Abstract
The COP9 signalosome (CSN), an eight-subunit protein complex, is conserved in all higher eukaryotes. CSN intersects the ubiquitin–proteasome pathway, modulating signaling pathways controlling various aspects of development. We are using Drosophila as a model system to elucidate the function of this important complex. Transcriptome data were generated for four csn mutants, sampled at three developmental time points. Our results are highly reproducible, being confirmed using two different experimental setups that entail different microarrays and different controls. Our results indicate that the CSN acts as a transcriptional repressor during development of Drosophila, resulting in achronic gene expression in the csn mutants. ‘Time shift' analysis with the publicly available Drosophila transcriptome data indicates that genes repressed by the CSN are normally induced primarily during late embryogenesis or during metamorphosis. These temporal shifts are likely due to the roles of the CSN in regulating transcription factors. A null mutation in CSN subunit 4 and hypomorphic mutations in csn5 lead to more severe defects than seen in the csn5-null mutants strain, suggesting that CSN5 carries only some of the CSN function.
Keywords: COP9 signalosome, Drosophila, heterochrony, JAB1, transcriptome analysis
Introduction
The COP9 signalosome (CSN) is a highly conserved protein complex that in higher eukaryotes consists of eight subunits named CSN1 to CSN8 (Deng et al, 2000; Cope and Deshaies, 2003; Wei and Deng, 2003; Schwechheimer, 2004). With its discovery in Arabidopsis, the complex was originally described as a repressor of light-dependent growth in plants (Wei et al, 1994; Chamovitz et al, 1996). Subsequent identification and characterization of the CSN from mammalian cells, insects and yeast highlighted the complex as a general modulator of signal transduction (Seeger et al, 1998; Wei et al, 1998; Freilich et al, 1999; Mundt et al, 1999).
The CSN is involved in diverse cellular and developmental processes such as DNA repair, cell cycle regulation, MAPK signalling, hormone signalling, axonal guidance, and embryogenesis. (Doronkin et al, 2002; Maytal-Kivity et al, 2002, 2003; Mundt et al, 2002; Oron et al, 2002; Suh et al, 2002; Wee et al, 2002; Busch et al, 2003; Cope and Deshaies, 2003; Liu et al, 2003; Lykke-Andersen et al, 2003; Nielsen, 2003; Serino and Deng, 2003; Yan et al, 2003; Harari-Steinberg and Chamovitz, 2004). In regulating these processes, the CSN has been shown to impact transcription, protein translation and protein degradation.
The most studied CSN function is regulation of protein degradation. The CSN intersects the ubiquitin–proteasome pathway at a number of junctions: The CSN or CSN subunits directly interact with the proteasome (Kwok et al, 1999; Peng et al, 2003). The CSN also interacts with E3-ubiquitin ligases, removing Nedd8, a ubiquitin-like modifier, from cullin-based E3s (Lyapina et al, 2001; Cope et al, 2002; Wolf et al, 2003), thereby regulating ligase activity. This deneddylation activity resides in the JAMM/MPN+ domain of CSN5 (Cope et al, 2002). The CSN also mediates phosphorylation and deubiquitination of ubiquitin–proteasome pathway substrates, and as a consequence alters their stability and subcellular localization (reviewed in Harari-Steinberg and Chamovitz, 2004).
Certain CSN activities impinge on transcriptional regulation. Through its regulation of protein degradation, the CSN affects the stability (and activity) of transcription factors such as HY5 in plants and cJUN and p53 in animals (Naumann et al, 1999; Osterlund et al, 1999; Bech-Otschir et al, 2001). CSN subunits also directly affect transcriptional activity as corepressors or coactivators. For example, CSN2 was originally discovered as a corepressor of the thyroid hormone receptor, where as CSN5 acts as a coactivator of AP-1 target genes (Dressel et al, 1999; Akiyama et al, 2003; Groisman et al, 2003; Tenbaum et al, 2003). Moreover, CSN associates with chromatin through CSA and RNA polymerase II (Groisman et al, 2003), suggesting that it may mediate gene expression directly.
To better understand the function of CSN in a multicellular animal, we initiated a study in Drosophila melanogaster (Freilich et al, 1999). The Drosophila CSN is highly similar in subunit composition, subunit sequence, and biochemical properties to its human counterpart, and is essential for Drosophila development (Freilich et al, 1999; Oron et al, 2002). Loss-of-function mutants in Drosophila CSN4 and CSN5 display overlapping phenotypes in oogenesis and larval development. However, each mutant also has specific defects. For example, csn5null mutants develop large melanotic masses in their hemolymph, whereas csn4null mutants never show these masses, but instead display multiple abnormalities consistent with defects in steroid hormone signalling (Oron et al, 2002; Harari-Steinberg et al, 2007). Work in Schizosaccharomyces pombe also showed that not all csn mutants are equal (Mundt et al, 2002). This lack of complete phenotypic overlap is different from the case in Arabidopsis where loss-of-function mutants in different CSN subunits are phenotypically indistinguishable (Kwok et al, 1996; Ma et al, 2003; Dohmann et al, 2005). The lack of complete phenotypic overlap between the csn mutants can be explained by at least two non-exclusive hypotheses. First, each subunit can have different roles within the CSN complex, perhaps binding different interacting proteins. Second, each subunit can have distinct roles independent of the CSN.
Indeed, the exact forms of a ‘functional' CSN are not clear. While some of the eight subunits are present only on a ∼500 kDa ‘core complex-dependent' form, some are present also in forms independent of the eight-subunit complex (which we refer to as ‘complex-independent' forms). In Drosophila, while both CSN4 and CSN5 are detected in both complex-dependent and complex-independent forms, CSN4 is essential for CSN core complex stability, while CSN5 is not (Oron et al, 2002). We therefore refer to CSN4 as a ‘core subunit' and to CSN5 as peripheral. Several studies have suggested various types of equilibria between these different forms of subunits (Tsuge et al, 2001; Oron et al, 2002; Fukumoto et al, 2005). This dynamic nature has consequences for experiment interpretation. It is still unclear in many cases which of the activities and phenotypes attributed to CSN reside in the complex as a whole and which reside in the individual subunits (or other complexed forms of the subunits).
One major caveat with many developmental studies employing mutants in CSN is the severe phenotypes of the mutants. The obvious severe morphological phenotypes observed, such as constitutive photomorphogenesis in Arabidopsis seedlings, embryo lethality in mice or larval lethality in Drosophila, may mask more subtle phenotypes arising from more specific roles for the complex. Furthermore, such strong phenotypes may be downstream indirect consequences of earlier perturbations resulting from the lack of CSN function.
Transcriptome analysis could be useful in overcoming these limitations for several reasons. First, transcriptome analysis will identify global changes in gene transcription, and not only those that lead to obvious visible morphological changes. Second, transcriptome analysis at early developmental stages can identify changes in gene transcription that precede overt morphological changes. Third, such analysis could be useful for elucidating the underlying molecular basis for observed morphological phenotypes. The fact that in Drosophila, as opposed to Arabidopsis, mutations in different CSN subunits cause different morphological and molecular phenotypes, makes this model system ideal for studying questions regarding subunit-specific functions. The analysis reported here implies that the neddylation is only a part of CSN function. Developmental timing has a major effect on the expression profiles of the mutants, and our results suggest that mutations in CSN lead to a heterochronic shift in regulation of gene expression. We also show that genes involved in hormone signalling are misexpressed in CSN mutants in a development-dependant manner.
Results
Transcription profiling of CSN mutants
Ma et al (2003) showed that in Arabidopsis, mutations in CSN1 and CSN8 lead to essentially identical transcriptomes, which correlates well with their identical morphological phenotypes. However, the non-overlapping phenotypes of the Drosophila mutants in csn4 and csn5, and the different structural roles of these proteins in the CSN complex (Table I), suggest that the transcriptomes of Drosophila csn4 and csn5 mutants should be different. Furthermore, as these mutations are obviously pleiotropic, we reasoned that there could be numerous misregulated pathways that are not morphologically noticeable, and that any transcriptional misregulation could be developmentally specific. To clarify these issues, we initiated a global analysis of transcription profiles on our available CSN mutants.
Table 1.
