Abstract
Cell adhesion and motility are very dynamic processes that require the temporal and spatial coordination of many cellular structures. ADP-ribosylation factor 6 (Arf6) has emerged as master regulator of endocytic membrane traffic and cytoskeletal dynamics during cell movement. Recently, a novel Arf6-binding protein known as FIP3/arfophilin/eferin has been identified. In addition to Arf6, FIP3 also interacts with Rab11, a small monomeric GTPase that regulates endocytic membrane transport. Both Arf6 and Rab11 GTPases have been implicated in regulation of cell motility. Here we test the role of FIP3 in breast carcinoma cell motility. First, we demonstrate that FIP3 is associated with recycling endosomes that are present at the leading edge of motile cells. Second, we show that FIP3 is required for the motility of MDA-MB-231 breast carcinoma cells. Third, we demonstrate that FIP3 regulates Rac1-dependent actin cytoskeleton dynamics and modulates the formation and ruffling of lamellipodia. Finally, we demonstrate that FIP3 regulates the localization of Arf6 at the plasma membrane of MDA-MB-231 cells. Based on our data we propose that FIP3 affects cell motility by regulating Arf6 localization to the plasma membrane of the leading edge, thus regulating polarized Rac1 activation and actin dynamics.
Keywords: Cell motility, Rab11, Rab11-FIP3, Endosomes, Actin cytoskeleton, Arf6
Introduction
Cell adhesion and motility are dynamic processes requiring both temporal and spatial coordination of many cellular structures. A key regulator of cell motility is the actin cytoskeleton. Cell motility requires continuous rearrangement of the cytoskeleton at the leading and retracting edges of moving cells. Much of this rearrangement is modulated by the coordinated action of small Ras-like GTPases of the Rac/Rho family. The prototype Rho family members, RhoA, Rac1, and Cdc42 are known for their specific effects on the actin cytoskeleton (Hall, 1998; Mackay and Hall, 1998). In particular, in migratory cells Rac1 stimulates the formation of the broad, actin-rich, membrane protrusions at the cell edges, called lamellipodia (Braga et al., 1997; Palacios and D’Souza-Schorey, 2003; Takaishi et al., 1997).
Vectorial cell migration also necessitates the extension of lamellipodia or filopodia in the direction of movement. It has been suggested that the generation of lamellipodia or filopodia requires massive directional influx of membrane (Nabi, 1999). This membrane influx may be achieved via targeted delivery of the membranes to the leading edge from the Golgi apparatus and/or endosomes. Interestingly, endosomes also are involved in several other processes that require massive influx of plasma membrane such as phagocytosis and cytokinesis (Cox et al., 2000; Riggs et al., 2003; Skop et al., 2001; Wilson et al., 2005).
Despite the involvement of cytoskeleton and endocytic membrane traffic in regulation of cell motility, we are only beginning to understand the machinery coordinating both of these processes. Recently, ADP-ribosylation factor 6 (Arf6) has emerged as master regulator of both vesicular transport and cytoskeleton dynamics (Donaldson and Honda, 2005; Sabe, 2003; Turner and Brown, 2001). Since both of these processes are important in regulation of cell motility, it is not very surprising that multiple studies have suggested that Arf6 plays a key role in regulation of cell movement. For example, Arf6 has been implicated in regulation of endocytic integrin recycling, as well as endocytosis of E-cadherin, a cell-cell adhesion molecule (D’Souza-Schorey et al., 1998; Dunphy et al., 2006; Palacios and D’Souza-Schorey, 2003; Powelka et al., 2004). It has been reported that Arf6 binds and regulates phosphatidylinositol 4-phosphate 5-kinase Iγ (PI4P-5K), as well as, phospholipase D (PLD) thus regulating the production of phosphatidylinositol (4,5)-bisphosphate (PI(4,5)P2) (Hernandez-Deviez et al., 2004; Hiroyama and Exton, 2005; Honda et al., 1999; Powner et al., 2002; Powner and Wakelam, 2002). Since PI(4,5)P2 regulates clathrin-mediated endocytosis, it is likely that Arf6 may regulate AP-2 and clathrin coat recruitment to the endocytic pits (Krauss et al., 2003; Paleotti et al., 2005). In addition, at the lateral membrane of epithelial cells Arf6 binds and recruits Nm23-H1 nucleoside diphosphate kinase, thereby regulating endocytosis of E-cadherin molecules (Palacios et al., 2002). Arf6 was also implicated in regulation of the endocytosis pathway that is used to internalize β1-integrin (for review see (Donaldson, 2003)).
In addition to regulating endocytosis and the recycling of integrins and cell-cell adhesion molecules, Arf6 is also known for its role in regulating the actin cytoskeleton. Over-expression of constitutively active Arf6 or its activator ARNO has been shown to induce the formation of large lamellipodia and stimulate cell motility in epithelial cells (Boshans et al., 2000; Radhakrishna et al., 1999; Santy and Casanova, 2001) by activating Rac1 GTPase and regulating actin dynamics (Radhakrishna et al., 1999; Santy and Casanova, 2001; Santy et al., 2005). Furthermore, it has been reported that Arf6 may regulate the redistribution of Rac1 from endosomes to the plasma membrane (Radhakrishna et al., 1999). Alternatively, Arf6 may regulate the recruitment of the Rac1 GEF, DOCK180-ELMO complex (Santy et al., 2005) or by binding to POR1, a Rac1-interacting protein (D’Souza-Schorey et al., 1997). It was recently shown that Arf6 regulates recruitment and activation of Rac GEF Kalirin (Koo et al., 2007). Finally, the increase in PI(4,5)P2 levels also can effect actin dynamics by regulating actin capping proteins.
Recently, an Arf-binding protein, named arfophilin, has been identified (Shin et al., 1999, 2001). While originally identified as an Arf5-binding protein, arfophilin was later shown to prefer Arf6 over Arf5 in vitro and in vivo (Fielding et al., 2005). Interestingly, arfophilin is identical to Rab11-FIP3/Eferin, which was independently identified as a Rab11-binding protein (Hales et al., 2001; Prekeris et al., 2001). Based on sequence homology Rab11-FIP3 belongs to a recently described FIP family of Rab11-binding proteins and will be referred to here as FIP3 (Hales et al., 2001; Lindsay et al., 2002; Prekeris et al., 2000, 2001).
The discovery that FIP3 can simultaneously bind to Rab11 and Arf6 GTPases has raised the possibility that endosome-associated FIP3 may play a role in regulating endocytic transport and cytoskeleton dynamics during cell motility. Rab11 GTPase is well known for its role in regulating endocytic membrane traffic. Furthermore, it has been suggested that both Rab11 and Arf6 are involved in regulating the same β1-integrin recycling pathway (Parsons et al., 2005). In this study we investigated the role of FIP3 in regulating motility of MDA-MB-231 breast carcinoma cells. First, we demonstrate that FIP3 is associated with Rab11-containing recycling endosomes that are present at the leading edge of motile cells. Second, we show that FIP3 is required for the motility of MDA-MB-231 breast carcinoma cells. Third, we demonstrate that while having no effect on integrin recycling, FIP3 regulates the polarized Rac1-dependent actin cytoskeleton dynamics, as well as formation and ruffling of the lamellipodia. Finally, we demonstrate that Arf6 binding to FIP3 is required for Arf6 targeting to the plasma membrane of the leading edge. Based on these data, we propose that FIP3 regulates polarized Rac1 activation by mediating the targeting of Arf6 to the leading edge of the moving breast cancer cells.
Materials and methods
Reagents and plasmids
Cell culture reagents were obtained from Invitrogen (Carlsbad, CA) unless otherwise specified. Most chemicals were from Sigma-Aldrich (St. Louis, MO) or Fisher (Pittsburgh, PA). Mouse monoclonal anti-HA antibody was purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Transferrin conjugated to Alexa488 (Tf-Alexa488) and phalloidin conjugated to Texas Red were purchased from Molecular Probes (Eugene, OR). Mouse monoclonal anti-transferrin receptor and rabbit polyclonal anti-Rab11 were purchased from Zymed laboratories (South San Francisco, CA). Mouse monoclonal anti-EEA1 and mouse monoclonal anti-Lamp1 antibodies were obtained from BD Biosciences (San José, CA). Rabbit polyclonal anti-Rac1 antibody was purchased from Cytoskeleton (Denver, CO). The polyclonal rabbit anti-FIP3 antibody was described previously (Wilson et al., 2005). The polyclonal goat anti-FIP3 antibody was generated using recombinant FIP3 as described previously (Meyers and Prekeris, 2002). Mouse anti-human EGF-R antibody (conjugated to Alexa647; clone 13A9), anti-β1-integrin (conjugated to APC) and anti-α5-integrin antibody (conjugated to PE) were generous gifts from Dr. Andrew Peden. Generation of the FIP3-GFP construct was as described previously (Wilson et al., 2005).
FIP3-YFP, CFP-Rab11a, HA-Arf6-27N, Arf6-CFP, myc-Rab11a-S25N and FIP3-GFP-I737E were previously described (Wilson et al., 2005; Fielding et al., 2005; Peden et al., 2004). FIP3-YFP-I737E and Arf6-CFP-W168L/L169V were generated using point-directed mutagenesis (BD Biosciences).
