CGNPs remain proliferation competent for 6 hours after Shh withdrawal or inhibition (Kenney and Rowitch, 2000).
Recent evidence suggests that IRS1 may have nuclear as well as cytoplasmic functions (Chen et al., 2005; Morelli et al., 2004). To determine the cellular localization of IRS1, we cultured CGNPs on coverslips with or without Shh for 24 hours. We then carried out immunostaining for IRS1 and p27, a predominantly nuclear protein associated with CGNP differentiation (Uziel et al., 2005). As shown in Figure 1C, Shh-treated CGNP cultures contained populations of cells expressing either p27 (green) or IRS1 (red) and expression of IRS1 and p27 is mutually exclusive in individual cells. When images were merged with blue DAPI (nuclear) staining, we observed IRS1 expression in the nucleus and cytoplasm, which was confirmed by confocal microscopy (Figure 1C, right panel). These results suggest that IRS1 may perform signaling functions in the cytoplasm, and may also play roles in regulating transcription or DNA repair, nuclear functions previously ascribed to IRS1 (Reiss 2006; Chen 2005). The role played by nuclear versus cytoplasmic IRS1 in proliferating CGNPs remains to be determined.
In our hands, mixed CGNP cultures contain up to 10% GFAP-positive cells. Previously it has been shown that the only Shh-responsive cells and BrdU-incorporating cells in mixed cultures are CGNPs, not those which stain positive for glial markers such as GFAP or O4 (Wechsler-Reya and Scott, 1999). In vivo staining of post-natal day 7 mouse cerebella for GFAP and IRS1 shows that these two markers do not co-localize (Figure 1D left panel). To confirm that increased IRS1 expression occurs in CGNPs, not glia, cultures were treated with Shh for 24 hours and then immunostained for GFAP and IRS1 (Figure 1D, middle and right panels) or BrdU and IRS1 (Figure 1E, right panel). BrdU-incorporating cells were also Zic1-positive, confirming their identity as CGNPs (Supplementary Figure 2). Our results demonstrate that increased IRS1 expression occurs in cells that have incorporated BrdU, both in culture and in vivo and is excluded from GFAP-expressing glial cells (Figure 1D and E)(Aruga et al., 2002). Thus, IRS1 is up-regulated in proliferating CGNPs.
In vivo, by post-natal day 15 IRS1 levels are reduced although some expression remains in the EGL in conjunction with the proliferation marker PCNA (Supplementary Figure 2). We found that IRS2 was restricted to Purkinje neurons and is excluded from the EGL (Supplementary Figure 2). Our observation that IRS2 is not found in CGNPs, but rather located in Purkinje neurons, the cells which provide Shh to CGNPs, suggests that a reported requirement for IRS2 in CGNP proliferation (Schubert et al., 2003) may be attributed to Purkinje cell defects (see Supplementary Figure 2). Indeed we detected reduced Shh signaling, and reduced levels of IRS1 protein in the EGL of IRS2-null neonatal mouse cerebella (Supplementary Figure 3).
Shh signaling stabilizes IRS1 protein levels without altering IRS1 transcripts
Canonical Shh signaling occurs through the action of Gli and N-myc transcription factors resulting in the up-regulation of target mRNA transcripts. In order to determine whether Shh signaling affects IRS1 protein levels by increasing IRS1 transcripts we used RT-PCR analysis for IRS1, Cyclin D2 and Actin (Figure 2A, B). RNA was collected for RT-PCR or quantitative PCR from CGNPs treated with or without Shh for 24 hours. Levels of Cyclin D2, an indirect target of Shh signaling (Kenney and Rowitch, 2000) are increased in Shh-treated CGNPs (Figure 2A). However, levels of IRS1 are constant regardless of treatment indicating that Shh does not affect IRS1 transcription (Figure 2A, B). These results are in agreement with previous work performed in non-neural cell types showing that IRS1 protein levels can change without changes in IRS1 transcription (Nemoto et al., 2006; Renstrom et al., 2005; Rice et al., 1993).
Figure 2.
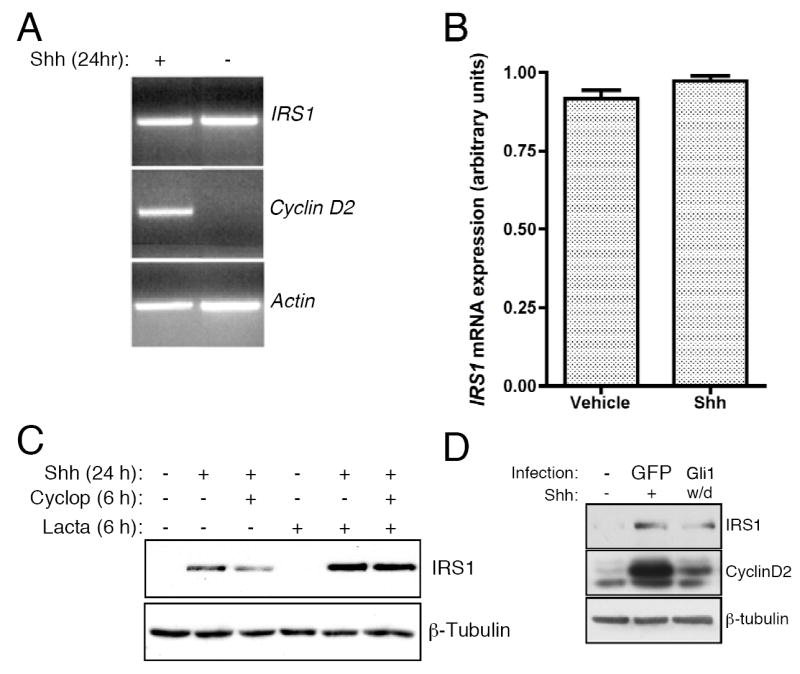
Shh signaling stabilizes IRS1 protein. (A) RNA was collected from Shh-treated CGNPs and RT-PCR analysis was conducted for IRS1, cyclin D2 and β-actin to verify equal RNA input. (B) qPCR was used to quantify levels of IRS1 expression. Expression levels of IRS1, depicted as arbitrary units, do not change in response to Shh treatment. (C) CGNPs were treated with Shh, cyclopamine or lactacystin (10 μM) for indicated times. Levels of IRS1 protein are stabilized in the presence of the lactacystin and cyclopamine. (D) Western blot analysis of IRS1 protein levels in CGNPs infected with Gli1 retroviruses and subsequently cultured with out Shh for 36 hours. Gli1 infection can sustain proliferation as indicated by cyclin D2 levels and as previously reported (Oliver et al., 2003). IRS1 was present but at lower levels than in CGNPs infected with GFP-expressing retroviruses and treated with exogenous Shh, suggesting the existence of Gli-mediated and non-Gli-mediated mechanisms promoting IRS1 protein accumulation.
