Abstract
Although microarray analysis has provided information regarding the dynamics of gene expression during development of the mouse lung, no extensive correlations have been made to the levels of corresponding protein products. Here, we present a global survey of protein expression during mouse lung organogenesis from embryonic day E13.5 until adulthood using gel-free two-dimensional liquid chromatography coupled to shotgun tandem mass spectrometry (MudPIT). Mathematical modeling of the proteomic profiles with parallel DNA microarray data identified large groups of gene products with statistically significant correlation or divergence in coregulation of protein and transcript levels during lung development. We also present an integrative analysis of mRNA and protein expression in Nmyc loss- and gain-of-function mutants. This revealed a set of 90 positively and negatively regulated putative target genes. These targets are evidence that Nmyc is a regulator of genes involved in mRNA processing and a repressor of the imprinted gene Igf2r in the developing lung.
Keywords: development, lung, microarray, mouse, Nmyc, proteomics
Introduction
The mouse lung primordia are specified from the ventral foregut endoderm at the 7–8 somite stage of the early embryo. Specification occurs in responses to Fgf signals arising from the cardiac mesoderm via Fgfr1 and 2 in the underlying definitive endoderm (Serls et al, 2005). At E9.5, two buds of lung epithelium protrude into the surrounding splanchnic mesoderm in response to Fgf10 signaling (Bellusci et al, 1997b; Arman et al, 1999; Sekine et al, 1999; Desai et al, 2006). The pseudoglandular stage follows, during which the trachea, mainstem bronchi and five major lung lobes are established. The majority of stereo-specific branching of lung bronchi occurs up to day 16.5. The branching process is thought to be developmentally hard-wired, wherein multiple signaling factors coordinately establish morphogenic signaling centers where branching occurs. This process involves interplay of morphogenic signals from the mesenchyme (Fgf10, Bmp4) and the epithelium (Shh, Bmp4) as well as contributions from Tgf-beta family members (reviewed in Cardoso and Lu, 2006). Vascular endothelial growth factor (Vegf), a potent inducer of vascular development (Kalinichenko et al, 2001; Ng et al, 2001), is expressed in the epithelium and promotes development of the vasculature in synchrony with the bronchi and bronchiole branching, such that major veins and arteries follow the branching pattern of the bronchi. Terminal bronchi develop over 24 h during the canalicular stage beginning at E16.5. This process involves the terminal budding of the epithelium into sacs, which are in tight apposition with the mesoderm-derived vasculature. The period from E 17.5 until postnatal day 5 is the terminal sac stage, which is characterized by an increase in the number of terminal sacs and capillaries. The increase in capillary plexus appears to take place by ‘intussusceptive angiogenesis', a process by which slender transcapillary tissue pillars extend at regular intervals between adjacent capillaries (Burri and Tarek, 1990; Patan et al, 1992).
The cells forming the terminal sacs next differentiate into Type I and Type II alveolar cells. The majority of gas exchange is mediated by Type I cells, whereas the Type II cells secrete lung surfactant proteins. A subset of the Type II cells appear to be stem cells for the generation of more alveolar Type I and Type II cells. Despite this detailed knowledge of the main signaling processes involved in mediating branching (reviewed in Chuang and McMahon, 2003; Warburton et al, 2003; Bartram and Speer, 2004; Cardoso and Lu, 2006) and morphology of lung development, little is known about the gene products that are downstream of these developmental signals.
Genetically engineered mouse mutant models using gene knockout (Kimura et al, 1996; Sekine et al, 1999), conditional alleles (Eblaghie et al, 2006), hypomorphic alleles (Moens et al, 1992, 1993) or transgene misexpression (Bellusci et al, 1997a) have all been used to gain insight into the genetic pathways controlling lung morphogenesis. However, the majority of these analyses used a small number of cell-specific markers that provided limited information regarding the full complement of molecular components underlying individual phenotypes, and did not reveal the full complexity of the developmental synthesis. To circumvent these limitations, three groups have published global microarray expression analyses of normal lung development, examining expression over time and comparing the proximal and distal parts of the lung (Mariani et al, 2002; Bonner et al, 2003; Lu et al, 2004a). However, the expression levels and localization of the corresponding cognate proteins are still largely unknown.
Large-scale analysis of the proteome in an unbiased manner is increasingly possible through developments in the application of tandem mass spectrometry (MS) for highly sensitive protein characterization. The advent of gel-free proteomic profiling in particular (Wolters et al, 2001) has facilitated large-scale, high-throughput shotgun peptide sequencing to systematically investigate the proteome of mammalian cells and tissues (Washburn et al, 2001; Florens et al, 2002; Koller et al, 2002). The accurate determination of protein relative abundance by replicate analysis, in combination with spectral counting methods, has made proteomic datasets amenable to comparative analysis using many of the same tools and techniques developed for microarray studies (Liu et al, 2004; Zybailov et al, 2005; Kislinger et al, 2006).
Our group has previously reported the development of a proteomics investigation strategy for mouse (PRISM) based on MudPIT, which markedly increases the depth of coverage via sample prefractionation into organellar compartments (Kislinger et al, 2003). Unlike other proteomic screening methods, our fractionation procedure has the advantage that both tissue and subcellular specificity can be assayed from the same sample preparation (Kislinger et al, 2003, 2006). We have reported the application of PRISM to exhaustively assess the molecular composition of adult mouse tissues (brain, heart, kidney, liver, lung and placenta) (Kislinger et al, 2006). We have also developed a principled mathematical model to compare protein and microarray expression profiles recorded in parallel (Kislinger et al, 2006).
Here, we report the application of these techniques to a time-course study of lung organogenesis at well-defined stages of development. Comparison of the proteomic data with mRNA expression profiles revealed a large number of gene products with coordinately regulated expression. There was also a smaller group of gene products with expression profiles suggestive of post-translational or post-transcriptional control. Having established a baseline of protein profiles during normal lung development, we analyzed the proteome and transcriptome of lungs from mice homozygous for a hypomorphic allele of Nmyc, which encodes a basic helix–loop–helix leucine-zipper protein shown to be critical for lung development (Moens et al, 1992, 1993; Okubo et al, 2005). We compared the resulting proteomic and transcriptomic patterns with an Nmyc gain-of-function microarray data set published recently (Okubo et al, 2005). Mining of the cognate expression profiles identified plausible direct targets of Nmyc regulation, including factors involved in mRNA splicing, nuclear export and localization, which had not previously been linked to Nmyc. Taken together, our findings support the view that Nmyc is a key regulator of the transcriptional processing environment of the undifferentiated lung epithelium.
Results
Efficient identification of proteins with restricted spatial and temporal expression
We collected lung samples at six time points (E13.5, E16.5, E18.5, P2, P14 and P56) from timed mating of ICR mice. These time points cover many of the major phases of lung development including pseudoglandular, canalicular, terminal sac, alveologenesis and septation and growth towards fully developed adult lungs. Sufficient tissues were collected at all times to fractionate and analyze in duplicate (see Materials and methods). Although we and others have shown that this level of replicate sampling by shotgun LC-MS does not achieve saturation of detection (Durr et al, 2004; Kislinger et al, 2006), the quantity of tissues at the earliest time point (E13.5) was limiting, with over 200 lungs required for two replicates of all three fractions; nevertheless, coverage is estimated to be between 75 and 80% (Kislinger et al, 2006). A total of 36 MudPIT analyses were performed, generating over 1.5 million MS/MS spectra. These were sequence mapped against a combined mouse and human protein database obtained from SwissProt/Trembl (EBI release 40) using SEQUEST (Eng et al, 1994). Combined data sets have been used by us and others to increase the protein identification rate of species with incomplete sequence coverage (Schirmer et al, 2003; Kislinger et al, 2006). Redundant cross species hits were removed by reciprocal BLAST, ensuring only unique proteins are reported (Schirmer et al, 2003; Kislinger et al, 2006). High-scoring peptide matches were then collated into candidate parent proteins.
The data set was then stringently filtered by accepting only those proteins with two or more independently collected MS/MS spectra, each with a predicted ⩾95% confidence likelihood as determined using the STATQUEST probability model (Kislinger et al, 2003, 2006). This resulted in the identification of 3330 proteins (Supplementary Table I). As our search database contained an equal number of reversed decoy sequences, we were able to estimate the false discovery rate on average to be ∼2.5% (ranging from 4.6 to 0.7% per fraction), similar to previous studies (Kislinger et al, 2003, 2006). The entire proteomic data set has been deposited in the public GEO database at NCBI under the series GSE6108.
Quantification of protein levels
Protein relative abundance was estimated using the spectral count method, which reflects the ratio of all matching MS/MS spectra above the specified statistical filter for any given protein per fraction (Liu et al, 2004). This technique was shown to be linear over at least two orders of magnitude (Liu et al, 2004). Furthermore, Washburn and colleagues (Zybailov et al, 2005) have shown that the spectral counts quantitative method performed as well or better than stable isotope labeling-based quantitation methods when determining relative change calls. This method, however, was not useful for the calculation of absolute protein abundance. As we wished to detect the relative changes of protein expression as a function of developmental time, absolute protein abundance is not necessary and the spectral counting method is sufficient.