Summary of the genetic and biochemical analysis of the Drosophila CSN
| Mutant | Visible phenotypes |
Biochemical phenotypes |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Lethal stage (hours AED) | DNA damage sensitivity | Oocyte defects | Axonal differentiation | Axonal guidance | Melanotic tumors | Molting defects | CSN complex | CSN5 monomer | CSN7 monomer | |
| WT | — | No | No | Normal | Normal | No | No | Present | Present | Present |
| csn4null | 96 | Yes | Yes | Defective | ND | No | Yes | Absent | Present | Present |
| csn5null | >120a | Yes | Yes | Defective | ND | Yes | No | Present | Absent | Present |
| csn51 | 96–120 | Yes | Yes | Normal | Defective | No | Yes1 | Present | Absent | Absent |
| csn53 | 96–120 | Yes | Yes | Normal | Defective | No | Yes1 | Present | Absent | Absent |
Based on (Freilich et al, 1999; Oron et al, 2002; Suh et al, 2002). AED=after egg deposition. 1—see Figure 7.
csn5null continue to live during a prolonged third larval instar but never pupariate.
Two rounds of microarray analyses were carried out. In the first round, the spotted microarrays used contain roughly half of the Drosophila estimated coding sequences (GEO GPL4285). Four mutants (csn4null, csn5null, csn51, csn53) in two CSN subunits (CSN4 and CSN5) were examined. The two null mutants are true genomic null alleles arising from gene deletion generated by imprecise excisions of P-elements contained in the genes (Freilich et al, 1999; Oron et al, 2002). Both csn51 and csn53 contain missense mutations in the CSN5 gene, maintain a CSN5 protein and were defined molecularly and phenotypically as hypomorphic mutants (Suh et al, 2002). mRNA populations were isolated from mutant larvae at three time points, 60, 72, and 96 h after egg deposition (AED), and compared with mRNA populations from age-matched wild-type (wt) larvae. The rationale for choosing these time points is outlined in Table II. Four biological repeats that included dye reversals were conducted for each experiment. The resulting data was standardized and analyzed using the FDR procedure (Reiner et al, 2003). The expression levels of 1362 genes, representing ∼20% of the genes on the microarray, were found to change significantly (FDR<0.05- and >1.85-fold change) in at least one experiment (Table III). This first round of experimentation is referred to below as ‘6k' reflecting the number of genes on the chip.
Table 2.
Rationale for time points sampled
| Time point | Rationale |
|---|---|
| 60 h AED (mid L2) | Mutant larvae molt from L1 to L2 concurrently with wt (at 48 h AED). There are no apparent differences in body size, phenotype, or behavior between the mutants and wt. |
| 72 h AED (L2/L3 transition) | Differences between csn4null and csn5null mutant larvae are first observed during the second molt at the L2/L3 transition (∼72 h AED), where csn4null larvae are unable to molt properly and display a double mouth hook phenotype. csn51 and csn53 larvae are smaller than csn5null larvae. |
| 96 h AED (mid L3) | Growth arrest of csn4null, csn51, and csn53 larvae, with obvious differences in body size observed relative to csn5null and WT. csn5null larvae develop melanotic capsules. |
AED=after egg deposition.
Table 3.
Summary of genes identified in the different experimental conditions using the 6k or 12k chip (shown in parentheses)
| Mutant | Hours AED | No. of regulated genes >1.85-fold, FDR<0.05 |
||
|---|---|---|---|---|
| Up | Down | Total | ||
| csn4null | 60 | 176 (493) | 61 (210) | 273 (703) |
| 72 | 191 | 179 | 370 | |
| 96 | 187 | 190 | 377 | |
| csn51 | 60 | 281 | 100 | 381 |
| 72 | 139 | 198 | 337 | |
| 96 | 292 | 263 | 555 | |
| csn53 | 60 | 293 (739) | 95 (297) | 388 (1036) |
| 72 | 122 | 153 | 275 | |
| 96 | 209 | 193 | 402 | |
| csn5null | 60 | 37 (131) | 41 (187) | 78 (318) |
| 72 | 108 | 52 | 160 | |
| 96 | 94 | 85 | 179 | |
The second round of microarray experiments differed from the first both in the microarray used and in experimental design. The spotted microarrays used in this second round of experiments contain 12 144 cDNAs representing most of the Drosophila estimated coding sequences (GEO GPL1908). Three mutants (csn4null, csn5null, csn53) and the wt were examined at 60 h AED. Each sample was hybridized together with a common reference (CR) comprised of a pooled wt RNA sample from multiple larval stages. Four biological repeats including dye reversals were conducted for each experiment. The resulting data were standardized and analyzed using the FDR procedure (Reiner et al, 2003). Differentially expressed genes were determined by comparing mutant versus CR to wt versus CR. The expression levels of 2816 genes, representing ∼20% of the genes on the microarray, were found to change significantly (FDR<0.05- and >1.85-fold change) in at least one experiment (Table III). This second round of experimentation is referred to below as ‘12k', reflecting the number of genes on the microarray.
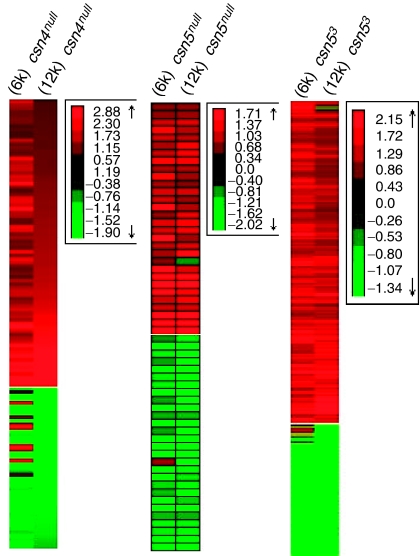
We first determined the overlap between differentially expressed genes detected in the 6k and 12k experiments. The genes on the 6k microarray used in the first round of experimentation are also present on the 12k microarray used in the second round of experimentation. Using a strict cutoff of FDR <0.05 for both sets of experiments, ∼60% of the genes detected in the first round of experimentation were also detected in the second round. This is the minimal overlap based on genes with >1.85-fold change in gene expression in both experimental sets. When these limitations were relaxed such that we consider genes with >1.5-fold change if the same gene has a 1.85-fold change in the second experiment, the overlap is much greater (∼90%). As seen in Figure 1, not only are the same genes identified but also their direction of differential expression remains the same, that is genes found to be upregulated on the 6k microarray were also detected as upregulated in the 12k microarray. This overlap, based on experiments carried out at different times with Drosophila grown in different laboratories and at different times, and with different experimental designs shows a high robustness of the results.
Figure 1.
Comparison of the ‘6k' and ‘12k' microarray analysies. Heat map analysis of genes present on both chips. The results show genes with >1.85-fold differential expression and FDR<0.05 at 60 h AED in at east one of the experimental setups.
Mutations in one CSN subunit do not affect expression of other CSN subunits
As an initial validation of the arrays and experimental conditions, we examined the expression levels of genes encoding CSN subunits (Figure 2A). As expected, csn4 and csn5 are not expressed in all experiments using csn4null or csn5null, respectively. No other genes encoding CSN subunits are misregulated in these strains, nor are any of the csn genes significantly misregulated in the two hypomorphic csn5 alleles examined. In addition to showing that mutations in one CSN subunit do not significantly affect the expression of the mRNAs encoding the other subunits, these results validate the experimental setup by showing that expected down-regulated genes defined by the null mutation are detected.
Figure 2.
General expression profile analyses of csn mutants. Expression profiles of four csn mutants were analyzed. For (B) and (C), only genes in experiments using the 6k chip, with >1.85-fold differential expression and FDR<0.05 were considered. (A) Heat map of the genes encoding CSN subunits. The genes encoding CSN3, 6 and, 8 are not present on the 6k chip and are marked with the hatched box. (B) Dendrogram showing expression profile relatedness following hierarchical clustering of the expression profiles of the four mutants at three time points. (C) Venn diagram showing numbers of common differentially expressed genes between csn4null, csn5null, and csn53.