To determine whether Shh stabilizes IRS1 protein levels by inhibiting its degradation, we asked how co-treatment of CGNPs with the Shh inhibitor cyclopamine and the proteasome inhibitor lactacystin affected IRS1 protein levels. In the presence of the Shh inhibitor cyclopamine, IRS1 levels declined as expected (Figure 2C). Reduction in IRS1 protein levels was prevented when lactacystin was also present, suggesting that inhibition of Shh causes IRS1 to be targeted for degradation (Figure 2C). Moreover, in addition to preventing IRS1 turnover, Shh may also affect IRS1 mRNA translation, since IRS1 transcripts are present in non-Shh-treated cells but lactacystin treatment did not induce IRS1 protein accumulation, indicating that the IRS1 mRNA is not being translated in CGNPs that have not been exposed to Shh. However, this remains to be conclusively determined, as current methodologies for examining Shh-responsive mRNA translation have not yet been refined for use with such limited starting material as primary CGNP cultures.
Although Shh signaling does not activate IRS1 transcription, it is possible that the Shh transcriptional target Gli1 can regulate IRS1 protein. To determine whether Gli activity can promote accumulation of IRS1 in CGNPs, we infected CGNP cultures with a Gli1 retrovirus. After the 2 hour infection period, the viral supernatent was withdrawn and replaced with fresh CGNP medium lacking Shh. We examined IRS1 protein levels 36 hours after infection. As shown in Figure 2D, IRS1 was present in Gli-infected cultures, albeit not at levels as high as in cultures treated with Shh. These results suggest that Gli1 can promote IRS1 protein accumulation, and that there are also Gli-independent mechanisms that synergize with Gli1 to achieve the full IRS1 accumulation response to exogenous Shh signaling. Future studies will determine whether Gli affects IRS1 stability and/or mRNA translation, and will identify non-Gli mediators of IRS1 accumulation.
Shh signaling stabilizes IRS1 by down-regulating S6 kinase
Several signaling proteins have been shown to play roles in IRS1 degradation, including suppressor of cytokine signaling (SOCS) signaling, retinoic-acid mediated protein kinase C activation, and mTOR, which mediates IRS1 phosphorylation on S636/639, to promote its degradation (del Rincon et al., 2004; Haruta et al., 2000; Ishizuka et al., 2007; Shah and Hunter, 2006). Since CGNPs are not cultured in the presence of cytokines or retinoic acid, we asked whether Shh signaling affected mTOR-regulated IRS1 turnover by reducing IRS1 phosphorylation and/or activity of S6 kinase, the mTOR substrate shown to directly phosphorylate IRS1 (Shah and Hunter, 2006). We treated CGNPs with Shh and cyclopamine for increasing periods of time, and then carried out western blot analysis for total IRS1 and S636/639-phosphorylated IRS1. As expected, IRS1 protein levels declined over time in the presence of cyclopamine (Figure 3A). However, the ratio of PS636/639IRS1 to total IRS1 increases, indicating Shh inhibition increases IRS1 phosphorylation on this destabilization-associated site.
Figure 3.
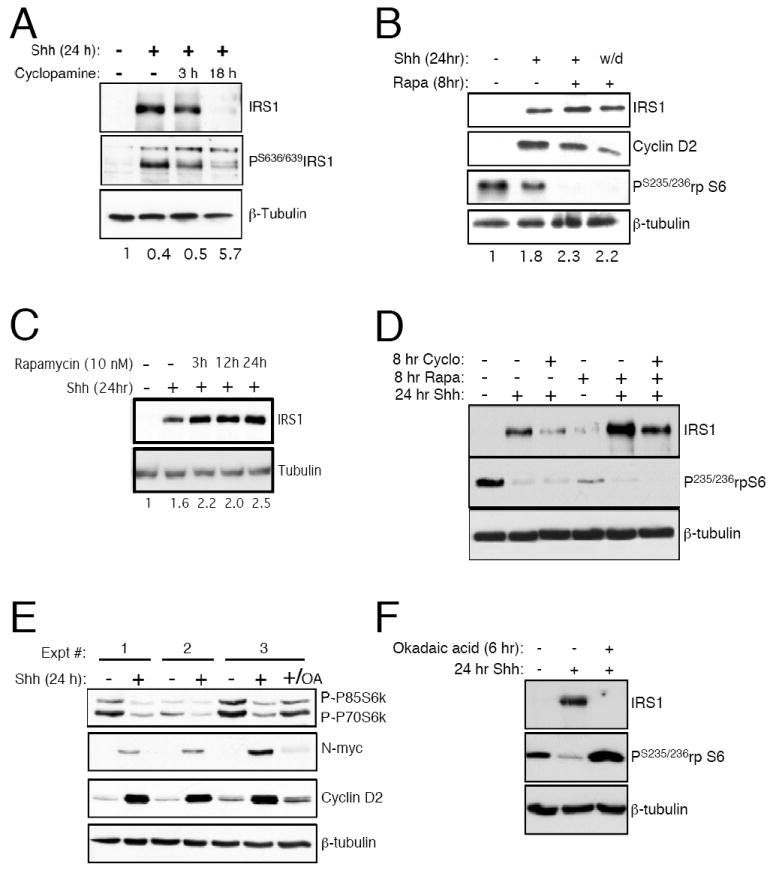
Shh signaling inhibits S6 kinase activation and hence IRS1 degradation. (A) Treatment of CGNPs with 1 μg/mL cyclopamine after increasing periods of time reduced IRS1 protein levels and increased detectable phosphorylated IRS1. The value under each lane represents the ratio of phosphorylated IRS1 to IRS1 as measured by densitometry. (B) CGNPs were treated with Shh and/or 10 nm rapamycin (Rapa) as indicated. The final lane represents CGNPs from which Shh was removed at the time of rapamycin addition. Rapamycin in combination with Shh causes an accumulation of IRS1 and rapamycin prevents reduction in IRS1 when Shh is removed. The value under each lane represents densitometric measurement of the IRS1 signal using the vehicle:tubulin value set to 1. (C) Treatment of CGNPs with rapamycin in the presence of Shh caused an accumulation of IRS1 over time. The value under each lane represents densitometric measurement of the IRS1 signal using the vehicle:tubulin value set to 1. (D) Shh, cyclopamine, and rapamycin were given to CGNPs as indicated above the lanes. Cyclopamine caused reduction in IRS1 levels, while rapamycin caused additional IRS1 accumulation in the presence of Shh. Rapamycin partially rescued the cyclopamine-mediated reduction of IRS1 in Shh treated cells. (E) Treatment with Shh reduces the phosphorylation of S6K and hence its activation. Inhibition of PP2A with okadaic acid (OA, 100 nM) restores S6K activation and reduces N-myc as well as cyclin D2 levels in CGNPs. (F) Treatment with OA restores S6 phosphorylation and reduces IRS1 levels even in the presence of Shh.