Cluster analysis of protein expression data
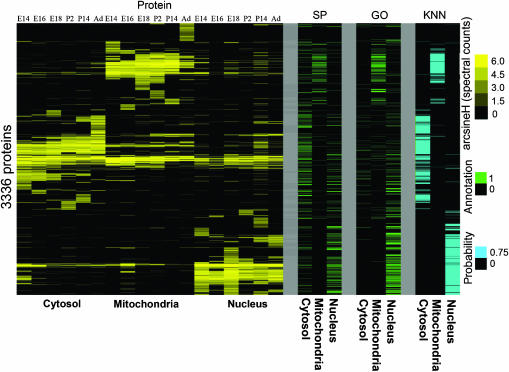
Two-dimensional hierarchical clustering of the entire data set (Figure 1) resolved the proteins back to their organelle(s) of origin. There was a statistically higher overlap of identified proteins in adjacent chronological time points (data not shown). This was observed strongest in the cytosolic fraction, where the embryonic fractions grouped away from the postnatal fractions (Figure 1). The nuclear and mitochondrial fractions also show some separation of the early embryonic from later embryonic and postnatal time points, although to a lesser degree. These trends imply that the protein data have captured cell biological and developmental relationships. Consistent with this, we also observed statistically significant enrichment of proteins with Gene Ontology (GO) functional and cellular location terms specific to each of the three subcellular compartments (Figure 1; Table I and Supplementary Table II).
Figure 1.
Double hierarchical cluster of all protein expression and subcellular fractions over six time points of lung development. Proteins are clustered along the horizontal axis with time point subcellular fractions along the vertical. Protein data are scaled as arcsine(H) in yellow, with black representing no data measured. Unguided double hierarchical clustering grouped subcellular fractions together in subclusters. Within the organelle subcluster, the developmental stages were generally separated into early and later developmental time points. This figure shows that both cellular and developmental information are captured by protein profiling. Clustering parameters were average linkage using an uncentered correlation similarity metric.
Table 1.
Enrichment of gene ontology terms to proteins clusters
| Number | P-value | GO ID | Term |
|---|---|---|---|
| Enriched to nuclear fraction | |||
| 469 | 0 | 5634 | Nucleus |
| 224 | 0 | 6351 | Transcription, DNA dependent |
| 65 | 0 | 6325 | Establishment and/or maintenance of chromatin architecture |
| 40 | 0 | 5681 | Spliceosome complex |
| 34 | 0 | 6281 | DNA repair |
| Enriched to mitochondrial fraction | |||
| 221 | 0 | 5739 | Mitochondrion |
| 65 | 0 | 6118 | Electron transport |
| 36 | 0.0001 | 5795 | Golgi stack |
| 34 | 0 | 5759 | Mitochondrial matrix |
| 32 | 0 | 5743 | Mitochondrial inner membrane |
| 30 | 0 | 6631 | Fatty acid metabolism |
| 16 | 0.0001 | 5789 | Endoplasmic reticulum membrane |
| Enriched to cytsolic fraction | |||
| 106 | 0.0013 | 5856 | Cytoskeleton |
| 102 | 0 | 5829 | Cytosol |
| 34 | 0 | 6512 | Ubiquitin cycle |
| 31 | 0 | 502 | Proteasome complex (sensu Eukaryota) |
| 25 | 0.0183 | 7264 | Small GTPase-mediated signal transduction |
The lung contains many specialized cellular structures such as microvilli and cilia, as well as several subcellular structures such as lamellar bodies and secretory vacuoles (Ten Have-Opbroek, 1991). The fractionation conditions utilized here will not separate these structures specifically and the detection of their component proteins will be suboptimal. Nevertheless, we do detect proteins such as ciliary dynein heavy chain 5 and 7 and subunits of vaculolar ATP synthase (subunits A, B, D and G), the retromer complex (Vps29 and Vps35) and the ESCRT-II complex (Vps36). This suggests that protein components of these specialized structures should be detectable if more optimized fractionation protocols are used.
Proteomics as a predictor of cellular location by KNN
Large-scale proteomics experiments that employ cellular fractionation can be used to annotate proteins to cellular locations (Foster et al, 2006; Kislinger et al, 2006). Similar to our previous work, nearly two-thirds (2100/3336) of the proteins were uniquely identified to single subcellular fractions (Supplementary Figure 1). We next employed machine learning to assign a probability score that a protein's observed subcellular location was the correct location.
We have previously shown the utility of K-nearest neighbor (KNN) machine learning as a method for refined prediction of protein subcellular localization (Kislinger et al, 2006). In our current study, we used a Dense KNN derivative (Liu et al, 2003) since it exhibited better performance (data not shown). The following three sets of data were required for this analysis: a training set with positive and negative examples of the desired outcome (e.g. known nuclear localized proteins), a test set with positive and negative examples that had not been used for training and the prediction set of the unknowns to be determined. For each subcellular fraction, independent training (total set size was 860 proteins, with 358 cytosolic positives, 187 mitochondrion positives and 315 nuclear positives) and test data sets were generated using SwissProt/TREMBL (SP) annotation current as of Uniprot release 7.0 (Supplementary Table III). A summary of set sizes is displayed in Table II. As these sets are not balanced, having more negative than positive examples, we used a 10-fold cross-validation procedure to ensure there was no bias during training. We used area under the curve (AUC) to derive the optimal value of K, which proved to be 15 for the KNN learner in all training sets (data not shown).
Table 2.
Summary of KNN prediction for subcellular location
| Fraction | Total number of traininga | Number of training predicted >0.75a | % | Number predicted >0.75a | % | Number of novel predictionsb | % |
|---|---|---|---|---|---|---|---|
| Cytosol | 358 | 224 | 63 | 511 | 23 | 455 | 24 |
| Mitochondrion | 187 | 142 | 76 | 238 | 11 | 180 | 10 |
| Nucleus | 315 | 248 | 79 | 539 | 24 | 366 | 19 |
| Total | 860 | 614 | 71 | 1288 | 58 | 1001 | 53 |
| Number no prediction | 245 | 28 | 942 | 42 | 889 | 47 |
Excludes 148 proteins with annotation to cytosol and nucleus or mitochondrion.
Excludes all test set proteins with GO annotation for any of the three fractions.
Receiver operator characteristic (ROC) curves for the training sets showed a high degree of specificity and sensitivity (Supplementary Figure 2). The specificity, sensitivity and precision were calculated by a confusion matrix (Lu et al, 2004b). Across all compartments, the mean sensitivity was 0.82 (varying from 0.75 to 0.91), precision was 0.84 (0.83–0.86) and specificity was 0.94 (0.92–0.95). To gauge the robustness of these predictions, we used a set of proteins with GO cellular location terms for cytosol, mitochondria and nucleus that did not have established SP localization terms as the test set. The ROC curves for the test data set are displayed along with the training ROC curves (Supplementary Figure 2) and were largely comparable, even considering the different annotation sources of the training and test data sets. The entire data set, along with spectral counts, predicted localizations and SP and GO annotations is provided in Supplementary Table IV, whereas a summary is provided in Table II.
Of the 2230 proteins in the prediction set, nearly 60% (1288) had predictions with >75% confidence; among these, nearly 80% (1001) represented novel localizations as they lacked relevant GO and SP annotations. Setting the prediction at a threshold of 75%, we then clustered the complete set of protein fractions along with the putative SP and GO annotations and our predictions for visual inspection. Good overall agreement was apparent in the resulting clustergram (Figure 2). Interesting patterns were also observed after clustering even with those proteins with lower confidence predictions (Figure 2), such as a large group of mitochondria-localized proteins. Closer inspection of this cluster indicated that the majority of these proteins occurred at a single time point of the mitochondrial fractions and that they were likewise coenriched with plasma membrane, ER, Golgi, as well as many extracellular/secreted proteins.
Figure 2.
Protein distributions in subcellular fractions are predictive of protein subcellular location in vivo. Shown is the same double hierarchical plot as in Figure 1, with subcellular location annotation for nucleus, cytosol and mitochondria from SwissProt (SP) and GO. Also shown are the predictions (P>0.75) of K-nearest neighbors machine learning on SP annotations. All predictions less than 75% are black. Note the high degree of correspondence between the subcellular fractions protein abundance, annotations and predictions.
As a further test of these predictions, we constructed and sequence confirmed 12 expression plasmids with C-terminal fusions to GFP, consisting of five nuclear, five mitochondrial and two cytosolic predicted localized proteins. Each of these proteins had a prediction greater than 75% and had no literature confirmation of their cellular location by direct in vivo or in vitro analysis. The plasmids were transfected into human embryonic kidney cells (293T) cells and imaged 24 h post infection by spinning disk confocal microscopy. Hoechst and Mito Tracker dye were used as a counter stain and to assess colocalization (Figure 3). Three of five nuclear, three of five of mitochondrial and both cytosolic proteins localized to the predicted organelle. Interestingly, each of the three nuclear proteins clearly labeled different regions of the nucleus indicating that we have sampled a range of sub-organellar compartments.
Figure 3.
Colocalization of GFP fusions of predicted localized proteins. Protein fusions with GFP expressed in 293T cells imaged with spinning disk confocal microscopy. Left column is the GFP channel (green), center column is the merge of Hoechst (blue) and MitoTracker CMXRos dye (red), with the nuclei and mitochondria labeled respectively, and the right panel is the merged image. In all cases, clear colocalization of predicted compartment is observed. Note the variety of localization patterns in the nucleus, indicating that a range of nuclear proteins were identified and tested.
Integration with public genome-scale data sets
The integration of multiple large-scale data sets could be used to mine information for hypothesis building. Three large-scale data sets in particular are the mutant phenotype data from Jackson Laboratories Informatics (www.informatics.jax.org), protein interaction networks from OPHID (Brown and Jurisica, 2005) and mRNA profiles of lung development (Mariani et al, 2002).
