Abstract
This work presents the genome sequencing of the lager brewing yeast (Saccharomyces pastorianus) Weihenstephan 34/70, a strain widely used in lager beer brewing. The 25 Mb genome comprises two nuclear sub-genomes originating from Saccharomyces cerevisiae and Saccharomyces bayanus and one circular mitochondrial genome originating from S. bayanus. Thirty-six different types of chromosomes were found including eight chromosomes with translocations between the two sub-genomes, whose breakpoints are within the orthologous open reading frames. Several gene loci responsible for typical lager brewing yeast characteristics such as maltotriose uptake and sulfite production have been increased in number by chromosomal rearrangements. Despite an overall high degree of conservation of the synteny with S. cerevisiae and S. bayanus, the syntenies were not well conserved in the sub-telomeric regions that contain lager brewing yeast characteristic and specific genes. Deletion of larger chromosomal regions, a massive unilateral decrease of the ribosomal DNA cluster and bilateral truncations of over 60 genes reflect a post-hybridization evolution process. Truncations and deletions of less efficient maltose and maltotriose uptake genes may indicate the result of adaptation to brewing. The genome sequence of this interspecies hybrid yeast provides a new tool for better understanding of lager brewing yeast behavior in industrial beer production.
Key words: Saccharomyces pastorianus, beer, genome, interspecies hybrid, larger yeast
1. Introduction
Beer is one of the most common alcoholic beverages, and ∼1.7 × 1011 L are produced annually worldwide.1 Brewing yeasts are mainly divided into two groups, ale brewing yeasts and lager brewing yeasts, according to their use for the production of ales and lagers, respectively. Lager beers are fermented by using lager brewing strains at low temperatures, ranging from 8 to 15°C, and they represent the largest part of the beer market. Therefore, most of the research on beer yeast has been conducted on lager brewing yeast.
Ale and lager brewing yeasts belong to the Saccharomyces genus. The first lager brewing yeast that was studied in cultures derived from a single cell was classified as Saccharomyces carlsbergensis, whereas ale brewing yeast was classified as Saccharomyces cerevisiae.2,3 According to subsequent more sophisticated taxonomic techniques such as DNA–DNA hybridization studies, the species S. carlsbergensis was later included in the species Saccharomyces pastorianus,4 and therefore, lager brewing yeast is now classified as S. pastorianus.3 On the basis of the results of DNA–DNA hybridization studies, lager brewing yeast was described as an allopolyploid interspecies hybrid between S. cerevisiae and Saccharomyces bayanus;5 this finding was later supported by some Southern hybridization studies using S. cerevisiae gene probes.6,7 Single-chromosome transfer from lager brewing yeast to S. cerevisiae and subsequent genetic analyses also showed that lager brewing yeast is a species hybrid between S. cerevisiae and another Saccharomyces species, possibly represented by S. bayanus.8–10 As expected in the case of an interspecies hybrid, two divergent orthologous genes were found in this yeast for almost all the genes tested: in most cases, they were very similar to the corresponding S. cerevisiae and S. bayanus genes.10–13 However, a significant sequence divergence was found between a part of the S. bayanus-type MET2 gene of the lager brewing yeast and the corresponding S. bayanus sequence, suggesting that the Sb-type sub-genome is slightly different from S. bayanus.14 Furthermore, because only a few genes or chromosomes have been identified and analyzed in the lager brewing yeast so far, the overall picture of its genome (e.g. genome size and chromosome number and precise structure) has not been clarified yet.
Lager brewing yeast mitochondrial DNA (mtDNA) show a similar map of restriction sites and the same gene order as S. bayanus, and it differs from S. cerevisiae mtDNA, suggesting that in lager brewing yeast mtDNA has been inherited from the S. bayanus parental species.15
There are several biological features that are characteristic of lager brewing yeast, such as a proper degree and timing of flocculation and the efficient ability to assimilate maltose and maltotriose at low temperature.16,17 Another characteristic of lager brewing yeast is the ability to produce a significant amount of sulfite, which is important for the flavor stability of beer in several ways, including antioxidant activity. Lager brewing yeasts produce a larger amount of sulfite [>4 parts per million (ppm)] than ale brewing yeasts (<2 ppm).18 However, the complete understanding of all these characteristics has been limited so far, due to the lack of a full genome sequence of this yeast.
Here, we report the entire sequence of the hybrid genome of the lager brewing yeast S. pastorianus strain Weihenstephan 34/70, and compare it with the genome sequences of other Saccharomyces species in order to explore the genetic basis of the characteristics of lager brewing yeast relevant to beer production. The sequence of a lager beer production strain has implications for understanding not only the biological characteristics of lager brewing yeasts but also the post-hybridization evolutionary history of hybrid yeast and species. We believe that the comprehensive analysis of lager brewing yeasts using this sequence will be highly beneficial to the brewing industry in terms of facilitating beer quality control, promoting the development of breeding programs for the construction of new yeast cultures and the development of new alcoholic products.
