Abstract
Bacterial virulence determinants can be identified, according to the molecular Koch’s postulates1, if inactivation of a gene associated with a suspected virulence trait results in a loss in pathogenicity. This approach is commonly used with genetically tractable organisms. However, the current lack of tools for targeted gene disruptions in obligate intracellular microbial pathogens seriously hampers the identification of their virulence factors. Here we demonstrate an approach to studying potential virulence factors of genetically intractable organisms, such as Chlamydia. Heterologous expression of Chlamydia pneumoniae CopN in yeast and mammalian cells resulted in a cell cycle arrest, presumably owing to alterations in the microtubule cytoskeleton. A screen of a small molecule library identified two compounds that alleviated CopN-induced growth inhibition in yeast. These compounds interfered with C. pneumoniae replication in mammalian cells, presumably by ‘knocking out’ CopN function, revealing an essential role of CopN in the support of C. pneumoniae growth during infection. This work demonstrates the role of a specific chlamydial protein in virulence. The chemical biology approach described here can be used to identify virulence factors, and the reverse chemical genetic strategy can result in the identification of lead compounds for the development of novel therapeutics.
Chlamydia pneumoniae, a human respiratory pathogen, is associated with atherosclerosis and has been linked to heart disease and stroke2. This obligate intracellular pathogen resides in host cells within vacuoles referred to as inclusions3. Chlamydia usurp various host cellular processes to promote virulence4–9, presumably through the actions of proteins that they directly secrete into host cells and/or express on the outer surface of the inclusion membrane10–13.
The yeast Saccharomyces cerevisiae is an established model system that can be used to identify and characterize bacterial virulence proteins14. The underlying premise of this system is that many bacterial virulence proteins target cellular processes conserved from yeast to mammals. Indeed, expression of numerous bacterial virulence proteins in yeast inhibits growth owing to targeting of conserved eukaryotic cellular processes15. We expressed five probable C. pneumoniae virulence proteins in yeast. Three of these proteins, CopN, CP1062 and CP0833, are putative substrates of the C. pneumoniae type III system, a specialized secretion systemthat directly translocates proteins from the bacterial cytosol into host cells. During an infection, CopN is detected on the inclusion membrane, CP0833 in the host cell cytosol, and CP1062 at both16. Whereas CP0679 encodes a putative serine/threonine kinase17, CP0358 encodes a serine/threonine protein phosphatase. As such, both encode potential virulence factors.
Expression of CopN and CP1062 severely inhibited yeast growth. This growth inhibition was alleviated when expression levels of CP1062 but not CopN were lowered (Fig. 1a). CopN inhibited yeast growth regardless of whether the protein was expressed on its own or fused to GFP (green fluorescent protein). This inhibitory activity was also observed with expression of CopN from Chlamydia psittaci B577 (Chlamydia abortus), but not with expression of more distally related CopN homologues, including CopN of Chlamydia trachomatis, YopN of Yersinia enterocolitica and PopN of Pseudomonas aeruginosa.
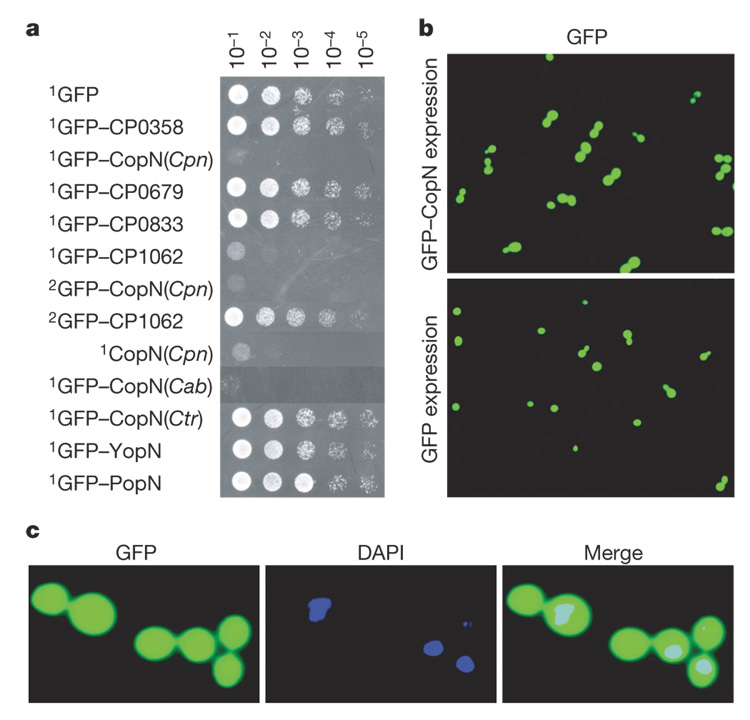
Figure 1. CopN expression inhibits yeast growth and results in the accumulation of large-budded yeast.
a, Serial dilutions of yeast that conditionally express the designated proteins were spotted on inducing media and grown for 48 h. Genes encoding the proteins were cloned on either a high (1) or low (2) copy number plasmid. Cpn, C. pneumoniae; Cab, C. abortus; Ctr, C. trachomatis. b, Image of yeast expressing GFP–CopN or GFP, visualized 6 h post-induction. c, Enlarged image of yeast 6 h postinduction of GFP–CopN. The yeast were fixed and stained with DAPI to visualize the nuclei.
Expression of GFP–CopN resulted in the accumulation of large-budded yeast (Fig. 1b, top panel). By 6 h post-induction of expression, 90% of the GFP–CopN-expressing yeast, but only 22% of GFP-expressing yeast (Supplementary Fig. 1), appeared as large-budded cells. Yeast normally undergo nuclear division coincident with the formation of large-budded cells, but the majority of the large-budded GFP–CopN-expressing yeast (91%) contained only a single nucleus, which was present in only one of the two buds (Fig. 1c).
To determine whether the large-budded yeast had undergone DNA replication, we quantified their DNA content using flow cytometry (FACS). Exponentially growing haploid yeast expressing GFP–CopN or GFP were synchronized at G1and then released to progress through the cell cycle. In both cases, a predominant 2N DNA peak was observed after 3 h, indicating that the majority of the yeast had progressed through S phase and completed DNA replication (Fig. 2a). However, while GFP-expressing yeast continued to proceed through the cell cycle, those expressing GFP–CopN arrested at this point. Thus, yeast expressing GFP–CopN arrest at the G2/M phase of the cell cycle.