Mutant phenotypes data
The Jackson Labs Informatics group maintains an up-to-date database of all published mouse mutant lines and scores their phenotypes with hierarchically related annotation terms similar to the framework used by GO. We cross-referenced our protein data to the list of mutant lines and annotated a number of genes with known lung mutant phenotypes. The phenotypes did not generally correlate with the putative organellar localization of the detected proteins (data not shown, Supplementary Table V). For example, broad terms such as ‘abnormal lung morphology' were found to span proteins from all three subcellular factions (Supplementary Table V). Similarly, there was little correlation for time and phenotype as many genes expressed throughout development of the lung are critical to development at different stages. Therefore, by assessing the coexpression of known interacting partners of these proteins with known mutant phenotypes, we thought it possible to uncover connections between these seemingly unrelated proteins.
Protein interaction networks
Examination of protein interaction networks may make it possible to predict the participation of other physically interacting proteins in developmental pathways based on spatial-temporal coexpression. To investigate this, we examined a set of 10 proteins identified in this study known to have the phenotype ‘abnormal alveolar morphology': ADA_MOUSE, CD81_MOUSE, DHI1_MOUSE, G3BP_MOUSE, PRDX6_MOUSE, FBLN5_MOUSE, PSPC_MOUSE, MO4L1_MOUSE, CO2A1_MOUSE and EGFR_MOUSE. An interaction map based on the human homologues of these 10 proteins in the OPHID database (http://ophid.utoronto.ca) (Brown and Jurisica, 2005) (see Materials and methods) returned a list of 161 known interactions for ADA_MOUSE, CD81_MOUSE, G3BP_MOUSE and EGFR_MOUSE, whereas the remaining proteins had no known interactions in the database. The interactions were then filtered for those proteins likewise detected in our proteomic screening, revealing a highly interconnected map of 46 proteins linking three of the alveoli phenotype gene products (CD81_MOUSE, G3BP_MOUSE and EGFR_MOUSE) (Figure 4). Consistent with the biological significance of this network, both EGFR and G3BP mutants have similar strain-dependent alveolar phenotypes, supporting the functionality of the inferred link between them through their interacting partners (Threadgill et al, 1995; Zekri et al, 2005). G3bp is linked to Egfr via Paxi (paxillin) and Tln1 (talin-1), both of which are involved in focal adhesion complexes, suggesting that this structure may be involved in alveologenesis.
Figure 4.
Proteins with similar phenotypes when deleted in vivo form a highly interconnected protein interaction network. Shown is the protein–protein interaction network of four proteins all annotated to have abnormal alveoli when deleted in vivo. Only proteins identified in this study are shown in the interaction network. The four nucleating proteins are all ovals, whereas interactions are shown as circles. Lines connect proteins with annotated interactions based on data deposited at OPHID. Note that three of the four form a connected network of proteins, possibly explaining their common alveolar phenotype.
Correlation of protein and microarray data
Microarray studies have shown the process of development to involve highly dynamic gene regulation. Two microarray data sets have shown this to be the case for lung development (Mariani et al, 2002; Bonner et al, 2003). The data set from the study published by You and co-workers (Bonner et al, 2003) has not been made publicly available and will not be considered in this paper. Conversely, Mariani et al (2002) released a complete data set covering 12 time points in mouse lung development recorded using the Affymetrix Mu11K A and B chip sets, allowing a high-resolution view of the temporal control of transcription during embryonic to prenatal lung organogenesis on through adulthood.
To ascertain the relationship between transcription and translation during lung development, we mapped (see Materials and methods) our protein data set against the lung development microarray data of Mariani et al (2002). A set of 1383 protein probe pairs were generated, with protein levels monitored by spectral counts across each time point and organelle, while mRNA levels were likewise estimated based on probe intensity (Supplementary Table VI). The combined data set was then clustered based on the gene expression patterns so as to reveal the overall degree of correlation as well as clusters of discordance in the global expression patterns (Supplementary Figure 3).
Previous correlation analyses of protein and mRNA, represented by either microarray or SAGE data, utilized simple Pearson or Spearman correlation (Gygi et al, 1999; Griffin et al, 2002). Although these simpler methods revealed correlated gene product relationships, they lacked a robust noise model or method for determination of confidence of the correlation. We opted, instead, for a probabilistic approach to model the relationship between protein and mRNA (Kislinger et al, 2006). This has the advantage of treating the microarray data with a Gaussian noise model and the protein data as a Poisson distribution. As equivalent data points are required for the correlation analysis, we created a table of protein data with all cell fraction data summed for each time point and paired this with microarray data that matched these time points. The strength of the relationship between microarray and protein data was determined and assigned a correlation score, while a confidence score (P-value) was assigned by permutation testing of randomized data. This allowed division of the data into significantly (or insignificantly) correlating (‘inliers') and non-correlating (‘outliers') gene product pairs.
Of the 1383 protein microarray data pairs, 643 were deemed to be significant inliers, 30 were called significant outliers, 699 as insignificant inliers and 11 as insignificant outliers (Figures 5A–D and Supplementary Table VI). For the significant inliers, we used K-means clustering (where K=7 was based on a figure of merit calculation; Yeung et al, 2001) to reveal correlated subgroups exhibiting no apparent change in expression or with dynamically changing expression levels from E13.5 to adult (Figure 5A). We next examined these clusters for evidence of functional coherence based on enrichment for common GO functional terms (Figure 5A). For example, a cluster of protein/mRNA pairs with correlated decreasing expression, Cluster 4 (Figure 5A), was enriched in genes associated with DNA replication, chromosome organization, and biogenesis (P<0.01 in all cases), genes that are critical to tissue and cellular development and differentiation (Supplementary Table VII). A cluster with relatively constant high expression, Cluster 7 (Figure 5A), was enriched in components of the ribosome and general metabolic/catabolic processes such as glycolysis (P<0.01 in all cases), as might be expected for housekeeping enzymes required for cellular homeostasis and growth (Supplementary Table VIII).
Figure 5.
Correlation analysis of protein and microarray data for lung developmental time course. Math modeling identified four subsets in the data sets: (A) significant inliers, (B) insignificant inliers, (C) insignificant outliers and (D) significant outliers. Whereas inlier denotes a positive correlation score, outliers denote a negative correlation score and significant indicates that the correlation is statistically different from the background (noise) model, whereas insignificant is not. Each subset was then either clustered by K-means (A, B) or simply visualized due to the lower information content. Note that the significant inliers contains many clear patterns of dynamic gene expression and those with static expression throughout development. The significant outliers shows several genes with off set protein and mRNA expression, where the mRNA increases before the protein.
Does poor correlation reflect post-transcriptional/post-translational control?
We identified a small significant outlier group, wherein the transcript and protein levels were statistically discordant. Many of these outliers showed clear peaks of protein expression from E18 to P2, whereas the corresponding mRNA peaked much earlier at E14 and E16. This would suggest that there is extensive temporal lag in the translation of some mRNAs or accumulation of significant protein.
The insignificant inliers were subclustered by K means (K=5, based on figure of merit calculation), which revealed highly regular mRNA expression but seemingly divergent protein profiles (Figure 5B). Although not statistically coherent, this group was nevertheless found to be enriched for mitochondrial proteins, which have previously been shown to have poor correlation at the mRNA level (Mootha et al, 2003; Kislinger et al, 2006). Also detected were numerous membrane proteins associated with the broad GO term establishment of localization, such as components of the clathrin coat adapter complex, which can be explained at least in part by biased, incomplete proteomic coverage analysis of components associated with the membrane and cytoskeletal systems.
Proteomic analysis of Nmyc mutant lungs
Mice homozygous for a null allele of Nmyc die mid-gestation, precluding simple analysis of its molecular function during the late stages of lung development (Sawai et al, 1991; Charron et al, 1992). However, Nmyc is essential for normal lung formation as pups homozygous for a hypomorphic allele fail to breath and die in the early postnatal period (Moens et al, 1992, 1993). In this allele, the neomycin resistance gene was inserted into the first intron of N-myc with a splice accepter, in such a way that alternative splicing around this insertion resulted in the generation of a normal N-myc transcript in addition to a mutant truncated transcript. The lungs form these mice exhibited reduced branching and were smaller than wild-type or heterozygous littermates (Moens et al, 1992, 1993). Moreover, a conditional deletion of the gene in lung epithelium produces a profound lung phenotype, resulting in large sac like structures with a thin mesenchyme (Okubo et al, 2005).
For this study, pups derived from crosses of mice heterozygous for the hypomorphic Mycntm1Jrt allele (back crossed onto ICR mice for several generations and herein referred to as Nmyc9a) were surgically delivered at E18.5. The pups were scored for color and rhythmic breathing, since homozygous Nmyc9a mutants fail to breathe properly. Pups that failed to breathe and quickly became listless were killed and their lungs removed for proteomic analysis and tail clipping taken for PCR genotyping. Enough material was collected to perform three technical replicate MudPIT analyses of cytosolic and nuclear fractions and two repeat runs of the mitochondrial fraction. As a wild-type control we compared the Nmyc9a protein profiles with our lung development protein data set. To equalize the mutant and wild-type data sets, further experimental replicates of the nuclear and cytosolic fractions for healthy normal E18.5 lungs from normal ICR crosses (i.e. identical to those used to profile normal development) were analyzed. These data were merged with the original E18.5 duplicates from the developmental profile and the added replicates correlated well with the previous replicates in both cell fractions (data not shown).