2. Materials and methods
2.1. Strain and sequence information
The lager brewing strain S. pastorianus Weihenstephan 34/70 was provided by Fachhochschule Weihenstephan, Freising, Germany. Genome sequences and annotations for S. cerevisiae S288C, S. bayanus CBS 7001, Saccharomyces castellii CBS 4309, Saccharomyces kudriavzevii CBS 8840, Saccharomyces mikatae CBS 8839 and Saccharomyces paradoxus CBS 432 were obtained from the Saccharomyces Genome Database (SGD; http://www.yeastgenome.org/) (30 August 2006). Genome sequences for S. cerevisiae YJM789 and RM11-1a were obtained from the GenBank database (accession no. AAFW02000000) and from the Broad Institute (http://www.broad.mit.edu/annotation/genome/saccharomyces_cerevisiae/Home.html), respectively.
2.2. Genome sequencing and assembly
A total of 331 798 clone ends were sequenced by the whole genome shotgun method and from cosmid libraries (Supplementary methods and Supplementary Table S1). The sequences were assembled into contigs (continuous blocks of uninterrupted sequence) and supercontigs (contigs linked by paired forward–reverse reads from the same plasmid) using the Phred19 and PCAP20 software packages.
2.3. Phylogenetic analysis
A NCBI-BLAST blastp search within S. cerevisiae S288C open reading frames (ORFs) was carried out with default parameters to find potential paralogs ORFs (query; S. cerevisiae ORFs DB; S. cerevisiae ORFs). The ORFs aligned against ORFs with high alignment lengths (match length of at least 50 amino acid) and high identities (≥60%) were listed as potential paralogous ORFs. An S. cerevisiae S288C orthologous ORFs set was prepared by removing the potential paralogous ORFs, and it was used for identification of orthologous ORFs.
For eight yeast strains (S. pastorianus Weihenstephan 34/70, S. cerevisiae YJM789, RM11-1a, S. bayanus CBS 7001, S. castellii CBS 4309, S. kudriavzevii CBS 8840, S. mikatae CBS 8839 and S. paradoxus CBS 432), the complete set of all predicted ORFs starting with a methionine and having a length of at least 50 amino acids were gathered.
Orthologous ORF sets for each strain were gathered by picking up bidirectional best hits. As for lager brewing yeast, each orthologous ORF of S. cerevisiae S288C was aligned to the two different ORFs of this yeast: the best matching ORFs were considered as class1 orthologous ORFs, whereas the second best matching ORFs (with scores >60) were considered as class2 orthologous ORFs. Pairwise distances between two strains were calculated using the Kimura two-parameter model using all the aligned ORFs, and phylogenetic trees were constructed using the UPGMA method. The relative distance of the closest species was computed using the neighbor-joining method.
2.4. Gene finding and annotations
ORFs that start with a methionine (ATG codon) and consist of more than 50 amino acids (150 bp) were predicted from the supercontigs of lager brewing yeast. Two sets of ORF data were used as references for gene finding and annotations. One consisted of S. bayanus CBS 7001 ORFs, which were also re-annotated from the genome sequence reported (GenBank accession no. AACG02000000). The other was a set of S. cerevisiae S288C annotated ORFs (SGD; http://www.yeastgenome.org/). Each ORF from a given species was used as a query to compare with ORFs from the two other species using NCBI-BLAST blastp. Blast results with scores above 60 were considered as significant hits for each ORF and used for the following steps. First, clusters of the ORF were generated. The 2930 clusters of the lager brewing yeast were composed of two types of ORF, and each ORF cluster was annotated with the functional role according to the descriptions of the S. cerevisiae ORF. In addition, the two sets of ORF in the lager brewing yeast were classified as Sc-type ORF, having higher identity to the S. cerevisiae ORF, and as Sb-type ORF, having higher identity to S. bayanus ORF. Subsequently, the remaining clusters, which had more than two S. cerevisiae ORFs, were annotated by preferentially keeping matches within gene order (synteny) over non-syntenic matches in the supercontigs. The remaining ORFs that were not annotated by the above methods were annotated to best-hit homologs with an identity of ≥40% and marked as non-orthologs. Of the non-orthologs, the ORFs having no homologies with S. cerevisiae S288C or S. bayanus CBS 7001 ORFs were defined as specific for the lager brewing yeast. These ORFs were annotated to the best-hit homologs and their functions were assigned or predicted by BLASTP against the non-redundant database using three thresholds: i.e. an E-value of ≤1 × 10−5, an identity of ≥40% and a match length of at least 60% of both protein lengths, and the domains by Pfam using one threshold, i.e. an E-value of ≤1 × 10−5. Six mtDNA contigs were obtained and assembled into a circular contig by closing the gaps by further sequencing using specific PCR primers across each gap. Gene annotation of the lager brewing yeast mtDNA genome was carried out using a variation of NCBI-BLAST blastn and blastx against mtDNA genomes of S. cerevisiae and manual curation. The annotation of tRNA genes was performed using the program tRNAscan-SE 1.21.