Cell culture and immunofluorescence microscopy
MDA-MB-231 and NIH-3T3 cells were cultured as described previously (Parsons et al., 2005). For immunofluorescence microscopy, cells were fixed with 4% paraformaldehyde for 15 min, permeabilized with 0.4% saponin, and non-specific sites were blocked with phosphate-buffered saline containing 0.2% bovine serum albumin and 1% fetal bovine serum. After incubation with specific antibodies, samples were extensively washed and mounted in VectaShield (Vector laboratories). Cells were imaged with an inverted Zeiss Axiovert 200M microscope. Image processing was performed using Intelligent Imaging Innovations three-dimensional rendering and exploration software.
RNA interference (RNAi)
Rab11a/b and FIP3#1 were depleted with siRNA oligonucleotides as described previously (Junutula et al., 2004; Wilson et al., 2005). siRNAs were co-transfected into MDA-MB-231 cells using Lipofectamine2000 (Gibco BRL) which allows >90% transfection efficiency of siRNA. Transfected cells were incubated for either 48 or 72 hours and analyzed for Rab11a/b or FIP3 expression by Western blotting (Supplemental Figure 1). Remaining cells were used for flow cytometry, fluorescence microscopy, cell attachment, cell spreading and cell motility studies. In most FIP3 siRNA experiments, the effect of siRNA treatment was confirmed with a second siRNA#2 oligo (5’-aaggcgtgtgctggagctgga-3’).
Transferrin and anti-EGF-R antibody uptake assays
For uptake assays, mock or FIP3 siRNA-treated MDA-MB-231 cells were incubated with 20 μg/ml Tf-Alexa488 at 4°C for 30 min. Cells were then incubated at 37°C for 30 min in the continuous presence of 20 μg/ml Tf-Alexa488 (pre-incubation at 4°C was required to generate sufficient signal for detection by flow cytometry). Cells were washed and the experiment completed by pelleting and resuspending the cells in 3% paraformaldehyde. Cell-associated Tf-Alexa488 was determined by flow cytometry analysis.
To analyze the presence of EGFR and TfR at the plasma membrane, mock or FIP3 siRNA-treated MDA-MB-231 cells were incubated with 20 μg/ml Tf-Alexa488 or anti-EGF-R conjugated to Alexa647 at 4°C for 60 min. Cells were washed and the experiment completed by pelleting and resuspending the cells in 3% paraformaldehyde. Cell-associated Tf-Alexa488 and anti-EGFR-Alexa647 were determined by flow cytometric analysis.
Flow cytometry analyses were performed using a Cytomics™ FC 500 flow cytometer (Beckman Coulter) equipped with 488-nm and 647-nm lasers at the University of Colorado Cancer Center Flow Cytometry Core Facility.
Cell motility assays
Motility of MDA-MB-231cells was measured by modified Boyden chamber assays. Briefly, mock or RNAi-treated cells were resuspended in Opti-Mem media and added to the top chamber of 8 μm pore size Transwell filter insets (6.5 mm filters, Corning Inc., Corning, NY) at 100,000 cells/filter. NIH-3T3 cell conditioned medium was added to the bottom chamber. After 8 hours, cells remaining on the upper surface of the filter were removed with a cotton swab. Cells on the bottom side were stained with 0.1% crystal violet. The dye was extracted into 10% sodium deoxycholate and quantitated by measuring the optical density at 590 nm. Under these conditions, about 60% of cells usually migrate to the bottom chamber. When transfected with FIP3-GFP or FIP3-GFP-I737E cells were incubated for 18 hours and then plated on Transwell filter insets at 100,000 cells/filter. After 8 hours, cell migration was estimated by counting transfected cells (as determined by presence of GFP fluorescence) on the upper or bottom surfaces of the filter. The total number of cells was calculated by adding transfected cells from both surfaces. The values presented are the percentage of transfected cells that migrated to the bottom surface.
For single-cell velocity studies, MDA-MB-231 or NIH-3T3 cells were plated on fibronectin-coated (at 20 μg/ml) glass coverslips in the presence of NIH-3T3 cell conditioned medium. Cells were then mounted on a PH2 heated platform fitted with a TC-344B dual automatic temperature controller (Warner Instruments). Cells were imaged at 37°C on an inverted microscope (Zeiss Axiovert 200M) using a 40× dry lens. Images were acquired and analyzed using Intelligent Imaging Innovations 3D rendering and exploration software. Time-lapse images (exposure 500 ms) were taken every 12 min. For each series 20 images were taken. To quantitate cell motility, the speeds of ten randomly chosen cells from two different time-lapse series were analyzed. The center of the nucleus was used as a reference point, and travel distance was measured between each frame. The speed was the expressed as distance in mm traveled per 1 hour.
For kymography, five randomly chosen MDA-MB-231 cells for each treatment condition were imaged at 37°C using a 63× oil lens. All images were taken in the presence of NIH-3T3 cell-conditioned medium. Time-lapse images (exposure 300 ms) were taken every 5 s. For each series 100 images were taken. Three kymographs from the most dynamic edge were collected from every cell. Kymographs were analyzed using Intelligent Imaging Innovations 3D rendering and exploration software as previously described (Hinz et al., 1999). Three main measurements were obtained: ruffling frequency, ruffle retraction velocity, and ruffle retraction distance. For the definitions of these measurements see Supplemental Figure 4.
For wound healing cell motility assays, MDA-MB-231 cells were plated at 100% confluence on fibronectin-coated (at 20 μg/ml) glass coverslips. The surface of the coverslip was scratched with a p200 pipette tip and incubated at 37°C for varying amounts of time. Cells were then fixed with 4% paraformaldehyde, mounted and imaged on an inverted microscope (Zeiss Axiovert 200M) using a 10× dry phase lens.
Cell spreading assay
Mock or FIP3 siRNA-treated MDA-MB-231 cells were placed for 1 or 3 hours onto glass coverslips coated with collagen. After the allotted time, cells were fixed and stained as described above using rhodamine-conjugated phalloidin. Images were acquired using an inverted microscope (Zeiss Axiovert 200M) with 63× oil immersion lens. The cell edges were outlined based on rhodamine-phalloidin staining. The outlined surface areas were calculated using Intelligent Imaging Innovations 3D rendering and exploration software. To evaluate the polarization of cell spreading, the ratio between length and width of randomly chosen mock, FIP3 siRNA-treated or Rip11/FIP5 siRNA-treated cells (after 3 hours of spreading) were measured. For each cell, the length was measured as a distance along the longest cell axis. The width was measured along the line (at the widest point) at a 90° angle from the length axis.
Reverse transcriptase polymerase chain reaction (RT-PCR)
Total RNA was extracted from 2 × 107 MDA-MB-231 or MCF-7 cells using TRIzol (Invitrogen) according to the manufacturer’s protocol. Reverse transcription to cDNA was performed with SuperScript III (Invitrogen) using random hexamers. PCR was performed using Taq polymerase (Invitrogen). All reactions were performed for 40 cycles with the following parameters: 94°C, 60 s; 55°C, 60 s; 72°C, 90 s. Primers for amplification of Rab25 and Rab11a were previously reported and are expected to generate 759-bp (Rab11a) or 532-bp (Rab25) PCR fragments (Cheng et al., 2006).
Rac GTPase activation assays
MDA-MB-231 cells were grown on 10-cm dishes. Cells were either mock-transfected or transfected with FIP3 siRNAs. Cells were incubated for 72 hours and then quickly rinsed in ice-cold phosphate-buffered saline. Cells were lysed in the reaction buffer (20 mM Tris-HCl, pH 7.4, 20 mM NaCl, 5 mM MgCl2 and 0.5 % Triton X-100) with protease inhibitors. Lysates were centrifuged for 5 min at 10,000 rpm at 4°C. To determine the level of activated Rac1, lysates were incubated with GST-PAK(CRIB) beads, washed, and bound Rac1-GTP was analyzed by immuoblotting.
Fluorescence energy transfer (FRET) time-lapse microscopy
FRET between FIP3-YFP and Arf6-CFP was measured as described previously (Schonteich et al., 2007). Briefly, cells were co-tranduced with proteins tagged with YFP and CFP. Live cells were then mounted on a PH2 heated platform fitted with a TC-344B dual automatic temperature controller (Warner Instruments). Cells co-expressing YFP and CFP-tagged proteins were randomly chosen and imaged at 37°C using an inverted microscope (Zeiss Axiovert 200M) with a 63× oil immersion lens. Corrected FRET (cFRET) was calculated using Intelligent Imaging Innovations three-dimensional rendering and exploration software as described previously using the equation cFRET = FRET - 0.22 × CFP - 0.037 × YFP (Schonteich et al., 2007). In all cases background cFRET was calculated by random sampling outside the cell and subtracted from images. Only cells expressing similar amounts of CFP- and YFP-tagged proteins (YFP/CFP=0.5-2.0) were included in the FRET analysis.