IRS1 phosphorylation on S636/639 occurs downstream of mTOR in 293HEK cells (Tzatsos and Kandror, 2006). In order to determine if mTOR signaling has an affect on IRS1 protein levels in response to Shh, we treated CGNPs with rapamycin, a compound that inhibits the mTOR:Raptor complex, in the presence of or after the withdrawal of Shh. Consistent with previous reports (Hartley and Cooper, 2002), we observed that in the presence of rapamycin, IRS1 levels accumulated (Figure 3B,C) without any affect on cell survival based on activated caspase 3 levels (data not shown). Interestingly, levels of IRS1 were stabilized after treatment with rapamycin even when Shh was removed at the time of rapamycin addition (Figure 3B, lane 4). This suggests that inhibiting mTOR can promote IRS1 stabilization in CGNPs. To further investigate the relationship between Shh signaling and the mTOR pathway, we treated CGNPs with cyclopamine and rapamycin and examined IRS1 levels. While treatment with rapamycin increased IRS levels compared to Shh alone, we observed only partial recovery of IRS1 protein when CGNPs were treated with cyclopamine and rapamycin (Figure 3D). This result suggests that inhibition of mTOR is not the sole mechanism through which Shh mediates IRS1 accumulation. For example, Shh may also regulate IRS1 mRNA translation in an mTOR-independent manner. The results may also indicate that mTOR is regulated in part by signaling through Smoothened.
S6K1, a major target of mTOR, is required for IRS1 phosphorylation in cells and can also directly phosphorylate IRS1 (Easton et al., 2006; Um et al., 2004). Stabilization of IRS1 protein in CGNPs after treatment with rapamycin could occur directly by the inhibition of mTOR or indirectly by inhibiting mTOR’s downstream target S6K1. Since both mTOR and S6K1 can phosphorylate IRS1 we wanted to determine the affects of Shh on the activation of S6K. Western blot analysis (Figure 3E) shows that S6K phosphorylation, an indicator of its activation by mTOR, is reduced in Shh-treated CGNPs. Consistent with reduced S6K activity in Shh-treated CGNPs, we also observed reduced phosphorylation of the S6K substrate ribosomal protein S6 in Shh-treated CGNPs (Figure 3B, D).
S6K de-phosphorylation is mediated by protein phosphatase 2A (Peterson et al., 1999; Petritsch et al., 2000), a positive regulator of N-myc stability (Sjostrom et al., 2005) and we can inhibit PP2A by the addition of okadaic acid (OA). As expected PP2A inhibition destabilized N-Myc (Figure 3E) (Sjostrom et al., 2005). OA treatment also rescued S6K phosphorylation in the presence of Shh (Figure 3E, final lane). In addition OA treatment not only rescues S6K activity, as determined by phosphorylation of its substrate ribosomal protein S6, but it also blocks Shh-mediated IRS1 stabilization (Figure 3F). These results suggest that Shh signaling inhibits S6K1 activity, thereby promoting stabilization of IRS1 protein.
Alteration of IRS1 levels modulates CGNP proliferation in vitro
To investigate a role for IRS1 in CGNP proliferation, we used lentiviruses expressing small hairpin RNAs (shRNAs) targeting IRS1. We found that of six shRNAs tested by transfection into a murine cell line, all six effectively knocked down IRS1 (data not shown). Shh-treated CGNPs infected with pooled lentiviruses expressing shRNA against IRS1 had reduced IRS1 protein levels (Figure 4A). We did not observe compensatory up-regulation of IRS2, nor did we detect effects of IRS1 knock down on other members of the IGF pathway (Figure 4A). Consistent with results shown in Figure 1, neither Shh treatment nor IRS1 knockdown affected Akt phosphorylation. However, levels of cyclin D2 are decreased in response to shRNA treatment, suggesting that reduction of IRS1 protein affects cell cycle progression (Figure 4A).
To determine whether shRNA-mediated IRS1 knock down impairs CGNP proliferation, we first assayed these viruses on PN5 cerebellar slices. We infected 300 μm cerebellar sections with IRS1 shRNA lentiviruses, then treated the slices with medium containing Shh or Shh vehicle (“Shh-“). After 48 hours the sections were pulsed with BrdU for 4 hours, fixed, sectioned and stained for BrdU incorporation. Treatment with exogenous Shh increases levels of BrdU incorporation (Figure 4B, right panels) as well as EGL thickness as previously reported (Wechlser-Reya and Scott, 1998). Infection of the slice cultures with shRNA lentiviruses in conjunction with Shh leads to reduced BrdU staining (Figure 4B, bottom right panel). Importantly, changes in proliferation in response to Shh and/or shRNA lentiviurus occurred in the EGL where CGNPs reside during their proliferation phase.
To quantify the effects of shRNA treatment on proliferation, we measured BrdU incorporation in control or shRNA lentivirus-infected dissociated CGNPs. Forty-eight hours after infection the CGNPs were pulsed with BrdU for 2 hours prior to fixation and immunofluorescent stained for BrdU incorporation or the proliferation mareker Ki67 (Supplementary Figure 4). We found a significant reduction in BrdU-positive cells in Shh-treated CGNPs infected with shRNA lentiviruses targeting IRS1 compared to Shh-treated alone (Figure 4C). To confirm that effects of IRS1 knock down are specifically attributed to CGNPs, Percoll purified cultures comprising 98% CGNPs were treated with shRNAs with or without Shh. As in the mixed culture system there was a significant decrease in proliferation after exposure to IRS1-specific shRNAs (Figure 4 D). CGNPs infected with lentiviruses targeting GFP did not show reduced proliferation (Supplementary Figure 4D). Vehicle-treated cells did not proliferate under any conditions. These results demonstrate that IRS1 is a critical mediator of Shh-mediated CGNP proliferation.
IGF-induced PI-3K and Akt activation are essential for neuronal survival (Dudek et al., 1997). Although we do not see changes in the activation of Akt in response to Shh (Figure 1A) or with treatment with IRS1 shRNA (Figure 4A), knock down of IRS1 may still impact CGNP survival. To determine whether proliferation decreases in IRS1 knock down CGNPs reflect compromised survival, we performed immunostaining for cleaved caspase-3. Knock down of IRS1 in the presence or absence of Shh did not affect cell survival in mixed or Percoll purified cultures (Figure 4E,F; Supplementary Figure 4) suggesting that the function of IRS1 in CGNPs promotes proliferation and not cell survival.