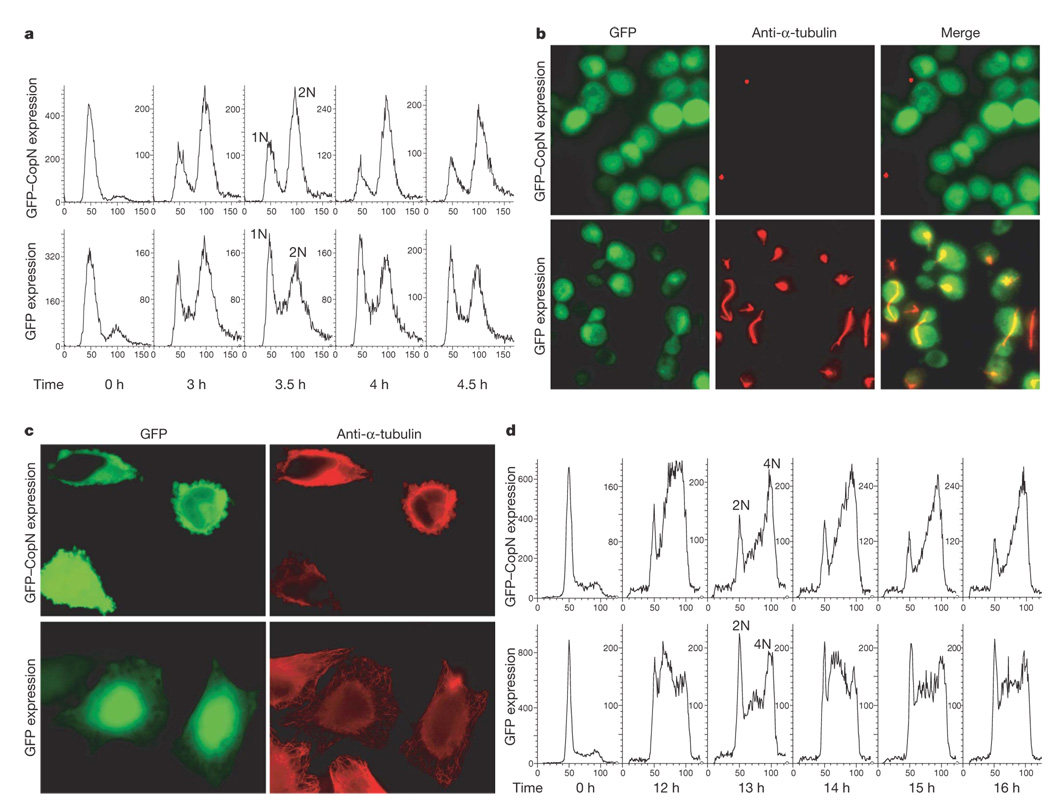
Figure 2. CopN expression induces a cell cycle arrest in both yeast and mammalian cells due to disruption of microtubules.
a, d, FACS analyses of theDNAcontent of a, yeast and d, 293 cells expressing GFP–CopN or GFP at the designated time points. The peaks labelled as 1N or 2N in yeast and 2N or 4N in 293 cells indicate the DNA content. b, c, Images of b, yeast 6 h postinduction of expression and c, HeLa cells 12 h post-transfection for transient expression of GFP–CopN or GFP. In both cases, cells were fixed and stained with anti-α-tubulin antibodies (red).
Disruption of yeast microtubules can prevent formation of the spindle apparatus, which is required for mitosis, resulting in the accumulation of large-budded 2N yeast. Thus, we examined the integrity of the spindle apparatus of CopN-expressing yeast. Remarkably, no spindles were detected in GFP–CopN-expressing yeast (Fig. 2b, top panels). GFP-expressing yeast displayed normal spindles at the appropriate point in the cell cycle (Fig. 2b, bottom panels). Thus, CopN expression results in a G2/M cell cycle arrest due to disruption of spindle apparatus.
We next investigated whether the activity of CopN was conserved from yeast to mammals. We examined the structural integrity of the microtubule network in GFP–CopN-expressing epithelial cells (HeLa cells). As shown in Fig. 2c, 12 h post-transfection, microtubule networks were disrupted in GFP–CopN-expressing cells. In contrast, a characteristic radial array of microtubules was observed in GFP-expressing cells.
Disruption of the microtubules in mammalian cells can also result in a G2/M cell cycle arrest. To test whether CopN confers such a phenotype, we established stable cell lines that conditionally express GFP–CopN or GFP. FACS analyses were performed to examine the effect of CopN expression on cell cycle progression for 16 h after release from G1 synchronization. Delay of cell cycle progression in the CopN-expressing cells was first observed at 12 h when the cells started to accumulate at the G2/M transition. This 4N peak continued to accumulate over the next four hours (Fig. 2d, top panel). In contrast, GFP-expressing cells continued to progress through the cell cycle (Fig. 2d, bottom panel). These results demonstrate that expression of CopN also induces a G2/M cell cycle arrest in mammalian cells.
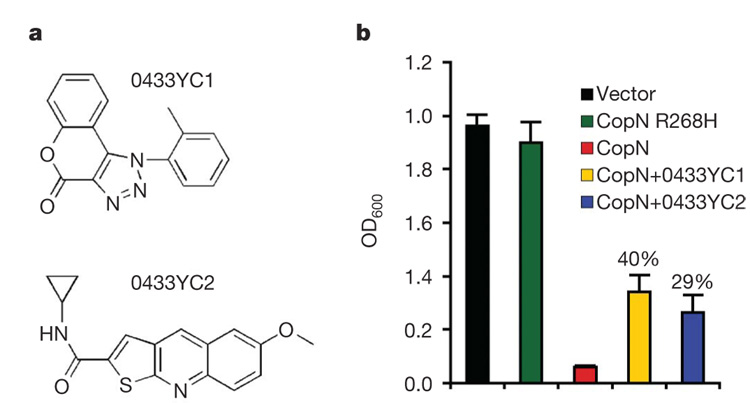
Genetic tools to create C. pneumoniae that do not express CopN are currently unavailable. To circumvent this limitation, we screened for small molecule inhibitors of CopN activity. Specifically, we screened a library of ~40,000 small molecules for those that alleviated yeast growth inhibition due to CopN expression. Two compounds, 0433YC1 and 0433YC2 (Fig. 3a), were found to reproducibly restore growth of CopN-expressing yeast to levels 40% and 29%, respectively, of yeast expressing an inactive CopN allele (CopN R268H) (Fig. 3b). At concentrations used in the screen, these compounds did not affect growth of wild-type yeast (data not shown).