Results
FIP3 is localized at the leading edge and centrosomes of breast cancer cells
The formation and extension of the leading edge is a key step in mediating the movement of cells. Multiple proteins that regulate cell motility, including Arf6 GTPase, are enriched at the leading edge of the moving cell. We therefore first sought to localize FIP3 in motile cells. To that end, we stained MDA-MB-231 breast cancer cells with anti-FIP3 antibody. As shown in Figure 1A-C, a small population of FIP3 is present at the leading edge of the cell, as marked by the actin cytoskeleton. A larger amount of FIP3 is present in multiple small punctate structures exhibiting peri-centriolar localization (see Supplemental Fig. 3). This is consistent with previous work demonstrating that FIP3 associates with the centrosomes during mitosis (Hickson et al., 2003; Wilson et al., 2005). Note that some anti-FIP3 staining can also be observed in the nucleus. This staining appears to be non-specific, since it persists in FIP3 siRNA-treated cells (Supplemental Fig. 1C and D).
Fig. 1.
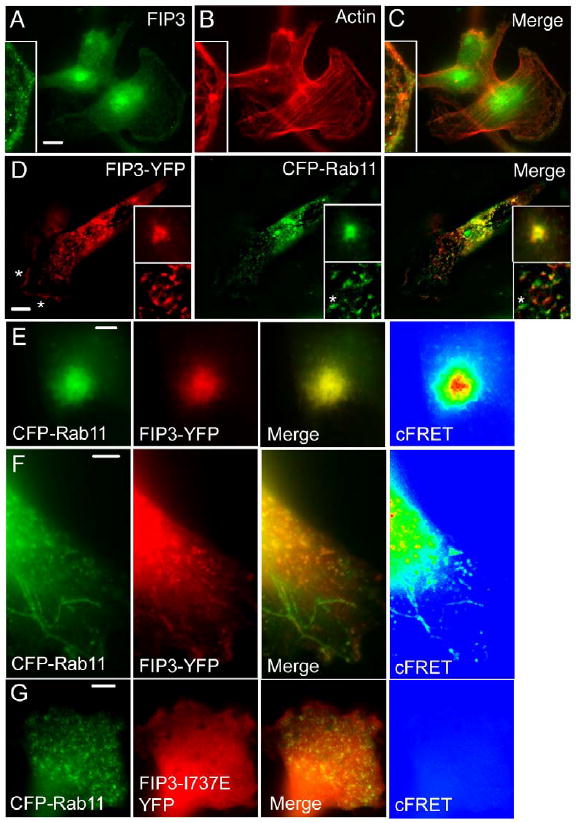
Sub-cellular localization of FIP3 in MDA-MB-231 cells. (A-C) MDA-MB-231 cells were plated on collagen-coated coverslips, fixed with paraformaldehyde and stained with anti-FIP3 antibodies (A) or rhodamine-labeled phalloidin (B). (C) Merged image. (D) MDA-MB-231 cells were transfected with FIP3-YFP (red) and CFP-Rab11a (green). Yellow in the merged image represents the overlap between CFP-Rab11a and FIP3-YFP. Asterisks mark the leading edge of the cell. Top inset is the peri-centriolar FIP3-YFP and CFP-Rab11a. Bottom inset is a magnified image of FIP3-YFP and CFP-Rab11a-containing endosomes. Asterisk in bottom inset marks an endosome that is positive for CFP-Rab11a but not FIP3-YFP. (E-G) MDA-MB-231 cells were co-transfected with FIP3-YFP/CFP-Rab11 (E, F) or FIP3-YFP-I737E/CFP-Rab11 (G). Then cells were fixed and imaged. (E) Peri-centriolar FIP3YFP/CFP-Rab11. (F, G) Peripheral endosomes positive for CFP-Rab11 and FIP3-YFP or FIP3-I737E-YFP. Yellow in merge panels represents the overlap between FIP3-YFP, FIP3-YFP-I737E and CFP-Rab11. cFRET images were generated as described in Materials and methods. Note that images in (E) were taken at the centrosomal level of the cell, while images in (F, G) were taken at the foot of the cell to visualize the leading edge. Bars: 7 μm (A-C), 5 μm (D), 1 μm (E), 2 μm (F, G).
Given that FIP3 is a Rab11-binding protein, we determined whether the FIP3-positive structures described above also contained Rab11. MDA-MB-231 cells were co-transfected with FIP3-YFP and CFP-Rab11a. Both the FIP3-positive cytoplasmic punctate structures and the peri-centriolar structures were found to also contain Rab11, suggesting that FIP3 is recruited to these organelles by binding to Rab11 (Fig. 1D). Indeed, it was previously reported that FIP3-I737E, a mutant that does not bind to Rab11, exhibits primarily cytosolic distribution (Eathiraj et al., 2006; Shiba et al., 2006; Wilson et al., 2005). Similarly, in MDA-MB-231 cells FIP3-I737E also exhibited primarily cytosolic distribution (Fig. 1G and Fig. 6D). Interestingly, while all FIP3-positive organelles contain CFP-Rab11, the converse is not true. Some of the CFP-Rab11-positive organelles did not contain FIP3-YFP (Fig. 1D, middle and right panel, insets; also see Fig. 1F), suggesting that Rab11 also exists in complexes with other FIPs. Consistent with that idea, we have previously shown that Rab11 forms mutually exclusive complexes with various FIP molecules (Meyers and Prekeris, 2002).
Fig. 6.

FIP3-I737E inhibits actin ruffling at the leading edge. MDA-MB-231 cells were transfected with FIP3-GFP (A-C) or FIP3-GFP-I737E (D, E) and stained with rhodamine-phalloidin. Asterisks mark the leading edge. Bars: 5 μm.
To further confirm that FIP3 and Rab11 form complexes in peri-centriolar as well as peripheral organelles we performed FRET analysis of cells co-expressing FIP3-YFP and CFP-Rab11. As shown in Figure 1E, FRET was observed between FIP3-YFP and CFP-Rab11 in peri-centriolar organelles. Similarly, FRET was detected in peripheral organelles (Fig. 1F). In contrast, no FRET was observed in peripheral organelles (Fig. 1G) or peri-centriolar structures (data not shown) when CFP-Rab11 and FIP3-I7373E-YFP were co-expressed.
FIP3 and Rab11 have been shown to associate with recycling endosomes in HeLa cells (Hickson et al., 2003; Horgan et al., 2004; Wilson et al., 2005). To confirm that the peri-centriolar structures in MDA-MB-231 cells are also recycling endosomes, we immunostained MDA-MB-231 cells with anti-FIP3 and various endocytic and Golgi markers. Staining MDA-MB-231 cells with anti-FIP3 and anti-mannosidase II (a Golgi marker) antibodies revealed that there is no colocalization between FIP3 and mannosidase II (data not shown). Next we tested whether FIP3-containing organelles could be early endosomes. As shown in Supplemental Figure 2, there is no colocalization between FIP3 and EEA1, the marker for early endosomes. No colocalization was also observed between FIP3 and Lamp1, suggesting that FIP3 was also not present on lysosomes (Supplemental Figure 2, middle row). Finally, we sought to determine whether these structures correspond to recycling endosomes. To label the recycling endosomes, we stained MDA-MB-231 cells with anti-transferrin receptor (TfR) antibodies, since at a steady state the majority of TfR is located in recycling endosomes. Consistent with the suggestion that FIP3 is localized in recycling endosomes, FIP3-containing organelles were also positive for TfR (Supplemental Figure 2, bottom panels).
Rab11 and FIP3 regulate the motility of breast cancer cells
Since a subset of FIP3-containing endosomes is present at the leading edge of MDA-MB-231 cells, we hypothesized that FIP3 and Rab11 may be important modulators of cell migration. Here we set out to directly test the role of Rab11 and FIP3 in motility. Besides Rab11, Rab25 has been implicated in regulation of breast cancer cell motility (Cheng et al., 2006; Cheng et al., 2004). Rab25 binds to all FIP family members (including FIP3) with an affinity similar to Rab11a and Rab11b. However, while FIP3, Rab11a and Rab11b are ubiquitously expressed, Rab25 expression is limited to epithelial cells (Goldenring et al., 1993). MDA-MB-231 cells do not express Rab25 (Cheng et al., 2006) (also see Supplemental Figure 1). Therefore, in this study we have focused only on Rab11a and Rab11b GTPases.
To establish the importance of Rab11 in this model, we transfected MDA-MB-231 cells with a dominant negative Rab11a mutant (S25N) and measured cell motility in Boyden chamber motility assays. As shown in Figure 2A, myc-Rab11a-S25N (Rab11a and Rab11b dominant negative mutant) reduced cell motility by 40%. A similar extent of inhibition was observed when levels of Rab11a/b were reduced by siRNA (Fig. 2A and Supplemental Figure 1A).
Fig. 2.

