Abstract
Cellular signal transduction pathways are usually studied following administration of an external stimulus. However, disease-associated aberrant activity of the pathway is often due to misregulation of the equilibrium state. The transcription factor NF-κB is typically described as being held inactive in the cytoplasm by binding its inhibitor, IκB, until an external stimulus triggers IκB degradation through an IκB kinase-dependent degradation pathway. Combining genetic, biochemical, and computational tools, we investigate steady-state regulation of the NF-κB signaling module and its impact on stimulus responsiveness. We present newly measured in vivo degradation rate constants for NF-κB-bound and -unbound IκB proteins that are critical for accurate computational predictions of steady-state IκB protein levels and basal NF-κB activity. Simulations reveal a homeostatic NF-κB signaling module in which differential degradation rates of free and bound pools of IκB represent a novel cross-regulation mechanism that imparts functional robustness to the signaling module.
Keywords: cross-regulation, homeostasis, mathematical modeling, robustness, signaling module
Introduction
Cellular signal transduction pathways mediate responses to extracellular and intracellular signals, such as changing environmental and metabolic conditions, pathogen assault, and developmental cues. Many signaling pathways control the activity of transcription factors that regulate cognate target genes (Brivanlou and Darnell, 2002). For immediate early transcriptional responses (not requiring induced synthesis), such regulation may involve the reversible phosphorylation of the transcription factor to induce dimerization or nuclear translocation (e.g. the Stat, IRF, AP-1 transcription factor families). An alternate means of pathway activation involves stabilization of the transcriptional effector, as in the case of the genotoxic response regulator p53, the hypoxia response factor HIF-1α, or the developmentally regulated coactivator β-catenin. Thus, signaling in response to stimulus involves alterations of the homeostatic rates of synthesis and degradation found in unstimulated cells.
In contrast, the cellular abundance of the transcription factor NF-κB does not change dramatically during signaling. NF-κB is the critical mediator of cellular responses to a large number of physiological stimuli, including inflammatory cytokines, developmental signals, pathogens, and cellular stresses (Figure 1A) (Hoffmann and Baltimore, 2006). Although inflammatory signaling leads to transient NF-κB activity that is dynamically regulated by feedback mechanisms, elevated constitutive levels of active NF-κB are associated with chronic inflammatory diseases and many types of cancer (Karin, 2006).
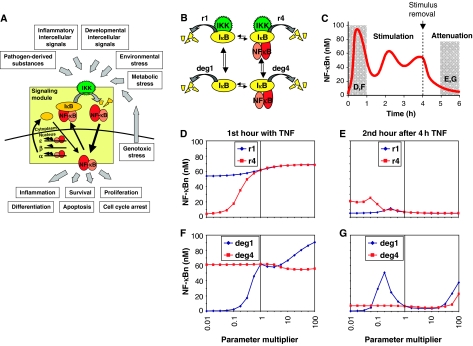
Figure 1.
Exploring the relative importance of IκB degradation mechanisms by computational parameter sensitivity analysis. (A) Schematic of the NF-κB signaling module and its physiological importance in the transduction of diverse inflammatory, developmental, and stress signals. (B) Illustration of the four IκB degradation pathways within the NF-κB signaling module. deg1 and deg4 are IKK-independent degradation rate constants for free and bound IκBα. r1 and r4 are IKK-dependent degradation rate constants for free and bound IκBα. (C) Computational simulation of NF-κB activation over a 6-h time course. TNF stimulation begins at time 0, and is removed at 4 h. Mean activity in the first hour of stimulation and the second hour after removal of the stimulus (shaded in gray) were used to create the plots in (D–F) and (G). (D–G) Graphs showing the average nuclear NF-κB (y axis) during the first hour (D, F) or during the second hour after 4 h (E, G) of TNF stimulation for different values (x axis) of the IKK-dependent (D, E) or -independent (F, G) degradation rate constants of free (blue line) and bound (red line) IκB.