Figure 3. The small molecule inhibitors 0433YC1 and 0433YC2 alleviate yeast growth inhibition due to CopN expression.
a, Structures of compounds 0433YC1 (ChemDiv 5947-0064) and 0433YC2 (ChemDiv C303-0665). b, Growth of yeast (mean + s.e.m., n = 4) expressing either GFP, an inactive allele of GFP–CopN (R268H), or GFP–CopN in the presence and absence of 0433YC1 or 0433YC2 at 12.5 µg ml−1. The percentages shown indicate the rate of restoration of growth in the presence of compounds relative to yeast expressing the inactive CopN allele. Student’s t-test was performed between CopN-expressing yeast treated with 0433YC1 (P = 0.004) or 0433YC2 (P = 0.02) and untreated control.
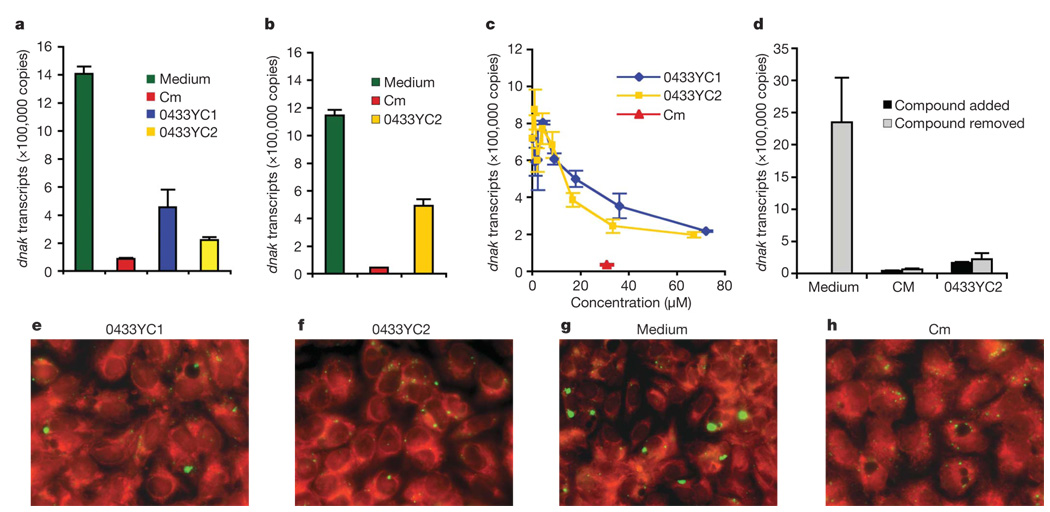
To investigate the role of CopN during a C. pneumoniae infection, the two inhibitors were used to essentially create ‘functional knockouts’ of CopN. Treatment of infected buffalo green monkey kidney (BGMK) cells18 with either 0433YC1 or 0433YC2 at 10 µg ml−1 for 72 h resulted in a significant reduction in the replication of C. pneumoniae (Fig. 4a). The presence of the compounds in the media led to a decrease in dnaK transcription by 68–84% as compared to dnaK levels present in host cells grown in untreated media19–21. Similarly, the addition of 0433YC2 inhibited replication in Hep-2 cells (Fig. 4b). Both inhibitors interfered with the intracellular replication of C. pneumoniae in a dose-dependent manner (Fig. 4c). No toxic effect on BGMK cells was observed when either compound was added at 20 µg ml−1 as assayed by either monitoring mitochondrial dehydrogenase activity or by microscopic examination of cell morphology (data not shown). Removal of 0433YC2 from the media of infected BGMK cells after 72-h treatment did not lead to an immediate recovery of C. pneumoniae growth (Fig. 4d). Neither of the compounds inhibited replication of C. trachomatis in BGMK cells (Supplementary Fig. 2). This result is perhaps not surprising, given that the expression of CopN from C. trachomatis did not inhibit yeast growth (Fig. 1a). Immunofluorescence microscopy revealed that the compounds also inhibited the development of C. pneumoniae inclusions observed within the infected BGMK cells (Fig. 4e, f and Supplementary Fig. 3). Infected cells treated with the compounds essentially lacked large inclusions (Fig. 4e, f) characteristic of C. pneumoniae growth in host cells (2.3–5 µm in diameter) (Fig. 4g). Rather, small inclusions were observed in cells incubated with the more potent compound 0433YC2 (8.8 inclusions per cell and 0.4–0.6 mm in diameter). These small inclusions resembled those seen in cells treated with chloramphenicol (9.2 inclusions per cell and 0.4–0.6 µm; Fig. 4h), an antimicrobial agent active against Chlamydia.
Figure 4. The CopN inhibitors 0433YC1 and 0433YC2 inhibit C. pneumoniae replication in host cells.
a–d, Growth of C. pneumoniae in host cells monitored by quantification of dnaK transcription by real-time PCR. Host cells were infected for one hour at a multiplicity of infection (m.o.i.) of 10:1 followed by incubation in fresh media containing the inhibitors. Each of the assays was repeated 3–10 times. Student’s t-test was performed between treated and untreated cells. a, BGMK cells treated with compounds 0433YC1 (P = 0.0004) and 0433YC2 (P = 0.000001) at 10 µg ml−1. Standard growth medium and that plus chloramphenicol (Cm) were used as growth and inhibition controls. b, Hep-2 cells treated with 0433YC2 (P = 0.0016) at 10 µg ml−1. c,BGMK cells treated with compounds at the designated concentrations (µM) on the×axis. d, BGMK cells were first treated with chloramphenicol or 0433YC2 for 72 h. The compounds were then removed and dnaK levels were determined after an additional 48 h. Data in a–d are presented as total copy number of dnaK transcripts per well of the 24-well plate (mean + s.e.m.). e–h, Immunofluorescent images of BGMK cells infected with C. pneumoniae at an m.o.i. of 10:1. The C. pneumoniae inclusions are stained with anti-Chlamydia-LPS antibody (green) and the host cell is counterstained red. Cells were treated with e, 0433YC1, f, 0433YC2, g, media control, or h, chloramphenicol.