2.5. Identification of chromosomal rearrangements
Supercontigs, in which either the gene synteny was not conserved with respect to the S. cerevisiae genome or both Sc- and Sb-type ORFs were included, were considered as candidates for chromosomal rearrangements. The regions containing the cosmid clones, which had an Sc-type sequence at one end and an Sb-type sequence at the other end, were considered as candidates for the chromosomal translocations. These putative translocations were confirmed by PCR amplification across the breakpoints using genomic DNA from the lager brewing yeast as a template and by sequencing analyses of the amplicons.
2.6. Optical mapping
Optical mapping (opmap; OpGen, Inc., Madison, WI, USA) was performed as reported earlier21 using XhoI restriction endonuclease. To validate and align the assembled sequence by opmap, the predicted lager brewing yeast chromosomal sequences were constructed on the basis of information provided by the assembled sequences, gene annotations and chromosomal rearrangements identified above.
2.7. Data deposition
This Whole Genome Shotgun project has been deposited at DDBJ/EMBL/GenBank under the project accession ABPO00000000. The version described in this paper is the first version, ABPO10000000. The accession number for the mitochondrial genome is EU852811.
3. Results and discussion
3.1. Genome assembly
We generated 7.8-fold redundant coverage in paired end sequences from the whole genome shotgun plasmids and additional cosmid clones. The sequences were assembled into 3184 contigs and 796 supercontigs using Phred19 and PCAP20 to yield a draft consensus sequence (Table 1). The sequence has a long-range continuity (N50 supercontig length of 108 kb) and 0.26 kb sequence gaps; such a sequence is short compared with the average gene length. The sum of the supercontig lengths including gaps is 23.4 Mb, not including the ribosomal DNA (rDNA) locus.
Table 1.
Genome assembly
| Sequence coverage (fold) | 7.8 |
| Assembled sequence (Mb)a | 22.5 |
| Assembled sequence, including gaps (Mb)b | 23.4 |
| Number of contigs | 3184 |
| Number of supercontigs | 796 |
| N50 supercontig length (kb) | 108 |
| Gaps per 100 kb | 13.2 |
| Average gap length (bp) | 258 |
aSum of contig lengths.
bSum of supercontig length, including contig lengths and estimated gap sizes between consecutive contigs.
3.2. Phylogenetic relationship
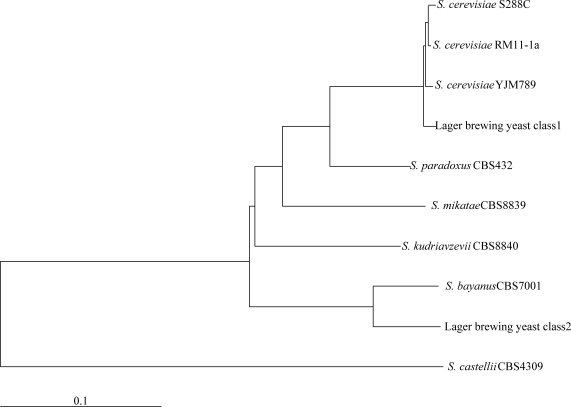
Previous works based on the sequences of the rDNA internal transcribed spacers (ITS1, ITS2) have led to the definition of a phylogenetic relationship between the Saccharomyces genus in which lager brewing yeast is closely related to S. bayanus.22 However, this definition has led to the construction of inaccurate phylogenetic trees because of the hybrid nature of lager brewing yeast. In addition, single gene phylogenies are much more error-prone than multiple gene phylogenies.23 Recently, whole genome data from Saccharomyces yeasts have provided a greater resolving power by allowing trees to be constructed based on concatenated sets of genes.23 Using this approach, we studied the relationship between lager brewing yeast and the other Saccharomyces yeasts.
A set of 2080 ORFs among class1 and 2 of the lager brewing yeast and eight Saccharomyces strains were compared and concatenated for phylogenetic analysis (see Materials and methods). As expected, the constructed phylogenetic tree showed that class1 and 2 were close to S. cerevisiae and S. bayanus, respectively (Fig. 1). This result supported the hypothesis that the lager brewing yeast is a hybrid between the two species, S. cerevisiae and S. bayanus at genomic level. However, evolutionarily, the set of class2 ORFs of the lager brewing yeast was clearly farther apart from S. bayanus CBS 7001 in comparison with the class1 ORFs and the S. cerevisae strains.
Figure 1.
Phylogenetic tree of class 1 and 2 of the larger brewing yeast and eight yeast species. Branch lengths denote the number of substitutions per base.
To explain the distance between the class2 ORFs of the lager brewing yeast and S. bayanus, it is important to highlight that the S. bayanus species is taxonomically very complex. It contains at least two sub-groups of strains:24,25 S. bayanus var. bayanus represented by a variety of strains not currently isolated from natural environment and S. bayanus var. uvarum, commonly known as Saccharomyces uvarum, represented by strains that are typically and abundantly isolated from the enological environment. The only S. bayanus strain that has currently been fully sequenced and can therefore be used as a reference in comparative genomic studies is strain CBS 7001, which belong to the S. uvarum group.26 It is reasonable to conclude that this strain is not an ideal reference for studying the lager brewing yeast genome; it is simply the best approximation available so far. Some of the main differences between this yeast and the corresponding sub-genome of the lager brewing yeast are described in this study.