NF-κB activity is inhibited by association with the inhibitor proteins, IκBα, IκBβ, or IκBɛ, which mask its nuclear localization sequence and inhibit its DNA-binding activity. The regulated metabolism of IκB proteins—their synthesis and degradation—critically controls NF-κB signaling (Ghosh et al, 1998). Synthesis of IκB proteins is a highly regulated process, with at least two isoforms, IκBα and IκBɛ, being subject to NF-κB-inducible synthesis, thereby providing negative feedback (Scott et al, 1993; Kearns et al, 2006). Stimulus-induced IκB degradation is controlled by the IκB kinase (IKK), which phosphorylates two N-terminal serines. This leads to IκB polyubiquitination and degradation via the 26S proteasome, thus liberating NF-κB for nuclear translocation (Ghosh et al, 1998; Yaron et al, 1998).
These processes were described in a mathematical model of the IKK-IκB-NF-κB signaling module to recapitulate NF-κB activation in response to TNF stimulation (Hoffmann et al, 2002). Its construction relied on rate constants available in the literature from a diverse set of experiments. As no isoform-specific data were available, rate constants pertaining to IκBβ and IκBɛ were assumed to be the same as those measured for IκBα. Although this model accurately recapitulates NF-κB signaling in response to TNF, in the unstimulated state the estimated IκB levels were found to be unexpectedly high (Lipniacki et al, 2004). In fact, the vast majority of IκB were calculated to be in the free form, contradictory to experimental studies showing that free IκBα accounts for less than 15% of the total cellular IκB (Rice and Ernst, 1993).
Despite our detailed understanding of stimulus-induced NF-κB signaling, there is less clarity about the mechanisms mediating IκB turnover in the absence of external stimulation. Early studies reported that basal turnover of IκB, unlike its induced degradation, does not require the IKK-targeted serines, the C-terminal PEST domain, or poly-ubiquitination of IκB (Krappmann et al, 1996), whereas others found robust C-terminal phosphorylation and poly-ubiquitination (Pando and Verma, 2000).
By distinguishing between NF-κB-bound and free IκB pools using an IκB interaction mutant, the half-life of bound IκB was found to be five-fold longer than that of free IκB in unstimulated cells (Pando and Verma, 2000). However, free IκB is a poorer substrate for IKK than NF-κB-bound IκB (Zandi et al, 1998), although it is routinely used as a substrate to measure IKK activity in vitro. Free IκB turnover was proposed to involve casein kinase 2 (CK2)-mediated phosphorylation of the C-terminal domain and ubiquitination (Schwarz et al, 1996; Bren et al, 2000), but others suggested that CK2 is involved in inducible degradation of NF-κB-bound IκB (Kato et al, 2003), or that ubiquitination was not required (Krappmann et al, 1996; Alvarez-Castelao and Castano, 2005).
Given these contradictory results in the literature, the lack of data on two of the three IκB isoforms, and the poor fit of computational simulations of the NF-κB signaling module in cells not exposed to TNF, we generated genetic tools—mouse knockout cell lines—to isolate cleanly the endogenous-free and -bound IκB protein pools and probe their degradation with kinase knockouts and pharmacological inhibitors. In addition, we used computational modeling (i) to identify which constitutive degradation rate constants play a critical role in determining stimulus responsiveness, (ii) to determine new biochemical rate constants based on our experimental results, (iii) to confirm the validity of the new parameters by simulating the cellular steady state, and (iv) to reveal the control of IκB degradation by NF-κB as a cross-regulatory mechanism.
Results and discussion
IKK-dependent and -independent degradation of IκBs determine NF-κB signaling
Four degradation rate constants govern the in vivo half-life of IκB proteins (Figure 1B). An IκB molecule can exist in either the free or NF-κB-bound form. Both forms may be degraded in an IKK-dependent manner (we denote the IκBα rate constants of these processes r1 and r4, respectively), but are also subject to constitutive degradation in an IKK-independent manner (with the rate constants denoted as deg1 and deg4). These mechanisms are described as first-order rate constants in our mathematical model of NF-κB signaling (Hoffmann et al, 2002).
To explore the functional significance of each IκB degradation rate constant in NF-κB signal transduction, we performed simulations of TNF signaling after altering one of the four rate constants (simultaneously for the IκBα, β, and ɛ isoforms) with a parameter multiplier ranging from 0.01 to 100. For each parameter multiplier, we calculated the average nuclear NF-κB level in response to TNF during the early phase (during the first hour of stimulation) and the later attenuation phase (during the second hour after a 4 h stimulation) (Figure 1C). By plotting the calculated NF-κB activity against its parameter multiplier, we can interpret the sensitivity of the system to each rate constant for two critical features of the NF-κB response to a transient TNF stimulus: activation and attenuation of NF-κB activity.