Taken together, our data demonstrate that CopN is required to support the intracellular growth of C. pneumoniae and plays an essential virulence role in a cell culture model of infection22. By using the CopN small molecule inhibitors identified in yeast, we were able to fulfil the molecular Koch’s postulates to identify the first chlamydial protein required for virulence of this obligate intracellular organism. This strategy can be extended to study candidate virulence factors from other pathogens, especially those, like Chlamydia species, that are genetically intractable. For example, more than 30 C. trachomatis candidate virulence proteins inhibit yeast growth23.
The expression of candidate virulence proteins in yeast can also result in new insights into the roles of these proteins in pathogenesis15. Our observation that heterologous expression of CopN in both yeast and mammalian cells affected the formation of microtubule structures and caused a cell cycle phase-specific cell division block is intriguing, and suggests that CopN directly or indirectly targets microtubules during the course of an infection. Infection with Chlamydia has been observed to delay host cell division when high titres of Chlamydia are found in the host cells4,24. Thus, if CopN does induce a cell cycle block, this could potentially divert resources of the infected cell to favour the multiplication of Chlamydia, a strategy used by other bacterial pathogens to facilitate infection25,26.
CopN is a member of a family of proteins common to pathogenic organisms including C. psittaci B577 (C. abortus), C. trachomatis, Yersinia species and P. aeruginosa. Expression of only CopN from C. pneumonia and its closest homologue from C. psittaci (Supplementary Fig. 4) inhibited yeast growth (Fig. 1a). Yersinia YopN has been implicated to regulate type III secretion by controlling access to the secretory channel. Following contact of the bacteria with a eukaryotic cell, YopN is translocated along with other effectors into the cytoplasm of the host cell27,28. Our results indicate that the limited homology of CopN to YopN may account for the lack of yeast toxicity of YopN and that CopN may play multiple roles in pathogenesis, including regulation of secretion and modification of the host microtubule network. There is a precedent for this phenomenon, as Shigella IpaB and Salmonella SipB, both components of the type III secretion system, are also delivered into host cells where they interact with caspase-1 to trigger apoptosis. In contrast, their Yersinia homologue, YopB, a component of the translocon, is not known to target caspase-1 (refs 29, 30).
Interestingly, the small molecule inhibitors of CopN that inhibited infection with C. pneumoniae did not inhibit infection with C. trachomatis, implying that the small molecule inhibitors target a function of CopN not shared among its more distal relatives. Thus, the specificity of our compounds for C. pneumoniae suggests that we have identified lead compounds for the development of therapeutics that specifically target C. pneumoniae and its closer relatives.
METHODS SUMMARY
The effect of expression of chlamydial proteins on yeast (W303) was determined by measuring growth 48 h after the cultures were spotted onto solid inducing media. Cellular activities of CopN were examined using immunofluorescence microscopy and flow cytometry (FACS). The high-throughput yeast growth suppression screen with a 40,000 compound chemical library (Supplementary Table 1) used RDY0433 expressing GFP–CopN. RDY0433 is a yeast strain that does not express PDR1 and PDR3, two major drug efflux pumps. The effect of compounds on growth of C. pneumoniae following infection of BGMK cells with C. pneumoniae strain AR39 was determined by immunofluorescence staining and by measurements of copies of dnaK transcripts using RT–PCR. Statistical analysis was performed with Student’s t-test.
METHODS
Plasmids and expression constructs
Original plasmid vectors and derived expression constructs are summarized in Supplementary Table 2. For yeast expression, the open reading frames of the C. pneumoniae genes (CP0358, CP0433, CP0679, CP0833 and CP1062) were PCR amplified from C. pneumoniae AR39 chromosomal DNA prepared as described21, and cloned by the Gateway technology (Invitrogen) into the yeast high-copy plasmid pDSTY1, a gateway-adapted 2µ-based pFUS14, which created expression constructs pY1(CP0358), pY1(CP0433), pY1(CP0679), pY1(CP0833) and pY1(CP1062). The same strategy was used for the cloning of CopN from C. trachomatis L2 genomic DNA (provided by Z. R. Balsara and M. N. Starnbach at Harvard Medical School), YopN from Y. entercolitica pYVe227 plasmid DNA (provided by V. T. Lee currently at University of Maryland), and PopN from P. aeruginosa PAO1 genomic DNA. This cloning allows for generation of N-terminal GFP fusion proteins under the control of the GAL10 promoter. The fragments containing the GAL10 promoter, GFP fusion gene and the ADH terminator from pFUS, pY1(CP0433) and pY1(CP1062) constructs were subcloned into the centromere-based (cen) pRS313 (refs 14, 31) through homologous recombination-mediated DNA replacement to make low-copy versions of GAL10–GFP–CopN and GAL10–GFP–CP1062 expression constructs pRS(0433) and pRS(1062). Integrating versions of the of GAL10–GFP (vector control) and GAL10–GFP–CopN were made by deleting the 2µ replication origin from the backbone of pFUS and pY1(0433) constructs which then gave rise to pYGFP/int and pY0433/int to target integration at the yeast chromosomal LEU2 locus. The high-copy (2µ) plasmid vector pDSTY3 is the non-GFP version of pDSTY1 modified by deleting the GFP open reading frame, and was used to create pY3(0433) construct for expressing pure CopN protein in the yeast. For transient mammalian expression, the CP0433 open reading frame was cloned by the Gateway technology into the vector pDEST53 (Invitrogen) to create pM53(CP0433) where expression of the GFP–CopN fusion protein is driven from a constitutive CMV promoter. The GFP expression construct pM53(GFP) was made by the removal of the att cassette containing the chloramphenicol resistance gene and ccdB gene from pDEST53 following restriction digestion with NotI and PacI, blunt-end treatment, and self-ligation. For integration and ‘stably’ regulated mammalian expression, the genes for GFP and GFP–CopN fusion protein were PCR amplified from the pM53(CP0433), and inserted into the EcoRV and NotI sites of the pcDNA5/FRT/TO (Invitrogen) to create pM5to(GFP) and pM5to(0433) for targeted chromosomal integration; thereafter expression of the GFP or GFP–CopN is regulated by a tetracycline-inducible CMV promoter. The pOG44 vector (Invitrogen) was used for the Flp recombinase expression in mammalian cells.