FIP3 is required for the motility of MDA-MB-231 cells (modified Boyden chamber assays; see Materials and methods). (A) MDA-MB-231 cells were either mock transfected (control) or transfected with Rab11a/b siRNA or myc-Rab11a-S25N. After 72 hours of incubation the motility of MDA-MB-231 cells was measured (B) MDA-MB-231 cells were either mock transfected or transfected with either FIP3 siRNA#1 or Rip11/FIP5 siRNA. After 72 hours of incubation the motility of cells was measured in the presence or absence of varying concentrations of EGF. (C) To measure the plasma membrane levels of TfR and EGFR, mock- or FIP3 siRNA#1-transfected MDA-MB-231 cells were incubated at 4°C in the presence of 1 μg/ml anti-EGFR-Alexa647 antibody and 10 μg/ml Tf-Alexa488. Cells were then washed, fixed and the levels of cell-bound anti-EGFR-Alexa647 antibody and Tf-Alexa488 were determined by flow cytometry. (D) To measure the endocytosis of TfR, mock- or FIP3 siRNA#1-transfected MDA-MB-231 cells were incubated at 37°C for 30 min in the presence of 10 μg/ml Tf-Alexa488. Cells were washed and the levels of internalized Tf-Alexa488 were determined by flow cytometry. (E) MDA-MB-231 cells were mock transfected or transfected with either FIP3-GFP or FIP3-GFP-I737E. After 16 hours of incubation the motility of cells was measured in the presence of 7.5 ng/ml EGF. The concentration dependency data in (B) is the mean of two experiments. The other data shown are the means and standard deviations of at least three independent experiments. The p values were calculated using a t-test.
To determine whether FIP3 is also involved in regulating cell motility, we tested the effect of FIP3 siRNA on basal and EGF-induced cell motility. It has been well documented that EGF stimulates the motility of various cancer cells. Consistent with this, the addition of EGF stimulated the motility of MDA-MB-231 cells in a concentration-dependent manner (Fig. 2B). Interestingly, FIP3 siRNA#1 (for siRNA#1 specificity see Supplemental Figure 1B-D) reduced basal cell motility and almost completely blocked the EGF-dependent enhancement of cell motility (Fig. 2B). The effect on cell motility appears to be specific for FIP3, since Rip11/FIP5 (the other FIP family member) did not have any effect on basal or EGF-induced cell motility (Fig. 2B). The effect of FIP3 siRNAs on EGF-induced cell motility raises the possibility that FIP3 may regulate EGF receptor (EGF-R) traffic to the plasma membrane, thus inhibiting EGF signaling. Rab11 is known for its role in plasma membrane receptor recycling. Thus, we tested the effect of FIP3 knockdown on the levels of EGF-R at the plasma membrane. As shown in Figure 2C, FIP3 siRNA#1 did not decrease the EGF-R level at the plasma membrane, rather we observed a small, although not statistically significant, increase in EGF-R levels (Fig. 2C). Since it is well established that Rab11 regulates plasma membrane protein recycling, it is possible that FIP3 does not directly regulate cell motility, but rather affects constitutive endocytic recycling, thereby indirectly affecting membrane dynamics at the leading edge of the cell. Recently published data have suggested that FIP3 may regulate the sub-cellular distribution of recycling endosomes, although no effect on TfR endocytic recycling kinetics was observed (Horgan et al., 2007; Inoue et al., 2008). Nevertheless, in order to confirm that FIP3 has no effect on endocytic recycling dynamics, we measured the plasma membrane level of TfR. As shown in Figure 2C, FIP3 siRNA#1 had no effect on plasma membrane levels of TfR. Furthermore, FIP3 siRNA#1 did not affect the uptake of Tf-Alexa488, suggesting that FIP3 is not required for constitutive endocytic recycling (Fig. 2D).
To determine whether the Rab11–FIP3 protein complex regulates cell motility, we transfected MDA-MB-231 cells with FIP3-GFP or FIP3-GFP-I737E. As shown in Figure 2E, the FIP3-GFP-I737E mutant significantly reduced cell motility as determined by a modified Boyden chamber assay. FIP3-GFP-I737E does not bind to Rab11, thus resulting in cytosolic distribution of FIP3 (Wilson et al., 2005). However, FIP3-I737E can still bind to Arf6 (Fielding et al., 2005), suggesting that localized Arf6 binding to endosome-associated FIP3 may be required for cell motility.
While Boyden chamber motility assays are commonly used to test the ability of cells to move, the inhibition of cell migration could be the result of many possible primary effects. Thus, to further understand the effect of FIP3 on cell movement, we have tested cell motility using microscopy-based assays. First, we used a wound-healing assay to examine the movement of mock- or FIP3 siRNA-treated MDA-MB-231 cells. As shown in Figure 3A, FIP3 knockdown reduced the ability of MDA-MB-231 cells to migrate into the “wound”. The effect was specific to FIP3 siRNA, since Rip11/FIP5 knockdown did not have any effect on cell motility (data not shown). Second, we used time-lapse microscopy to image individual cell movement. As shown in Figure 3B and C, FIP3 knockdown inhibited cell motility (p < 0.05). Interestingly, cells treated with FIP3 siRNA seem to be less polarized, since their lamellipodia were much broader as compared to mock cells (Fig. 3B and C). Furthermore, the lamellipodia of untreated MDA-MB-231 cells were more dynamic, since cells often could be observed rapidly relocating lamellipodia from one side of the cell to another, thus rapidly changing direction of movement (Fig. 3B). Similar rapid translocation of lamellipodia to a different side of the cell was almost never observed in FIP3 siRNA-treated cells (Fig. 3C). As the result, FIP3 siRNA-treated cells moved much slower and rarely changed the direction of movement (Fig. 3C). To confirm that FIP3 may regulate cell motility, we also transfected NIH-3T3 cells with either GFP or FIP3-GFP-I373E constructs and measured cell motility using single-cell velocity studies. Consistent with the data from MDA-MB-231 cells FIP3-GFP-I737E inhibited the motility of NIH-3T3 cells (control 21.4 ± 3.2 mm/h, GFP only 20.2 ± 4.1 mm/h, FIP3-GFP-I737E 13.6 ± 2.8 mm/h).
Fig. 3.

FIP3 knockdown inhibits motility of MDA-MB-231 cells. (A) Mock-, FIP3 siRNA#1- or FIP3 siRNA#2-treated MDA-MB-231 cells were plated on glass coverslips coated with 20 μg/ml fibronectin and grown to confluence. The surface of the coverslip was then scratched and cells were incubated for varying amounts of time. Cells were then fixed and imaged by phase contrast. (B) Mock- or FIP3 siRNA#1-treated MDA-MB-231 cells were plated on glass coverslips coated with 20 μg/ml fibronectin. Cell motility was analyzed by time-lapse microscopy. Images were taken every 12 min for 240 min. Shown are the still images taken from the time-lapse series. The numbers in the left panels are the average speed and standard deviation obtained from ten different mock- or FIP3 siRNA-treated cells. Bars: 10 μm.
To further understand the role of FIP3 in lamellipodia dynamics, we analyzed time-lapse series of mock- or FIP3 siRNA-treated MDA-MB-231 cells by kymography (Supplemental Figure 4). As shown in Figure 4A, E and F (also see Supplemental movie 1), the lamellipodia of mock-treated MDA-MB-231 cells were very dynamic. Continuous membrane waves (or ruffles) could be observed at the leading edge of the cell. In contrast, while FIP3 siRNA-treated cells exhibited some membrane ruffling, the ruffling was much less dynamic (Fig. 4B, E and F, Supplemental movie 2). While the ruffle retraction distance did not decrease, FIP3 knockdown significantly reduced the frequency of ruffling (Fig. 4F and Supplemental movie 2). The ruffle retraction velocity also appeared to be reduced. The transfection of MDA-MB-231 cells with FIP3-GFP-I737E also had a similar effect on lamellipodia dynamics, with a reduction in ruffling frequency and ruffle retraction velocity (Fig. 4C-F and Supplemental movie 3). In contrast, the lamellipodia dynamics of the cells expressing GFP only was not different from mock or untreated cells (data not shown). In summary, the cell motility and kymography analyses suggest that FIP3 is involved in regulating lamellipodia polarization and dynamics during cell movement.
Fig. 4.

FIP3 knockdown inhibits lamellipodia ruffling. Lamellipodia dynamics was analyzed in mock-, FIP3 siRNA#1- or FIP3-GFP-I737E-treated MDA-MB-231 cells. (A-D) Single time-frame images (for full time-lapse series, see Supplemental movies 1-3). Representative kymographs are shown in (E). The lines in (A-C) show the area used to generate the kymographs. (F) Quantitation of ruffle frequency, ruffle retraction distance and ruffle retraction velocity obtained from the kymographs (for definitions see Supplemental Figure 4). The data shown are the means and standard deviations from 15 kymographs obtained from five randomly chosen cells (three separate kymographs from every cell). *p<0.01. **p<0.05. Bars: 4 μm.