We first examined the impact of changes in IKK-dependent IκB degradation rate constants on NF-κB activation. During the first hour of TNF stimulation, the amount of nuclear NF-κB calculated by the model is fairly insensitive to even drastic changes in the IKK-dependent degradation rate of free IκB (Figure 1D, blue line). In contrast, slowing down the IKK-induced degradation of NF-κB-bound IκB severely dampens NF-κB activity (Figure 1D, red line). During the attenuation phase, the amount of nuclear NF-κB predicted by the model was similarly found to be insensitive to changes in IKK-dependent degradation of free IκB (Figure 1E, blue line), but slowing the IKK-dependent degradation rate of bound IκB results in a loss of attenuation (Figure 1E, red line).
The IKK-independent IκB degradation rates control the stimulus-independent turnover of IκB proteins, and thus maintain a resting state equilibrium of IκB levels. Examining whether these IKK-independent degradation rates play a role in determining the cellular responsiveness to inflammatory stimuli revealed that during the first hour of TNF stimulation the signaling module is dramatically more sensitive to the basal turnover rate of free IκB (Figure 1F, blue line) than of bound IκB (Figure 1F, red line). Furthermore, our simulations predicted that a more stable free IκB results in a loss of attenuation, whereas the basal turnover rate of the bound IκBs had no effect (Figure 1G).
In sum, our computational simulations revealed that two of the four possible degradation pathways play a particularly important role in controlling NF-κB signaling. Whereas much is known about the stimulus-responsive IKK-mediated degradation pathway, the IKK-independent degradation mechanism of free IκB has received surprisingly little experimental attention. Given the importance of these degradation rate constants in our computational analysis, we set out to examine them in more detail experimentally.
NF-κB regulation of IκB protein turnover and synthesis
To measure experimentally in vivo degradation rate constants for NF-κB-bound IκB proteins, we used the ribosomal inhibitor cycloheximide (CHX) to reduce the synthesis of new IκB proteins (by 85%; Supplementary Figure S1A), and examined the amount of nuclear NF-κB DNA-binding activity via electrophoretic mobility shift assay (EMSA). Treatment of wild-type MEFs with CHX over a 60 h time course induced nuclear NF-κB activity that corresponds to 25–35% of peak TNF-induced NF-κB activity (Figure 2A, Supplementary Figure S1B). To determine the relative contributions of each NF-κB-bound IκB isoform (α, β, and ɛ) to CHX-mediated NF-κB activation, we used a panel of IκB double-knockout MEFs, which contain only one IκB isoform. These cells were previously used to determine the degradation rate constants for each IκB isoform by TNF-induced NF-κB activation, which revealed that upon IKK activation, IκBα was degraded most rapidly, followed by IκBɛ, and then IκBβ (Hoffmann et al, 2002). Interestingly, we find the same trend in stimulus-independent degradation, where IκBβ is the most stable and IκBα is the least stable (Figure 2B and Supplementary Figure S1C).
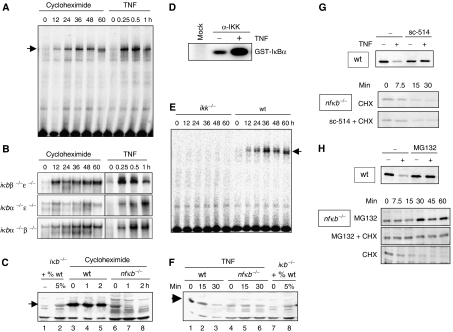
Figure 2.