Random mutagenesis
Mutagenesis of the gene CP0433 was carried out with the GeneMorph II Random Mutagenesis kit following the manufacture’s instruction (Stratagene). The target DNA was pY1(0433) and primers were the universal attB primers (Invitrogen). The pool of mutagenized PCR products and the BglII and BsiWI (within theCP0433 insert)-linearizedpY1(0433)were used to co-transform the yeast wherein the mutagenized CP0433 gene fragments were incorporated into the yeast expression vector pDSTY1 through in vivo homologous recombination and gap repair. The growth of resulting yeast transformants was selected on inducing selective medium plate supplemented with 2% galactose, and the plasmids were recovered for sequencing analysis.
Yeast strains and growth assay
The yeast strains for episomal and integrative expression of bacterial genes were created by transformation of the yeast strain W303a using different plasmid expression constructs (Supplementary Table 2) and a lithium acetate method32. Yeast growth assays were conducted and the yeast growth rates of individual strains were compared as described14. Briefly, saturated overnight cultures of the strains of interest were grown in non-inducing selective synthetic media supplemented with 2%raffinose. Each culture was normalized to OD600 = 1 and then serial tenfold dilutions (5 µl) were spotted onto inducing media. The plates were incubated at 30 °C and photographs of the plates were taken 48 h after plating.
Yeast microscopy and immunofluorescence
Yeast strains carrying the plasmid constructs of interest were grown overnight in non-inducing selective synthetic media supplemented with 2% raffinose. Yeast cells were diluted to OD60050.5–0.6 and grown for an additional 1 h. Then 2% galactose was added to induce expression of the fusion protein. For examination of budding morphology and nuclear division, yeast cells sampled at the designated time points were resuspended in mounting media containing DAPI (Sigma). For immunofluorescence observation of microtubules, yeast cells were fixed in 3.7% formaldehyde, and stained with rat anti-α-tubulin antibody YOL1/34 (SeroTec) and the secondary antibody Texas Red dye-conjugated donkey anti-rat IgG (H+L, Jackson ImmunoResearch Laboratory) followed by DAPI (Sigma) staining of DNA as described33. All microscopic observations were performed on an inverted Nikon Eclipse TE2000-U microscope. Images were generated by using MetaMorph software, converted to .tif format, and then transferred into Adobe Photoshop CS2 Version 9.0.2 (Adobe Microsystems) where they were re-sized, contrast-enhanced, pseudocoloured, and/or merged.
Yeast cell synchronization and flow cytometry
The yeast strains Y0433 and YGFP, derivatives of W303a carrying integrating versions of the GFP–CP0433 fusion gene and the GFP gene for integrative expression of the GFP–CopN fusion protein and GFP, were grown to early-log phase in non-inducing selective synthetic media supplemented with 2% raffinose at 30 °C. Then α-factor (10 µg ml−1, Zymo Research) was added to synchronize the yeast cells at G1 phase for a total of 3 h as described34. At 1.5 h after addition of the α-factor, 2% galactose was added to induce protein expression. The yeast cells were released from α-factor-arrest 1.5 h after the galactose was added to the media where designated as ‘0 h’ point, and continued to grow in inducing selective synthetic media supplemented with 2% galactose at 30 °C. Yeast were harvested at 30 min intervals and fixed in 70% ethanol. For flow cytometric analysis of DNA contents, DNA of the fixed yeast cells was stained using the fluorescence marker propidium iodide (PI, Sigma) as previously described35. Samples were analysed on the FACScan flow cytometer using ModFit software. The distribution of fluorescence intensity from individual cells is presented as histograms, with the x axis showing fluorescence intensity whereas the y axis shows cell number.
Mammalian cell transfection and immunofluorescence
HeLa cells were grown in DMEM medium supplemented with 10% fetal bovine serum (Invitrogen) in an incubator at 37 °C, 5% CO2. Chemical transfection of HeLa cells was performed with GeneJuice transfection reagent according to the manufacturer’s instruction (Novagen). Transfected HeLa cells were cultured for 12 h. All immunostaining procedures were performed at room temperature according to the online protocol of the Mitchison laboratory authored by A. Desai at Harvard Medical School (mitchison.med.harvard.edu/protocols). Cells were rinsed in BRB80 (80mM PIPES, 1mM EGTA and 1mM MgCl2, pH 6.8) and fixed for 10 min in 0.5% glutaradehyde in BRB80. Cell membranes were permeabilized for 15 min with a solution of 1% Triton X-100 in PBS (12mM phosphate, 137mM NaCl and 3mM KCl, pH 7.4). Free aldehydes were quenched three times with NaBH4 (1 mg ml−1, Sigma) in PBS for 10 min each. Fixed cells were rinsed three times with PBST (PBS + 0.1% Triton X-100) and blocked in 1% bovine serum albumin (BSA) in PBST for 20 min. All subsequent rinses between antibody incubations were performed using PBST. All antibodies were diluted in 1% BSA in PBST. For immunofluorescence of microtubules, cells were incubated for 60 min in 1/8,000 mouse anti-α-tubulin primary antibody (B-5-1-2, Sigma) followed by a 60-min incubation in 1/2,000 Alexa Fluor 594-conjugated goat anti-mouse IgG (H+L) secondary antibody (Invitrogen). For labelling DNA, cells were incubated in DAPI (10 mg ml−1, Sigma) for 20 min. After the coverslips had been washed three times with PBS and once with deionized water, they were mounted and observed on an inverted Nikon Eclipse TE2000-U microscope. Images were generated by using MetaMorph software, converted to .tif format, and then transferred into Adobe Photoshop CS2 Version 9.0.2 (Adobe Microsystems) where they were re-sized, contrast-enhanced, pseudocoloured and/or overlaid.