3.3. Chromosomal structure
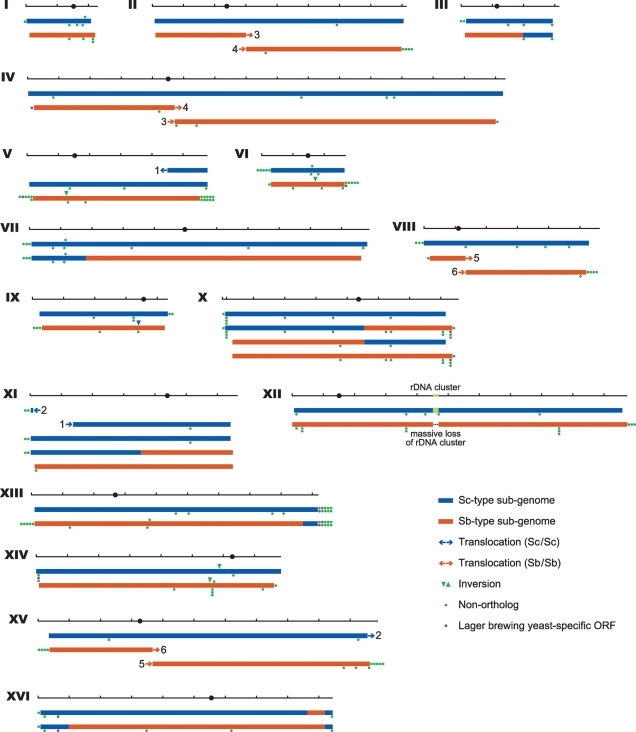
After clarifying that lager brewing yeast was a hybrid between S. cerevisiae and S. bayanus, we annotated the ORFs of the lager brewing yeast and produced a large-scale alignment of genomic regions by aligning the lager brewing yeast genome with the S. cerevisiae S288C and S. bayanus CBS 7001 genomes (see Materials and methods). The Sb-type ORFs showed clearly lower sequence identities with the corresponding S. bayanus CBS 7001 ORFs (average 92.7%) in comparison with the Sc-type ORFs and the S. cerevisae S288C (99.2%) (Supplementary Table S2), and it was consistent with the phylogenetic analysis. Most of the supercontigs were classified into two groups, i.e. those consisting entirely of Sc-type ORFs and those consisting entirely of Sb-type ORFs (Supplementary Table S2); therefore, the two sub-genomes were designated Sc- and Sb-type, respectively. Except for the sub-telomeric regions, large regions of the Sc- and the Sb-type sub-genomes shared conserved synteny with the S. cerevisiae and S. bayanus genomes (Fig. 2). On the sub-telomeric regions, where syntenies were not well conserved, ∼59% of non-orthologs (111/187) were found. It is in these sub-telomeric regions where many important genes for beer brewing (sugar transporters; HXT and MAL families and lectin-like protein involved in flocculation; FLO families etc.) were found (Supplementary Table S2).
Figure 2.
Chromosomal structure of lager brewing yeast. The types of lager brewing yeast chromosomes are summarized. The black horizontal bars indicate the S. cerevisiae genome with tick marks for every 100 kbp. Matching pairs of translocations between chromosomes with different numbers are indicated by the orientation of the arrowheads with their common numbering. The locations of chromosomal rearrangements (inversions and deletions) relative to the S. cerevisiae genome are indicated by colored triangles, and the locations and numbers of non-orthologs and lager brewing yeast-specific genes are indicated by colored dots. The non-orthologs that are split into more than two genes by stop codon or frameshift mutation are counted as one, and the genes whose characteristics are deemed dubious are not counted. The positions of these marks, whether they are shown above or below the chromosome bars, do not indicate the strand in which those rearrangements or genes are located in the lager brewing yeast genome.