Experimental studies of degradation pathways of NF-κB-bound and -free IκB proteins. (A) NF-κB activity as measured by EMSA of nuclear extracts from wild-type cells treated with 10 μg/ml CHX or 1 ng/ml TNF for indicated times. (B) NF-κB activity as measured by EMSA of nuclear extracts from ikbβ−/−ɛ−/−, ikbα−/−ɛ−/−, or ikbα−/−β−/− cells treated with 10 μg/ml CHX or 1 ng/ml TNF. (C) Western blot for IκBα in CHX-treated wild-type and nfkb−/− cells. The first two lanes show iκbα−/−β−/−ɛ−/− extract and iκbα−/−β−/−ɛ−/− extract mixed with 5% wild-type extract to show the protein level of IκBα in the nfkb−/− cells was approximately 5% that in the wild-type cells at time zero. (D) Cytoplasmic extracts of wild-type cells were immunoprecipitated with IKKγ antibody and subject to an in vitro kinase assay. In the ‘mock' lane, no antibody was added during the IP. (E) NF-κB activity as measured by EMSA of nuclear extracts from ikkα−/−β−/− or wild-type MEFs treated with 10 μg/ml CHX. (F) Western blot for IκBα of protein extracts from TNF (1 ng/ml)-treated wild-type or rela−/−crel−/−nfkb1−/− cells. (G) Western blots for IκBα of protein extracts from TNF-treated wild-type cells (top panel) in the presence or absence of the IKK-inhibitor sc-514. Bottom panels show Western blots for IκBα of protein extracts from CHX (10 μg/ml)-treated cells in the presence or absence of sc-514. (H) Western blots for IκBα of protein extracts from wild-type cells treated with TNFα (1 ng/ml) with or without the presence of the proteasome inhibitor MG132 (top panel). Western blots for IκBα of protein extracts from nfkb−/− cells treated with 10 μg/ml CHX, 10 μM MG132, or both.
To investigate the stability of the unbound, or ‘free', IκB proteins in resting cells, we generated crel−/−rela−/−nfkb1−/− (termed ‘nfkb−/−') MEFs deficient in the three NF-κB proteins known to interact with the classical IκB proteins: RelA, c-Rel, and p50. Western blots revealed a dramatic reduction in the amount of total IκB protein level in these cells compared to wild type (Figure 2C, compare lanes 3 and 6). A dilution series of wild-type protein extract with ikbα−/−β−/−ɛ−/− extract showed that the amount of IκBα in the nfkb−/− cells was approximately one-twentieth the amount in wild-type cells, and that this ratio is probably even lower for IκBβ and IκBɛ (Supplementary Figure S2A). No decrease in IκB levels was detected in MEFs deficient in the NF-κB proteins RelB and nfκb2 p52, which are non-canonical NF-κB proteins that do not bind canonical IκB proteins.
Strikingly, the level of IκBα mRNA in the nfkb−/− cells was only two-fold lower than in wild type, with even smaller differences in IκBβ and IκBɛ mRNA levels (Supplementary Figure S2B), suggesting that differential protein stability may account for different IκB protein levels in wild-type and nfkb−/− cells. Indeed, treating nfkb−/− cells with CHX resulted in rapid decreases of IκBα protein, whereas it remains stable in the wild-type cells beyond 2 h (Figure 2C). These results suggest that NF-κB has a regulatory role not only in controlling IκBα transcription, but also in stabilizing IκB proteins.
We next investigated whether the dramatically different half-life of free and bound IκB proteins may be due to different mechanisms governing their degradation.
IKK phosphorylation is a key mediator of the stimulus-induced degradation of NF-κB-bound IκB proteins, yet it is unclear if and how IKK may participate in the basal degradation of bound IκB. We first performed a kinase assay to examine the IKK activity of immunoprecipitated IKK complex from wild-type MEFs. Surprisingly, even in resting cells, a substantial amount of basal activity associated with the IKK complex was detectable (Figure 2D). In cells lacking the IKK catalytic subunits, IKKα and IKKβ, no activation of NF-κB upon CHX treatment (Figure 2E) was observed, indicating that IKK-dependent phosphorylation is required for the basal turnover of NF-κB-bound IκB proteins.
We sought to determine if IKK activity is involved in the turnover of free IκB proteins as well. IP-IKK kinase assays determined that the basal and inducible IKK activities are intact in the nfkb−/− cell line (Supplementary Figure S3A). Treatment of wild-type cells with TNF led to the rapid degradation of IκBα, but did not affect the levels of IκBα in the nfkb−/− cells (Figure 2F and Supplementary Figure S3B), suggesting that the inducible IKK activity is not involved in the degradation of free IκBα. Further, the IKK inhibitor, sc-514, which diminishes the TNF-induced degradation of IκBα in wild-type cells, did not have an effect on the basal turnover of free IκB in nfkb−/− cells (Figure 2G). In contrast, the proteasome inhibitor MG132 prevented not only TNF-induced degradation of IκBα in wild-type cells, but also led to accumulation of free IκB in nfkb−/− cells, and prevented its degradation when cells were cotreated with CHX (Figure 2H).