Slightly more proteins were identified in the wild-type lungs (1808) versus the Nmyc9a lungs (1509) (Supplementary Figure 4A), presumably due to perturbed, development in the mutant. Overall, 882 proteins were uniquely identified in wild-type lungs and 500 in Nmyc9a lungs (Supplementary Figure 4A). The nuclear fraction of wild-type lungs had the largest discrepancy as compared with Nmyc9a lungs (443 versus 126 uniquely identified proteins) (Supplementary Figure 4A). This data set is deposited at GEO under the series GSE6108.
Characterization of error in protein abundance by spectral counts
Before assessing the apparent differences in relative levels of protein abundance between the Nmyc9a and wild-type lungs, we performed an analysis of the normal distribution of ratios of spectral counts for replicates to assign a background error model for spurious variance in quantitation (see Materials and methods). As represented graphically in Supplementary Figure 4B, we determined that a fold expression ratio of greater than three-fold up or down explains most of the residual variance (∼90% of the proteins are within this level of reproducibility), irrespective of the spectral count intensity, indicating that even potentially lower abundance proteins (low spectral counts) are amenable to accurate determination of differential expression. We calculated the relative levels of those proteins detected in either Nmyc9a or wild-type at E18.5 in at least two of three replicates of the cytosolic and nuclear fractions, and in two of two of the mitochondrial samples.
Altered expression of proteins in Nmyc9a lungs
Of the 700 proteins quantified between Nmyc9a and wild-type lungs, 170 proteins showed significantly (>three-fold) increased expression in Nmyc9a lungs, including 77 that were only detected in Nmyc9a, whereas 182 had decreased expression (<0.33-fold), of which 42 were not detected in Nmyc9a (Supplementary Figure 4C; Supplementary Table IX). The overall distribution of spectral counts recorded for the quantified group of proteins had a similar profile to the entire data set (Supplementary Figure 4D).
A comparison of the proteins with increased or decreased expression with wild-type mice was made against the entire developmental protein profiling data set. Of 315 proteins differentially regulated (37 proteins were not observed in the developmental data set), nearly 60% were found to be localized to the cytosol, over 23% to the nuclear and 17% to the mitochondrial fractions. The majority of proteins with decreased expression in Nmyc9a lungs were proteins with constitutive expression profiles when compared with our lung development data set (Supplementary Figure 4E). These proteins were enriched for GO molecular functional terms such as mRNA splicing and mRNA transport (P<0.01). Conversely, the majority of proteins with increased expression in Nmyc9a lungs were proteins with dynamic expression profiles when compared with our lung development data set (Supplementary Figure 4F). These proteins were enriched for GO molecular function terms such as cell adhesion and anatomical structure development (P<0.01).
Microarray analysis of Nmyc9a mutant lungs
To allow for a comparison of the protein data with corresponding mRNA patterns, total RNA was isolated from six mutant Nmyc9a lungs and six wild-type control lungs at E18.5 and submitted for microarray expression analysis by Affymetrix MOE430v2 chip (deposited at GEO, GSE6079). To facilitate identifying possible direct targets genes of Nmyc, we utilized another previously published microarray data set of stage-matched lungs misexpressing Nmyc under the human lung surfactant protein C promoter (SftpC-Nmyc) (Okubo et al, 2005) (GEO, GSE6077). In total, 441 proteins could be paired with 1217 probe sets from both the Affymetrix MOE430 v2 chip and the MOE430A chip (Supplementary Table X), allowing a three-way comparison. Within this set, 257 proteins (paired to 451 probe sets) displayed differential mRNA and/or protein abundance in either one or both of the loss- and gain-of-function Nmyc mutants (Supplementary Table X).
Gene products wherein both the mRNA and protein exhibited depressed expression in Nmyc9a lungs but increased levels in SftpC-Nmyc lungs are strong candidates for being direct targets of Nmyc transcriptional regulation. Conversely, as Nmyc has also been reported to function as a transcriptional repressor, we also looked for mRNA and protein showing increased abundance in the Nmyc9a allele and decreased mRNA in the SftpC-Nmyc lungs. This coordinate mining of expression profiles revealed a list of 63 positive and one negative potential targets of Nmyc regulation.
The positively regulated list was highly enriched (P<0.001) in gene products associated with mRNA metabolism, including splicing, nuclear export and localization, and contained nucleolin and lactate dehydrogenase (Ldh), which are both known targets of Nmyc (Patel et al, 2004), suggesting that the other members in this group are genuine. The negatively regulated target was spectrin beta 2.
It was noted that one of the redundant probe sets for nucleolin showed a decrease in expression in the Nmyc9a lungs, although it did not test as significant by the Affymetrix statistical change call. However, the protein did show a significant change in expression as did the same probe set in SftpC-Nmyc lungs. We extended our mining to include those with increased or decreased mRNA expression in Nmyc9a lungs, even if it did not test as significant, as long as there was a significant change in the Nmyc9a protein or SftpC-Nmyc mRNA. The rationale is that the hypomorphic Nmyc9a allele may not necessarily induce as robust a change in gene regulation as observed with a null allele. This extended mining strategy revealed a further 22 potentially positively regulated genes and four more potentially negatively regulated genes. The combined results from the initial and extended mining strategy are all displayed in Supplementary Table XI. The potentially directly regulated data set is graphically displayed as a heat map of mRNA or protein ratios (Figure 6). More complex regulatory patterns were suggested by hierarchical clustering of all the significantly differentially regulated gene products detected in all the data sets (data not shown; Supplementary Table X). This second positively regulated set contained Nmyc downstream-regulated gene 1 (Ndrg1) and was also significantly enriched in mRNA processing gene products (P<0.01). Intriguingly, one positively regulated gene, hepatoma-derived growth factor (Hdgf), was detected in the cytoplasm of both Nymc9a and wild-type lungs, while nuclear localized Hdgf was only detected in the wild-type lung. Hdgf has been shown to be cytoplasmic and to translocate to the nucleus where it can activate cell growth (Kishima et al, 2002). The potentially negatively regulated data set included insulin-like growth factor 2 receptor (Igf2r), mannose receptor C type 2 (Mrc2) and signal transducer and activator of transcription 3 (Stat3).
Figure 6.
mRNA and protein regulation in Nmyc loss- and gain-of-function mutant E18.5 lungs. Combined protein and microarray that tested as being significantly regulated was separated into direct and indirect classes based on the expected profiles of positive, where both transcript and protein are reduced in the loss of function mutant and the transcript is increased in the gain of function mutant and negative, with the opposite profile to positive. Based on these profiles, 90 protein microarray pairs were extracted and are organized with the potential positive targets followed by the four negative targets at the bottom of the figure.
Discussion
Data mining can lead to genes with developmental roles
As more large-scale microarray, and increasingly, protein data sets are assembled and published, there is a growing resource of information on lung development. A common criticism of large-scale screens is that they lack a hypothesis or are overly broad in scope. However, by combining resources, one is able to ask focused questions of the data sets and select promising candidate genes for mechanistic studies. We have presented several examples throughout this work, including comparison of protein and mRNA profiles and protein interaction databases.
Machine learning was able to learn the profiles of known nuclear, mitochondrial and cytosolic localized proteins and calculate probabilities for proteins with no cellular location annotation. Expression of GFP fusions of proteins with novel cellular location prediction held up in the majority of cases (eight of 12 fusion proteins expressed) when expressed in 293T cells. Aside from failure of the KNN algorithm prediction, failure of the protein to localize as predicted could be due to the differences in the in vivo cellular environment of lungs versus in vitro cell culture. As well, the presence of GFP on the C-terminus may affect the ability of the proteins to be properly localized.
In an examination of the interaction partners of lung proteins with known alveolar developmental phenotypes, we were able to link three into an interaction cluster. In this analysis, G3bp was found to be linked to Paxi (paxillin) and Tln1 (talin-1), both of which are involved in focal adhesion complex linking the cytoskeleton and plasma membrane as well as responding to signaling environments. Paxi and Tln1 interact with integrin alpha 5 (Ita5) and Tenascin (Tena) linking them to Egfr. Null mutations of either paxillin or talin-1 result in early embryonic lethality: talin-1-null embryos have an earlier phenotype at gastrulation (Monkley et al, 2000), whereas paxillin-null embryos exhibit a later phenotype with absent hearts and abnormal somites (Hagel et al, 2002). For this reason, the role of talin-1 or paxillin in alveologenesis is as yet unexplored, but perhaps by use of conditional alleles of paxillin or talin-1 their function in alveologenesis may be uncovered. Nevertheless, our data suggest a possible role for G3bp regulation of the focal adhesion complex in the development of the alveolar structures. The focal adhesion complex is also involved in vascular development and it is possible that the observed protein expression is derived from the lung vascular tissue; however, there is a tight association with vascular and alveolar development and a defect in either can affect the other.
Dynamic gene regulation during development
With so much expression data at hand, we could examine the relationship between the mRNA and corresponding protein levels. In our current analysis, the microarray data set was generated on an older chip containing fewer probe sets (∼11 000 compared with the current Affymetrix MOE430 2 chip with ∼40 000) such that there were many proteins with no corresponding mRNA probe set. There are several caveats to this analysis. First, mammalian genes are often differentially spliced to produce specific variants, which can confound attempts to map to the cognate protein product. It is also possible that differences are a reflection of variables in the genetic background of the mice used for this study and those for the microarray. Despite these caveats, there were still over 1300 pairs of gene products with sufficient information for correlative analysis.