In addition to the sub-telomeric rearrangements, several internal rearrangements were found in comparison with S. cerevisiae S288C (Fig. 2). One and three inversions were found in Sc- and Sb-type sub-genomes, respectively (Fig. 2). The Sc-type sub-genome showed an inversion of 30 kb (570–600 kb of Sc-type chromosome XIV) (Fig. 2 and Supplementary Table S3A). The sequence of S. cerevisiae RM11-1a27 and YJM78928 revealed that the same region was also inverted in comparison with S. cerevisiae S288C. Two of the inversions found in the Sb-type sub-genome (117–133 and 164–181 kb of Sb-type chromosomes V and VI, respectively) (Fig. 2 and Supplementary Table S3A) have also been identified in S. bayanus CBS 7001.26 The remaining inversion spans within the interval between 553 and 560 kb on the Sb-type chromosome XIV. Two translocations were found in the Sc-type sub-genomes. A novel chromosome comprising a combination of Sc-type chromosomes V and XI created by the translocation present in the Ty element was detected, and a fragment corresponding to 32 kb of the Sc-type chromosome XV of S. cerevisiae S288C was also found to be translocated to the Sc-type chromosome XI inside an identical area of 34 bp located between FRE2 and FRE3 (Table 2). On the other hand, two reciprocal translocations (Sb-types II–IV and VIII–XV) were found in the Sb-type sub-genome (Table 2); these translocations have been reported previously in lager brewing yeast6,7 as well as in S. bayanus.26 Furthermore, the genome of S. bayanus CBS 7001 has three reciprocal translocations (right arm of II and left arm of II, VI–X and II–XIV).26 However, the gene syntenies of the corresponding regions in the Sb-type sub-genome are the same as those of S. cerevisiae (Supplementary Table S3C). Eight chimera chromosomes between the Sc- and the Sb-type sub-genomes were found in the lager brewing yeast, indicating a post-hybridization event (Fig. 2). Although the breakpoints of the chromosomal translocations in the Saccharomyces lineage seem to be associated to Ty elements or highly similar pairs of ribosomal protein genes,29 those in the lager brewing yeast were found between the orthologous non-ribosomal protein genes, with the exception of chromosome III (Table 2). One was a reciprocal translocation between the orthologs, ScYJR009C (ScX) and SbYJR009C (SbX), which share the 96.1% sequence identity over an 840 bp homology region (Supplementary Fig. S1). The other seven translocations occurred by non-reciprocal recombinations. The non-reciprocal translocation between ScIII and SbIII was found to be at the MAT-locus, which involved a 604 bp homologous region. The remaining six non-reciprocal translocations (ScVII/SbVII, ScXI/SbXI, ScXIII/SbXIII, ScXVI/SbXVI, ScXVI/SbXVI and ScXVI/SbXVI) occurred between their orthologs ORFs (YGL173C, YKL045W, YMR302C, YPL240C, YPR160W and YPR191W), which share 81–87% sequence identity (Table 2 and Supplementary Fig. S1). The sequencing of these translocation products showed that exchange occurred inside their identical short regions (8–23 bp) (Supplementary Fig. S1).
Table 2.
List of chromosomal translocations
| Translocation | Breakpoint |
|---|---|
| Sc–Sc-type chromosomal translocation | |
| ScV/ScXI | ScV Ty LTR > ScXI Ty LTR |
| ScXI/ScXV | ScYKL220C (FRE2) > ScYOR381W(FRE3) |
| Sb–Sb-type chromosomal translocations | |
| SbII/SbIV | SbYBR031W (RPL4A) > SbYDR012W (RPL4B) |
| SbII/SbIV | SbYDR012W (RPL4B) > SbYBR031W (RPL4A) |
| SbVIII/SbXV | SbYOR018W-SbYOR019W > SbYHR014W-SbYHR015W |
| SbVIII/SbXV | SbYHR014W-SbYHR015W > SbYOR018W-SbYOR019W |
| Sc–Sb-type chromosomal translocations | |
| ScIII/SbIII | SbMAT locus > ScMAT locus |
| ScVII/SbVII | ScYGL173C (KEM1) > SbYGL173C (KEM1) |
| ScX/SbX | ScYJR009C (TDH2) > SbYJR009C (TDH2) |
| ScX/SbX | SbYJR009C (TDH2) > ScYJR009C (TDH2) |
| ScXI/SbXI | ScYKL045W (PRI2) > SbYKL045W (PRI2) |
| ScXIII/SbXIII | SbYMR302C (YME2) > ScYMR302C (YME2) |
| ScXVI/SbXVI | ScYPL240C (HSP82) > SbYPL240C (HSP82) |
| ScXVI/SbXVI | ScYPR160W (GPH1) > SbYPR160W (GPH1) |
| ScXVI/SbXVI | SbYPR191W (QCR2) > ScYPR191W (QCR2) |
Translocations usually occur between sequences at identical positions on homologous chromosomes in order to repair DNA for chromosome disjunction.30,31 Abbreviated translocations have been shown to be common in S. cerevisiae, but most of them are mediated through long regions of homologous Ty or sub-telomeric Y′ element recombination.32 However, in the present study, we observed that most of the chromosomal translocations between the Sc- and the Sb-type chromosomes occurred between the short homologous regions of orthologous genes, which showed 81–96% sequence divergences. Sequence divergence acts as a potent barrier for translocations. The translocations found in the lager brewing yeast are rare since the recombination rates for diverged sequences are 2.2 × 10−9 (82% sequence divergence) and 5.4 × 10−8 (96%) in Saccharomyces yeast.33 The frequencies are increased in double-strand breaks (DBSs). When DSBs were induced by ionizing radiation in the diploid S. cerevisiae genome, a translocation was found between the short homology region (26 bp) of the two homologs (90.7% sequence identity).34 Thus, it seems that the translocations found between Sc- and Sb-type chromosomes were mediated by the fortuitous appearance of broken chromosomes ends produced by DSBs in either of the two orthologs.