FIP3 regulates the actin cytoskeleton at the leading edge of breast cancer cells
Our motility assays suggest that FIP3 regulates lamellipodia dynamics in the moving cell. The actin cytoskeleton is known to be a major driving force in lamellipodia expansion and/or retraction. This raises the intriguing possibility that FIP3 may regulate actin cytoskeleton dynamics at the leading edge of motile cells. Consistent with this possibility, it has been reported that nuclear fallout (Nuf) protein, the putative Drosophila homologue of FIP3, regulates the cortical actin cytoskeleton during the cellularization of Drosophila embryos (Riggs et al., 2003; Rothwell et al., 1998, 1999). The next series of experiments were designed to test this hypothesis. First, mock- or FIP3 siRNA-treated MDA-MB-231 cells were stained with rhodamine-conjugated phalloidin. The majority of mock-treated MDA-MB-231 cells (83% from 250 cells counted) displayed polarized leading edges that were rich in actin ruffles containing FIP3-positive endosomes (Fig. 5A, C and D). In marked contrast, FIP3 siRNA-treated MDA-MB-231 cells usually lacked well-developed polarized leading edges (14% from 250 cells counted) and actin ruffling at the leading edge (Fig. 5B and E), suggesting that FIP3 may regulate leading edge formation and cell motility by modulating the actin cytoskeleton. To test whether FIP3 also regulates the actin cytoskeleton in other cell types, we stained actin in mock-, FIP3 siRNA- or Rip11/FIP5 siRNA-treated HeLa cells (Fig. 5F-H). Unlike MDA-MB-231 cells, HeLa cells do not form large lamellipodia. Nevertheless, FIP3 siRNA treatment also decreased actin ruffling at the edges of the cells. This effect was specific to FIP3, as RCP/FIP1 or Rip11/FIP5 siRNAs did not affect actin ruffling, although Rip11/FIP5 knockdown did seem to induce filopodia formation in HeLa cells (data not shown and Figure 5H). To test whether FIP3 and Rab11 binding is required for the regulation of the actin cytoskeleton, we transfected cells with either FIP3-GFP or FIP3-GFP-I737E. As shown in Figure 6, FIP3-GFP-I737E over-expression also inhibited actin ruffling at the leading edge. To confirm that FIP3 is required for lamelipodia formation and/or stability, we have tested the spreading of MDA-MB-231 cells on collagen-coated glass coverslips. As shown in Figure 7A, after a one-hour incubation, mock-treated (or Rip11/FIP5 siRNA-treated) cells started polarizing by forming lamellipodia extensions at distinct plasma membrane sites (see arrows). In contrast, cells depleted of FIP3 showed little polarization and spread out in a “pancake” fashion. Furthermore, cells treated with FIP3 siRNA had more prominent stress fibers as compare to the mock cells (Fig. 7A, left column). The difference between mock or FIP3-depleted cells was even more prominent after a three-hour incubation (Fig. 7A, right column). The mock- or Rip11/FIP5 siRNA-treated cells were almost completely spread out and in many cases had well-formed polarized lamellipodia with actin ruffles at the leading edge. FIP3 siRNA-treated cells lacked a polarized lamellipodium. Indeed, the ratio between length and width of FIP3 siRNA-treated cells was 1.23 ± 0.1 (for comparison, mock-transfected cells: 2.13 ± 0.31), suggesting the diminished development and/or maintenance of polarized lamellipodia (Fig. 7A). In addition, after three hours of incubation FIP3-depleted cells were less spread out as compared to the mock of Rip11/FIP5-depleted cells (Fig. 7B), although it remains unclear whether that is a direct result of decrease in the rate of cell spreading, since after 1 hour of incubation, the area occupied by mock or FIP3 siRNA-treated cells were not significantly different (data not shown).
Fig. 5.

FIP3 regulates the actin cytoskeleton at the leading edge of cells. (A-E) Mock- or FIP3 siRNA#1-treated MDA-MB-231 cells were plated on collagen-coated coverslips, fixed and stained with anti-FIP3 antibodies (D) and rhodamine-phalloidin (A-C, E). (F-H) Mock-, FIP3 siRNA#1- or Rip11/FIP5 siRNA-treated HeLa cells were plated on collagen-coated coverslips, fixed and stained with rhodamine-phalloidin. Bars: 10 μm (A, B, F-H), 2 μm (C, D), 5 μm (E).
Fig. 7.

FIP3 is required for cell spreading and lamellipodia formation. (A, B) To measure cell spreading, mock, FIP3 siRNA#1- or Rip11/FIP5 siRNA-treated MDA-MB-231 cells were plated on collagen-coated coverslips and incubated for either 1 or 3 h at 37°C. Cells were then gently washed with phosphate-buffered saline, fixed and stained with rhodamine-phalloidin (A). Arrows point to forming lamellipodia in mock- and Rip11/FIP5 siRNA-treated cells. Bars: 10 μm. The numbers on the right column panels are the means and standard deviations of length/width ratio obtained from several randomly chosen cells; n is the number of cells analyzed. The length/width ratio of FIP3 siRNA-treated cells is significantly different from mock (p<0.05). (B) Quantitation of cell spreading after 3 hours of incubation. The data shown are the mean and standard deviation of the spreading area measured in randomly chosen cells; n equals the number of cells used for each condition. (C) Lysates obtained from mock- or FIP3 siRNA#1-treated cells were incubated with glutathione beads coated with either GST alone or GST-GGA3 fusion protein. Beads were then washed and bound protein was eluted with 1% SDS. Lysates (upper panels) or bead eluates (lower panels) were then blotted for the presence of FIP3 or Arf6. The numbers shown are the mean (normalized against mock) and standard deviation derived from three independent experiments.
FIP3 regulates localization of Arf6 at the plasma membrane of the leading edge
Since Arf6 is well known for its role in regulating actin ruffling, it is possible that FIP3 may affect Arf6 activation, perhaps by regulating its binding to Arf6 GAPs and/or GEFs. To test this, we have measured the activation of endogenous Arf6 in mock- of FIP3 siRNA-treated cells. GTP-bound Arf6 was detected by a glutathione bead pull-down assay using GST fused to Arf-binding protein GGA3 (Santy and Casanova, 2001). As shown in Figure 7C, FIP3 knockdown did not affect either total Arf6 or activated Arf6 levels.
While our data demonstrate that FIP3 does not mediate Arf6 activation, FIP3 may still affect Arf6-dependent actin ruffling by regulating the targeting of Arf6 to the leading edge of the cell. Arf6 localization to the lamellipodia is known to result in localized activation of Rac1, thus regulating actin ruffling at the leading edge of the cell. Consistent with this, in MDA-MB-231 breast cancer cells Arf6-CFP is enriched at the lamellipodia where it colocalizes with actin ruffles (Fig. 8A-C). To test whether FIP3 binding is required for Arf6-CFP localization to the actin ruffles, we tested the sub-cellular localization of Arf6-CFP-W168L/L169V. We have previously shown that FIP3 binds to the C-terminal end of Arf6 (Schonteich et al., 2007). We have also shown that W168 and L169 play a key role in Arf6 binding to FIP3 in vitro and in vivo while having no effect on Arf6 activation or GTPase activity (Schonteich et al., 2007). As shown in Figure 8D-F, Arf6-CFP-W168L/L169V does not localize to the actin ruffles at the leading edge, but instead is present on internal organelles. These organelles are likely late endosomes and lysosomes, since Arf6-CFP-W168L/L169V colocalizes with Lamp1 (Fig. 8G-E).
Fig. 8.

FIP3 regulates Arf6 targeting to the plasma membrane of the leading edge. MDA-MB-231 cells were transfected with either Arf6-CFP (A-C) or Arf6-CFP-W168L/L169V (D-I). Cells were fixed and either stained with rhodamin-phalloidin or anti-Lamp1 antibodies. Bars: 5 μm.
FIP3 is required for activation of Rac1 GTPase
Arf6 is known to affect the actin cytoskeleton by inducing the activation of Rac1 GTPase, thus stimulating the formation and dynamics of the leading edge (Radhakrishna et al., 1999; Santy and Casanova, 2001; Santy et al., 2005). Since our data demonstrate that FIP3 mediates Arf6 localization to the leading edge, it is possible that FIP3 may regulate Arf6-dependent activation of Rac1 at the leading edge of moving cells. To test this, we transfected mock- or FIP3 siRNA-treated cells with a constitutively active Rac1 mutant (Rac1-L61). As shown in Figure 9C, over-expression of myc-Rac1-L61 inhibited FIP3 siRNA-induced stress fiber formation and reversed FIP3 siRNA-induced inhibition of actin ruffling (compare to untransfected mock and FIP3 siRNA cells in Figure 9A and B). The effect was specific to the constitutively active Rac1 mutant, since myc-Rac1-N17 (GDP-locked mutant)-expressing FIP3 siRNA-treated cells still displayed prominent stress fibers (Fig. 9E). Interestingly, over-expression of myc-Rac1-L61 did not fully rescue the phenotype of FIP3 siRNA-treated cells. While over-expression of myc-Rac1-L61 induced actin ruffling, the ruffling can now be observed along the entire edge of the cell and is not polarized as in untreated cells. That data is consistent with our data showing that FIP3 is required for polarized targeting of Arf6 to the leading edge of the cell. Thus, localized Arf6-dependent activation of Rac1 by FIP3 may be required for polarized lamellipodia formation and/or stability.
Fig. 9.