Mutations in csn4 and csn5 lead to unique but overlapping expression profiles
We first examined the overall gene expression overlap between the mutants at different time points using the first round of experiments with the 6k microarray, to determine if the phenotypic differences are manifested in transcriptome differences. The entire data set of 1362 genes was subjected to hierarchical clustering analysis. Several trends are obvious from this analysis. First, Figure 2B shows a dendrogram based on the gene expression profile clustering. The clades center on developmental time points rather than individual mutants; for example, the expression profile of csn4null at time 96 is more closely related to csn5null at 96 than it is to the profile for csn4null at time 72 or time 60. This indicates that different changes occur throughout development, but these changes are similar among the different mutants.
Second, the expression profiles of csn51 and csn53 are very closely related (>80% overlap). csn51 and csn53 are both hypomorphic alleles: csn51 is mutated at the carboxyl border of the MPN domain and csn53 contains a mutation in the middle of the MPN domain (Suh et al, 2002). However, both point mutations lead to essentially identical morphological phenotypes and similar transcriptome profiles. Because of this overlap, in some of the subsequent analyses, including the second round of microarray analysis (‘12k'), we used only one of these mutants for comparison.
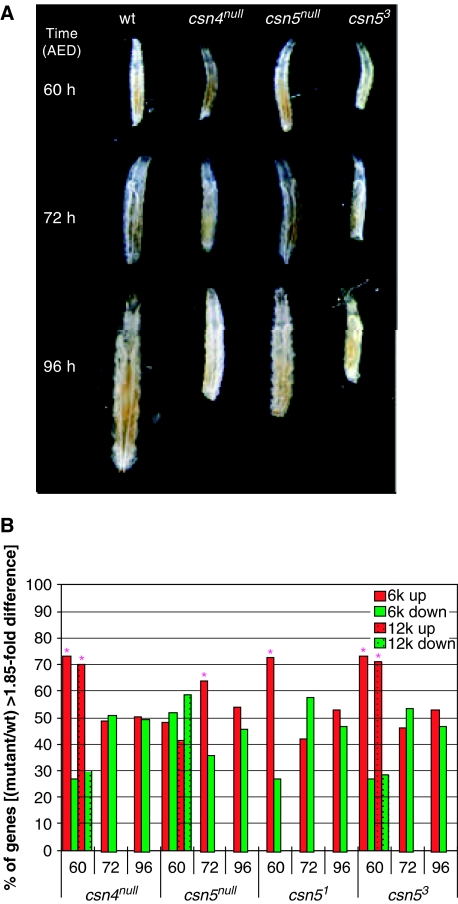
Third, the expression profiles of csn4null, csn5null, and csn53 partially overlap, consistent with each mutation affecting CSN activity leading to mutual underlying molecular effects, while subunits also have additional functions that are not shared with the entire CSN (Figure 2C). While fewer genes are misregulated in csn5null relative to the other mutants, most of the expression profile of csn5null is shared with both csn4null and csn53. The difference between csn5null and the other mutants is especially obvious at 60 h AED. At this time point, where the mutants are developmentally indistinguishable from the wt (Figure 3A), csn4null, csn51, and csn53 have >280 misregulated genes each, while csn5null shows only 75 misregulated genes (Table III).
Figure 3.
(A) Morphology of csn mutants and wt at 60, 72, and 96 h AED. (B) Distribution of up- and downregulated genes in the mutants at the time points checked. The stars designate time points with more up- than downregulated genes. Only genes with >1.85-fold differential expression and FDR<0.05 were considered.
The CSN represses temporal regulation of gene expression
We examined the distribution of up- and downregulated genes in the mutants at the different time points. As seen in Table III and Figure 3B, for mutants csn4null, csn51, and csn53, the number of up- and downregulated genes was more or less equal at 72 and 96 h AED, while at 60 h AED, in both experimental sets, there were more up- than downregulated genes. Very few genes are misregulated at all in csn5null at 60 h AED (Table III), although a similar skew toward upregulated genes is also noticed in csn5null at 72 h as opposed to 96 h AED.
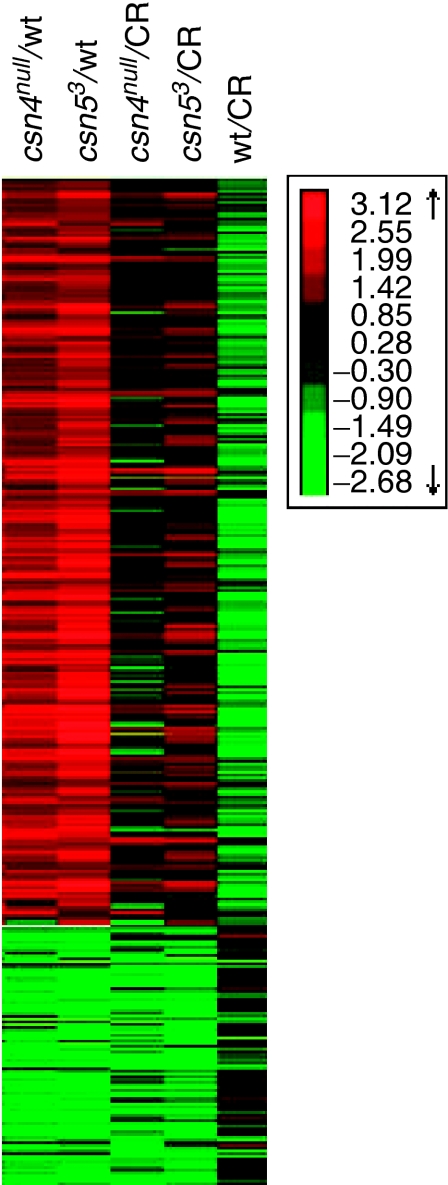
At 60 h AED, all four mutants are morphologically indistinguishable from each other and from wt in terms of body size and behavior (Figure 3A). We therefore reasoned that at this time point the underlying molecular differences between the mutant and wt larvae represent more primary effects of the mutations. We further hypothesized that similar to the role of the Arabidopsis CSN as a general repressor of environmentally induced gene expression, the prevalence of up- rather than downregulated genes in the mutants at 60 h AED indicates that the Drosophila CSN is a general repressor of various developmental/temporal cues that induce gene expression. In absence of the CSN repressor activity, as in the mutants, these genes would be expressed achronically. If this hypothesis is true, then groups of genes that are upregulated (derepressed) in the mutants at 60 h AED should be induced at different developmental stages in the wt. In other words, these genes would normally be repressed at 60 AED relative to other developmental stages (before or after 60 AED) where the transcription is induced. As the experimental setup of the 12k experiment included a common control, we could check the behavior in the wt of the genes that are misregulated at T60 in the mutants. This could not be determined in the first round of experimentation (6k) owing to experimental design. As seen in Figure 4, most of the genes that are upregulated in the mutants at T60 are normally downregulated in the wt at T60 relative to the common control.
Figure 4.
Genes upregulated in the mutants at T60 are normally downregulated in the wt. The first two columns on the left show the expression levels of genes in csn4null and csn53 relative to wt, such that the red represents genes upregulated and the green downregulated genes. The next two columns show the expression levels of genes in csn4null and csn53 relative to the common reference. The first column on the right shows the expression levels of the same genes in wt relative to the common reference. Note that most of the genes identified as upregulated in the mutants in the first two columns are green (downregulated) in the wt relative to the common reference.
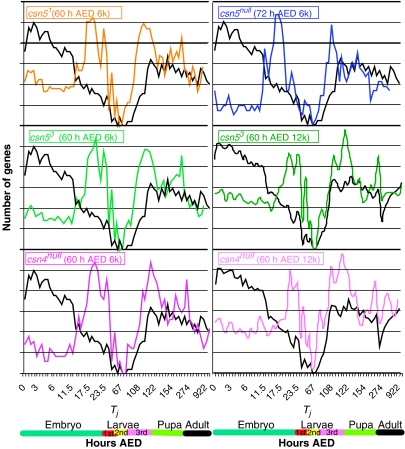
More specifically, we further hypothesized that these genes would normally be upregulated early in development and repressed at 60 AED, but not repressed in the mutants; alternatively, they would normally be upregulated later in development, but derepressed early in the mutants. To test this hypothesis, we used the available data sets from the developmental time-course expression profiling of wt Drosophila (Arbeitman et al, 2002) (Figure 5). A total of 54 and 33% of the genes in the 6k and 12k micrroarrays respectively that are upregulated in the csn mutants at 60 h AED are present in the Arbeitman et al (2002) data set. Briefly, we searched for the normal time of induction of genes that were derepressed in the csn mutants at 60 AED. We mined for time points (T) at which the misregulated genes (g) at 60 h AED (T60) in the csn mutants fit the following criterion: gnwt(Tj)>2gnwt(T60), where Tj is the normal time of induction of a gene n that is upregulated at T60 in the mutant. Of the genes upregulated in the mutants at T60, >85% fit the requirements above, that is, are upregulated in the wt at a different developmental time relative to T60 (Figure 6A). Similarly, of the genes upregulated in csn5null at T72, >85% fit the same requirements. A total of 81% of the genes identified as time shifted in the 6k experiment were also identified in the 12 k experiment (Supplementary Table I).