In order to determine if IRS1 over-expression can maintain CGNP proliferation in the absence of Shh, we infected CGNPs with a retrovirus expressing IRS1. Ectopic expression of IRS1 in CGNPs did not result in alteration of other members of the IGF pathway (Figure 5A). Ectopic IRS1 expression in Shh-treated cells did not result in increased cyclin D2 levels. However, we observed that over-expression of IRS1 maintained cyclin D2 expression when Shh was removed (Figure 5A). To determine if IRS1-driven cyclin D2 expression in the absence of exogenous Shh is associated with activation of intracellular Shh pathway components, we assayed expression levels of Gli1 and Gli2. As shown in Figure 5B, RT-PCR analysis of these transcription factors demonstrates that IRS1 does not induce their expression. However expression of N-Myc, a well-characterized Shh signaling target (Kenney et al., 2003), is increased in response to IRS1 over-expression. These results suggest that Shh-mediated activation of N-myc may be a result of IRS1 stabilization, and that IRS1 does not act upstream of Gli1.
We next determined the effect of IRS1 expression on CGNP proliferation by staining BrdU-pulsed CGNPs infected with IRS1-expressing retroviruses. We see an increase in BrdU staining as well as increased Ki67, a proliferation marker, in Shh-treated CGNPs compared to untreated cells as expected (Supplementary Figure 5A). Ki67 expression is maintained in the absence of Shh when cells are infected with IRS1 expressing retrovirus before Shh withdrawal. To confirm these results and to quantify the affects of IRS1 expression on proliferation, CGNPs were pulsed with BrdU as described. CGNPs from which Shh was removed after infection with IRS1 have significantly more BrdU incorporation compared to untreated alone (Figure 5C). Similar results were obtained with IRS1-infected Shh-treated cells were exposed to cyclopamine, indicating that IRS1 effects on CGNP proliferation are Smoothened-independent (data not shown). However, in comparison to Shh-treated, non-IRS1-infected CGNPs, BrdU incorporation is reduced indicating that other components of the Shh signaling pathway are necessary to maintain full CGNP proliferation in vitro. Treatment of IRS1-infected CGNPs with cyclopamine yielded similar results (data not shown), indicating that sustained proliferation in IRS1-infected, Shh withdrawn CGNPs is not a result of residual Shh in the medium.
The increase in proliferation levels in IRS1 over-expressing, non-Shh-treated cells does not appear to result from a cell survival advantage as levels of activated caspase-3 remain the same in all treatment groups (Figure 5D, Supplementary Figure 5B). Over-expression of IRS1 followed by on-going Shh treatment did not promote increased proliferation. We speculate that this is because IRS1 is a large scaffolding protein and inducing supra-normal levels may lead to formation of non-functional complexes due to limiting levels of other components. Taken together, our results suggest that through IRS1 up-regulation, the Shh signaling pathway may in effect be hijacking mitogenic effectors of IGF signaling. It is also possible that IRS1 in CGNPs may have additional, IGF-independent functions contributing to proliferation.
IRS1 in Shh-mediated mouse medulloblastoma
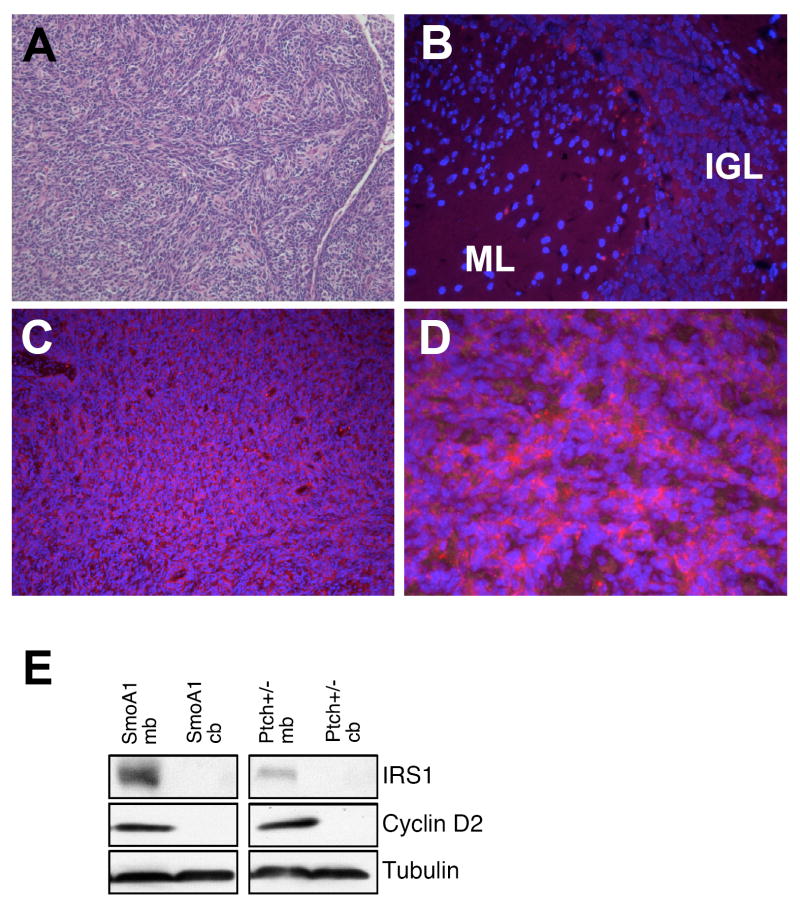
Aberrant IRS1 expression has been associated with several types of cancer including medulloblastoma (Del Valle et al., 2002; Waters et al., 1993). Since we see a role for IRS1 in mediating CGNP proliferation, we looked at IRS1 protein in two mouse models of medulloblastoma. Both the Patched+/- and Neuro-D2-SmoA1 mice form spontaneous medulloblastoma as a result of aberrant activation of the Shh signaling pathway (Berman et al., 2002; Goodrich et al., 1997; Hallahan et al., 2004). We found that tumors in both mice strains showed elevated IRS1 levels compared to adjacent normal brain tissue (Figure 6 and data not shown). Tumor lysates from these mice also show increased IRS1 levels compared to non-tumor cerebellar tissue, which correlates with increased cyclin D2 levels (Figure 6E).
Figure 6.
IRS1 is present in Shh-mediated mouse medulloblastoma. (A) Hematoxylin and eosin staining of a Neuro-D2-SmoA1 medulloblastoma depicts a desmoplastic medulloblastoma. (B) Adjacent normal cerebellar areas in the SmoA1 tumor model show very little IRS1 (red) immunostaining. (C-D) Increased IRS1 protein can be seen within the tumor at both 10X and 63X magnification (blue=DAPI). (E) Western blot analysis for both Patched +/- and SmoA1 medulloblastoma models show increased IRS1 expression coinciding with increased cyclin D2 expression.
DISCUSSION
Normal CGNP proliferation is dependent upon Shh and IGF signaling and both signaling pathways are implicated in medulloblastoma, a brain tumor for which CGNPs are a proposed cell-of-origin. We found that Shh specifically up-regulates the IGF receptor-interacting scaffolding protein IRS1 without altering levels of other members of the IGF pathway, including IRS2, Gab1 or activated Akt. Furthermore, IRS1 protein levels depend upon constant Shh signaling in vitro and IRS1 is found in proliferating CGNPs in vivo suggesting that IRS1 appears at the right time and place to play roles in Shh-dependent CGNP proliferation. IRS1 protein is also seen in mouse medulloblastoma in conjunction with high levels of cyclin D2 expression suggesting that IRS1 may also be playing a role in disease. Such a role for IRS1 is supported by our experiments showing that IRS1 over-expression in CGNPs is capable of sustaining proliferation even in the absence of Shh while knock down of IRS1 blocks proliferation in the presence of Shh.