After taking into account the reliability of the experimental measurements, we were able to cross-compare the protein and microarray patterns using a more robust statistical modeling method. The high-confidence correlated gene product group (significant inliers) revealed subgroups exhibiting either dynamic or stable expression profiles during lung organogenesis enriched with anticipated gene products. The more interesting groups of gene products were the non-correlating groups as these may be enriched for genes that undergo post-transcriptional or post-translational control. We note several data pairs that appear to have a delayed translation; however, it is not possible without further experimental evidence to conclude if this is a post-translational or post-transcriptional mechanism.
In the list of insignificant correlating pairs, we note many groups of proteins enrich to general terms such as localization, metabolism and mitochondria. Mitochondrial proteins have been shown by us and others to have a poor correlation to mRNA levels, and there is evidence that the mitochondrial localized protein cytochrome c is under post-transcriptional control (Kawai et al, 2006). It will be difficult to further test post-transcriptional or post-translational events in our data sets due to the use of in vivo tissue. The treatment of tissue in vivo with transcription and translation inhibitors to test rates of production and turnover of mRNA and protein would be difficult. It may be possible to substitute a cell-based system or organ culture system, but the behavior of mRNA and protein regulation in vitro may not faithfully recapitulate in vivo dynamics.
Other model organisms such as Drosophila have many genes known to be regulated at the post-transcriptional and post-translational levels (reviewed in Lipshitz and Smibert, 2000). Future studies in these model systems may help establish better mathematical models, which are needed to facilitate prediction of post-transcriptional or post-translational control mechanisms from combined proteomics and microarray data.
Differential expression of proteins in Nmyc mutant lungs
The ability to test the developmental role of a gene by loss and gain of function mutants is a powerful tool. It is therefore of great value to have the ability to measure the molecular changes in protein and transcript expression to more fully characterize the biochemical function of a gene in development. We used the available gain- and loss-of-function alleles of the transcription factor Nmyc to study its role in lung development in great molecular detail. Nmyc is a basic helix–loop–helix leucine-zipper transcription factor known to function as a transcriptional activator and a repressor (Cole and McMahon, 1999; Patel et al, 2004). Nmyc has been noted to be overexpressed in many cancer cell types and to confer a selective advantage for rates of cell division when overexpressed (Brodeur et al, 1985; Schwab et al, 1985). Mice homozygous for null alleles of Nmyc die in early gestation, while a hypomorphic allele has been shown to cause a severe lung phenotype (Sawai et al, 1991; Charron et al, 1992; Moens et al, 1992, 1993). Recently, a conditional allele of Nmyc has been used to completely delete it from the lung epithelium, revealing an even stronger phenotype in the lung (Okubo et al, 2005). Misexpression of Nmyc with the SftpC promoter has revealed a role in maintaining the undifferentiated state of multipotent epithelial progenitor cells (Okubo et al, 2005).
By combining the complementary hypomorphic and misexpression microarray data sets and the protein data of the hypomorphic allele, we mined gene products with opposite regulation in the two different Nmyc genotypes compared with wild-type. This data set included several known direct targets of Nmyc regulation and is now a great source for future studies involving transcription factor motif identification and chromatin immunoprecipitation. Of the potential positive targets of Nmyc, we noted some correspondence to previously described target proteins involved in ribosome biogenesis, protein synthesis and DNA replication (Boon et al, 2001) and reviewed in Patel et al (2004), including Nucleolin (Murakami et al, 1991a, 1991b). Nmyc has also been shown to regulate genes involved in the cell cycle (Boon et al, 2001; Okubo et al, 2005).
We have observed a large number of gene products that are potentially positively regulated targets of Nmyc. These gene products were enriched in mRNA processing factors that spanned a wide range of activities. For example, there were two ATP-dependent helicases, four factors annotated as involved in mRNA splicing, one suggested to be involved in the regulation of the U4/U6 × U5 tri-snRNP complex, one potential translation elongation factor and two potential mRNA nuclear export factors. The exact role of these factors cannot be discerned as many have only electronic annotation based on the presence of protein domains with associated functions, such as RNA binding motifs. These novel mRNA processing factors may regulate a specific subset of mRNAs and may not be part of the general splicing/translation machinery. It will be of great interest to further characterize and identify the targets of these mRNA processing factors.
Our data suggest that Nmyc maintains the undifferentiated state of the distal epithelial cells of the lung not by directly regulating other multipotentcy genes but largely through the positive regulation of the mRNA processing factors that may maintain the active translation and splicing of multipotentcy genes. This is partially supported by our observation that proteins with an increased expression in Nmyc9a were enriched for structural and cell adhesion functions that are typically more robustly expressed at later stages in development or in association with mature differentiated cell types.
We also identified a small number of potential negative targets of Nmyc regulation with a variety of functions. One target was Stat3, a TF often activated in a variety of human cancers, including lung (reviewed in Bromberg, 2002), although previous specific deletion of Stat3 in the lung epithelium resulted in no obvious developmental defect (Hokuto et al, 2004). Igf2r also appeared to be negatively regulated. Igf2r is a negative regulator of Igf2 signaling and embryos with a null mutation in Igf2r are 30% larger and present defects in alveologenesis, a process which generates terminally differentiated epithelial cell types (Wang et al, 1994). Igf2r is an imprinted gene (Barlow et al, 1991), regulated by the expression of the non-coding transcript Air (Sleutels et al, 2002). The question of whether Nmyc has a role on the imprinting status of Igf2r may be interesting to investigate in future work. Mrc2, which also appears negatively regulated, is involved in regulation of extracellular matrix, specifically collagen (East et al, 2003) and matrix metalloproteinases (Engelholm et al, 2001). ECM remodeling is a process that is dynamically regulated during lung development (reviewed in Suki et al, 2005) and is correlated with regulation of cell signaling environments and cell differentiation.
In conclusion, Nmyc appears to play a role in regulating the environment of the undifferentiated epithelium. Internally Nmyc may positively regulate many novel mRNA processing factors, ranging from splicing, nuclear export and translation elongation factors. Nmyc may also negatively regulate a suppressor of Igf2 signaling and the deposition of collagen and secretion of MMP involved in ECM remodeling.
Improvements to detection limits and sample preparation enable analysis of embryonic tissues
Although the basic MudPIT procedure has undergone substantive improvements in terms of instrumentation, analytical methodologies and analysis algorithms, the procedure still does not achieve the apparent detection sensitivity or coverage reported for DNA microarray technologies. Nevertheless, it has still afforded an in-depth probing of the proteome of mammalian tissue systems (Washburn et al, 2001; Florens et al, 2002; Koller et al, 2002). Using high-throughput screening techniques such as tandem MS, a biological system like lung can potentially be interrogated to identify its core components and their respective interactions and modifications (Washburn et al, 2001; Wolters et al, 2001; Ballif et al, 2004).
The application of proteomics to developmental biology presents several critical challenges, however. The first issue pertains to the often limiting amounts of material that can be gathered for proteomic analysis. In our case, this limited our MudPIT analysis to only two technical replicates, which, although below the theoretical saturation of detection, putatively achieve >75% coverage of the detectable proteome. This imposes constraints on the analysis of the data. Reassuringly, the majority of the protein profiles were not deemed to be statistically uncorrelated with the corresponding mRNA transcript levels as measured by microarray.
Another challenge is the accurate assessment of changes in protein abundance over time. The use of spectral counting as a means to measure relative protein levels between samples appears to have worked well in this study. With the development of more sensitive techniques and instrumentation, it should be possible to increase our sampling efficiency and overall confidence in protein quantitation for small sample sizes.
Materials and methods
Mouse tissues
Mouse tissues were derived from ICR timed mated mice. Timing of conception was designated by assigning noon of the day of plug formation as day 0.5. Pregnant animals were killed and uteri dissected to obtain individual embryos from which lung tissue was derived. A minimum of 0.5 g or more lung tissue was used in all cases, which required approximately 50 pairs of lungs at E13.5, 24 at E16.5 and at later stages, we homogenized lungs from a minimum of two litters (∼24 pairs of lungs) but isolated protein extracts from an aliquot of the homogenate equivalent to 4 g of lung tissue. Protein extracts were made form four separate isolations of lung tissues. Nmyc9a mice back crossed to ICR and heterozygous for the hypomorphic allele were timed mated and litters delivered on day 18.5. The uterus was removed and the pups dissected from the uterus. The yolk sac and amnion were carefully removed. The placental vein and artery were left intact while the pup was dried with Kim wipes, then they were cut and the pups moved under a heat lamp for warmth. The pups' mouths and noses were frequently blotted to remove any mucus that was expelled. They were also gently rubbed to stimulate breathing. After 30 min, pups that had failed to breath or had become listless were killed, a tail clipping removed for genotyping as described in Moens et al (1992) and lungs removed and processed for protein. Geneotyping was performed using the following primer pairs: for Nmyc wild-type allele NmycA GGT AGT CGC GCT AGT AAG AGC and NmycB GGC GTG GGC AGC AGC TCA AAC and for Nmyc9a hypomorphic allele NmycB and NmycC (neo) GGA GAA CCT GCG TGC AAT CC. Lung samples were processed individually into nuclear, mitochondria and cytosol fractions and stored at −70°C until confirmed by genotyping to be homozygous for the mutant allele and then pooled before preparation for MS.