Figure 5.
Overview of time-shift analysis, comparing the data presented here with that shown in Arbeitman et al (2002).
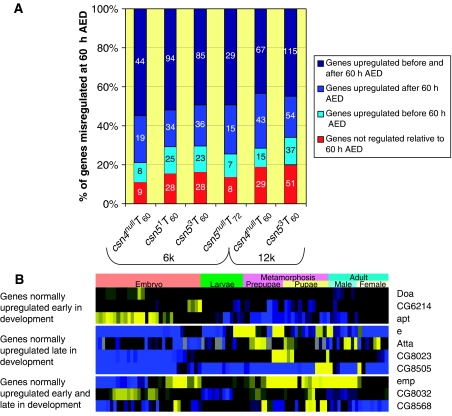
Figure 6.
Genes upregulated at 60 h AED in the mutants are putatively time shifted. (A) Percentage of upregulated genes that are putatively time shifted in the first (6k) and second (12k) rounds of microarray analysis, and their predicted normal time of induction. The number in each bar is absolute number of genes. (B) Snapshot of wt developmental time-course data (Based on data in Arbeitman et al, 2002) for genes putatively time shifted in the csn mutants. Representative time-shifted genes that were identified in both the first and second sets of experimentation are shown for each category (see Supplementary information for complete lists). Yellow shows upregulation while blue shows downregulation.
Figure 6B shows the normal expression pattern in the wt of selected genes that are upregulated at T60 in the mutants. Three classes of genes are identified. The first class comprises genes normally upregulated before T60. A clear example here is apontic (apt), which encodes a Myb–homeodomain transcription factor involved in a number of early developmental processes including heart and central nervous system development. apt is strongly induced during embryogenesis and repressed in later development. apt levels are highly induced in csn4null, csn51, and csn53 at T60. Interestingly, although apt is not induced in csn5null at T60, it is induced in this mutant at T72, providing further evidence for a delay in phenotypic development in csn5null relative to the other mutants. The second class contains genes that are normally upregulated later in development, as seen in CG8505. CG8505 encodes a structural component of the cuticle and is normally induced only during late metamorphosis. CG8505 levels are increased in csn4null, csn51, and csn53 at T60. The third class of genes is induced in both early and late development, relative to T60. An example of this class, epithelial membrane protein (emp) is involved in apoptosis connected to autophagy and defense responses. emp is normally induced in late embryogenesis and during metamorphosis, but is induced in csn4null, csn51, and csn53 at T60, and in csn5null at T72.
To determine if CSN-dependent repression of gene expression is time specific, we plotted the number of genes identified in each time point as a function of Tj (Figure 7). As a control, we sampled random groups of genes from the data set. In agreement with our hypothesis, we find that transcript levels of genes upregulated in the mutants normally increase in the wt predominantly at one of two time windows—either during late embryogenesis (16–24 h) or late larval/early metamorphosis (96–126 h). The transcript levels of genes upregulated in csn5null at T72 normally increase in the wt also in these same two time windows. Random sampling indicated that these windows are enriched above a 95% confidence interval, and shows that in the wt, the largest group of genes is normally induced, relative to T60, very early in development (<12 h AED).
Figure 7.
Time-shift analysis. For each mutant, the wt expression levels of putatively time-shifted genes identified in Figure 6A were compared relative to their expression levels at T60 such that at T60, the relative value=1 (base line). The y-axis shows the number of genes upregulated (e.g. relative value >1) at a particular time point. The black lines represent the average of >100 random gene groups that were sampled. Note that for csn5null, the analysis was carried out using T72. The total number of genes in each analysis is different as detailed in Figure 6A and is presented here as normalized.
The ‘time-shifted' genes were analyzed for two GO criteria, ‘biological process' and ‘molecular function' (Table IV). Several GO definitions are statistically more prevalent among the time-shifted genes. While a more detailed GO analysis of the entire data set will be presented elsewhere, several trends are obvious from this initial analysis. First, >70% of the ‘time-shifted' genes with a known GO definition are clearly involved in some aspect of development or signaling (Supplementary Table I). Second, several of the GO groups identified as significantly enriched are closely associated with cellular stress responses including oxidoreductase activity, electron transport and the antibacterial humeral response. Indeed, this last group is especially interesting in light of the melanotic tumor phenotype of csn5null (Oron et al, 2002) and the implication of Arabidopsis CSN in plant defense responses (Azevedo et al, 2002; Liu et al, 2002). Consistent with problems in molting (see Figure 8), genes involved in cuticle formation (chitin metabolism and binding) are also enriched in the time-shifted genes. One other group worth mentioning are genes involved in amine metabolism. These genes could potentially affect a wide range of responses, including timing of gene expression, through their essential function in dopamine biosynthesis (De Luca et al, 2003; Jordan et al, 2006).
Table 4.
Gene ontology classifications for ‘time-shifted' genes
| GO category | No. of genes (array) | % of array | No. of genes (time shift) | % of time-shifted genes | P-value | |
|---|---|---|---|---|---|---|
| Biological process | ||||||
| 6k | Chitin metabolism | 23 | 0.44 | 6 | 4.26 | 6.75E-06 |
| N-acetylglucosamine metabolism | 25 | 0.48 | 6 | 4.26 | 3.66E-05 | |
| Glucosamine metabolism | 25 | 0.48 | 6 | 4.26 | 4.60E-05 | |
| Amino sugar metabolism | 25 | 0.48 | 6 | 4.26 | 4.60E-05 | |
| Amine metabolism | 111 | 2.13 | 12 | 8.51 | 1.21E-04 | |
| Cellular polysaccharide metabolism | 31 | 0.60 | 6 | 4.26 | 1.31E-04 | |
| Electron transport | 113 | 2.17 | 11 | 7.80 | 1.63E-04 | |
| Nitrogen compound metabolism | 115 | 2.21 | 12 | 8.51 | 1.63E-04 | |
| Polysaccharide metabolism | 31 | 0.60 | 6 | 4.26 | 2.30E-04 | |
| Antibacterial humoral response | 3 | 0.06 | 2 | 1.42 | 3.32E-03 | |
| Cellular carbohydrate metabolism | 103 | 1.98 | 8 | 5.67 | 5.28E-03 | |
| 12k | Electron transport | 203 | 5.86 | 18 | 20.22 | 5.58E-06 |
| Antibacterial humoral response | 13 | 0.59 | 5 | 10.64 | 2.25E-05 | |
| Amine metabolism | 178 | 4.54 | 15 | 14.56 | 9.17E-05 | |
| Catecholamine metabolism | 7 | 0.32 | 3 | 6.38 | 9.29E-04 | |
| Response to starvation | 2 | 0.06 | 2 | 2.25 | 3.60E-03 | |
| Amino-acid metabolism | 105 | 3.5 | 8 | 11.27 | 4.04E-03 | |
| Chitin metabolism | 52 | 2.36 | 5 | 10.64 | 5.98E-03 | |
| Nitrogen compound biosynthesis | 27 | 0.78 | 4 | 4.49 | 6.89E-03 | |
| N-acetylglucosamine metabolism | 57 | 2.59 | 5 | 10.64 | 8.54E-03 | |
| Glucosamine metabolism | 57 | 1.9 | 5 | 7.04 | 1.34E-02 | |
| Cellular polysaccharide metabolism | 62 | 2.07 | 5 | 7.04 | 1.83E-02 | |
| Polysaccharide metabolism | 62 | 1.79 | 5 | 5.62 | 2.51E-02 | |
| Molecular function | ||||||
| 6k | Pattern binding | 21 | 0.40 | 7 | 4.96 | 7.71E-06 |
| Oxidoreductase activity | 207 | 3.97 | 19 | 13.47 | 1.00E-05 | |
| Carbohydrate binding | 33 | 0.63 | 8 | 5.67 | 1.22E-05 | |
| Iron ion binding | 64 | 1.22 | 10 | 7.09 | 2.64E-05 | |
| Polysaccharide binding | 18 | 0.34 | 6 | 4.25 | 4.57E-05 | |
| Monooxygenase activity | 39 | 0.74 | 8 | 5.67 | 4.65E-05 | |
| Chitin binding | 16 | 0.30 | 5 | 3.54 | 3.97E-04 | |
| Tetrapyrrole binding | 34 | 0.65 | 6 | 4.25 | 7.13E-04 | |
| Heme binding | 34 | 0.65 | 6 | 4.25 | 8.97E-04 | |
| Oxidoreductase activity, acting on CH-OH group of donors | 27 | 0.53 | 5 | 3.55 | 2.04E-03 | |
| 12k | Oxidoreductase activity | 340 | 5.83 | 32 | 19.75 | 4.21E-06 |
| Monooxygenase activity | 63 | 1.69 | 13 | 11.5 | 1.48E-04 | |
| Iron ion binding | 108 | 4.27 | 17 | 19.54 | 1.76E-04 | |
| Tetrapyrrole binding | 58 | 1 | 11 | 6.79 | 7.17E-04 | |
| Heme binding | 58 | 1.55 | 11 | 9.73 | 1.14E-03 | |
| Structural constituent of cuticle | 40 | 0.69 | 6 | 3.7 | 3.44E-01 | |
| Oxidoreductase activity, acting on CH-OH group of donors | 45 | 1.21 | 6 | 5.31 | 7.45E-01 | |
| Pattern binding | 51 | 0.88 | 5 | 3.09 | 1.71E-02 | |
The entire group of ‘time-shifted' genes identified in Figure 6 were subjected to GO analysis using Babilomics FatiGO (Al-Shahrour et al, 2006). Only significantly enriched GO groups are shown.