In cell lines, it has been shown that IRS1 stability can be regulated by a negative feedback loop wherein S6K1 phosphorylates IRS1, targeting it for degradation by the proteasome (Easton et al., 2006). We observed that this feedback loop exists in primary CGNP cultures, and that Shh interferes with this process by suppressing S6 kinase activity, thereby stabilizing IRS1. Our results indicate that Shh suppresses S6 kinase activity by inhibiting its upsream regulator mTOR. Two previous studies have indicated interactions between the mTOR pathway and the hedgehog pathway, in that mTOR can regulate Indian hedgehog levels in chondrocytes and that mTOR inhibition impairs survival in epitheloid cells over-expressing Gli1 (Chanika Phornphutkul, 2008; Louro et al., 1999). These studies did not investigate how Shh signaling affects mTOR activity in primary neurons. Our data indicate that Shh-mediated mTOR inhibition is to some extent dependent upon Smoothened signaling, but the inability of cyclopamine to completely rescue S6K activity indicates existence of additional mechanisms. IRS1 regulation by stabilization instead of increased transcription has been reported in other cell types (Lee et al., 2003; Nemoto et al., 2006; Renstrom et al., 2005), but not in the setting of Shh signaling. Our study indicates a role for Shh-mediated IRS1 mRNA translation in addition to its stabilization in proliferating CGNPs.
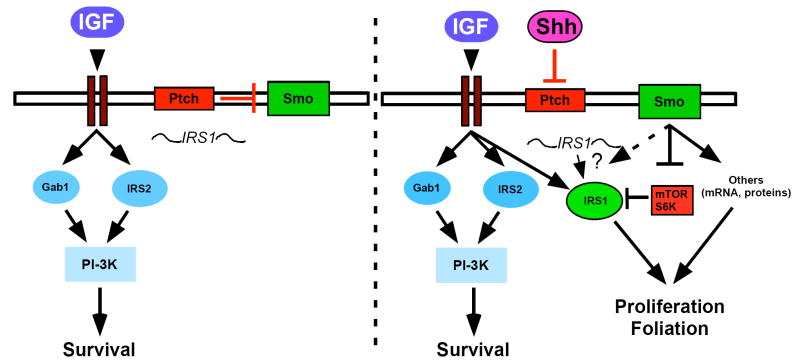
In addition to regulating IRS1 stability, Shh may also affect IRS1 mRNA translation, as the mRNA for IRS1 is present in un-treated CGNPs but the protein is not detectable, even upon the addition of lactacystin. The determine whether Shh influences laoding of IRS1 mRNA onto polysomes in CGNPs will be of future interest when techniques have evolved to make this experiment feasible. Currently, our results are consistent with a role for IGF signaling through Akt to promote survival (Dudek et al., 1997; Miller et al., 1997), coincident with stabilization of N-myc through GSK3β (Kenney et al., 2004). In the absence of Shh signaling, IGF signaling activates the PI-3K pathway leading to neuronal survival (Figure 7). Since levels of the IGF effectors IRS2 and Gab1 are unchanged in response to Shh, IGF survival signals may procede through these signaling molecules and not IRS1. In the presence of Shh, IGF signaling continues to send survival signals but can now also exert mitogenic effects through newly translated, stabilized IRS1, along with other factors important in mediating CGNP proliferation such as N-myc (Figure 7) (Kenney et al., 2003).
Figure 7.
Model suggesting how Shh mediates IRS1 expression during CGNP proliferation. In the absence of Shh IGF signaling sends survival cues through IRS2 and/or Gab1 and PI-3K signaling (left panel). IRS1 mRNA (squiggle) is present. In the presence of Shh, IRS1 is up-regulated by a mechanism that may involve both enhanced translation and protein stabilization. Shh stabilizes IRS1 protein by inhibiting mTOR-mediated activation of S6K, which is known to phosphorylate IRS1 leading to its degradation.
Our results demonstrate a role for IRS1 in mediating CGNP proliferation, and thus IRS1 may have a role in cell cycle progression in medulloblastoma. It has been shown that over-expression of IRS1 is sufficient to mediate transformation of mouse fibroblasts (D’Ambrosio et al., 1995). IRS1 has been found to have a role in several types of cancer including breast cancer while the IGF-pathway has been strongly linked to medulloblastoma (Dearth et al., 2007; Dearth et al., 2006a; Del Valle et al., 2002; Rao et al., 2004). One group has reported IRS1 expression in a JC-virus induced mouse medulloblastoma (Khalili et al., 2003), but a relationship between IRS1 and Shh-mediated medulloblastoma has not been reported. We report over-expression of IRS1 in two models of mouse Shh-induced medulloblastoma. This makes IRS1 an attractive candidate as a potential target for cancer therapies.
How IRS1 mediates CGNP proliferation remains unclear. Our data suggest that the effects of IRS1 do not occur through the activity of PI-3K. One possibility is that IRS1 increases CGNP survival by interacting with Bcl-2 (Ueno et al., 2000) however, modulation of IRS1 levels in vitro do not alter cell survival making this scenario unlikely. Recent studies in mammary tumors suggest that IRS1 interacts with proteins with known roles in proliferation, such as β-catenin (Dearth et al., 2006a). It remains to be determined whether this occurs in CGNPs and Shh-derived medulloblastomas. It is also possible that IRS1, a large scaffolding protein, has unknown interactors in Shh-stimulated CGNPs. Future studies exploring the specific mechanism through which IRS1 promotes Shh-stimulated CGNP proliferation may also identify novel targets for development of new treatments for medulloblastoma and other cancers.
Supplementary Material
Acknowledgments
We thank the Sontag Foundation and Alex’s Lemonade Stand Foundation for supporting these studies. This work was also supported by NIH 1R01NS061070-01 (AMK) and NRSA fellowship F32AG030888 (SP). Africa Fernandez-L receives fellowship support from the Spanish Ministry of Education. The contents of this manuscript are the sole responsibility of the authors and do not necessarily represent the official views of the NINDS, the NIA, the Sontag Foundation or Alex’s Lemonade Stand Foundation. We thank Tim Gershon for assistance with generating IRS1 retroviruses, and we thank William Hahn for advice concerning use of shRNA lentiviruses.