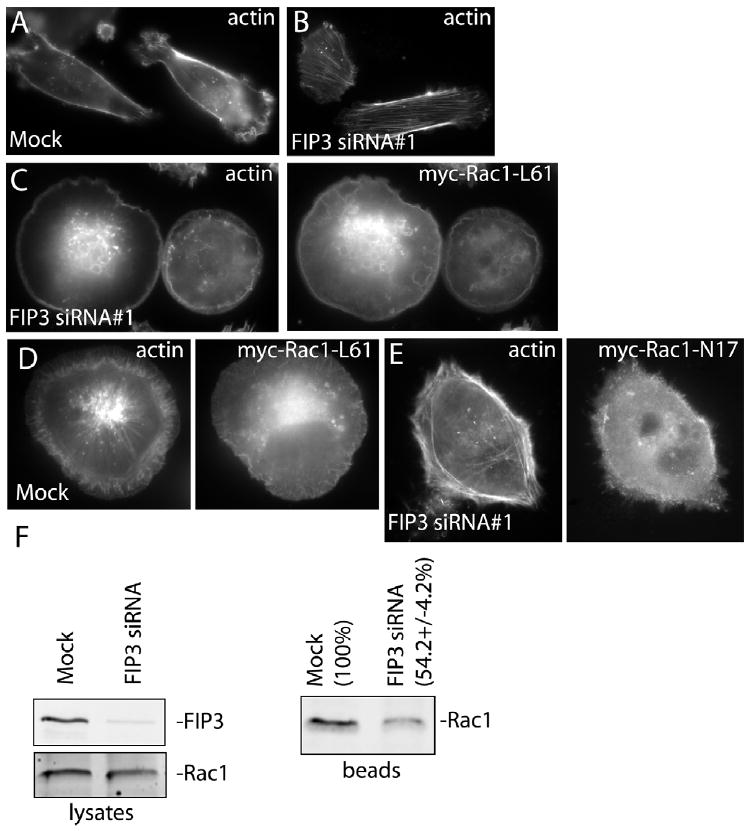
FIP3 regulates Rac1 activation. (C-E) Mock- or FIP3 siRNA#1-treated MDA-MB-231 cells were transfected either with myc-Rac1-L61 or myc-Rac1-N17 and stained with rhodamin-phalloidin and anti-myc antibodies. (A, B) For comparison, untransfected mock- or FIP3 siRNA#1-treated cells stained with rhodamin-phalloidin are also shown. (F) GST-PAK(CRIB) beads were incubated with Triton X-100 lysates from mock- or FIP3 siRNA-treated MDA-MB-231 cells. Beads were washed, and the levels of Rac1 (activated) bound to beads were determined by immunoblotting (right panel). The left two panels show the levels of total cellular FIP3 and Rac1 in the lysates used for the experiment. The numbers shown are the means (normalized against mock) and standard deviations derived from three independent experiments.
Our microscopy data suggests that FIP3 may be required for polarized activation of Rac1. To further test this possibility, we measured the levels of activated Rac1 in mock- or FIP3 siRNA#1-treated MDA-MB-231 cells using a GST-PAK(CRIB) pull-down assay. As shown in Figure 9E, FIP3 knockdown decreased the levels of activated Rac1 while having no effect on total cellular Rac1. In summary, our data suggest that FIP3 regulates the actin dynamics, at least in part, by modulating Arf6-dependent Rac1 activation at the leading edge of moving cells.
Discussion
Endocytic membrane transport plays an important role in regulating cell motility acting in tandem with the actin cytoskeleton. Consistent with this data, recycling endosomes have been shown to mediate the recycling of integrins (Fabbri et al., 2005; Powelka et al., 2004; Yoon et al., 2005). Endosomes may also be required for the delivery of the membrane to the extending lamellipodia/filopodia, providing a source of membrane influx. In this work, we have identified FIP3 as an important regulator of cell motility. Furthermore, we show that FIP3 modulates actin cytoskeleton dynamics, as well as lamellipodia polarization and ruffling, by regulating Arf6 targeting to the lamellipodia and Arf6-dependent activation of Rac1 GTPase. Based on these data, we suggest that endosome-associated FIP3 is a regulator of actin dynamics at the leading edge of motile cells.
The sub-cellular localization of proteins often serves as clues to their function. Consistent with its role in regulating cell motility, we have shown that the FIP3/Rab11 protein complex is present at the leading edge of breast cancer cells, where it could regulate cell motility. FIP3 or Rab11a/b knockdown by siRNA inhibited the motility of MDA-MB-231 cells. Similar effects on motility were also observed when cells were transfected with a Rab11-dominant negative mutant (Rab11-S25N). Interestingly, Rab11 only affected cell motility when both isoforms (Rab11a and Rab11b) were knocked down by siRNA (data not shown), consistent with a previously published report that FIPs can bind to both Rab11a and Rab11b isoforms with similar affinity (Junutula et al., 2004). Several studies have implicated Rab25 in regulating the motility of cells in ovarian and breast cancers (Caswell et al., 2007; Cheng et al., 2006; Cheng et al., 2004). Interestingly, all the FIPs (including FIP3) also bind to Rab25, since it is closely related to Rab11a and Rab11b. Since Rab25 is not expressed in MDA-MB-231 and HeLa cells (Cheng et al., 2006) it is not likely to be a factor in our studies (Supplemental Figure 1E). However, it is possible that a Rab25 and FIP3 interaction may play an important role in regulation of cell motility in other cells.
In addition to Rab11, FIP3 also binds to Arf6 GTPase (Fielding et al., 2005; Schonteich et al., 2007). Since Arf6 is well known for its role in activating Rac1 GTPase and regulating actin ruffling in motile cells (Santy and Casanova, 2001; Santy et al., 2005) we have tested whether FIP3 may affect actin dynamics at the leading edge of the cell. Remarkably, FIP3 knockdown by siRNA resulted in the loss of actin ruffles in both MDA-MB-231 and HeLa cells. Furthermore, FIP3 knockdown also resulted in a dramatic decrease in plasma membrane ruffling frequency, as well as loss of polarized extension of lamellipodia. Such data suggest that FIP3 actually regulates cell motility by regulating actin cytoskeleton dynamics.
Since Arf6 is a well-established modulator of actin ruffling at the leading edge, we tested whether FIP3 may regulate Arf6 activity at the lamellipodia of MDA-MB-231 cells. While FIP3 did not seem to have any effect on the activity of total cellular Arf6, blocking the interaction between Arf6 and FIP3 inhibited Arf6 accumulation at the leading edge of the cells. Thus, our data suggest that FIP3 regulates actin dynamics by mediating Arf6 targeting and enrichment at the leading edge of mobile cells. Consistent with that, FIP3 knockdown also affected the polarized formation of lamellipodia during cell spreading and motility. It is likely that localized delivery of FIP3-containing endosomes may provide the means of rapid and restricted activation of Rac1 and subsequent polarized formation of lamellipodia. Indeed, FIP3 depletion resulted in a decrease in Rac1 activation. Furthermore, over-expression of constitutively active Rac1 reverses the FIP3 knockdown-induced inhibition of actin ruffling. Interestingly, over-expression of constitutively active Rac1 resulted in non-polarized activation of actin ruffling, since actin ruffles could be observed along the edge of the entire cell. Thus, FIP3 appears to play an important role in restricted/localized activation of the Arf6-Rac1-actin ruffling pathway, thus allowing the directional motility of MDA-MB-231 cells.
It has been previously reported that Rab11 and Arf6 regulate the recycling of integrins during cell motility (Caswell and Norman, 2006; Pellinen and Ivaska, 2006; Powelka et al., 2004). Since FIP3 interacts with both, Rab11 and Arf6, it is possible that, in addition to regulating the actin cytoskeleton, FIP3 may also regulate integrin recycling. While additional work will be required to resolve this point, several studies have shown that FIP3 does not seem to affect the rates of protein recycling (Horgan et al., 2007; Inoue et al., 2008). Furthermore, recent work from Norman and colleagues has implicated FIP1/RCP, the other member of the FIP family, in regulating integrin recycling (Caswell et al., 2008).
In summary, we show that FIP3 regulates cell motility by mediating Arf6 targeting to the leading edge of the cell. The polarized Arf6 enrichment at the leading edge then results in localized activation of Rac1 and increased actin and plasma membrane ruffling.