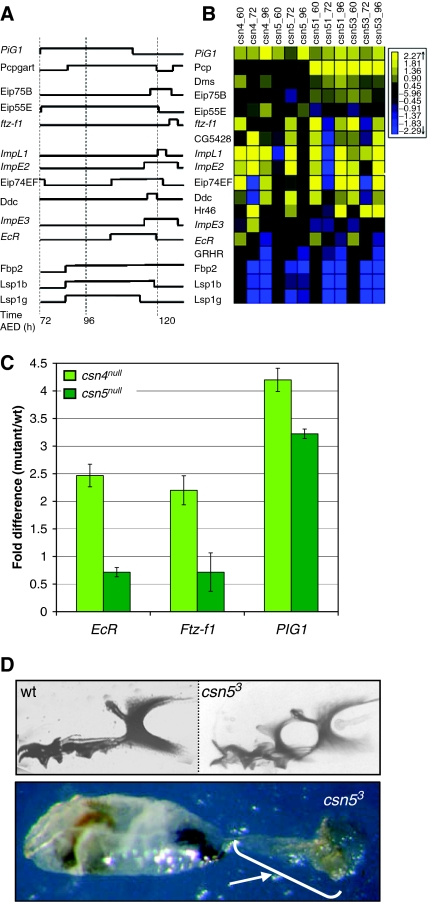
Figure 8.
Ecdysone-regulated genes are misexpressed in csn mutants. (A) wt expression pattern of ecdysone-related genes during larval development (adapted from Andres et al (1993). (B) Heat map of expression levels of the same genes in (A), plus additional genes. (C) Expression at 60 h AED of three ecdysone-regulated genes (EcR, βFtz-1, and PIG1) in two CSN mutants (csn4null and csn5null) was validated using QRT-PCR. Each bar shows the relative mRNA expression of mutant versus wt with associated standard errors. Data shown are averages of quadruplicate QRT-PCR measurements. Rp49 was used for normalization. (D) csn53 mutant L3 larvae contain a double set of mouth hooks. The top two panels show mouth hooks from wt (left) and csn53 (right) larvae dissected following the second molt (∼80 h AED). The bottom panel illustrates that csn53 larva are unable to undergo the second/third instar molt, resulting in an L2 cuticle (designated by the arrow) attached through the L2 mouth parts to the L3 mouth parts.
Ecdysone-regulated genes are misregulated in csn mutants
In light of the abnormal molting phenotype of csn4null (Oron et al, 2002) and that CSN2 was shown to interact with Drosophila Ecdysone receptor (EcR) (Dressel et al, 1999), we previously suggested that the CSN is involved in steroid hormone signaling in Drosophila development (Oron et al, 2002). To further explore this hypothesis, we examined groups of genes present on the 6k microarray that had been implicated in ecdysone action (Andres et al, 1993). Indeed, a group of genes implicated in ecdysone signaling are misexpressed in csn mutants (Figure 8B).
All mutants analyzed showed misregulation of this group of genes. However, although these genes are misexpressed in both csn4 and csn5 mutants, we find a difference in the onset of their misexpression. While in csn4null and the csn5 hypomorphs, a majority of these genes show abnormal expression already at 60 h AED, in csn5null, the misexpression is evident only at 72 h AED, as seen for Ftz-f1, ImpL1, and ImpE2 (Figure 8B). This raises the possibility that the reason that csn5null does not display the molting phenotypes clearly observed in csn4null is not because CSN5 and CSN4 regulate different pathways, as had been proposed, but rather because of different temporal disruption of the same pathways. That is, the loss of both CSN4 and CSN5 results in deregulation of ecdysone signaling, but this deregulation manifests earlier in csn4null than in csn5null.
To validate these results, RNA was isolated from larvae at 60 h AED from newly established cultures of each mutant and the wt and was used as a template for quantitative reverse transcription–PCR (RT–PCR) (Figure 8C). The transcript levels of three representative genes, EcR and ftz transcription factor 1 (ftz-f1), whose gene products physically interact with the CSN2 (Dressel et al, 1999) and Preintermolt Gene 1 (pig1) were found to be upregulated relative to the wt in csn4null at 60 AED and downregulated in csn5null, confirming the microarray results.
As the transcriptome of the csn5 hypomorphic mutants was similar to that of csn4null in general (Figure 2) and specifically with regard to the timing of ecdysone-related genes, we further examined these mutants for morphological changes. As seen in Figure 8D, csn53 larvae at 72 h AED display double mouth hook phenotypes similar to those seen in csn4null (Oron et al, 2002). This phenotype was not observed in csn5null. Thus, from the transcriptome results, we succeeded in predicting a morphological change that was confirmed experimentally.
Discussion
We present here the first genomic analysis of animal mutants in the COP9 signalosome. Using two different experimental setups, entailing two different cDNA arrays, different controls, and carried out with strains grown in different labs and at different times, we have obtained similar results indicating that (1) the CSN appears to act as a transcriptional repressor at 60 h AED, (2) the genes repressed by the CSN are normally induced at earlier or later developmental time points, (3) CSN5 carries only part of CSN functions, and (4) the hypomorphic mutations in csn5 are more severe than the null mutation.
A major difficulty in analysis of mutants in the COP9 signalosome is determining which of the many phenotypes arise directly from the lesions in the CSN, and which are pleiotropic downstream effects. Indeed, the severe phenotypes of mutants in the CSN in Drosophila, mouse, and Arabidopsis all converge on death. But mutations in CSN itself are not likely the direct cause of death, as null mutations in the CSN in several organisms including S. pombe, Caenorhabditis elegans, and Aspergillus nidulans are not lethal (Mundt et al, 1999; Maytal-Kivity et al, 2002; Busch et al, 2003). That mutation in CSN leads to accumulating downstream effects can be seen in the transcription profiles of later developmental stages. The number of misregulated genes increases with time in all mutants and the overlap between the mutants also increases. This is likely due to each mutant approaching developmental arrest and subsequent death.