An optical mapping (opmap)21 using XhoI restriction endonuclease was performed to determine the genome size and chromosomes number. The DNA from the lager brewing yeast yielded an optical map of 36 chromosomes of ∼26.1 Mb, not including the rDNA cluster (Table 3). This genome size and the number of chromosomes are more than twice those of S. cerevisiae S288C (16 chromosomes and 12.1 Mb, respectively). In order to validate the predicted chromosomal structures, 36 chromosomal DNA sequences were constructed by connecting supercontigs without gaps (as shown in Supplementary Table S2) and then compared with the opmap. All the 36 sequences showed clear one-to-one matches to the 36 corresponding chromosomes found by the opmap (Supplementary Fig. S2). The total size of the 36 chromosomal DNA sequences was 25.0 Mb, and hence, the coverage was 95.8% of the estimated genome size, as determined by opmap (Table 3). On the basis of these results, we conclude that the lager brewing yeast has 36 chromosomes, whereas its parental species, S. cerevisiae and S. bayanus, have 16 chromosomes each.
Table 3.
Opmap and comparison with the predicted chromosome
| Number of chromosomes found by the opmap | 36 |
| Total size of the opmap chromosomes (Mb) | 26.1 |
| Number of the predicted chromosomes | 36 |
| Total size of the predicted chromosomes (Mb) | 25.0 |
| Genome coverage (%) | 95.8 |
3.4. rDNA locus
The rDNA of S. cerevisiae is a region of ∼1.5 Mb, consisting of 150 tandem copies of a 9.1 kb repeat containing the ITS and is located on the right arm of chromosome XII. The lager brewing yeast we analyzed has two types of chromosomes XII and two types of ITS sequence: one is closely related to S. cerevisiae and the other to S. bayanus (Fig 2 and Supplementary Fig. S3). Strains S. pastorianus CBS 1538 and S. carlsbergensis CBS 1513, historical lager brewing yeasts, not currently used in beer manufacturing, show only the Sb-type ITS sequence. Comparative genomic hybridization (CGH) of these two strains with S. cerevisiae yeast DNA microarrays revealed, in fact, that the Sc-type chromosome XII has been lost in both strains.35 Although the length of the rDNA region on the Sc-type chromosome XII was estimated to be >350 kb, that on the Sb-type chromosome XII, according to the opmap and PCR amplification, was of 18 kb (Supplementary Fig. S2 and Table S3C). This result indicates that the rDNA region on the Sb-type chromosome XII decreased massively. Analysis carried out by genomic in situ hybridization with labeled rDNA sequences in the alloploid grass, Zingeria trichopoda, also revealed that the 45S rDNA from one of the parental genomes had undergone significant reduction.36
3.5. Structure of the mitochondrial genome
The mitochondrial DNA (mtDNA) of the lager brewing yeast was also sequenced and assembled into a circular contig. The size of the mtDNA was 70 578 bp, somewhat smaller than that of S. cerevisiae S288C (85 779 bp) and slightly larger than that of S. bayanus (64.3 kb) (Supplementary Fig. S4). The mtDNA of the lager brewing yeast was also found to contain several differences (gene order, number of exons and intron–exon boundaries of COX1, COB1 and 21SrRNA) in comparison with S. cerevisiae (Supplementary Fig. S4). On the other hand, it showed many aspects of similarity with the mtDNA of S. bayanus. The gene order was the same as that of S. bayanus,37 and it was completely consistent with the results of mapping obtained with restriction enzymes.15 In addition, all mtDNA genes exhibited a higher sequence similarity with the corresponding genes of S. bayanus than with those of S. cerevisiae (Supplementary Table S4). We concluded that the mtDNA of the lager brewing yeast was inherited from its Sb-type ancestor without shuffling between the Sc- and the Sb-type mtDNA genomes.
3.6. ORF deletion
Twenty-eight and 33 ORFs were found to have been shortened (truncated) by frameshift or stop codon mutation in the Sc- and Sb-type sub-genomes, respectively (Supplementary Table S5), whereas the ORF lengths between Saccharomyces sensu stricto are quite conserved.26 This can be expected as the result of a yeast having two functional copies of each gene that releases the evolutionary pressure on one copy. Of these ORFs, three were found to be essential by a large-scale deletion study in S. cerevisiae,38 indicating that they must have been truncated after the interspecies hybridization event. In addition, some ORFs involved in the characteristics of lager brewing yeast were also found to be truncated (see below).
3.7. Maltose and maltotriose uptake genes
Maltose and maltotriose are the most abundant fermentable sugars in wort, and the rate at which brewing yeast takes up these sugars can have a major impact on the fermentation performance. The α-glucoside transporter genes, with nine predicted transmembrane domains, are responsible for the uptake of maltose and maltotriose. Increasing the expression of the α-glucoside transporters have been shown to increase the rate of maltose fermentation.39 Three α-glucoside transporter genes in each Sc- and Sb-type sub-genome were found on seven loci (Table 4). LBYG03039 was identical to MTT1, which encodes an efficient maltotriose transporter in lager brewing yeasts,40 and was found to be on both chromosomes ScVII and SbVII (Table 4). This indicates that the gene copy number of MTT1 in the lager brewing yeast has increased thanks to the chromosomal translocation. Another α-glucoside transporter gene (MPH3), which was found in S. cerevisiae S288C, was not found in the lager brewing yeast genome. This is consistent with previous studies carried out with Southern blotting.41,42 LBYG09472 (Sc-type AGT1) and LBYG14145 (Sb-type MAL31) showed truncated forms having only two and three predicted transmembrane domains and are, hence, unlikely to be functional. In total, six α-glucoside transporter genes were found in seven chromosomal regions in the lager brewing yeast sequenced in this work, but two of these were truncated.