References
- Alcedo J, Ayzenzon M, Von Ohlen T, Noll M, Hooper JE. The Drosophila smoothened Gene Encodes a Seven-Pass Membrane Protein, a Putative Receptor for the Hedgehog Signal. Cell. 1996;86:221. doi: 10.1016/s0092-8674(00)80094-x. [DOI] [PubMed] [Google Scholar]
- Altman J, Bayer S. Development of the cerebellar system in relation to its evolution, structure, and functions. CRC Press; 1997. [Google Scholar]
- Aruga J, Inoue T, Hoshino J, Mikoshiba K. Zic2 Controls Cerebellar Development in Cooperation with Zic1. J Neurosci. 2002;22:218–225. doi: 10.1523/JNEUROSCI.22-01-00218.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman DM, Karhadkar SS, Hallahan AR, Pritchard JI, Eberhart CG, Watkins DN, Chen JK, Cooper MK, Taipale J, Olson JM, et al. Medulloblastoma growth inhibition by hedgehog pathway blockade. Science. 2002;297:1559–61. doi: 10.1126/science.1073733. [DOI] [PubMed] [Google Scholar]
- Cai D, Shen Y, De Bellard M, Tang S, Filbin MT. Prior exposure to neurotrophins blocks inhibition of axonal regeneration by MAG and myelin via a cAMP-dependent mechanism. Neuron. 1999;22:89–101. doi: 10.1016/s0896-6273(00)80681-9. [DOI] [PubMed] [Google Scholar]
- Phornphutkul Chanika, W K-Y, A V, C Q, G PA. mTOR signaling contributes to chondrocyte differentiation. Developmental Dynamics. 2008;237:702–712. doi: 10.1002/dvdy.21464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Wu A, Sun H, Drakas R, Garofalo C, Cascio S, Surmacz E, Baserga R. Functional significance of type 1 insulin-like growth factor-mediated nuclear translocation of the insulin receptor substrate-1 and beta-catenin. J Biol Chem. 2005;280:29912–20. doi: 10.1074/jbc.M504516200. [DOI] [PubMed] [Google Scholar]
- Chen JK, Taipale J, Young KE, Maiti T, Beachy PA. Small molecule modulation of Smoothened activity. Proceedings of the National Academy of Sciences. 2002;99:14071–14076. doi: 10.1073/pnas.182542899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corrales JD, Blaess S, Mahoney EM, Joyner AL. The level of sonic hedgehog signaling regulates the complexity of cerebellar foliation. Development. 2006;133:1811–21. doi: 10.1242/dev.02351. [DOI] [PubMed] [Google Scholar]
- Corrales JD, Rocco GL, Blaess S, Guo Q, Joyner AL. Spatial pattern of sonic hedgehog signaling through Gli genes during cerebellum development. Development. 2004;131:5581–90. doi: 10.1242/dev.01438. [DOI] [PubMed] [Google Scholar]
- D’Ambrosio C, Keller S, Morrione A, Lienhard G, Baserga R, Surmacz E. Transforming potential of the insulin receptor substrate 1. Cell Growth Differ. 1995;6:557–62. [PubMed] [Google Scholar]
- Dahmane N, Ruiz-i-Altaba A. Sonic hedgehog regulates the growth and patterning of the cerebellum. Development. 1999;126:3089–100. doi: 10.1242/dev.126.14.3089. [DOI] [PubMed] [Google Scholar]
- Dearth R, Cui X, Kim H, Hadsell D, Lee A. Oncogenic transformation by the signaling adaptor proteins insulin receptor substrate (IRS)-1 and IRS-2. Cell Cycle. 2007;6:705–713. doi: 10.4161/cc.6.6.4035. [DOI] [PubMed] [Google Scholar]
- Dearth RK, Cui X, Kim H-J, Kuiatse I, Lawrence NA, Zhang X, Divisova J, Britton OL, Mohsin S, Allred DC, et al. Mammary Tumorigenesis and Metastasis Caused by Overexpression of Insulin Receptor Substrate 1 (IRS-1) or IRS-2. Mol Cell Biol %R 10.1128/MCB.00260-06. 2006a;26:9302–9314. doi: 10.1128/MCB.00260-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dearth RK, Cui X, Kim HJ, Kuiatse I, Lawrence NA, Zhang X, Divisova J, Britton OL, Mohsin S, Allred DC, et al. Mammary tumorigenesis and metastasis caused by overexpression of insulin receptor substrate 1 (IRS-1) or IRS-2. Molecular & Cellular Biology. 2006b;26:9302–14. doi: 10.1128/MCB.00260-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Rincon S, Guo Q, Morelli C, Shiu H, Surmacz E, Miller WJ. Retinoic acid mediates degradation of IRS1 by the ubiquitin-proteasome pathway, via a PKC-dependant mechanism. Oncogene. 2004;23:9269–9279. doi: 10.1038/sj.onc.1208104. [DOI] [PubMed] [Google Scholar]
- Del Valle L, Enam S, Lassak A, Wang JY, Croul S, Khalili K, Reiss K. Insulin-like growth factor I receptor activity in human medulloblastomas. Clin Cancer Res. 2002;8:1822–30. [PubMed] [Google Scholar]
- Dudek H, Datta SR, Franke TF, Birnbaum MJ, Yao R, Cooper GM, Segal RA, Kaplan DR, Greenberg ME. Regulation of neuronal survival by the serine-threonine protein kinase Akt. Science. 1997;275:661–5. doi: 10.1126/science.275.5300.661. [DOI] [PubMed] [Google Scholar]
- Easton JB, Kurmasheva RT, Houghton PJ. IRS-1: Auditing the effectiveness of mTOR inhibitors. Cancer Cell. 2006;9:153. doi: 10.1016/j.ccr.2006.02.027. [DOI] [PubMed] [Google Scholar]
- Goodrich LV, Milenkovic L, Higgins KM, Scott MP. Altered neural cell fates and medulloblastoma in mouse patched mutants. Science. 1997;277:1109–13. doi: 10.1126/science.277.5329.1109. [DOI] [PubMed] [Google Scholar]
- Hahn H, Wojnowski L, Specht K, Kappler R, Calzada-Wack J, Potter D, Zimmer A, Muller U, Samson E, Quintanilla-Martinez L. Patched target Igf2 is indispensable for the formation of medulloblastoma and rhabdomyosarcoma. J Biol Chem. 2000;275:28341–4. doi: 10.1074/jbc.C000352200. [DOI] [PubMed] [Google Scholar]
- Hallahan AR, Pritchard JI, Hansen S, Benson M, Stoeck J, Hatton BA, Russell TL, Ellenbogen RG, Bernstein ID, Beachy PA, et al. The SmoA1 Mouse Model Reveals That Notch Signaling Is Critical for the Growth and Survival of Sonic Hedgehog-Induced Medulloblastomas. Cancer Res. 2004;64:7794–7800. doi: 10.1158/0008-5472.CAN-04-1813. [DOI] [PubMed] [Google Scholar]