Based on our new biochemical data, we revised the parameter values governing degradation of IκB proteins within the NF-κB signaling module and incorporated these into our mathematical model (now termed model 1.1). Half-lives for free IκB proteins were determined to be 5–10 min (Figures 2H and Supplementary Figure S2, allowing us to calculate the respective first order rate constants (deg1-3). Our data highly constrained IKK-independent degradation rate constants (deg4-6) for NF-κB-bound IκB proteins (Figure 2E). While previous studies suggested that NF-κB stabilized free IκB degradation by a factor of 5 (Pando and Verma, 2000), our new measurements (Table I) indicate an NF-κB effect of 2000-fold with respect to the IKK-independent degradation of IκB proteins. This large discrepancy likely lies in the facts that (i) we have used a clean genetic system to isolate free endogenous IκB from NF-κB proteins, and have thus obtained a much faster degradation rate for free IκB and (ii) we have determined that degradation of NF-κB-bound IκB proteins can only occur through an IKK-involving mechanism and have thus drastically decreased the IKK-independent degradation rate of bound IκB.
Table 1.
Rate constants for IKK-independent IκB degradation: a role for NF-κB
| Rate constants (s−1) | NF-κB effect | ||
|---|---|---|---|
| IκBα | |||
| Free | deg1:2 × 10−3 | 2000 | |
| Bound | deg4:1 × 10−6 | ||
| IκBβ | |||
| Free | deg2:3 × 10−3 | 3000 | |
| Bound | deg5:1 × 10−6 | ||
| IκBɛ | |||
| Free | deg3:3 × 10−3 | 3000 | |
| Bound | deg6:1 × 10−6 | ||
New rate constants governing the uninduced IKK-independent degradation of IκB proteins derived from experiments in this paper and used in an updated version of the mathematical model. The ratio of IκB degradation rate constants in the presence and absence of NF-κB is defined as the ‘NF-κB effect'.
After incorporation of the new rate constants in Table I, we performed model fitting as described previously (Hoffmann et al, 2002) to obtain new degradation rate constants for IKK-induced degradation of NF-κB-bound IκB proteins (r4-6; Supplementary Table SI). As IKK-mediated phosphorylation of IκB is five-fold more efficient when NF-κB is present (Zandi et al, 1998), we divided the newly determined r4-6 by 5 to determine IKK-induced degradation of free IκB proteins (Supplementary Table SI, see Supplementary information for rate constant derivations). Including eight-fold differential IKK association rate constants (Zandi et al, 1998), the combined NF-κB effect on IKK-mediated degradation of free and bound IκB proteins is almost 50-fold.
Our results emphasize that NF-κB determines the degradation mechanism of IκB proteins. When bound to NF-κB, IκB turnover is slow and dependent on the basal activity of IKK. In contrast, when not bound to NF-κB, IκB degradation is rapid and independent of IKK activity.
Cross-regulation between IκB proteins via half-life control by NF-κB
We compared the steady-state levels of IκB predicted for unstimulated cells by the previous version of the model (referred to as model 1.0) (Hoffmann et al, 2002) and the new version of the model that incorporates the new rate constants of Tables I and Supplementary Table I (model 1.1) (Figure 3A, white and gray bars, respectively). The new degradation rate constants result in predictions of a much smaller pool of free IκB protein, as well as less total cellular IκB protein. The simulation results produced with the new model (1.1) are therefore in much better agreement with experimental observations (Rice and Ernst, 1993) than those with the previous model. Although a previous study (Lipniacki et al, 2004) lowered the IκB synthesis rate to correct the model-predicted ratio of free to bound IκB protein in the steady state, our new data indicate that the rapid free IκB degradation necessitates a high synthesis rate.
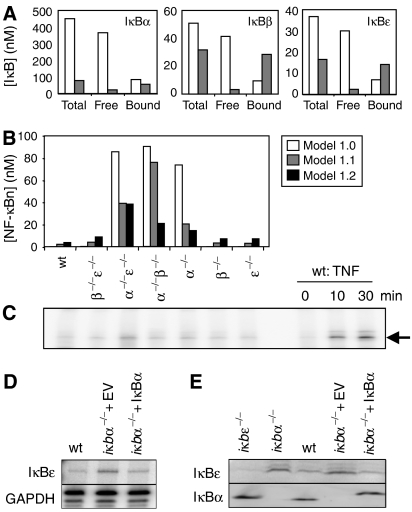
Figure 3.