Table 4.
Genes related to lager brewing yeast characteristics
| Gene | Type | Location | Length (LBYG) (aa) | Predicted Protein | Systematic Name | Standard Name | Length (S288C) (aa) | Identity (%) |
|---|---|---|---|---|---|---|---|---|
| α-glucoside transporter genes | ||||||||
| LBYG00616 | Sc | ScII right sub-telomere | 614 | α-glucoside transporter | YBR298C | MAL31 | 614 | 98 |
| LBYG03039 | Sb | ScVII right sub-telomere SbVII right sub-telomere |
615 | α-glucoside transporter | YBR298C | MAL31 | 614 | 91 |
| LBYG13187 | Sb | SbXV-VIII left sub-telomere | 610 | α-glucoside transporter | YGR289C | AGT1 | 616 | 85 |
| LBYG00811-00812a | Sc | ScIV left sub-telomere | 602 | α-glucoside transporter | YDL247W | MPH2 | 609 | 94 |
| LBYG09472a | Sb | SbV right sub-telomere | 231 (tr) | α-glucoside transporter | YBR298C | MAL31 | 614 | 80 |
| LBYG14145a | Sc | Unkown | 321 (tr) | α-glucoside transporter | YGR289C | AGT1 | 616 | 96 |
| Genes related to the sulfite production pathway | ||||||||
| LBYG00610 | Sc | ScII right sub-telomere | 183 (tr) | High affinity sulfate permease | YBR294W | SUL1 | 859 | 100 |
| LBYG09125 | Sb | SbIV-II right sub-telomere | 295 (tr) | High affinity sulfate permease | YBR294W | SUL1 | 859 | 83 |
| LBYG04711-04712a | Sc | ScXII | 893 | High affinity sulfate permease | YLR092W | SUL2 | 893 | 99 |
| LBYG11823 | Sb | ScXII | 893 | High affinity sulfate permease | YLR092W | SUL2 | 893 | 87 |
| LBYG03911 | Sc | ScX SbX-ScX |
511 | ATP sulfurylase | YJR010W | MET3 | 511 | 100 |
| LBYG11111 | Sb | SbX ScX-SbX |
511 | ATP sulfurylase | YJR010W | MET3 | 511 | 89 |
| LBYG04364 | Sc | ScXI ScV-ScXI |
202 | Adenylylsulfate kinase | YKL001C | MET14 | 202 | 100 |
| LBYG11543 | Sb | SbXI ScXI-SbXI |
202 | Adenylylsulfate kinase | YKL001C | MET14 | 202 | 97 |
| LBYG07555 | Sb | mosaic-ScXVI mosaic-SbXVI |
261 | 3′-phosphoadenylsulfate reductase | YPR167C | MET16 | 261 | 92 |
| LBYG07205 | Sc | mosaic-ScXVI | 458 | Plasma membrane sulfite pump | YPL092W | SSU1 | 458 | 99 |
| LBYG13862 | Sb | mosaic-SbXVI | 458 | Plasma membrane sulfite pump | YPL092W | SSU1 | 458 | 79 |
α-Glucoside transporter genes and genes related to the sulfite production pathway are shown. In Saccharomyces sensu stricto yeast, sulfite is produced in the methionine and cysteine biosynthesis pathway. Sulfate is imported into a cell by sulfate transporters, Sul1p and Sul2p and is converted into APS (adenylylsulfate) by ATP sulfurylase, Met3p. APS is phosphorylated, yielding PAPS (3′-phosphoadenylyl-sulfate) by adenylylsulfate kinase, Met14p. Sulfite is yielded by the reduction of PAPS by 3′-phosphoadenylylsulfate reductase, Met16p. Excess sulfite is passed out of the cell by a sulfite transporter, Ssu1p.
The Tr in brackets indicates the truncated genes. The genes were aligned against S. cerevisiae gene using Blastp, and each S. cerevisiae gene with the highest score is shown. Non-truncated and truncated protein sequence identities were calculated by the global alignment tool (needle) and local alignment tool, respectively, which were provided by EMBOSS. Genes that were found in lager brewing yeast but in which the specific chromosome could not be decided are shown as ‘unknown’.
aNote that the genes were confirmed by PCR amplification and sequencing (Supplementary Sequence S1).
Integration of an intact AGT1 gene containing its promoter region from S. cerevisiae into a lager brewing yeast caused a reduction in sugar assimilation rates during lager beer fermentation in comparison with the parental strain (H. Hatanaka et al., manuscript in preparation). These results indicate that adaptation to brewing conditions resulted not only in the increase of the copy number of efficient α-glucoside transporter genes but also in the inactivation of less efficient genes, since the capacity of membrane-localized proteins may have been limited.