Can such analysis help us in elucidating the contribution of individual subunits to CSN function? Comparison of the profiles of the null mutants in csn4 and csn5 with the point mutations in csn5 can help answer this question. While fewer genes are misregulated in csn5null relative to the other mutants, about two-thirds of the expression profile of csn5null is shared with csn4null, csn51, and csn53. This suggests that part of the molecular phenotype of csn4null, csn51, and csn53 is due to the absence of CSN5 activity, presumably its deneddylase activity. However, as CSN4 is essential for CSN complex integrity, while CSN5 is not (Oron et al, 2002; Dohmann et al, 2005), the additional genes misregulated in csn4null could represent functions dependent on the entire CSN complex (which remains intact in csn5null) but not connected to CSN5-mediated deneddylation. As the overlap between csn4null, and csn51, and csn53 extends beyond those genes shared with csn5null, that is, that these mutants affect the expression of genes not dependent on CSN5 itself, this suggests that the point mutations in csn5 affect the entire CSN complex, leading to a molecular and physical phenotype that mimics loss of the entire complex.
As the Arabidopsis CSN is a negative regulator of transcription factor stability, we hypothesized that a lack of CSN activity in Drosophila would lead to transcription factor stabilization and increase in target gene activation. The first evidence supporting this was the larger number of up- than downregulated genes early in development before any obvious morphological difference between the mutants and wt. This enrichment of upregulated genes was not detected at later time points, further indicating that the transcription profiles at these times reflect mainly secondary effects of loss of CSN function. As a number of transcription factors are induced at T60 in the mutants (e.g. dei1, ken, Mef2, ftz-f1, bun, Rel, and apt), it is likely that some of the expression profile of the mutants in later development is due to induction of target genes for these transcription factors.
Comparison of the upregulated genes with the developmental transcriptional data provided further evidence for the CSN as a repressor of developmentally regulated transcription. The time-shift analysis developed here revealed that the vast majority of the genes upregulated in the csn mutants early in development are normally induced at other specific stages. The largest group of genes is normally induced at late embryogenesis. That lack of CSN function should predominantly affect genes regulated at late embryogenesis is especially interesting considering that CSN subunits are maternally contributed. Both CSN4 and CSN5 are maternally contributed to the egg and this contribution abates by late embryogenesis (Oron et al, 2002). The overlap between loss of maternally contributed CSN and normal induction pattern of misregulated genes suggests that the increase in gene derepression reflects a primary response caused by the mutations.
Maternally contributed CSN is a potential stumbling block in the analysis of our results. The use of germline clones to remove the maternal contribution was not useful as this resulted in similar oogenic arrest at stages 5–6 for csn4null, csn5null, and csn51 (Oron et al, 2002). While maternally contributed CSN is depleted by late embryogenesis, we do not know the exact timing of this depletion, and indeed, a slight difference in persistence of the maternally contributed subunit could have drastic developmental effects. Certainly, exact timing of changes in transcriptional regulation has major phenotypic consequences. For example, while both csn4null and csn5null show misregulation of ecdysone-associated genes, only csn4null shows an ecdysone-related phenotype, probably due to the earlier appearance of misregulation of these genes.
However, if differences in maternal contribution were the major cause of the phenotypic differences, we would expect that maternally contributed CSN5 would persist in the null background as in the hypomorphic backgrounds, leading to similar timing of phenotypic changes, which is not the case. Alternatively, the csn5null mutant larvae display mutant phenotypes later than the hypomorphic csn5 and csn4null mutants not because of the difference in maternal contribution but rather because of the persistence of a CSN5-less CSN complex, which we hypothesize maintains partial function. csn4null larvae lack the entire CSN complex and thus this partial function would be missing. Apparently, the mutated CSN5 protein in the hypomorphic alleles also disrupts CSN function.
This possibility is supported by the morphological and transcriptome phenotypes of csn51 and csn53, which are much more similar to csn4null than csn5null. These mutants maintain a CSN that contains the mutated CSN5 (Oron et al, 2002), but apparently the point mutations in CSN5 have a negative effect on the entire CSN, such that a mutated CSN5 in the complex is worse than a lack of CSN5. However, this is not a classic dominant-negative effect, as heterozygotes for the point mutation (i.e. csn51/+) are completely wt in phenotype, consistent with a hypomorph, and any effect is only seen once the normal CSN is not present. Biochemically, while these point mutations do not affect CSN complex stability, they do lack CSN-mediated Cul1 deneddylation activity, similar to the null mutants (not shown), suggesting some type of neomorphic action, and influence the stability of the complex-independent forms of other CSN subunits. For example, these mutants lack the complex-independent forms of both CSN5 and CSN7 (Oron et al, 2002). We thus conclude that the phenotypes detected are likely consequences of primary lesions and not simply mistiming of maternal genes.
This has major implications for our understanding of CSN function. The most celebrated function of the CSN is as a deneddylase or deubiquitinase. Both functions are dependent on CSN5, which carries the catalytic activity (Cope et al, 2002). However, if these were the only, or even most central, roles for the CSN, we would expect that the transcriptome phenotype of the csn5null mutant would be very similar to that of the csn4null mutant. As csn4null and the hypomorphic csn5 mutants affect more genes, we conclude that the entire CSN has additional functions that are not dependent on CSN5.
In trying to understand the specific roles of individual subunits, we looked at transcriptome overlap. The lack of complete overlap between mutants could be due to several factors: (1) it could represent true differences between mutants; (2) it could represent differences in the strains not connected to the CSN lesion; and (3) it could be a result of inherent error in the transcriptome analysis. While our results discussed above clearly point to phenotypic differences arising from differences in effects on timing of gene expression, finding clear and specific differences in gene regulation aside from timing between the mutants is not trivial. When the experiments were repeated with different controls and with strains grown at different times and places, we found a very large overlap between profiles for the same mutants, indicating a robustness of our results. However, the drift in results is large enough to explain a major part of the transcriptome differences between any two mutants. Determining subunit-specific effects will necessitate an expanded analysis that includes more mutants, to find out specific differences between CSN subunits.
Materials and methods
Fly stocks and growth conditions
wt and mutant strains were maintained on standard medium and all experiments were performed at 25°C. Fly stocks: w1118 was used as wt. Three mutant alleles of csn5 and one of csn4 were utilized. csn5null and csn4null were generated by imprecise P-element excision, resulting in protein-null mutations (Freilich et al, 1999; Oron et al, 2002). csn51 and csn53 alleles were generated by EMS mutagenesis and encode the missense mutations E160V and T100I respectively (Suh et al, 2002). csn5 and csn4 mutations were balanced over TM3, Ser GFP and CyO, GFP respectively, to permit sorting of homozygous mutant larvae.
Transcriptional profiling
For all microarray experiments, larvae were staged from egg deposition, collected in 2-h time windows following hatching and raised at a density of 50 per vial at 25°C. Age-matched mutants and wt larvae were collected at 60, 72, and 96 h time points AED. Total RNA was extracted with TRIzol reagent (Invitrogen) followed by RNeasy (Qiagen) clear up. cRNA targets were generated and coupled to either Cy3 or Cy5 fluorophores according to ‘FHCRC Genomics Resource DNA Array Laboratory' protocols. Hybridization and scanning were performed by the FHCRC Genomics Resource DNA Array Laboratory. Expression profiles were performed using spotted microarrays (GEO GPL4285=‘6k' and GEO GPL1908=‘12k'). Microarray images were quantified using GenePix Pro software (Axon Instruments). Statistical analysis was conducted in R (R 2.2.1—The R Development Core Team). Raw expression readings were log-transformed using the R VSN package (Huber et al, 2002); within-array expression measurement biases were removed by applying LOWESS normalization to expression log ratios; and the FDR was controlled by computing FDR adjusted P-values (Reiner et al, 2003) to test for differential expression of each gene in each CSN mutant and comparing them to 0.05. Data were generated from four or five independent replicates (two with one dye orientation and two/three with the reversed dye orientation).
Heat maps and clustering analysis were performed with EXPANDER (EXPression Analyzer and DisplayER) gene expression analysis and visualization tool Version 2 (Shamir et al, 2005). GO analysis employed Babelomics (Al-Shahrour et al, 2006).
Time-shift analysis
The definition and mathematical properties of a ‘time shift' as well as the algorithms and heuristics employed are described in detail in Tuller et al (2005).
The expression data have been deposited in the GEO database under accession number GSE7303.