An improved model of the homeostatic NF-κB signaling module. (A) Model calculations of IκBα, β, and ɛ protein levels (nM) in unstimulated cells for total IκB, free IκB, or NF-κB-bound IκB. Model 1.0 predictions are white bars, model 1.1 predictions are gray bars. (B) Model calculations of NF-κB activity (nM) in unstimulated wild-type and ikb−/− cells predicted by model version 1.0 (white bars), version 1.1 (gray bars), and version 1.2 (black bars). (C) NF-κB activity in untreated cells as measured by EMSA of nuclear extracts from the iκb−/− cell genotype labeled above each lane. The last three lanes are controls for the NF-κB band and are nuclear extracts from wild-type cells treated with TNF (1 ng/ml). (D) RNase protection assay showing levels of IκBɛ mRNA in untreated wild-type cells, iκbα−/− cells with empty vector control, or iκbα−/− cells expressing a retroviral iκbα transgene. GAPDH is used as a loading control. (E) Western blots for IκBɛ and IκBα in resting cells. The cell genotype is listed above each lane.
Next, we examined the consequences of the new degradation parameters on constitutive NF-κB activity in a series of IκB knockout cell lines. The previous model predicts that the removal of IκBα results in high levels of nuclear NF-κB activity in unstimulated cells (Figure 3B), which does not match with our experimental observations (Figure 3C). The differential degradation rates of bound and unbound IκB protein may result in molecular compensation among the IκB isoforms; upon deletion of a single IκB isoform, the newly available NF-κB may act to stabilize the remaining IκB isoforms, resulting in the cytoplasmic retention of NF-κB. Indeed, model 1.1 predicts a lower level of NF-κB activity in unstimulated knockout cells than version 1.0 (Figure 3B, compare white and gray bars). EMSA results (Figure 3C) confirm the new predictions, indicating that functional IκB compensation via differential half-life control indeed exists.
In the case of iκbα−/−β−/− cells where IκBɛ is the only isoform present, our model predicts a markedly higher level of NF-κB activity than seen experimentally. However, we have recently characterized an NF-κB-inducible IκBɛ mRNA synthesis mechanism (Kearns et al, 2006). Incorporation of this feedback mechanism into the model (referred to as model 1.2) indeed lowers the predicted basal NF-κB levels (Figure 3B, black bars) to levels that are in good agreement with the EMSA results. We measured IκBɛ mRNA levels and found that they are indeed upregulated in iκbα−/− cells compared to wild type (Figure 3D), resulting in higher IκBɛ protein levels (Figure 3E). To determine whether this effect was the result of homeostatic regulation within the NF-κB signaling module, we used a retroviral transgene to reconstitute IκBα expression in iκbα−/− cells. Indeed, we found that IκBɛ upregulation was reversible, confirming that even in resting cells constitutive NF-κB activity plays a role in transcriptional regulation of its inhibitors to controls its own steady-state activity.
Homeostatic control via distinct IκB degradation pathways
Our analysis of the NF-κB signaling module in unstimulated cells reveals a highly dynamic homeostatic state that is controlled by multiple synthesis and degradation mechanisms of the regulatory IκB proteins. As such we find that NF-κB itself has two roles in regulating its own basal activity. NF-κB binding to IκB proteins removes them from this rapid degradation pathway, and sensitizes them to a slow degradation mechanism that is dependent on basal IKK activity. Second, constitutive NF-κB activity also impacts transcription rates of IκBα and IκBɛ, thus providing for negative feedback even in the absence of an external stimulus.
Our studies identify the free IκB protein degradation pathway as a major determinant of constitutive NF-κB and of stimulus responsiveness of the NF-κB signaling module. Given this hitherto unappreciated importance, determining the enzymatic and potentially regulatory mechanisms of the free IκB degradation pathway is critical for understanding the regulation of NF-κB in diverse physiological and pathological settings.