- Hartley D, Cooper GM. Role of mTOR in the degradation of IRS-1: Regulation of PP2A activity. Journal of Cellular Biochemistry. 2002;85:304–314. doi: 10.1002/jcb.10135. [DOI] [PubMed] [Google Scholar]
- Hartmann W, Koch A, Brune H, Waha A, Schuller U, Dani I, Denkhaus D, Langmann W, Bode U, Wiestler OD, et al. Insulin-like growth factor II is involved in the proliferation control of medulloblastoma and its cerebellar precursor cells. Am J Pathol. 2005;166:1153–62. doi: 10.1016/S0002-9440(10)62335-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haruta T, Uno T, Kawahara J, Takano A, Egawa K, Sharma PM, Olefsky JM, Kobayashi M. A Rapamycin-Sensitive Pathway Down-Regulates Insulin Signaling via Phosphorylation and Proteasomal Degradation of Insulin Receptor Substrate-1. Mol Endocrinol %R 10.1210/me.14.6.783. 2000;14:783–794. doi: 10.1210/mend.14.6.0446. [DOI] [PubMed] [Google Scholar]
- Hatten M, Heintz N. Mechanisms of neural patterning and specification in the developing cerebellum. Annu Rev Neurosci. 1995;18:385–408. doi: 10.1146/annurev.ne.18.030195.002125. [DOI] [PubMed] [Google Scholar]
- Ho K, Scott M. Sonic hedgehog in the nervous systme: functions, modifications and mechanisms. Curr Opin Neurobiol. 2002a;12:57–63. doi: 10.1016/s0959-4388(02)00290-8. [DOI] [PubMed] [Google Scholar]
- Ho KS, Scott MP. Sonic hedgehog in the nervous system: functions, modifications and mechanisms. Curr Opin Neurobiol. 2002b;12:57–63. doi: 10.1016/s0959-4388(02)00290-8. [DOI] [PubMed] [Google Scholar]
- Ishizuka K, Usui I, Kanatani Y, Bukhari A, He J, Fujisaka S, Yamazaki Y, Suzuki H, Hiratani K, Ishiki M, et al. Chronic tumor necrosis factor-alpha treatment causes insulin resistance via insulin receptor substrate-1 serine phosphorylation and suppressor of cytokine signaling-3 induction in 3T3-L1 adipocytes. Endocrinology. 2007;148:2994–3003. doi: 10.1210/en.2006-1702. [DOI] [PubMed] [Google Scholar]
- Kenney AM, Cole MD, Rowitch DH. Nmyc upregulation by sonic hedgehog signaling promotes proliferation in developing cerebellar granule neuron precursors. Development. 2003;130:15–28. doi: 10.1242/dev.00182. [DOI] [PubMed] [Google Scholar]
- Kenney AM, Rowitch DH. Sonic hedgehog promotes G(1) cyclin expression and sustained cell cycle progression in mammalian neuronal precursors. Mol Cell Biol. 2000;20:9055–67. doi: 10.1128/mcb.20.23.9055-9067.2000. In Process Citation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenney AM, Widlund HR, Rowitch DH. Hedgehog and PI-3 kinase signaling converge on Nmyc1 to promote cell cycle progression in cerebellar neuronal precursors. Development. 2004;131:217–28. doi: 10.1242/dev.00891. [DOI] [PubMed] [Google Scholar]
- Khalili K, Del Valle L, Wang JY, Darbinian N, Lassak A, Safak M, Reiss K. T-antigen of human polyomavirus JC cooperates with IGF-IRS singaling system in cerebellar tumors of the childhood-medulloblastomas. Anticancer Res. 2003;23:2035–41. [PubMed] [Google Scholar]
- Knoepfler P, Kenney A. Neural precursors cycling at sonic speed: N-Myc pedals, GSK-3 breaks. Cell Cycle. 2006;5:47–52. doi: 10.4161/cc.5.1.2292. [DOI] [PubMed] [Google Scholar]
- Lee AV, Gooch JL, Oesterreich S, Guler RL, Yee D. Insulin-Like Growth Factor I-Induced Degradation of Insulin Receptor Substrate 1 Is Mediated by the 26S Proteasome and Blocked by Phosphatidylinositol 3’-Kinase Inhibition. Mol Cell Biol %R 10.1128/MCB.20.5.1489-1496.2000. 2000;20:1489–1496. doi: 10.1128/mcb.20.5.1489-1496.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee AV, Zhang P, Ivanova M, Bonnette S, Oesterreich S, Rosen JM, Grimm S, Hovey RC, Vonderhaar BK, Kahn CR, et al. Developmental and Hormonal Signals Dramatically Alter the Localization and Abundance of Insulin Receptor Substrate Proteins in the Mammary Gland. Endocrinology %R 10.1210/en.2002-221103. 2003;144:2683–2694. doi: 10.1210/en.2002-221103. [DOI] [PubMed] [Google Scholar]
- Louro ID, McKie-Bell P, Gosnell H, Brindley BC, Bucy RP, Ruppert JM. The zinc finger protein GLI induces cellular sensitivity to the mTOR inhibitor rapamycin. Cell Growth Differ. 1999;10:503–16. [PubMed] [Google Scholar]
- Marino S. Medulloblastoma: developmental mechanisms out of control. Trends Mol Med. 2005;11:17–22. doi: 10.1016/j.molmed.2004.11.008. [DOI] [PubMed] [Google Scholar]
- Mill P, Mo R, Hu MC, Dagnino L, Rosenblum ND, Hui CC. Shh controls epithelial proliferation via independent pathways that converge on N-Myc. Dev Cell. 2005;9:293–303. doi: 10.1016/j.devcel.2005.05.009. [DOI] [PubMed] [Google Scholar]
- Miller TM, Tansey MG, Johnson EM, Jr, Creedon DJ. Inhibition of phosphatidylinositol 3-kinase activity blocks depolarization- and insulin-like growth factor I-mediated survival of cerebellar granule cells. J Biol Chem. 1997;272:9847–53. doi: 10.1074/jbc.272.15.9847. [DOI] [PubMed] [Google Scholar]
- Morelli C, Garofalo C, Sisci D, del Rincon S, Cascio S, Tu X, Vecchione A, Sauter ER, Miller WH, Jr, Surmacz E. Nuclear insulin receptor substrate 1 interacts with estrogen receptor alpha at ERE promoters. Oncogene. 2004;23:7517–26. doi: 10.1038/sj.onc.1208014. [DOI] [PubMed] [Google Scholar]
- Nemoto T, Yokoo H, Satoh S, Yanagita T, Sugano T, Yoshikawa N, Maruta T, Kobayashi H, Wada A. Constitutive activity of glycogen synthase kinase-3B: Positive regulation of steady-state levels of insulin receptor substrates-1 and -2 in adrenal chromaffin cells. Brain Research. 2006;1110:1–12. doi: 10.1016/j.brainres.2006.06.053. [DOI] [PubMed] [Google Scholar]