Supplementary Material
Acknowledgments
We thank Dr. Gwyn Gould and Dr. Kathryn Howell for critical reading of the manuscript. We also thank Dr. Andrew Peden for the anti-EGF-R, anti-β1 integrin and anti-α5-integrin antibodies and Dr. Gary Bokoch for providing the GST-PAK(CRIB) construct. Finally, we thank Dr. Heide Ford and Erica McCoy for all the technical help. This work was supported in part by a grant from NIH-NIDDK (R01 DK064380 to R. Prekeris).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Boshans RL, Szanto S, van Aelst L, D’Souza-Schorey C. ADP-ribosylation factor 6 regulates actin cytoskeleton remodeling in coordination with Rac1 and RhoA. Mol Cell Biol. 2000;20:3685–3694. doi: 10.1128/mcb.20.10.3685-3694.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braga VM, Machesky LM, Hall A, Hotchin NA. The small GTPases Rho and Rac are required for the establishment of cadherin-dependent cell-cell contacts. J Cell Biol. 1997;137:1421–1431. doi: 10.1083/jcb.137.6.1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caswell PT, Norman JC. Integrin trafficking and the control of cell migration. Traffic. 2006;7:14–21. doi: 10.1111/j.1600-0854.2005.00362.x. [DOI] [PubMed] [Google Scholar]
- Caswell PT, Spence HJ, Parsons M, White DP, Clark K, Cheng KW, Mills GB, Humphries MJ, Messent AJ, Anderson KI, McCaffrey MW, Ozanne BW, Norman JC. Rab25 associates with alpha5beta1 integrin to promote invasive migration in 3D microenvironments. Dev Cell. 2007;13:496–510. doi: 10.1016/j.devcel.2007.08.012. [DOI] [PubMed] [Google Scholar]
- Caswell PT, Chan M, Lindsay AJ, McCaffrey MW, Boettiger D, Norman JC. Rab-coupling protein coordinates recycling of alpha5beta1 integrin and EGFR1 to promote cell migration in 3D microenvironments. J Cell Biol. 2008;183:143–155. doi: 10.1083/jcb.200804140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng JM, Ding M, Aribi A, Shah P, Rao K. Loss of RAB25 expression in breast cancer. Int J Cancer. 2006;118:2957–2964. doi: 10.1002/ijc.21739. [DOI] [PubMed] [Google Scholar]
- Cheng KW, Lahad JP, Kuo WL, Lapuk A, Yamada K, Auersperg N, Liu J, Smith-McCune K, Lu KH, Fishman D, Gray JW, Mills GB. The RAB25 small GTPase determines aggressiveness of ovarian and breast cancers. Nat Med. 2004;10:1251–1256. doi: 10.1038/nm1125. [DOI] [PubMed] [Google Scholar]
- Cox D, Lee DJ, Dale BM, Calafat J, Greenberg S. A Rab11-containing rapidly recycling compartment in macrophages that promotes phagocytosis. Proc Natl Acad Sci USA. 2000;97:680–685. doi: 10.1073/pnas.97.2.680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Souza-Schorey C, Boshans RL, McDonough M, Stahl PD, Van Aelst L. A role for POR1, a Rac1-interacting protein, in ARF6-mediated cytoskeletal rearrangements. EMBO J. 1997;16:5445–5454. doi: 10.1093/emboj/16.17.5445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Souza-Schorey C, Boettner B, Van Aelst L. Rac regulates integrin-mediated spreading and increased adhesion of T lymphocytes. Mol Cell Biol. 1998;18:3936–3946. doi: 10.1128/mcb.18.7.3936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson JG. Multiple roles for Arf6: sorting, structuring, and signaling at the plasma membrane. J Biol Chem. 2003;278:41573–41576. doi: 10.1074/jbc.R300026200. [DOI] [PubMed] [Google Scholar]
- Donaldson JG, Honda A. Localization and function of Arf family GTPases. Biochem Soc Trans. 2005;33:639–642. doi: 10.1042/BST0330639. [DOI] [PubMed] [Google Scholar]
- Dunphy JL, Moravec R, Ly K, Lasell TK, Melancon P, Casanova JE. The Arf6 GEF GEP100/BRAG2 regulates cell adhesion by controlling endocytosis of beta1 integrins. Curr Biol. 2006;16:315–320. doi: 10.1016/j.cub.2005.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eathiraj S, Mishra A, Prekeris R, Lambright DG. Structural basis for Rab11-mediated recruitment of FIP3 to recycling endosomes. J Mol Biol. 2006;364:121–135. doi: 10.1016/j.jmb.2006.08.064. [DOI] [PubMed] [Google Scholar]
- Fabbri M, Di Meglio S, Gagliani MC, Consonni E, Molteni R, Bender JR, Tacchetti C, Pardi R. Dynamic partitioning into lipid rafts controls the endo-exocytic cycle of the alphaL/beta2 integrin, LFA-1, during leukocyte chemotaxis. Mol Biol Cell. 2005;16:5793–5803. doi: 10.1091/mbc.E05-05-0413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fielding AB, Schonteich E, Matheson J, Wilson G, Yu X, Hickson GR, Srivastava S, Baldwin SA, Prekeris R, Gould GW. Rab11-FIP3 and FIP4 interact with Arf6 and the exocyst to control membrane traffic in cytokinesis. EMBO J. 2005;24:3389–3399. doi: 10.1038/sj.emboj.7600803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldenring JR, Shen KR, Vaughan HD, Modlin IM. Identification of a small GTP-binding protein, Rab25, expressed in the gastrointestinal mucosa, kidney, and lung. J Biol Chem. 1993;268:18419–18422. [PubMed] [Google Scholar]
- Hales CM, Griner R, Hobdy-Henderson KC, Dorn MC, Hardy D, Kumar R, Navarre J, Chan EK, Lapierre LA, Goldenring JR. Identification and characterization of a family of Rab11-interacting proteins. J Biol Chem. 2001;276:39067–39075. doi: 10.1074/jbc.M104831200. [DOI] [PubMed] [Google Scholar]
- Hall A. G proteins and small GTPases: distant relatives keep in touch. Science. 1998;280:2074–2075. doi: 10.1126/science.280.5372.2074. [DOI] [PubMed] [Google Scholar]
- Hernandez-Deviez DJ, Roth MG, Casanova JE, Wilson JM. ARNO and ARF6 regulate axonal elongation and branching through downstream activation of phosphatidylinositol 4-phosphate 5-kinase alpha. Mol Biol Cell. 2004;15:111–120. doi: 10.1091/mbc.E03-06-0410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickson GRX, Matheson J, Riggs B, Maier VH, Fielding AB, Prekeris R, Sullivan W, Barr FA, Gould GW. Arfophilins are dual Arf/Rab11 binding proteins that regulate recycling endosome distribution and are related to Drosophila nuclear fallout. Mol Biol Cell. 2003;14:2908–2920. doi: 10.1091/mbc.E03-03-0160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinz B, Alt W, Johnen C, Herzog V, Kaiser HW. Quantifying lamella dynamics of cultured cells by SACED, a new computer-assisted motion analysis. Exp Cell Res. 1999;251:234–243. doi: 10.1006/excr.1999.4541. [DOI] [PubMed] [Google Scholar]
- Hiroyama M, Exton JH. Localization and regulation of phospholipase D2 by ARF6. J Cell Biochem. 2005;95:149–164. doi: 10.1002/jcb.20351. [DOI] [PubMed] [Google Scholar]
- Honda A, Nogami M, Yokozeki T, Yamazaki M, Nakamura H, Watanabe H, Kawamoto K, Nakayama K, Morris AJ, Frohman MA, Kanaho Y. Phosphatidylinositol 4-phosphate 5-kinase alpha is a downstream effector of the small G protein ARF6 in membrane ruffle formation. Cell. 1999;99:521–532. doi: 10.1016/s0092-8674(00)81540-8. [DOI] [PubMed] [Google Scholar]
- Horgan CP, Walsh M, Zurawski TH, McCaffrey MW. Rab11-FIP3 localises to a Rab11-positive pericentrosomal compartment during interphase and to the cleavage furrow during cytokinesis. Biochem Biophys Res Commun. 2004;319:83–94. doi: 10.1016/j.bbrc.2004.04.157. [DOI] [PubMed] [Google Scholar]
- Horgan CP, Oleksy A, Zhdanov AV, Lall PY, White IJ, Khan AR, Futter CE, McCaffrey JG, McCaffrey MW. Rab11-FIP3 is critical for the structural integrity of the endosomal recycling compartment. Traffic. 2007;8:414–430. doi: 10.1111/j.1600-0854.2007.00543.x. [DOI] [PubMed] [Google Scholar]
- Inoue H, Ha VL, Prekeris R, Randazzo PA. Arf GAP ASAP1 interacts with Rab11 effector FIP3 and regulates pericentrosomal localization of transferrin receptor-positive recycling endosome. Mol Biol Cell. 2008;19:4224–4237. doi: 10.1091/mbc.E08-03-0290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junutula JR, Schonteich E, Wilson GM, Peden AA, Scheller RH, Prekeris R. Molecular characterization of Rab11 interactions with members of the family of Rab11-interacting proteins. J Biol Chem. 2004;279:33430–33437. doi: 10.1074/jbc.M404633200. [DOI] [PubMed] [Google Scholar]