Tissue fractionation
The tissue fractionation was performed as described (Cox and Emili, 2006; Kislinger et al, 2006). Briefly, lung tissue was rinsed twice in ice-cold phosphate-buffered saline and homogenized in ice-cold lysis buffer containing 250 mM sucrose, 50 mM Tris–HCl (pH 7.4), 5 mM MgCl2, 1 mM DDT and 1 mM PMSF using a tight fitting Teflon pestle. The lysate was centrifuged in a bench-top centrifuge at 800 g for 15 min; the supernatant served as source for cytosol, mitochondria and microsomes. The pellet, which contains the nuclei was rehomogenized in lysis buffer and centrifuged as above. The nuclei were resuspended in 2 M sucrose buffer (2 M sucrose, 50 mM Tris–HCl (pH 7.4), 5 mM MgCl2, 1 mM DDT and 1 mM PMSF) and pelleted by ultracentrifugation at 80 000 g in a SW40Ti (Beckman) for 35 min. Mitochondria were isolated from the crude cytoplasmic fraction by bench-top centrifugation at 6000 g for 15 min. The mitochondrial pellet was washed and pelleted twice in lysis buffer. The cytosolic fraction was obtained after removal of the microsomal fraction by ultracentrifugation at 100 000 g in a SW60Ti (Beckman) for 1 h.
Organelle protein extraction
Nuclear proteins were extracted by resuspending the isolated nuclei in five volumes of 20 mM HEPES (pH 7.9), 1.5 mM MgCl2, 0.42 M NaCl, 0.2 mM EDTA, 0.1% Triton X-100 and 25% glycerol for 30 min with gentle shaking at 4°C. The nuclei were lysed by 10 passages through an 18-gauge needle and debris were removed by microcentrifugation at 13 000 r.p.m. The supernatant served as nuclear fraction. Mitochondrial proteins were isolated by incubating the mitochondria in a hypotonic lysis buffer (10 mM HEPES, pH 7.9) for 30 min on ice, briefly sonicated and debris pelleted at 13 000 r.p.m. for 30 min.
Protein digestion and MudPIT analysis
An aliquot of 150 μg of total protein from each fraction was precipitated overnight with five volumes of ice-cold acetone followed by centrifugation at 13 000 r.p.m. for 20 min. The protein pellet was solubilized in 8 M urea, 50 mM Tris–HCl (pH 8.5) and 1 mM DTT for 1 h followed by carboxyamidomethylation with 5 mM iodoacetamide for 1 h at 37°C. The samples were then diluted to 4 M urea with 100 mM ammonium bicarbonate (pH 8.5) and digested overnight with endoproteinase Lys-C at 37°C. The next day, the mixture was further diluted to 2 M urea with 50 mM ammonium bicarbonate (pH 8.5) supplemented with CaCl2 to a final concentration of 1 mM and incubated overnight with Porozyme trypsin beads at 30°C with rotation. The resulting peptide mixture was solid phase-extracted with SPEC-Plus C18 cartridges according to the manufacture's instruction and stored at −70°C.
Mass spectrometry
A fully automated 12-cycle, 24 h MudPIT chromatographic procedure was set up essentially as described previously (Washburn et al, 2001). Briefly, an HPLC quaternary pump was interfaced with an LCQ DECA XP ion trap tandem mass spectrometer (ThermoFinnigan, San Jose, CA). A 100 μm inner diameter fused silica capillary microcolumn (Polymicro Technologies, Phoenix, AZ) was pulled to a fine tip using a P-2000 laser puller (Sutter Instruments, Novato, CA) and packed with 10 cm of 5-μm Zorbax Eclipse XDB-C18 resin (Agilent Technologies, Mississagua, Ontario, Canada) and then with 6 cm of 5-μm Partisphere strong cation exchange resin (Whatman). Samples were loaded manually onto separate columns using a pressure vessel. The chromatography was carried out as described by Wolters et al (2001).
Protein identification and validation
The SEQUEST program (a kind gift from J Eng and J Yates III) was used to search peptide spectra essentially as described previously using a minimally redundant mouse/human protein sequence database (EBI database) (Eng et al, 1994; Kislinger et al, 2003). The statistical confidence of identified proteins was validated by the use of an in-house algorithm (STATQUEST as described earlier; Kislinger et al, 2003).
Quantitative analysis and data clustering
The profiles were clustered based on the Spearman rank correlation metric with average linkage using the freeware program Cluster 3.0 based on the original Cluster program (Eisen et al, 1998). Hierarchical clustering was performed using average linkage and the uncentered correlation metric. K-means clustering also used uncentered correlation metric and K was determined by a figure of merit calculation (Yeung et al, 2001). The resulting clusters were visualized in heat-map format using Java TreeView based on the original TreeView (Eisen et al, 1998). Spectral counts were normalized essentially as previously described (Cox et al, 2005). Relative protein abundance was inferred using the normalized spectral counts as a semiquantitative metric after arcsine(H) transformation of the data. Arcsine(H) transformation has the advantage over log transformation as zeros transform into zeros. Further, for proteins with low spectral counts, statistically insignificant changes in counts across experimental conditions can give a false impression of large changes in expression levels when data has been transformed as log ratios.
Cloning and expression of GFP fusions
Plasmids with cDNAs of interest were ordered as transformed bacterial glycerol stocks from Open Biosystems (Pycr2, MMM1013-63668; Ssu72, MMM1013-65805; Sltm, MMM1013-66003; 0710008K08Rik, MMM1013-7513094; Serpina1b, MMM1013-7513091; A730098D12Rik, MHS1010-9205573; Tfg, MMM1013-7510551; Echdc2, MMM1013-7513730; Cyb5r3, MMM1013-7513899; Thrap3, EMM1002-8537; BC018371, MMM1013-9200793; Rbm3, MMM1013-98478344). Primers were designed to amplify the open reading frame from the ATG to the last amino-acid codon, removing the stop codon. Restriction sites were added to the primers to facilitate cloning into the GFP fusion vector (pEGFP-N1, Clonetech). PCR was preformed with a high-fidelity PCR kit (BD Biosciences, Advantage 2) and PCR products were TA cloned into pCR2.1-TOPO vector (Invitrogen). Sequence verified clones were then subcloned into the fusion vector and checked by restriction digestion and PCR to verify the insert. 293T were plated 24 h before transfection on to 35 mm glass bottom culture dishes (MatTek, P35G-0-10-C) to achieve 50−80% confluence by the following day. Cell were grown in DMEM (Gibco, 11960) supplemented with L-glutamine, sodium pyruvate, penicillin, streptomycin and 10% fetal bovine serum (Wisent, 080150, lot 11514). Approximately 1.5 μg of a plasmid containing a GFP fusion construct was transfected using FuGENE 6 (Roche, 11814443001) as per the product instructions. The cells were cultured for a further 24 h before imaging. Approximately 30 min before imaging the cells were labeled with 5 μl Hoechst 33342 and 0.4 μl Mito Tracker Red CMXRos dye (Molecular Probes, I34154) The cells were live imaged on a Zeiss Axiovert 200 M inverted microscope fitted with a Volocity spinning disk confocal system. Channels were sequentially scanned and collected for each fluorophore using a × 63 oil-immersion objective.
Mathematical modeling
SwissProt/Trembl (SPTR) IDs from the protein data set were mapped to the Mu11K chip set via the Affymetrix website (www.affymetrix.com/analysis/index.affx) in order to relate the two data sets. All redundantly matching microarray probe sets were included with the protein. The model for comparison of protein data and microarray data was as previously described (Kislinger et al, 2006). K-means clustering of the correlated data sets was selected after applying a figure of merit to determine the optimal rage of clusters (K) for each group.
Protein–protein interaction analysis
Human homologues of selected target genes were mapped to SwissProt (build 49.1) to query the protein–protein interaction database, OPHID (REF); (http://ophid.utoronto.ca). OPHID comprises 11 109 proteins and 57 081 interactions, of which 33 713 are known and 23 368 are predicted interologs from Mus musculus, D. melanogaster, Caenorhabditis elegans, and Saccharomyces cerevisiae. Visualization of the resulting network (49 proteins, 107 interactions) was achieved using NAViG@Tor (http://ophid.utoronto.ca/navigator).
KNN prediction of subcellular localization and annotation of organelle localization
The algorithm first gathered the K nearest neighbors for a given target protein based on the minimum Euclidean distance in the observed compartment profiles relative to a reference training set, and then predicted target localization based on a majority vote (Liu et al, 2003). As the training sets were unbalanced a 10-fold, cross-validation procedure was used as there were a greater number of negative than positive examples. The optimal number of neighbors (K=15) was chosen by testing a range values for K from 9 to 20. A confidence score indicating the probability of a prediction was generated based the proportion of votes for the winning class. Classifier specificity, sensitivity, accuracy and precision were evaluated by AUC analysis (Liu et al, 2003). AUC curves are the ratio of the perfect learner (i.e. where all positives are perfectly predicted and all negatives are excluded) and the actual learner. The actual learner was assessed with ROC plots. An AUC score of 1.0 would be a perfect learner, whereas 0.5 would be random guessing. Precision, sensitivity and specificity were calculated using previously published methods (Lu et al, 2004b).
Background error for protein quantitation
Triplicate analyses of E18 lung mitochondria were analyze to generate a replicate sample set of proteins that were detected in all three replicates and quantified by spectral counts. From the triplicates, a distribution of duplicates was generated using all combinations of pairs of triplicates (total of three combinations). The duplicates were then summed to get an experimental value. Ratios of the experimental values were calculated in all combinations (total of three combinations) to generate a distribution of ratios. Ratios were sorted into bins ranging from <0.2- to >5-fold by single-fold increments (1.1–2, 2.1–3, etc) and graphed to empirically determine the background error for determination of ratios in the replicate samples.