- Oliver TG, Grasfeder LL, Carroll AL, Kaiser C, Gillingham CL, Lin SM, Wickramasinghe R, Scott MP, Wechsler-Reya RJ. Transcriptional profiling of the Sonic hedgehog response: a critical role for N-myc in proliferation of neuronal precursors. Proc Natl Acad Sci U S A. 2003;100:7331–6. doi: 10.1073/pnas.0832317100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Packer RJ, Cogen P, Vezina G, Rorke LB. Medulloblastoma: clinical and biologic aspects. Neuro-oncol. 1999;1:232–50. doi: 10.1215/15228517-1-3-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson RT, Desai BN, Hardwick JS, Schreiber SL. Protein phosphatase 2A interacts with the 70-kDa S6 kinase and is activated by inhibition of FKBP12-rapamycinassociated protein. Proc Natl Acad Sci U S A. 1999;96:4438–42. doi: 10.1073/pnas.96.8.4438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petritsch C, Beug H, Balmain A, Oft M. TGF-beta inhibits p70 S6 kinase via protein phosphatase 2A to induce G(1) arrest. Genes Dev. 2000;14:3093–101. doi: 10.1101/gad.854200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Provias J, Becker L. Cellular and molecular patholgy of medulloblastoma. J Neurooncol. 1996;29:35–43. doi: 10.1007/BF00165516. [DOI] [PubMed] [Google Scholar]
- Rao G, Pedone CA, Valle LD, Reiss K, Holland EC, Fults DW. Sonic hedgehog and insulin-like growth factor signaling synergize to induce medulloblastoma formation from nestin-expressing neural progenitors in mice. Oncogene. 2004;23:6156–62. doi: 10.1038/sj.onc.1207818. [DOI] [PubMed] [Google Scholar]
- Reiss K. Insulin-like growth factor-I receptor - a potential therapeutic target in medulloblastomas. Expert Opin Ther Targets. 2002;6:539–44. doi: 10.1517/14728222.6.5.539. [DOI] [PubMed] [Google Scholar]
- Renstrom F, Bruen J, Eriksson J. Insulin receptor substrates-1 and -2 are both depleted but via different mechanisms after down-regulation of glucose transport in rat adipocytes. Endocrinology. 2005;146:3044–3051. doi: 10.1210/en.2004-1675. [DOI] [PubMed] [Google Scholar]
- Rice M, Turnbow M, Garner C. Insulin stimulates the degradation of IRS-1 in 3T3-L1 adipocytes. Biochem and Biophys res comm. 1993;190:961–967. doi: 10.1006/bbrc.1993.1143. [DOI] [PubMed] [Google Scholar]
- Riobo NA, Lu K, Ai X, Haines GM, Emerson CP., Jr Phosphoinositide 3-kinase and Akt are essential for Sonic Hedgehog signaling. PNAS %R 10.1073/pnas.0504337103. 2006;103:4505–4510. doi: 10.1073/pnas.0504337103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert M, Brazil DP, Burks DJ, Kushner JA, Ye J, Flint CL, Farhang-Fallah J, Dikkes P, Warot XM, Rio C, et al. Insulin receptor substrate-2 deficiency impairs brain growth and promotes tau phosphorylation. J Neurosci. 2003;23:7084–92. doi: 10.1523/JNEUROSCI.23-18-07084.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah O, Hunter T. Turnover of the active fraction of IRS1 involves raptor-mTOR and S6K1-dependent serine phosphorylation in cell culture models of tuberous sclerosis. Mol Cell Biol. 2006;26:6425–34. doi: 10.1128/MCB.01254-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjostrom SK, Finn G, Hahn WC, Rowitch DH, Kenney AM. The cdk1 complex plays a prime role in regulating N-myc phosphorylation and turnover in neural precursors. Dev Cell. 2005;9:327–38. doi: 10.1016/j.devcel.2005.07.014. [DOI] [PubMed] [Google Scholar]
- Tzatsos A, Kandror K. Nutrients suppress phosphatidylinositol 3-kinase/Akt signaling via raptor-dependent mTOR-mediated insulin receptor substrate 1 phosphorylation. Mol Cell Biol. 2006;26:63–76. doi: 10.1128/MCB.26.1.63-76.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueno H, Kondo E, Yamamoto-Honda R, Tobe K, Nakamoto T, Sasaki K, Mitani K, Furusaka A, Tanaka T, Tsujimoto Y, et al. Association of Insulin Receptor Substrate Proteins with Bcl-2 and Their Effects on Its Phosphorylation and Antiapoptotic Function. Mol Biol Cell. 2000;11:735–746. doi: 10.1091/mbc.11.2.735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Um SH, Frigerio F, Watanabe M, Picard F, Joaquin M, Sticker M, Fumagalli S, Allegrini PR, Kozma SC, Auwerx J, et al. Absence of S6K1 protects against age- and diet-induced obesity while enhancing insulin sensitivity. Nature. 2004;431:200. doi: 10.1038/nature02866. [DOI] [PubMed] [Google Scholar]
- Uziel T, Zindy F, Xie S, Lee Y, Forget A, Magdaleno S, Rehg JE, Calabrese C, Solecki D, Eberhart CG, et al. The tumor suppressors Ink4c and p53 collaborate independently with Patched to suppress medulloblastoma formation. Genes Dev. 2005;19:2656–2667. doi: 10.1101/gad.1368605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Obberghen E, Baron V, Delahaye L, Emanuelli B, Filippa N, Giorgetti-Peraldi S, et al. Surfing the insulin signaling web. Eur J Clin Invest. 2001;31:966–77. doi: 10.1046/j.1365-2362.2001.00896.x. [DOI] [PubMed] [Google Scholar]
- Wallace VA. Purkinje-cell-derived Sonic hedgehog regulates granule neuron precursor cell proliferation in the developing mouse cerebellum. Curr Biol. 1999;9:445–8. doi: 10.1016/s0960-9822(99)80195-x. [DOI] [PubMed] [Google Scholar]
- Waters SB, Yamauchi K, Pessin JE. Functional expression of insulin receptor substrate-1 is required for insulin-stimulated mitogenic signaling. J Biol Chem. 1993;268:22231–4. [PubMed] [Google Scholar]
- Wechsler-Reya RJ, Scott MP. Control of neuronal precursor proliferation in the cerebellum by Sonic Hedgehog. Neuron. 1999;22:103–14. doi: 10.1016/s0896-6273(00)80682-0. see comments. [DOI] [PubMed] [Google Scholar]
- Wetmore C. Sonic hedgehog in normal and neoplastic proliferation: insight gained from human tumors and animal models. Curr Opin Genet Dev. 2003;13:34–42. doi: 10.1016/s0959-437x(03)00002-9. [DOI] [PubMed] [Google Scholar]
- White M. The IRS-signaling system: A network of docking proteins that mediate insulin action. Mol and Cell Biochem. 1998;182:3–11. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.