Quantitative RT–PCR
Quantitative real-time reverse transcription–PCR (qPCR) reactions were carried out to validate changes in expression of three ecdysone-related genes (EcR, Ftz-f1, and PIG1). RNA was extracted from w1118, csn4null, and csn5null larvae as described above. cDNA synthesis was carried out using Superscript II (Invitrogen).
Four qPCR primer pairs were designed to generate intron-spanning products using Primer Express Version 1.5 software:
EcR: (F) 5′-GCCACTATTCCGCTACTACCTGA-3′ (R) 5′-ATATAACGGCCAACTGATTGTACG-3′; Ftz-f1: (F) 5′-GATCTGAAGGTCGACGACCAA-3′ (R) 5′-CGTTATGGATTCGATGATGCAG-3′; PIG1: (F) 5′-GAGTCCAGTGCCACGAACAGT-3′ (R) 5′-CGGGAACCATCGTCCACATC-3′; and RP49: (F) 5′-TAAGCTGTCGCACAAATGGC-3′ (R) 5′-ACCGATGTTGGGCATCAGATA-3′.
qPCR analysis was performed on an ABI Prism® 7000 Sequence Detection System (Applied Biosystems) using SYBR Green® I chemistry. Samples were in triplicate and normalized for RNA levels based on rp49 expression. Analysis was performed with the ABI Prism 7000 SDS software RQ study Application v1.1 using the Δ−ΔCt method, which determines fold changes in gene expression relative to a comparative sample (age matched w1118).
Supplementary Material
Supplementary Table I
Acknowledgments
This work would not have been completed without the invaluable assistance of Roni Elkon and Professor Ron Shamir in adapting Expander for use with Drosophila and The Genomics Resources Center at the Fred Hutchinson Cancer Research Center. We thank Professor Bernard Epel for important comments in developing this project and Ronen Globinski for assistance in organizing microarray data. This work was supported by grants from the Israel Academy of Sciences (to DAC), NIH R01 GM51186 (to BAE), and the Manna Center for Plant Biosciences and Constantiner Institute of Molecular Genetics (to EO).
References
- Akiyama H, Fujisawa N, Tashiro Y, Takanabe N, Sugiyama A, Tashiro F (2003) The role of transcriptional corepressor Nif3l1 in early stage of neural differentiation via cooperation with Trip15/CSN2. J Biol Chem 8: 8. [DOI] [PubMed] [Google Scholar]
- Al-Shahrour F, Minguez P, Tarraga J, Montaner D, Alloza E, Vaquerizas JM, Conde L, Blaschke C, Vera J, Dopazo J (2006) BABELOMICS: a systems biology perspective in the functional annotation of genome-scale experiments. Nucleic Acids Res 34: W472–W476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andres AJ, Fletcher JC, Karim FD, Thummel CS (1993) Molecular analysis of the initiation of insect metamorphosis: a comparative study of Drosophila ecdysteroid-regulated transcription. Dev Biol 160: 388–404 [DOI] [PubMed] [Google Scholar]
- Arbeitman MN, Furlong EE, Imam F, Johnson E, Null BH, Baker BS, Krasnow MA, Scott MP, Davis RW, White KP (2002) Gene expression during the life cycle of Drosophila melanogaster. Science 297: 2270–2275 [DOI] [PubMed] [Google Scholar]
- Azevedo C, Sadanandom A, Kitagawa K, Freialdenhoven A, Shirasu K, Schulze-Lefert P (2002) The RAR1 interactor SGT1, an essential component of R gene-triggered disease resistance. Science 14: 14. [DOI] [PubMed] [Google Scholar]
- Bech-Otschir D, Kraft R, Huang X, Henklein P, Kapelari B, Pollmann C, Dubiel W (2001) COP9 signalosome-specific phosphorylation targets p53 to degradation by the ubiquitin system. EMBO J 20: 1630–1639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busch S, Eckert SE, Krappmann S, Braus GH (2003) The COP9 signalosome is an essential regulator of development in the filamentous fungus Aspergillus nidulans. Mol Microbiol 49: 717–730 [DOI] [PubMed] [Google Scholar]
- Chamovitz DA, Wei N, Osterlund MT, von Arnim AG, Staub JM, Matsui M, Deng XW (1996) The COP9 complex, a novel multisubunit nuclear regulator involved in light control of a plant developmental switch. Cell 86: 115–121 [DOI] [PubMed] [Google Scholar]
- Cope G, Deshaies RJ (2003) COP9 signalosome: a multifunctional regulator of SCF and other cullin-based ubiquitin ligases. Cell 114: 663–671 [DOI] [PubMed] [Google Scholar]
- Cope GA, Suh GS, Aravind L, Schwarz SE, Zipursky SL, Koonin EV, Deshaies RJ (2002) Role of predicted metalloprotease motif of Jab1/Csn5 in cleavage of NEDD8 from CUL1. Science 15: 15. [DOI] [PubMed] [Google Scholar]
- De Luca M, Roshina NV, Geiger-Thornsberry GL, Lyman RF, Pasyukova EG, Mackay TF (2003) Dopa decarboxylase (Ddc) affects variation in Drosophila longevity. Nat Genet 34: 429–433 [DOI] [PubMed] [Google Scholar]
- Deng XW, Dubiel W, Wei N, Hofmann K, Mundt K, Colicelli J, Kato J, Naumann M, Segal D, Seeger M, Carr A, Glickman M, Chamovitz DA (2000) Unified nomenclature for the COP9 signalosome and its subunits: an essential regulator of development [letter] [published erratum appears in Trends Genet 2000 Jul;16(7):289]. Trends Genet 16: 202–203 [DOI] [PubMed] [Google Scholar]
- Dohmann EM, Kuhnle C, Schwechheimer C (2005) Loss of the CONSTITUTIVE PHOTOMORPHOGENIC9 signalosome subunit 5 is sufficient to cause the cop/det/fus mutant phenotype in Arabidopsis. Plant Cell 17: 1967–1978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doronkin S, Djagaeva I, Beckendorf SK (2002) CSN5/Jab1 mutations affect axis formation in the Drosophila oocyte by activating a meiotic checkpoint. Development 129: 5053–5064 [DOI] [PubMed] [Google Scholar]
- Dressel U, Thormeyer D, Altincicek B, Paululat A, Eggert M, Schneider S, Tenbaum SP, Renkawitz R, Baniahmad A (1999) Alien, a highly conserved protein with characteristics of a corepressor for members of the nuclear hormone receptor superfamily. Mol Cell Biol 19: 3383–3394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freilich S, Oron E, Kapp Y, Nevo-Caspi Y, Orgad S, Segal D, Chamovitz DA (1999) The COP9 signalosome is essential for development of Drosophila melanogaster. Curr Biol 9: 1187–1190 [DOI] [PubMed] [Google Scholar]
- Fukumoto A, Tomoda K, Kubota M, Kato JY, Yoneda-Kato N (2005) Small Jab1-containing subcomplex is regulated in an anchorage- and cell cycle-dependent manner, which is abrogated by ras transformation. FEBS Lett 579: 1047–1054 [DOI] [PubMed] [Google Scholar]
- Groisman R, Polanowska J, Kuraoka I, Sawada J, Saijo M, Drapkin R, Kisselev AF, Tanaka K, Nakatani Y (2003) The ubiquitin ligase activity in the DDB2 and CSA complexes is differentially regulated by the COP9 signalosome in response to DNA damage. Cell 113: 357–367 [DOI] [PubMed] [Google Scholar]
- Harari-Steinberg O, Cantera R, Denti S, Bianchi E, Oron E, Segal D, Chamovitz DA (2007) COP9 signalosome subunit 5 (CSN5) regulates the development of Drosophila immune system: effects on cactus, Dorsal and hematopoiesis. Genes Cells 12: 183–195 [DOI] [PubMed] [Google Scholar]
- Harari-Steinberg O, Chamovitz DA (2004) The COP9 signalosome: mediating between kinase signaling and protein degradation. Curr Protein PeptSci 5: 185–189 [DOI] [PubMed] [Google Scholar]
- Huber W, von Heydebreck A, Sultmann H, Poustka A, Vingron M (2002) Variance stabilization applied to microarray data calibration and to the quantification of differential expression. Bioinformatics 18 Suppl 1: S96–S104 [DOI] [PubMed] [Google Scholar]