- Koo TH, Eipper BA, Donaldson JG. Arf6 recruits the Rac GEF Kalirin to the plasma membrane facilitating Rac activation. BMC Cell Biol. 2007;8:29. doi: 10.1186/1471-2121-8-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krauss M, Kinuta M, Wenk MR, De Camilli P, Takei K, Haucke V. ARF6 stimulates clathrin/AP-2 recruitment to synaptic membranes by activating phosphatidylinositol phosphate kinase type Igamma. J Cell Biol. 2003;162:113–124. doi: 10.1083/jcb.200301006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsay AJ, Hendrick AG, Cantalupo G, Senic-Matuglia F, Goud B, Bucci C, McCaffrey MW. Rab coupling protein (RCP), a novel Rab4 and Rab11 effector protein. J Biol Chem. 2002;277:12190–12199. doi: 10.1074/jbc.M108665200. [DOI] [PubMed] [Google Scholar]
- Mackay DJ, Hall A. Rho GTPases. J Biol Chem. 1998;273:20685–20688. doi: 10.1074/jbc.273.33.20685. [DOI] [PubMed] [Google Scholar]
- Meyers JM, Prekeris R. Formation of mutually exclusive Rab11 complexes with members of the family of Rab11-interacting proteins regulates Rab11 endocytic targeting and function. J Biol Chem. 2002;277:49003–49010. doi: 10.1074/jbc.M205728200. [DOI] [PubMed] [Google Scholar]
- Nabi IR. The polarization of the motile cell. J Cell Sci. 1999;112:1803–1811. doi: 10.1242/jcs.112.12.1803. [DOI] [PubMed] [Google Scholar]
- Palacios F, D’Souza-Schorey C. Modulation of Rac1 and ARF6 activation during epithelial cell scattering. J Biol Chem. 2003;278:17395–17400. doi: 10.1074/jbc.M300998200. [DOI] [PubMed] [Google Scholar]
- Palacios F, Schweitzer JK, Boshans RL, D’Souza-Schorey C. ARF6-GTP recruits Nm23-H1 to facilitate dynamin-mediated endocytosis during adherens junctions disassembly. Nat Cell Biol. 2002;4:929–936. doi: 10.1038/ncb881. [DOI] [PubMed] [Google Scholar]
- Paleotti O, Macia E, Luton F, Klein S, Partisani M, Chardin P, Kirchhausen T, Franco M. The small G-protein Arf6GTP recruits the AP-2 adaptor complex to membranes. J Biol Chem. 2005;280:21661–21666. doi: 10.1074/jbc.M503099200. [DOI] [PubMed] [Google Scholar]
- Parsons M, Monypenny J, Ameer-Beg SM, Millard TH, Machesky LM, Peter M, Keppler MD, Schiavo G, Watson R, Chernoff J, Zicha D, Vojnovic B, Ng T. Spatially distinct binding of Cdc42 to PAK1 and N-WASP in breast carcinoma cells. Mol Cell Biol. 2005;25:1680–1695. doi: 10.1128/MCB.25.5.1680-1695.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peden AA, Schonteich E, Chun J, Jagath JR, Scheller RH, Prekeris R. The RCP-Rab11 complex regulates endocytic protein sorting. Mol Biol Cell. 2004;15:3530–3541. doi: 10.1091/mbc.E03-12-0918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellinen T, Ivaska J. Integrin traffic. J Cell Sci. 2006;119:3723–3731. doi: 10.1242/jcs.03216. [DOI] [PubMed] [Google Scholar]
- Powelka AM, Sun J, Li J, Gao M, Shaw LM, Sonnenberg A, Hsu VW. Stimulation-dependent recycling of integrin beta1 regulated by ARF6 and Rab11. Traffic. 2004;5:20–36. doi: 10.1111/j.1600-0854.2004.00150.x. [DOI] [PubMed] [Google Scholar]
- Powner DJ, Wakelam MJ. The regulation of phospholipase D by inositol phospholipids and small GTPases. FEBS Lett. 2002;531:62–64. doi: 10.1016/s0014-5793(02)03410-5. [DOI] [PubMed] [Google Scholar]
- Powner DJ, Hodgkin MN, Wakelam MJ. Antigen-stimulated activation of phospholipase D1b by Rac1, ARF6, and PKCalpha in RBL-2H3 cells. Mol Biol Cell. 2002;13:1252–1262. doi: 10.1091/mbc.01-05-0235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prekeris R, Klumperman J, Scheller RH. A Rab11/Rip11 protein complex regulates apical membrane trafficking via recycling endosomes. Mol Cell. 2000;6:1437–1448. doi: 10.1016/s1097-2765(00)00140-4. [DOI] [PubMed] [Google Scholar]
- Prekeris R, Davies JM, Scheller RH. Identification of a novel Rab11/25 binding domain present in Eferin and Rip proteins. J Biol Chem. 2001;276:38966–38970. doi: 10.1074/jbc.M106133200. [DOI] [PubMed] [Google Scholar]
- Radhakrishna H, Al-Awar O, Khachikian Z, Donaldson JG. ARF6 requirement for Rac ruffling suggests a role for membrane trafficking in cortical actin rearrangements. J Cell Sci. 1999;112:855–866. doi: 10.1242/jcs.112.6.855. [DOI] [PubMed] [Google Scholar]
- Riggs B, Rothwell W, Mische S, Hickson GR, Matheson J, Hays TS, Gould GW, Sullivan W. Actin cytoskeleton remodeling during early Drosophila furrow formation requires recycling endosomal components Nuclear-fallout and Rab11. J Cell Biol. 2003;163:143–154. doi: 10.1083/jcb.200305115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothwell WF, Fogarty P, Field CM, Sullivan W. Nuclear-fallout, a Drosophila protein that cycles from the cytoplasm to the centrosomes, regulates cortical microfilament organization. Development. 1998;125:1295–1303. doi: 10.1242/dev.125.7.1295. [DOI] [PubMed] [Google Scholar]
- Rothwell WF, Zhang CX, Zelano C, Hsieh TS, Sullivan W. The Drosophila centrosomal protein Nuf is required for recruiting Dah, a membrane associated protein, to furrows in the early embryo. J Cell Sci. 1999;112:2885–2893. doi: 10.1242/jcs.112.17.2885. [DOI] [PubMed] [Google Scholar]
- Sabe H. Requirement for Arf6 in cell adhesion, migration, and cancer cell invasion. J Biochem (Tokyo) 2003;134:485–489. doi: 10.1093/jb/mvg181. [DOI] [PubMed] [Google Scholar]
- Santy LC, Casanova JE. Activation of ARF6 by ARNO stimulates epithelial cell migration through downstream activation of both Rac1 and phospholipase D. J Cell Biol. 2001;154:599–610. doi: 10.1083/jcb.200104019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santy LC, Ravichandran KS, Casanova JE. The DOCK180/Elmo complex couples ARNO-mediated Arf6 activation to the downstream activation of Rac1. Curr Biol. 2005;15:1749–1754. doi: 10.1016/j.cub.2005.08.052. [DOI] [PubMed] [Google Scholar]
- Schonteich E, Pilli M, Simon GC, Matern HT, Junutula JR, Sentz D, Holmes RK, Prekeris R. Molecular characterization of Rab11-FIP3 binding to ARF GTPases. Eur J Cell Biol. 2007;86:417–431. doi: 10.1016/j.ejcb.2007.05.004. [DOI] [PubMed] [Google Scholar]
- Shiba T, Koga H, Shin HW, Kawasaki M, Kato R, Nakayama K, Wakatsuki S. Structural basis for Rab11-dependent membrane recruitment of a family of Rab11-interacting protein 3 (FIP3)/Arfophilin-1. Proc Natl Acad Sci USA. 2006;103:15416–15421. doi: 10.1073/pnas.0605357103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin OH, Ross AH, Mihai I, Exton JH. Identification of arfophilin, a target protein for GTP-bound class II ADP-ribosylation factors. J Biol Chem. 1999;274:36609–36615. doi: 10.1074/jbc.274.51.36609. [DOI] [PubMed] [Google Scholar]
- Shin OH, Couvillon AD, Exton JH. Arfophilin is a common target of both class II and class III ADP-ribosylation factors. Biochemistry. 2001;40:10846–10852. doi: 10.1021/bi0107391. [DOI] [PubMed] [Google Scholar]
- Skop AR, Bergmann D, Mohler WA, White JG. Completion of cytokinesis in C. elegans requires a brefeldin A-sensitive membrane accumulation at the cleavage furrow apex. Curr Biol. 2001;11:735–746. doi: 10.1016/s0960-9822(01)00231-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takaishi K, Sasaki T, Kotani H, Nishioka H, Takai Y. Regulation of cell-cell adhesion by rac and rho small G proteins in MDCK cells. J Cell Biol. 1997;139:1047–1059. doi: 10.1083/jcb.139.4.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner CE, Brown MC. Cell motility: ARNOand ARF6 at the cutting edge. Curr Biol. 2001;11:R875–877. doi: 10.1016/s0960-9822(01)00521-8. [DOI] [PubMed] [Google Scholar]
- Wilson GM, Fielding AB, Simon GC, Yu X, Andrews PD, Peden AA, Gould GW, Prekeris R. The FIP3-Rab11 protein complex regulates recycling endosome targeting to the cleavage furrow during late cytokinesis. Mol Biol Cell. 2005;16:849–860. doi: 10.1091/mbc.E04-10-0927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon SO, Shin S, Mercurio AM. Hypoxia stimulates carcinoma invasion by stabilizing microtubules and promoting the Rab11 trafficking of the alpha6beta4 integrin. Cancer Res. 2005;65:2761–2769. doi: 10.1158/0008-5472.CAN-04-4122. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