Stable cell lines and mammalian cell flow cytometry
Using Lipofectamine 2000 (Invitrogen), the Flp-In T-REx 293 cells (Invitrogen) were co-transfected with the plasmid construct pM5to(0433) or pM5to(GFP) along with the Flp recombinase-expressing plasmid pOG44 according to the manufacturer’s instructions (Invitrogen). Being selected in the presence of hygromycin (100 µg ml−1) and blasticidin (15 µg ml−1), individual colonies were tested for tetracycline (1 µg ml−1)-regulatable expression of the relevant constructs by fluorescence microscopy and western blotting. The resulting stable cell lines TR293-CopN and TR293-GFP capable of expressing CopN and GFP, respectively, were maintained in the constant selection of hygromycin (50 µg ml−1) and blasticidin (15 µg ml−1). For flow cytometric analysis of DNA contents, (5–10) × 105 cells were seeded in individual T-25 cell culture flasks. Following a 24-h incubation, the semi-confluent cells were synchronized in the G1 stage with amphidicolin (5 µg ml−1) for a total of 20 h. Removal of amphidicolin defined the ‘0 h’ time point. To ensure the presence of the protein at work immediately after release of the G1 synchronization, tetracycline (1 µg ml−1) was added to the cultures to initiate the protein expression at the −12 h point, and the post-synchronization induction continued for 18 h. Cells in one flask from each different cell line were collected at 1-h intervals starting at the 0-h point, and then fixed and permeabilized in 4 ml of 75% ethanol in PBS at −20 °C for at least 16 h. After one wash with PBS containing 1% BSA, the cell pellets were resuspended in 1 ml of solution containing propidium iodide (50 µg ml−1; Sigma), RNase (240 µg ml−1; Sigma) and Triton X-100 (0.01% v/v; Sigma). Cells were stained for at least 30 min in the dark before cell cycle analysis. The distribution of cells in the various phases of the cell cycle was analysed on a Becton-Dickinson FACScan flow cytometer using ModFit software.
Chemical library and high-throughput screen
The screening was performed at the Institute of Chemistry and Cell Biology (ICCB) at Harvard Medical School. The isogenic yeast strain RDY0433 capable of integratively expressing GFP–CopN was screened in the CopN-based yeast growth interference assay against a pilot library of 40,000 small-molecule compounds representing a diverse portion of the ICCB collection from multiple sources (Supplementary Table 1). The assay strains were constructed from the drug-sensitive strain RDY84 (MATa, pdr1DKAN, pdr3DHIS5+, ade2, trp1, his3, leu2, ura3, can1), a derivative of S. cerevisiae W303a lacking the major efflux pumps PDR1 and PDR3 (ref. 36). The screen assay was validated37 by a genetically introduced point mutation to create the mutant CopN R268H that completely eliminated the inhibitory growth effect of the wild type CopN. All primary screening was done in duplicate in 384-well plates (Costar, Corning). DMSO stock compounds were transferred using pin arrays from a stock solution of 5 mg ml−1 to the wells, each of which was pre-filled with 30 µl of inducing synthetic selective media containing 2% galactose. Then 10 µl of RDY0433 cells diluted to an OD600 of 0.16 in inducing synthetic media containing 2% galactose were added immediately to 384-well plates. The final volume in each well was 40 µl, which contained DMSO at a final concentration of 2% and compounds at a final concentration of about 12.5 mg ml−1. As a positive control, cells of the strain RDY0433(R268H) carrying integrated CopN R268H were similarly constructed, grown and diluted to the sameOD600 and then inoculated. The negative control was the assay strain RDY0433 without compounds. All plates were incubated at 30 °C for 40–42 h, and OD600 was read with a microtitre plate reader (Molecular Device). The effect of compounds was measured as a percentage of growth restoration using the following equation: percentage of growth restoration = [(ODt − ODn)/(ODp − ODn)] × 100, where ODt is OD600 of the well with the assay strain RDY0433 and test compounds, ODn is the median value of OD600 of the RDY0433 cells without compounds, and ODp is the median value of OD600 of RDY0433(R268H) cells without compounds. Compounds showing ≥10% of growth restoration in duplicate tests were scored as hits38. Hit compounds identified from the primary screening were confirmed by repeating the growth restoration assay in 96-well plates (Costar, Corning). The compounds were tested against other isogenic strains expressing different proteins that also elicit lethal phenotypes but are not related to CopN.
C. pneumoniae infection and immunofluorescence
C. pneumoniae strain AR39 (53592; ATCC) was cultured in BGMK cells and the inclusion forming units of partially purified elementary bodies (EBs) were determined as previously described21. Test of small molecule compounds on C. pneumoniae growth was performed in the BGMK cell culture in 24-well cell culture plates (Costar, Corning) in 5% CO2 at 37 °C, each well containing 1 ml of growth medium. The confluent monolayer BGMK cells were infected with EBs at a multiplicity of infection (m.o.i.) of 10 by centrifugation at 35 °C with 1,200g for 1 h, washed twice with Hanks’ balanced salt solution, and incubated in the fresh medium plus 0.2% DMSO, with or without compounds, for up to 72 h. Compounds were all used at a final concentration of 10 µg ml−1 except in the dose-dependence experiment where compounds were used at 0.3125, 0.625, 1.25, 2.5, 5, 10 and 20 µg ml−1 corresponding to 1.125, 2.25, 4.5, 9, 18, 36 and 72.1 µM for 0433YC1, and 1.0, 2.1, 4.2, 8.4, 16.8, 33.5 and 67 µM for 0433YC2. Chloramphenicol (10 µg ml−1)-treated and untreated BGMK cells were also prepared as the positive inhibition (no-growth) and negative inhibition (growth) controls, respectively. Cells from three or four wells as indicated for each concentration of compounds and from the positive and negative control wells were harvested for RNA extraction and subsequent RT–PCR. For visualization of chlamydial inclusions, the chlamydial infection and compound treatments were carried out following the same procedures in the BGMK cells cultured on coverslips in wells of 24-well plates. After incubation for 72 h, cells were washed with PBS, fixed with 100% methanol, and stained with FITC-conjugated mAb against chlamydial LPS of the Chlamydia Culture Confirmation System (Pathfinder, BIO-RAD). Hep-2 cells were grown and treated with the same procedures as used for BGMK cells.