Supplementary Material
Supplementary Tables
Supplementary Figures
Acknowledgments
We thank Eric Sat for assistance in genenotyping and breeding the Nmyc9a mice and Owen Tamplin for assistance with confocal microscopy. This work was supported by grants from the Canadian Institutes of Health Research to JR and grants from Canadian Institutes of Health Research, the McLaughlin Centre for Molecular Medicine, Genome Canada and the Ontario Genomics Institute (OGI) and the Natural Science and Engineering Council of Canada (NSERC) to AE. DAW was supported by a Canadian Institutes of Health Research postdoctoral fellowship, TK was supported in part by the Josef Schormüller Gedächtnisstiftung, BC by a Canadian Institutes of Health Research Doctoral fellowship and JR a distinguished investigator award from the CIHR. OPHID is supported by funding from Genome Canada through the Ontario Genomics Institute, the Younger and Firemen Foundations and IBM.
References
- Arman E, Haffner-Krausz R, Gorivodsky M, Lonai P (1999) Fgfr2 is required for limb outgrowth and lung-branching morphogenesis. Proc Natl Acad Sci USA 96: 11895–11899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballif BA, Villen J, Beausoleil SA, Schwartz D, Gygi SP (2004) Phosphoproteomic analysis of the developing mouse brain. Mol Cell Proteomics 3: 1093–1101 [DOI] [PubMed] [Google Scholar]
- Barlow DP, Stoger R, Herrmann BG, Saito K, Schweifer N (1991) The mouse insulin-like growth factor type-2 receptor is imprinted and closely linked to the Tme locus. Nature 349: 84–87 [DOI] [PubMed] [Google Scholar]
- Bartram U, Speer CP (2004) The role of transforming growth factor beta in lung development and disease. Chest 125: 754–765 [DOI] [PubMed] [Google Scholar]
- Bellusci S, Furuta Y, Rush MG, Henderson R, Winnier G, Hogan BL (1997a) Involvement of Sonic hedgehog (Shh) in mouse embryonic lung growth and morphogenesis. Development 124: 53–63 [DOI] [PubMed] [Google Scholar]
- Bellusci S, Grindley J, Emoto H, Itoh N, Hogan BL (1997b) Fibroblast growth factor 10 (FGF10) and branching morphogenesis in the embryonic mouse lung. Development 124: 4867–4878 [DOI] [PubMed] [Google Scholar]
- Bonner AE, Lemon WJ, You M (2003) Gene expression signatures identify novel regulatory pathways during murine lung development: implications for lung tumorigenesis. J Med Genet 40: 408–417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boon K, Caron HN, van Asperen R, Valentijn L, Hermus MC, van Sluis P, Roobeek I, Weis I, Voute PA, Schwab M, Versteeg R (2001) N-myc enhances the expression of a large set of genes functioning in ribosome biogenesis and protein synthesis. EMBO J 20: 1383–1393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodeur GM, Seeger RC, Schwab M, Varmus HE, Bishop JM (1985) Amplification of N-myc sequences in primary human neuroblastomas: correlation with advanced disease stage. Prog Clin Biol Res 175: 105–113 [PubMed] [Google Scholar]
- Bromberg J (2002) Stat proteins and oncogenesis. J Clin Invest 109: 1139–1142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown KR, Jurisica I (2005) Online predicted human interaction database. Bioinformatics 21: 2076–2082 [DOI] [PubMed] [Google Scholar]
- Burri PH, Tarek MR (1990) A novel mechanism of capillary growth in the rat pulmonary microcirculation. Anat Rec 228: 35–45 [DOI] [PubMed] [Google Scholar]
- Cardoso WV, Lu J (2006) Regulation of early lung morphogenesis: questions, facts and controversies. Development 133: 1611–1624 [DOI] [PubMed] [Google Scholar]
- Charron J, Malynn BA, Fisher P, Stewart V, Jeannotte L, Goff SP, Robertson EJ, Alt FW (1992) Embryonic lethality in mice homozygous for a targeted disruption of the N-myc gene. Genes Dev 6: 2248–2257 [DOI] [PubMed] [Google Scholar]
- Chuang PT, McMahon AP (2003) Branching morphogenesis of the lung: new molecular insights into an old problem. Trends Cell Biol 13: 86–91 [DOI] [PubMed] [Google Scholar]
- Cole MD, McMahon SB (1999) The Myc oncoprotein: a critical evaluation of transactivation and target gene regulation. Oncogene 18: 2916–2924 [DOI] [PubMed] [Google Scholar]
- Cox B, Emili A (2006) Tissue subcellular fractionation and protein extraction for use in mass-spectrometry-based proteomics. Nat Protoc 1: 1872–1878 [DOI] [PubMed] [Google Scholar]
- Cox B, Kislinger T, Emili A (2005) Integrating gene and protein expression data: pattern analysis and profile mining. Methods 35: 303–314 [DOI] [PubMed] [Google Scholar]
- Desai TJ, Chen F, Lu J, Qian J, Niederreither K, Dolle P, Chambon P, Cardoso WV (2006) Distinct roles for retinoic acid receptors alpha and beta in early lung morphogenesis. Dev Biol 291: 12–24 [DOI] [PubMed] [Google Scholar]
- Durr E, Yu J, Krasinska KM, Carver LA, Yates JR, Testa JE, Oh P, Schnitzer JE (2004) Direct proteomic mapping of the lung microvascular endothelial cell surface in vivo and in cell culture. Nat Biotechnol 22: 985–992 [DOI] [PubMed] [Google Scholar]
- East L, McCarthy A, Wienke D, Sturge J, Ashworth A, Isacke CM (2003) A targeted deletion in the endocytic receptor gene Endo180 results in a defect in collagen uptake. EMBO Rep 4: 710–716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eblaghie MC, Reedy M, Oliver T, Mishina Y, Hogan BL (2006) Evidence that autocrine signaling through Bmpr1a regulates the proliferation, survival and morphogenetic behavior of distal lung epithelial cells. Dev Biol 291: 67–82 [DOI] [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D (1998) Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA 95: 14863–14868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eng JK, McCormack AL, Yates JR III (1994) An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J Am Soc Mass Spectrom 5: 976–989 [DOI] [PubMed] [Google Scholar]
- Engelholm LH, Nielsen BS, Netzel-Arnett S, Solberg H, Chen XD, Lopez Garcia JM, Lopez-Otin C, Young MF, Birkedal-Hansen H, Dano K, Lund LR, Behrendt N, Bugge TH (2001) The urokinase plasminogen activator receptor-associated protein/endo180 is coexpressed with its interaction partners urokinase plasminogen activator receptor and matrix metalloprotease-13 during osteogenesis. Lab Invest 81: 1403–1414 [DOI] [PubMed] [Google Scholar]
- Florens L, Washburn MP, Raine JD, Anthony RM, Grainger M, Haynes JD, Moch JK, Muster N, Sacci JB, Tabb DL, Witney AA, Wolters D, Wu Y, Gardner MJ, Holder AA, Sinden RE, Yates JR, Carucci DJ (2002) A proteomic view of the Plasmodium falciparum life cycle. Nature 419: 520–526 [DOI] [PubMed] [Google Scholar]
- Foster LJ, de Hoog CL, Zhang Y, Xie X, Mootha VK, Mann M (2006) A mammalian organelle map by protein correlation profiling. Cell 125: 187–199 [DOI] [PubMed] [Google Scholar]
- Griffin TJ, Gygi SP, Ideker T, Rist B, Eng J, Hood L, Aebersold R (2002) Complementary profiling of gene expression at the transcriptome and proteome levels in Saccharomyces cerevisiae. Mol Cell Proteomics 1: 323–333 [DOI] [PubMed] [Google Scholar]
- Gygi SP, Rochon Y, Franza BR, Aebersold R (1999) Correlation between protein and mRNA abundance in yeast. Mol Cell Biol 19: 1720–1730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagel M, George EL, Kim A, Tamimi R, Opitz SL, Turner CE, Imamoto A, Thomas SM (2002) The adaptor protein paxillin is essential for normal development in the mouse and is a critical transducer of fibronectin signaling. Mol Cell Biol 22: 901–915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hokuto I, Ikegami M, Yoshida M, Takeda K, Akira S, Perl AK, Hull WM, Wert SE, Whitsett JA (2004) Stat-3 is required for pulmonary homeostasis during hyperoxia. J Clin Invest 113: 28–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalinichenko VV, Lim L, Stolz DB, Shin B, Rausa FM, Clark J, Whitsett JA, Watkins SC, Costa RH (2001) Defects in pulmonary vasculature and perinatal lung hemorrhage in mice heterozygous null for the Forkhead Box f1 transcription factor. Dev Biol 235: 489–506 [DOI] [PubMed] [Google Scholar]
- Kawai T, Lal A, Yang X, Galban S, Mazan-Mamczarz K, Gorospe M (2006) Translational control of cytochrome c by RNA-binding proteins TIA-1 and HuR. Mol Cell Biol 26: 3295–3307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura S, Hara Y, Pineau T, Fernandez-Salguero P, Fox CH, Ward JM, Gonzalez FJ (1996) The T/ebp null mouse: thyroid-specific enhancer-binding protein is essential for the organogenesis of the thyroid, lung, ventral forebrain, and pituitary. Genes Dev 10: 60–69 [DOI] [PubMed] [Google Scholar]
