Systems biology is the latest fashion in biology, driven by advances in technology that have provided us with a suite of ‘omics' techniques providing information or at least data, which is not quite the same thing, but can easily be confused with it at a whole range of levels of biomolecular organisation from genes through proteins to metabolites. The hope is that all of these, when combined in some way, will provide new insights into important biological processes, such as development, ageing, disease and so on, to the benefit of, among other things, the development of new medicines and treatment regimes. While it is easy to be cynical and deride all the hype surrounding this new ‘revolution' in biology as simply a way of extracting grants from gullible funding agencies and merely yet another panacea for reversing the perceived decline in innovation in pharmaceutical research, there is no doubt that some of the results coming out of some of this work are scientifically very beautiful. For me this is particularly the case where the results provide new insight into something which we have known about for a long time, but which we have forgotten in the rush to gene-based solutions to all our problems. One area where this is clearly the case is the rediscovery of the intimate relationship that animals have with the organisms inhabiting their guts. This is very well illustrated in a recent study published in Molecular Systems Biology where François-Pierre Martin, Jeremy Nicholson and colleagues describe the results of a top-down view of these microbial–mammalian interactions showing the effects of different gut flora on the metabolic profiles of the host organism (Martin et al, 2007). The symbiotic relationship that animals have with their gut microflora is the product of co-evolution and has led to the use of the term ‘superorganism', originally proposed by Lederberg (2000), to describe the complex web of interactions that occur between the host and the consortium of gut microorganisms that constitute the ‘microbiome'. While the importance of the gut flora is well recognised, it is often forgotten and the sheer complexity of the gut ecosystem has required the development of modern molecular biology techniques (many of the organisms present cannot be cultured and are only known as a result of their detection via ribosomal 16S rDNA (Gill et al, 2006)). We now know that these microbial populations are immensely complex, with many hundreds of different species cohabiting in the gut and forming a complex ecology.
While the effects that the microflora can have on the metabolic profiles of biofluids such as urine have been recognised for sometime (Phipps et al, 1998), the experiments described in the current paper show the metabolic effects of inoculating germ-free mice with human-derived flora or exposing them to conventional mice, enabling them to acquire a normal mouse microbiome (Figure 1). Using a range of state-of-the-art metabolic profiling techniques, the authors were able to show profound differences between normal and ‘re-conventionalised' mice on the one hand and the ‘humanised' mice on the other in their metabolite profiles for biofluids such as plasma and urine, and in the metabolite profiles of organs such as the liver. These differences were reflected in marked changes in bile acid and short-chain fatty acid profiles obtained via the analysis of ileal washes and caecum contents, respectively. The application of metabonomics in this way revealed specific metabotypes (metabolic phenotypes) associated with the different microflora. There are also changes in bile acid composition noted by the authors (a shift to taurine-conjugated bile acids in humanised mice compared to cholic acid in conventional and re-conventionalised mice). The authors show that these changes are correlated with the gut microbial composition and suggest that humanised mice are relatively unable to hydrolyse taurine-conjugated bile acids, with a consequent effects on the absorption of dietary fat. The role of the gut microflora in modulating host lipid metabolism has recently been highlighted (Backed et al, 2004; Ley et al, 2006), and changes in plasma lipid profiles that relate to the type of gut microflora that are present in the current study support the importance of this interaction, with a concomitant increase in lipid accumulation in the livers of humanised animals.
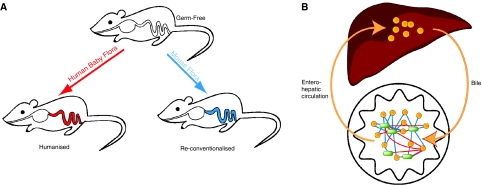
Figure 1.
Impact of the bacterial flora on host physiology. (A) Germ-free mice were either ‘humanised' by inoculation of human-derived flora (left red arrow) or ‘re-conventionalised' by exposure to normal non-germ-free mice (right blue arrow). (B) Bile acids (orange nodes) are produced in the liver, processed by the gut microflora (green nodes) and are recycled to the liver via the enterohepatic circulation. Analysis of the correlation between gut flora composition and bile acid composition reveals that in ‘re-conventionalised' mice, the correlation network is much denser (blue edges) than in ‘humanised' mice (red edges). These changes may underlie the profound changes observed in host lipid metabolism and illustrate the close adaptation of the flora to its natural host.
The authors show that both system-wide (i.e., whole organism) and organ-specific changes in metabolic profiles result from alterations in gut microbial populations. The implications of such studies are profound. If the microflora represent, as an increasing number of publications suggest, an important source of metabolic variability in the host, then studies that simply look at the host genome, for example, will miss a potentially major confounding factor (Nicholson et al, 2005). If in fact it is the gut flora that decides the amount of fat that the host absorbs from its diet, or modulates where that fat is distributed, then whether or not an individual is susceptible to, for example, type II diabetes or non-alcoholic liver disease may depend not only on the genetics and diet of the host but also critically on the microbiome. Increasingly, studies are showing significant effects on mammalian, including human, phenotypes resulting from differences in gut flora and frankly it does not require much imagination to see the profound consequences to human health, both positive and negative, that might result from changes in the normal gut microflora.
References
- Backed F, Ding H, Wang T, Hooper LV, Koh GY, Nagy A, Semenkovich CF, Gordon JI (2004) The gut microbiota as an environmental factor that regulates fat storage. Proc Natl Acad Sci USA 101: 15718–15723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill SR, Pop M, DeBoy RT, Eckburg PB, Turnbaugh PJ, Samuel BS, Gordon JI, Relman DA, Fraser-Liggett CM, Nelson KE (2006) Metagenomic analysis of the human distal gut microbiome. Science 312: 1355–1359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lederberg J (2000) Infectious history. Science 288: 287–293 [DOI] [PubMed] [Google Scholar]
- Ley R, Turnbaugh PJ, Klein S, Gordon J (2006) Microbial ecology: human gut microbes associated with obesity. Nature 444: 1022–1023 [DOI] [PubMed] [Google Scholar]
- Martin F-PJ, Dumas M, Wang Y, Legido-Quigley C, Tang H, Zirah S, Murphy GH, Cloarec O, lindon JC, Sprenger N, Fay LB, Kochhar S, Holmes E, Nicholson J (2007) A top-down systems biology view of microbiome–mammalian metabolic interactions in a mouse model. Mol Syst Biol 3:112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson JK, Holmes E, Wilson ID (2005) Gut microorganisms, mammalian metabolism and personalised healthcare. Nat Rev Microbiol 3: 431–438 [DOI] [PubMed] [Google Scholar]
- Phipps AN, Stewart J, Wright B, Wilson ID (1998) Effect of diet on the urinary excretion of hippuric acid and other dietary-derived aromatics in rat. A complex interaction between diet, gut microbiotia and substrate specificity. Xenobiotica 28: 527–537 [DOI] [PubMed] [Google Scholar]