RNA extraction and real-time RT–PCR
Total RNA was extracted from single inoculated wells by using the RNAqueous-Micro kit (Ambion) in accordance with the manufacturer’s instructions. The extracted RNAs were treated with DNase I included in the kit to eliminate the contaminating DNA. The DNA-free RNAs were confirmed by PCR without RT. RT was performed using the reverse primer specific for C. pneumoniae dnaK gene with the SuperScript III reverse transcriptase (Invitrogen) according to the manufacturer’s instructions. The resulting cDNAs were then subjected to the real-time PCR with primers specific for C. pneumoniae dnaK gene and with the use of the Platinum SYBR Green qPCR SuperMix-UDG kit (Invitrogen) following the manufacturer’s instructions on the ABI PRISM 7700 Sequence Detection System.
Acknowledgements
We thank members of the Lesser and Lory laboratories for discussions, N. Slagowski of the Lesser laboratory for assistance in the yeast growth assay, C. Shamu and the members of the ICCB Screening facility at Harvard Medical School for granting access to chemical compounds and assistance with screening, and R. Dorer of the laboratory of A. Murray at Harvard University for sharing the drug sensitive yeast strain. We thank B. Kaltenboeck at Auburn University, V. Lee, Z. Balsara and M. N. Starnbach at Harvard Medical School for sharing BGMK cells, Y. entercolitica pYVe227 plasmid DNA, and C. trachomatis L2 genomic DNA.
Footnotes
Full Methods and any associated references are available in the online version of the paper at www.nature.com/nature.
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Author Information Reprints and permissions information is available at www.nature.com/reprints.
References
- 1.Falkow S. Molecular Koch’s postulates applied to microbial pathogenicity. Rev. Infect. Dis. 1988;10 Suppl. 2:S274–S276. doi: 10.1093/cid/10.supplement_2.s274. [DOI] [PubMed] [Google Scholar]
- 2.Campbell LA, Kuo CC. Chlamydia pneumoniae – an infectious risk factor for atherosclerosis? Nature Rev. Microbiol. 2004;2:23–32. doi: 10.1038/nrmicro796. [DOI] [PubMed] [Google Scholar]
- 3.Hackstadt T. In: Chlamydia: Intracellular Biology, Pathogenesis, and Immunity. Stephens RS, editor. ASM Press; 1999. pp. 101–138. [Google Scholar]
- 4.Crocker TT, Pelc SR, Nielsen BI, Eastwood JM, Banks J. Population dynamics and deoxyribonucleic acid synthesis in HeLa cells infected with an ornithosis agent. J. Infect. Dis. 1965;115:105–122. doi: 10.1093/infdis/115.2.105. [DOI] [PubMed] [Google Scholar]
- 5.Hackstadt T, Scidmore MA, Rockey DD. Lipid metabolism in Chlamydia trachomatis-infected cells: Directed trafficking of Golgi-derived sphingolipids to the chlamydial inclusion. Proc. Natl Acad. Sci. USA. 1995;92:4877–4881. doi: 10.1073/pnas.92.11.4877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fan T, et al. Inhibition of apoptosis in Chlamydia-infected cells: Blockade of mitochondrial cytochrome c release and caspase activation. J. Exp. Med. 1998;187:487–496. doi: 10.1084/jem.187.4.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carabeo RA, Grieshaber SS, Fischer E, Hackstadt T. Chlamydia trachomatis induces remodeling of the actin cytoskeleton during attachment and entry into HeLa cells. Infect. Immun. 2002;70:3793–3803. doi: 10.1128/IAI.70.7.3793-3803.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carabeo RA, Mead DJ, Hackstadt T. Golgi-dependent transport of cholesterol to the Chlamydia trachomatis inclusion. Proc. Natl Acad. Sci. USA. 2003;100:6771–6776. doi: 10.1073/pnas.1131289100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Su H, et al. Activation of Raf/MEK/ERK/cPLA2 signaling pathway is essential for chlamydial acquisition of host glycerophospholipids. J. Biol. Chem. 2004;279:9409–9416. doi: 10.1074/jbc.M312008200. [DOI] [PubMed] [Google Scholar]
- 10.Hsia RC, Pannekoek Y, Ingerowski E, Bavoil PM. Type III secretion genes identify a putative virulence locus of Chlamydia. Mol. Microbiol. 1997;25:351–359. doi: 10.1046/j.1365-2958.1997.4701834.x. [DOI] [PubMed] [Google Scholar]
- 11.Fields KA, Mead DJ, Dooley CA, Hackstadt T. Chlamydia trachomatis type III secretion: Evidence for a functional apparatus during early-cycle development. Mol. Microbiol. 2003;48:671–683. doi: 10.1046/j.1365-2958.2003.03462.x. [DOI] [PubMed] [Google Scholar]
- 12.Peters J, Wilson DP, Myers G, Timms P, Bavoil PM. Type III secretion a la Chlamydia. Trends Microbiol. 2007;15:241–251. doi: 10.1016/j.tim.2007.04.005. [DOI] [PubMed] [Google Scholar]
- 13.Valdivia RH. Chlamydia effector proteins and new insights into chlamydial cellular microbiology. Curr. Opin. Microbiol. 2008;11:53–59. doi: 10.1016/j.mib.2008.01.003. [DOI] [PubMed] [Google Scholar]
- 14.Lesser CF, Miller SI. Expression ofmicrobial virulence proteins in Saccharomyces cerevisiae models mammalian infection. EMBO J. 2001;20:1840–1849. doi: 10.1093/emboj/20.8.1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Siggers KA, Lesser CF. The yeast Saccharomyces cerevisiae: A versatile model system for the identification and characterization of bacterial virulence proteins. Cell Host Microbe. 2008;4:8–15. doi: 10.1016/j.chom.2008.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lugert R, Kuhns M, Polch T, Gross U. Expression and localization of type III secretion-related proteins of Chlamydia pneumoniae. Med. Microbiol. Immunol. (Berl.) 2004;193:163–171. doi: 10.1007/s00430-003-0206-x. [DOI] [PubMed] [Google Scholar]