- Jordan K, Morgan T, Mackay TF (2006) Quantitative trait loci for locomotor behavior in Drosophila melanogaster. Genetics 174: 271–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwok SF, Piekos B, Misera S, Deng XW (1996) A complement of ten essential and pleiotropic arabidopsis COP/DET/FUS genes is necessary for repression of photomorphogenesis in darkness. Plant Physiol 110: 731–742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwok SF, Staub JM, Deng XW (1999) Characterization of two subunits of Arabidopsis 19S proteasome regulatory complex and its possible interaction with the COP9 complex. J Mol Biol 285: 85–95 [DOI] [PubMed] [Google Scholar]
- Liu C, Powell KA, Mundt K, Wu L, Carr AM, Caspari T (2003) Cop9/signalosome subunits and Pcu4 regulate ribonucleotide reductase by both checkpoint-dependent and -independent mechanisms. Genes Dev 14: 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Schiff M, Serino G, Deng X-W, Dinesh-Kumar SP (2002) Role of SCF ubiquitin-ligase and the COP9 signalosome in the N gene-mediated resistance response to tobacco mosaic virus. Plant Cell 14: 1483–1496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyapina S, Cope G, Shevchenko A, Serino G, Tsuge T, Zhou C, Wolf DA, Wei N, Deshaies RJ (2001) Promotion of NEDD-CUL1 conjugate cleavage by COP9 signalosome. Science 292: 1382–1385 [DOI] [PubMed] [Google Scholar]
- Lykke-Andersen K, Schaefer L, Menon S, Deng X-W, Miller JB, Wei N (2003) Disruption of the COP9 signalosome Csn2 subunit in mice causes deficient cell proliferation, accumulation of p53 and cyclin E, and early embryonic death. Mol Cell Biol 23: 6790–6797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L, Zhao H, Deng XW (2003) Analysis of the mutational effects of the COP/DET/FUS loci on genome expression profiles reveals their overlapping yet not identical roles in regulating Arabidopsis seedling development. Development 130: 969–981 [DOI] [PubMed] [Google Scholar]
- Maytal-Kivity V, Pick E, Piran R, Hofmann K, Glickman MH (2003) The COP9 signalosome-like complex in S. cerevisiae and links to other PCI complexes. Int J Biochem Cell Biol 35: 706–715 [DOI] [PubMed] [Google Scholar]
- Maytal-Kivity V, Piran R, Pick E, Hofmann K, Glickman MH (2002) COP9 signalosome components play a role in the mating pheromone response of S. cerevisiae. EMBO Rep 3: 1215–1221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mundt KE, Liu C, Carr AM (2002) Deletion mutants in COP9/signalosome subunits in fission yeast Schizosaccharomyces pombe display distinct phenotypes. Mol Biol Cell 13: 493–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mundt KE, Porte J, Murray JM, Brikos C, Christensen PU, Caspari T, Hagan IM, Millar JB, Simanis V, Hofmann K, Carr AM (1999) The COP9/signalosome complex is conserved in fission yeast and has a role in S phase. Curr Biol 9: 1427–1430 [DOI] [PubMed] [Google Scholar]
- Naumann M, Bech-Otschir D, Huang X, Ferrell K, Dubiel W (1999) COP9 signalosome-directed c-Jun activation/stabilization is independent of JNK. J Biol Chem 274: 35297–35300 [DOI] [PubMed] [Google Scholar]
- Nielsen O (2003) COP9 signalosome: a provider of DNA building blocks. Curr Biol 13: R565–R567 [DOI] [PubMed] [Google Scholar]
- Oron E, Mannervik M, Rencus S, Harari-Steinberg O, Neuman-Silberberg S, Segal D, Chamovitz DA (2002) COP9 signalosome subunits 4 and 5 regulate multiple pleiotropic pathways in Drosophila melanogaster. Development 129: 4399–4409 [DOI] [PubMed] [Google Scholar]
- Osterlund MT, Ang LH, Deng XW (1999) The role of COP1 in repression of Arabidopsis photomorphogenic development. Trends Cell Biol 9: 113–118 [DOI] [PubMed] [Google Scholar]
- Peng Z, Shen Y, Feng S, Wang X, Chitteti BN, Vierstra RD, Deng XW (2003) Evidence for a physical association of the COP9 signalosome, the proteasome, and specific SCF E3 ligases in vivo. Curr Biol 13: 504–505 [DOI] [PubMed] [Google Scholar]
- Reiner A, Yekutieli D, Benjamini Y (2003) Identifying differentially expressed genes using false discovery rate controlling procedures. Bioinformatics 19: 368–375 [DOI] [PubMed] [Google Scholar]
- Schwechheimer C (2004) The COP9 signalosome (CSN): an evolutionary conserved proteolysis regulator in eukaryotic development. Biochim Biophys Acta 1695: 45–54 [DOI] [PubMed] [Google Scholar]
- Seeger M, Kraft R, Ferrell K, Bech-Otschir D, Dumdey R, Schade R, Gordon C, Naumann M, Dubiel W (1998) A novel protein complex involved in signal transduction possessing similarities to 26S proteasome subunits. FASEB J 12: 469–478 [PubMed] [Google Scholar]
- Serino G, Deng X-W (2003) The COP9 signalosome: regulating plant development through control of proteolysis. Annu Rev Plant Biol 54: 165–182 [DOI] [PubMed] [Google Scholar]
- Shamir R, Maron-Katz A, Tanay A, Linhart C, Steinfeld I, Sharan R, Shiloh Y, Elkon R (2005) EXPANDER—an integrative program suite for microarray data analysis. BMC Bioinformatics 6: 232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh GSB, Poeck B, Chouard T, Oron E, Segal D, Chamovitz DA, Zipursky SL (2002) Drosophila JAB1/CSN5 acts in photoreceptor cells to induce glial cells. Neuron 33: 35–46 [DOI] [PubMed] [Google Scholar]
- Tenbaum SP, Juenemann S, Schlitt T, Bernal J, Renkawitz R, Munoz A, Baniahmad A (2003) Alien/CSN2 gene expression is regulated by thyroid hormone in rat brain. Dev Biol 254: 149–160 [DOI] [PubMed] [Google Scholar]
- Tsuge T, Matsui M, Wei N (2001) The subunit 1 of the COP9 signalosome suppresses gene expression through its N-terminal domain and incorporates into the complex through the PCI domain. J Mol Biol 305: 1–9 [DOI] [PubMed] [Google Scholar]
- Tuller T, Oron E, Makavy E, Chamovitz DA, Chor B (2005) Time-window analysis of developmental gene expression data with multiple genetic backgrounds. In Algorithms in Bioinformatics, Casadio R, Myers G (eds) Vol. 3692, pp 53–64. Berlin: Springer-Verlag [Google Scholar]
- Wee S, Hetfeld B, Dubiel W, Wolf DA (2002) Conservation of the COP9/signalosome in budding yeast. BMC Genet 3: 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei N, Chamovitz DA, Deng XW (1994) Arabidopsis COP9 is a component of a novel signaling complex mediating light control of development [published erratum appears in Cell 1994 Oct 7;79(1):following 179]. Cell 78: 117–124 [DOI] [PubMed] [Google Scholar]
- Wei N, Deng XW (2003) The COP9 signalosome. Annu Rev Cell Dev Biol 19: 261–286 [DOI] [PubMed] [Google Scholar]
- Wei N, Tsuge T, Serino G, Dohmae N, Takio K, Matsui M, Deng XW (1998) Conservation of the COP9 complex between plants and mammals and its relationship to the 26S proteasome regulatory complex. Curr Biol 8: 919–922 [DOI] [PubMed] [Google Scholar]
- Wolf DA, Zhou C, Wee S (2003) The COP9 signalosome: an assembly and maintenance platform for cullin ubiquitin ligases? Nat Cell Biol 5: 1029–1033 [DOI] [PubMed] [Google Scholar]
- Yan J, Walz K, Nakamura H, Carattini-Rivera S, Zhao Q, Vogel H, Wei N, Justice MJ, Bradley A, Lupski JR (2003) COP9 signalosome subunit 3 is essential for maintenance of cell proliferation in the mouse embryonic epiblast. Mol Cell Biol 23: 6798–6808 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table I