Owing to the dynamic nature of the IκB-NF-κB equilibrium, the majority of newly synthesized IκB is likely degraded before ever binding NF-κB. However, this is not unlike other signal transduction pathways that consume significant cellular resources for the maintenance of a dynamic homeostatic state. For example, the transcription factors p53, HIF-1α, and β-catenin are continually synthesized and degraded. Upon signaling, the respective degradation pathways are inhibited to allow for their nuclear accumulation and function (Ivan et al, 2001; Jaakkola et al, 2001; Moon, 2005). How may this energy-consuming process of maintaining a dynamic homeostasis benefit the cell? Future computational studies may suggest that homeostatic control of the NF-κB signaling module confers sensitivity to signals but ensures a very steady low equilibrium activity that is less likely to drift (D Barken, unpublished results). In addition, combined computational and experimental studies may demonstrate that such a dynamic equilibrium state sensitizes the signaling pathway to metabolic changes, such that stress conditions constitute an input signal that results in cellular responses (Ellen L O'Dea, unpublished results).
Materials and methods
Cells and reagents
Primary and 3T3 immortalized MEF were generated from E12.5–14.5 embryos and maintained as described previously (Hoffmann et al, 2002). rela−/−crel−/−nfkb1−/− MEFs were generated from E12.5-timed matings of rela+/−crel−/−nfkb1−/− mice and iκbα−/−β−/−ɛ−/− cells will be described elsewhere. ikk1−/−ikk2−/− cells were a kind gift from Inder Verma. iκbα−/− MEF lines reconstituted with pBabe-IκBα and empty vector control were a generous gift from Erika Mathes. Recombinant murine TNF was from Roche; CHX, sc-514, and MG132 from Sigma. RelA/p65 (sc-372), RelB (sc-226), cRel (sc-71), IκBα (sc-371), IκBβ (sc-946), and IκBɛ (sc-7156) antibodies were from Santa Cruz Biotechnology. Trans35S-methionine label was from MP Biomedicals.
Biochemical analysis
Whole-cell extracts were prepared in RIPA buffer and equivalent protein amounts subjected to immunoblot analysis using ECL-plus (Amersham/GE Healthcare). Nuclear extracts were prepared and used for electrophoretic EMSA as described (Hoffmann et al, 2002). Immunoprecipitation kinase assay performed as in Werner et al (2005). Signals were quantified using a phosphorimager (Molecular Dynamics) and ImageQuant software version 5.2 (GE Healthcare). Dilution series with knockout extracts assured that Western blot signals were in the linear range. Total cellular RNA was isolated with Trizol reagent (Invitrogen) and used for RNase protection assay as described in Kearns et al (2006).
Cells were labeled with 200 μCi/ml 35S-methionine label for indicated times. Whole-cell extracts were prepared in RIPA buffer and dried on filter paper. 35S-Met incorporation was measured by scintillation count and CHX-treated cells versus untreated cells were compared to measure the percentage of translational inhibition.
Computational modeling
The mathematical model of the IKK-IκB-NF-κB signaling module was described in Hoffmann et al (2002). This model (version 1.0) was used to generate Figure 1. Model version 1.1 includes the parameter values shown in Table I and baseline level of IKK of 1 nM. Simulations were performed in Matlab and Excel as described previously (Hoffmann et al, 2002) with extended equilibration times. A complete list of the parameter values can be found in the Supplementary information. Graphs were generated in Excel. The Matlab code file is available upon request, and the SBML code is available at the MSB website.
Supplementary Material
Supplementary Data
Structured Data
Acknowledgments
We thank Erika Mathes, Gouri Ghosh, and Betsy Komives for insightful discussions, and Gouri Ghosh for critical reading of the manuscript. We are grateful to Santa Cruz Biotechnology for antibodies. EO is supported by the Heme Training Grant, JDK by the Molecular Biophysics Training Grant, DB by the UCSD Bioinformatics Graduate Training Grant, and SLW by an NSF Graduate Fellowship. This study was supported by NIH grants GM72024, GM69013, and GM071573.