- 17.Verma A, Maurelli AT. Identification of two eukaryote-like serine/threonine kinases encoded by Chlamydia trachomatis serovar L2 and characterization of interacting partners of Pkn1. Infect. Immun. 2003;71:5772–5784. doi: 10.1128/IAI.71.10.5772-5784.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li D, et al. High-yield culture and purification of Chlamydiaceae bacteria. J. Microbiol. Methods. 2005;61:17–24. doi: 10.1016/j.mimet.2004.10.020. [DOI] [PubMed] [Google Scholar]
- 19.Khan MA, Potter CW, Sharrard RM. A reverse transcriptase-PCR based assay for in-vitro antibiotic susceptibility testing of Chlamydia pneumoniae. J. Antimicrob. Chemother. 1996;37:677–685. doi: 10.1093/jac/37.4.677. [DOI] [PubMed] [Google Scholar]
- 20.Cross NA, et al. Antimicrobial susceptibility testing of Chlamydia trachomatis using a reverse transcriptase PCR-based method. Antimicrob. Agents Chemother. 1999;43:2311–2313. doi: 10.1128/aac.43.9.2311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang J, et al. The quantity of nitric oxide released by macrophages regulates Chlamydia-induced disease. Proc. Natl Acad. Sci. USA. 2002;99:3914–3919. doi: 10.1073/pnas.062578399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moulder JW. The relation of basic biology to pathogenic potential in the genus Chlamydia. Infection. 1982;10 Suppl 1:S10–S18. doi: 10.1007/BF01640709. [DOI] [PubMed] [Google Scholar]
- 23.Sisko JL, Spaeth K, Kumar Y, Valdivia RH. Multifunctional analysis of Chlamydia-specific genes in a yeast expression system. Mol. Microbiol. 2006;60:51–66. doi: 10.1111/j.1365-2958.2006.05074.x. [DOI] [PubMed] [Google Scholar]
- 24.Horoschak KD, Moulder JW. Division of single host cells after infection with chlamydiae. Infect. Immun. 1978;19:281–286. doi: 10.1128/iai.19.1.281-286.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oswald E, Nougayrede JP, Taieb F, Sugai M. Bacterial toxins that modulate host cell-cycle progression. Curr. Opin. Microbiol. 2005;8:83–91. doi: 10.1016/j.mib.2004.12.011. [DOI] [PubMed] [Google Scholar]
- 26.Shafikhani SH, Engel J. Pseudomonas aeruginosa type III-secreted toxin ExoT inhibits host-cell division by targeting cytokinesis at multiple steps. Proc. Natl Acad. Sci. USA. 2006;103:15605–15610. doi: 10.1073/pnas.0605949103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee VT, Anderson DM, Schneewind O. Targeting of Yersinia Yop proteins into the cytosol of HeLa cells: One-step translocation of YopE across bacterial and eukaryotic membranes is dependent on SycE chaperone. Mol. Microbiol. 1998;28:593–601. doi: 10.1046/j.1365-2958.1998.00822.x. [DOI] [PubMed] [Google Scholar]
- 28.Day JB, Ferracci F, Plano GV. Translocation of YopE and YopN into eukaryotic cells by Yersinia pestis yopN, tyeA, sycN, yscB and lcrG deletion mutants measured using a phosphorylatable peptide tag and phosphospecific antibodies. Mol. Microbiol. 2003;47:807–823. doi: 10.1046/j.1365-2958.2003.03343.x. [DOI] [PubMed] [Google Scholar]
- 29.Chen Y, Smith MR, Thirumalai K, Zychlinsky A. A bacterial invasin induces macrophage apoptosis by binding directly to ICE. EMBO J. 1996;15:3853–3860. [PMC free article] [PubMed] [Google Scholar]
- 30.Guichon A, Hersh D, Smith MR, Zychlinsky A. Structure-function analysis of the Shigella virulence factor IpaB. J. Bacteriol. 2001;183:1269–1276. doi: 10.1128/JB.183.4.1269-1276.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mumberg D, Muller R, Funk M. Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene. 1995;156:119–122. doi: 10.1016/0378-1119(95)00037-7. [DOI] [PubMed] [Google Scholar]
- 32.Gietz RD, Woods RA. Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Methods Enzymol. 2002;350:87–96. doi: 10.1016/s0076-6879(02)50957-5. [DOI] [PubMed] [Google Scholar]
- 33.Miller RK. Monitoring spindle assembly and disassembly in yeast by indirect immunofluorescence. Methods Mol. Biol. 2004;241:341–352. doi: 10.1385/1-59259-646-0:341. [DOI] [PubMed] [Google Scholar]
- 34.Day A, Schneider C, Schneider BL. Yeast cell synchronization. Methods Mol. Biol. 2004;241:55–76. doi: 10.1385/1-59259-646-0:55. [DOI] [PubMed] [Google Scholar]
- 35.Zhang H, Siede W. Analysis of the budding yeast Saccharomyces cerevisiae cell cycle by morphological criteria and flow cytometry. Methods Mol. Biol. 2004;241:77–91. doi: 10.1385/1-59259-646-0:77. [DOI] [PubMed] [Google Scholar]
- 36.Dorer RK, et al. A small-molecule inhibitor of Mps1 blocks the spindle-checkpoint response to a lack of tension on mitotic chromosomes. Curr. Biol. 2005;15:1070–1076. doi: 10.1016/j.cub.2005.05.020. [DOI] [PubMed] [Google Scholar]
- 37.Tugendreich S, et al. A streamlined process to phenotypically profile heterologous cDNAs in parallel using yeast cell-based assays. Genome Res. 2001;11:1899–1912. doi: 10.1101/gr.191601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Perkins E, et al. Novel inhibitors of poly(ADP-ribose) polymerase/PARP1 and PARP2 identified using a cell-based screen in yeast. Cancer Res. 2001;61:4175–4183. [PubMed] [Google Scholar]