- Kishima Y, Yamamoto H, Izumoto Y, Yoshida K, Enomoto H, Yamamoto M, Kuroda T, Ito H, Yoshizaki K, Nakamura H (2002) Hepatoma-derived growth factor stimulates cell growth after translocation to the nucleus by nuclear localization signals. J Biol Chem 277: 10315–10322 [DOI] [PubMed] [Google Scholar]
- Kislinger T, Cox B, Kannan A, Chung C, Hu P, Ignatchenko A, Scott MS, Gramolini AO, Morris Q, Hallett MT, Rossant J, Hughes TR, Frey B, Emili A (2006) Global survey of organ and organelle protein expression in mouse: combined proteomic and transcriptomic profiling. Cell 125: 173–186 [DOI] [PubMed] [Google Scholar]
- Kislinger T, Rahman K, Radulovic D, Cox B, Rossant J, Emili A (2003) PRISM, a generic large scale proteomic investigation strategy for mammals. Mol Cell Proteomics 2: 96–106 [DOI] [PubMed] [Google Scholar]
- Koller A, Washburn MP, Lange BM, Andon NL, Deciu C, Haynes PA, Hays L, Schieltz D, Ulaszek R, Wei J, Wolters D, Yates JR III (2002) Proteomic survey of metabolic pathways in rice. Proc Natl Acad Sci USA 99: 11969–11974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipshitz HD, Smibert CA (2000) Mechanisms of RNA localization and translational regulation. Curr Opin Genet Dev 10: 476–488 [DOI] [PubMed] [Google Scholar]
- Liu H, Sadygov RG, Yates JR III (2004) A model for random sampling and estimation of relative protein abundance in shotgun proteomics. Anal Chem 76: 4193–4201 [DOI] [PubMed] [Google Scholar]
- Liu T, Moore A, Gray A (2003) Efficient exact k-NN and nonparametric classification in high dimensions. In Proceedings of Neural Information Processing Systems, Vol. 15 [Google Scholar]
- Lu J, Qian J, Izvolsky KI, Cardoso WV (2004a) Global analysis of genes differentially expressed in branching and non-branching regions of the mouse embryonic lung. Dev Biol 273: 418–435 [DOI] [PubMed] [Google Scholar]
- Lu Z, Szafron D, Greiner R, Lu P, Wishart DS, Poulin B, Anvik J, Macdonell C, Eisner R (2004b) Predicting subcellular localization of proteins using machine-learned classifiers. Bioinformatics 20: 547–556 [DOI] [PubMed] [Google Scholar]
- Mariani TJ, Reed JJ, Shapiro SD (2002) Expression profiling of the developing mouse lung: insights into the establishment of the extracellular matrix. Am J Respir Cell Mol Biol 26: 541–548 [DOI] [PubMed] [Google Scholar]
- Moens CB, Auerbach AB, Conlon RA, Joyner AL, Rossant J (1992) A targeted mutation reveals a role for N-myc in branching morphogenesis in the embryonic mouse lung. Genes Dev 6: 691–704 [DOI] [PubMed] [Google Scholar]
- Moens CB, Stanton BR, Parada LF, Rossant J (1993) Defects in heart and lung development in compound heterozygotes for two different targeted mutations at the N-myc locus. Development 119: 485–499 [DOI] [PubMed] [Google Scholar]
- Monkley SJ, Zhou XH, Kinston SJ, Giblett SM, Hemmings L, Priddle H, Brown JE, Pritchard CA, Critchley DR, Fassler R (2000) Disruption of the talin gene arrests mouse development at the gastrulation stage. Dev Dyn 219: 560–574 [DOI] [PubMed] [Google Scholar]
- Mootha VK, Bunkenborg J, Olsen JV, Hjerrild M, Wisniewski JR, Stahl E, Bolouri MS, Ray HN, Sihag S, Kamal M, Patterson N, Lander ES, Mann M (2003) Integrated analysis of protein composition, tissue diversity, and gene regulation in mouse mitochondria. Cell 115: 629–640 [DOI] [PubMed] [Google Scholar]
- Murakami T, Ohmori H, Gotoh S, Tsuda T, Ohya R, Akiya S, Higashi K (1991a) Down modulation of N-myc, heat-shock protein 70, and nucleolin during the differentiation of human neuroblastoma cells. J Biochem (Tokyo) 110: 146–150 [DOI] [PubMed] [Google Scholar]
- Murakami T, Ohmori H, Tsuda T, Higashi K (1991b) The effect of changes of N-myc products on the gene expression of heat-shock protein (hsp-70) and nucleolin during differentiation of neuroblastoma cells. Prog Clin Biol Res 366: 65–70 [PubMed] [Google Scholar]
- Ng YS, Rohan R, Sunday ME, Demello DE, D'Amore PA (2001) Differential expression of VEGF isoforms in mouse during development and in the adult. Dev Dyn 220: 112–121 [DOI] [PubMed] [Google Scholar]
- Okubo T, Knoepfler PS, Eisenman RN, Hogan BL (2005) Nmyc plays an essential role during lung development as a dosage-sensitive regulator of progenitor cell proliferation and differentiation. Development 132: 1363–1374 [DOI] [PubMed] [Google Scholar]
- Patan S, Alvarez MJ, Schittny JC, Burri PH (1992) Intussusceptive microvascular growth: a common alternative to capillary sprouting. Arch Histol Cytol 55 (Suppl): 65–75 [DOI] [PubMed] [Google Scholar]
- Patel JH, Loboda AP, Showe MK, Showe LC, McMahon SB (2004) Analysis of genomic targets reveals complex functions of MYC. Nat Rev Cancer 4: 562–568 [DOI] [PubMed] [Google Scholar]
- Sawai S, Shimono A, Hanaoka K, Kondoh H (1991) Embryonic lethality resulting from disruption of both N-myc alleles in mouse zygotes. New Biol 3: 861–869 [PubMed] [Google Scholar]
- Schirmer EC, Florens L, Guan T, Yates JR III, Gerace L (2003) Nuclear membrane proteins with potential disease links found by subtractive proteomics. Science 301: 1380–1382 [DOI] [PubMed] [Google Scholar]
- Schwab M, Varmus HE, Bishop JM (1985) Human N-myc gene contributes to neoplastic transformation of mammalian cells in culture. Nature 316: 160–162 [DOI] [PubMed] [Google Scholar]
- Sekine K, Ohuchi H, Fujiwara M, Yamasaki M, Yoshizawa T, Sato T, Yagishita N, Matsui D, Koga Y, Itoh N, Kato S (1999) Fgf10 is essential for limb and lung formation. Nat Genet 21: 138–141 [DOI] [PubMed] [Google Scholar]
- Serls AE, Doherty S, Parvatiyar P, Wells JM, Deutsch GH (2005) Different thresholds of fibroblast growth factors pattern the ventral foregut into liver and lung. Development 132: 35–47 [DOI] [PubMed] [Google Scholar]
- Sleutels F, Zwart R, Barlow DP (2002) The non-coding Air RNA is required for silencing autosomal imprinted genes. Nature 415: 810–813 [DOI] [PubMed] [Google Scholar]
- Suki B, Ito S, Stamenovic D, Lutchen KR, Ingenito EP (2005) Biomechanics of the lung parenchyma: critical roles of collagen and mechanical forces. J Appl Physiol 98: 1892–1899 [DOI] [PubMed] [Google Scholar]
- Ten Have-Opbroek AA (1991) Lung development in the mouse embryo. Exp Lung Res 17: 111–130 [DOI] [PubMed] [Google Scholar]
- Threadgill DW, Dlugosz AA, Hansen LA, Tennenbaum T, Lichti U, Yee D, LaMantia C, Mourton T, Herrup K, Harris RC, Barnard JA, Yuspa SH, Coffey RJ, Magnuson T (1995) Targeted disruption of mouse EGF receptor: effect of genetic background on mutant phenotype. Science 269: 230–234 [DOI] [PubMed] [Google Scholar]
- Wang ZQ, Fung MR, Barlow DP, Wagner EF (1994) Regulation of embryonic growth and lysosomal targeting by the imprinted Igf2/Mpr gene. Nature 372: 464–467 [DOI] [PubMed] [Google Scholar]
- Warburton D, Bellusci S, Del Moral PM, Kaartinen V, Lee M, Tefft D, Shi W (2003) Growth factor signaling in lung morphogenetic centers: automaticity, stereotypy and symmetry. Respir Res 4: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Washburn MP, Wolters D, Yates JR III (2001) Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat Biotechnol 19: 242–247 [DOI] [PubMed] [Google Scholar]
- Wolters DA, Washburn MP, Yates JR III (2001) An automated multidimensional protein identification technology for shotgun proteomics. Anal Chem 73: 5683–5690 [DOI] [PubMed] [Google Scholar]
- Yeung KY, Haynor DR, Ruzzo WL (2001) Validating clustering for gene expression data. Bioinformatics 17: 309–318 [DOI] [PubMed] [Google Scholar]
- Zekri L, Chebli K, Tourriere H, Nielsen FC, Hansen TV, Rami A, Tazi J (2005) Control of fetal growth and neonatal survival by the RasGAP-associated endoribonuclease G3BP. Mol Cell Biol 25: 8703–8716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zybailov B, Coleman MK, Florens L, Washburn MP (2005) Correlation of relative abundance ratios derived from peptide ion chromatograms and spectrum counting for quantitative proteomic analysis using stable isotope labeling. Anal Chem 77: 6218–6224 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Tables
Supplementary Figures