References
- Alvarez-Castelao B, Castano JG (2005) Mechanism of direct degradation of IkappaBalpha by 20S proteasome. FEBS Lett 579: 4797–4802 [DOI] [PubMed] [Google Scholar]
- Bren GD, Pennington KN, Paya CV (2000) PKC-zeta-associated CK2 participates in the turnover of free IkappaBalpha. J Mol Biol 297: 1245–1258 [DOI] [PubMed] [Google Scholar]
- Brivanlou AH, Darnell JE Jr (2002) Signal transduction and the control of gene expression. Science 295: 813–818 [DOI] [PubMed] [Google Scholar]
- Ghosh S, May MJ, Kopp EB (1998) NF-kappa B and Rel proteins: evolutionarily conserved mediators of immune responses. Annu Rev Immunol 16: 225–260 [DOI] [PubMed] [Google Scholar]
- Hoffmann A, Baltimore D (2006) Circuitry of nuclear factor kappaB signaling. Immunol Rev 210: 171–186 [DOI] [PubMed] [Google Scholar]
- Hoffmann A, Levchenko A, Scott ML, Baltimore D (2002) The IkappaB-NF-kappaB signaling module: temporal control and selective gene activation. Science 298: 1241–1245 [DOI] [PubMed] [Google Scholar]
- Ivan M, Kondo K, Yang H, Kim W, Valiando J, Ohh M, Salic A, Asara JM, Lane WS, Kaelin WG Jr (2001) HIFalpha targeted for VHL-mediated destruction by proline hydroxylation: implications for O2 sensing. Science 292: 464–468 [DOI] [PubMed] [Google Scholar]
- Jaakkola P, Mole DR, Tian YM, Wilson MI, Gielbert J, Gaskell SJ, Kriegsheim A, Hebestreit HF, Mukherji M, Schofield CJ, Maxwell PH, Pugh CW, Ratcliffe PJ (2001) Targeting of HIF-alpha to the von Hippel-Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science 292: 468–472 [DOI] [PubMed] [Google Scholar]
- Karin M (2006) Nuclear factor-kappaB in cancer development and progression. Nature 441: 431–436 [DOI] [PubMed] [Google Scholar]
- Kato T Jr, Delhase M, Hoffmann A, Karin M (2003) CK2 is a C-terminal IkappaB kinase responsible for NF-kappaB activation during the UV response. Mol Cell 12: 829–839 [DOI] [PubMed] [Google Scholar]
- Kearns JD, Basak S, Werner SL, Huang CS, Hoffmann A (2006) IkappaBepsilon provides negative feedback to control NF-kappaB oscillations, signaling dynamics, and inflammatory gene expression. J Cell Biol 173: 659–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krappmann D, Wulczyn FG, Scheidereit C (1996) Different mechanisms control signal-induced degradation and basal turnover of the NF-kappaB inhibitor IkappaB alpha in vivo. EMBO J 15: 6716–6726 [PMC free article] [PubMed] [Google Scholar]
- Lipniacki T, Paszek P, Brasier AR, Luxon B, Kimmel M (2004) Mathematical model of NF-kappaB regulatory module. J Theor Biol 228: 195–215 [DOI] [PubMed] [Google Scholar]
- Moon RT (2005) Wnt/beta-catenin pathway. Sci STKE, 2005, cm1 [DOI] [PubMed] [Google Scholar]
- Pando MP, Verma IM (2000) Signal-dependent and -independent degradation of free and NF-kappa B-bound IkappaBalpha. J Biol Chem 275: 21278–21286 [DOI] [PubMed] [Google Scholar]
- Rice NR, Ernst MK (1993) In vivo control of NF-kappa B activation by I kappa B alpha. EMBO J 12: 4685–4695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz EM, Van Antwerp D, Verma IM (1996) Constitutive phosphorylation of IkappaBalpha by casein kinase II occurs preferentially at serine 293: requirement for degradation of free IkappaBalpha. Mol Cell Biol 16: 3554–3559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott ML, Fujita T, Liou HC, Nolan GP, Baltimore D (1993) The p65 subunit of NF-kappa B regulates I kappa B by two distinct mechanisms. Genes Dev 7: 1266–1276 [DOI] [PubMed] [Google Scholar]
- Werner SL, Barken D, Hoffmann A (2005) Stimulus specificity of gene expression programs determined by temporal control of IKK activity. Science 309: 1857–1861 [DOI] [PubMed] [Google Scholar]
- Yaron A, Hatzubai A, Davis M, Lavon I, Amit S, Manning AM, Andersen JS, Mann M, Mercurio F, Ben-Neriah Y (1998) Identification of the receptor component of the IkappaBalpha-ubiquitin ligase. Nature 396: 590–594 [DOI] [PubMed] [Google Scholar]
- Zandi E, Chen Y, Karin M (1998) Direct phosphorylation of IkappaB by IKKalpha and IKKbeta: discrimination between free and NF-kappaB-bound substrate. Science 281: 1360–1363 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Data
Structured Data