3.8. Sulfite production pathway
Lager brewing yeasts produce relatively high levels of sulfite compared with ale brewing yeasts.18 Two distinct Sc- and Sb-type genes were found in the lager brewing yeast sulfite production pathway, with the exception of MET16 (Table 4). Lager brewing yeast, in fact, has only Sb-type MET16 because of a deletion on chromosome XVI in the area (from 861 to 920 kb) that includes the Sc-type MET16. Lager brewing yeast has four sulfate transporter genes: Sc-type-SUL1, SUL2 and Sb-type-SUL1, SUL2. The SUL1 gene sequences from S. cerevisiae encode an 859 amino acid polypeptide with nine predicted transmembrane domains, but Sc- and Sb-type SUL1 genes were found to encode truncated (183 and 295 amino acids) polypeptides with stop codon and frameshift mutations, respectively. These proteins are unlikely to be functional transporters because they have only one and three transmembrane domains. Five genes (Sc-MET3, Sb-MET3, Sc-MET14, Sb-MET14 and Sb-MET16) were found on two chromosomes due to chromosome translocations (Table 4), indicating that these gene loci were increased in copy number.
3.9. Lager brewing yeast-specific genes
Eight genes were found to be present in the lager brewing yeast but absent in both S. cerevisiae S288C and S. bayanus CBS 7001. Except for an YJM789 homolog (LBYG13 665), these were all located in sub-telomeric regions (Table 5). LBYG05796 (encoding a transcriptional regulatory protein) was also found in YJM789.28 LBYG11275 (encoding a melibiase), LBYG08543 (a fructose symporter), LBYG05774, (a tyrosine permease), LBYG05783 (an amidase) and LBYG09147 (an amidase) have been identified and characterized in lager brewing yeast, and some of their functions are specific of lager brewing yeast.43–46 Only one of the lager brewing yeast specific genes (LBYG09608) did not have any previous Saccharomyces homologs. According to the results retrieved by the PSORT II system (http://psort.hgc.jp/form2.html), this gene is predicted to encode a monocarboxylate transporter (MCT) with major facilitator superfamily, and localize in the plasma membrane or in the endoplasmic reticulum. Subsequent tests demonstrated that it is expressed at the middle and late stages of lager beer fermentation (data not shown).
Table 5.
Lager brewing yeast-specific genes
| Gene | Location | Length (aa) | Predicted Protein | E-value | Homolog (BLASTP) | Pfam Domains |
|---|---|---|---|---|---|---|
| LBYG11275 | SbX right sub-telomere ScX-SbX right sub-telomere |
471 | Melibiase | 0 | α-galactosidase (S. pastorianus) | Melibiase |
| LBYG08543 | SbIV-II left sub-telomere | 570 | Fructose symporter | 0 | fructose symporter (S. pastorianus) | Sugar (and other) transporter |
| LBYG05774 | SbXIII-ScXIII right sub-telomere ScXIII right sub-telomere |
557 | tyrosine permease | 0 | tyrosine permease (S. pastorianus) | Amino acid permease |
| LBYG05783 | ScXIII right sub-telomere SbXIII-ScXIII right sub-telomere |
598 | amidase | 0 | amidase homolog AMI1-A (S. pastorianus) | Amidase |
| LBYG09147 | SbV left sub-telomere | 395 | amidase | 0 | amidase homolog AMI1-B (S. pastorianus) | Amidase |
| LBYG13665 | SbXVI left arm | 372 | hypothetical protein | e−138 | hypothetical protein SCY_5476 (S. cerevisiae YJM789) | - |
| LBYG05796 | ScXIV left sub-telomere | 795 | transcriptional regulatory protein | 0 | hypothetical protein SCY_1426 (S. cerevisiae YJM789) | Fungal Zn(2)-Cys(6) binuclear cluster domain |
| LBYG09608 | SbVI right sub-telomere | 356 | Monocarboxylate transporter | 1e−55 | MFS monocarboxylate transporter, putative (Aspergillus clavatus NRRL 1) | Major Facilitator Superfamily |
Lager brewing yeast-specific genes that were found to be present only in the lager brewing yeast strain but absent in S. cerevisiae S288C and S. bayanus CBS 7001. The genes were identified by Blastp search against the non-redundant database and by Pfam search, and each description with the highest score or nearly the highest score is shown.
MCTs form a large family of proton-linked carriers that have the ability to transport monocarboxylates such as lactate, pyruvate or acetate across the plasma membrane, and they can be found in bacteria, protozoa, fungi, invertebrates, as well as in vertebrates.47 Pyruvate, acetate and lactate are by-products formed during yeast alcoholic fermentation. Therefore, LBYG09608 gene encoding MCT may be expressed at the middle and late stages of fermentation to control these monocarboxylates in the plasma membrane or in the endoplasmic reticulum.
3.10. Origins and evolution of lager brewing yeast
The genome sequencing of S. pastorianus Weihenstephan 34/70 confirmed at genomic level that lager brewing yeasts are hybrids between the two species S. cerevisiae and S. bayanus. The phylogenetic tree shown in Fig. 1, constructed on the basis of concatenated sets of genes of various Saccharomyces yeasts and the high sequence identities between Sc-type and S. cerevisiae ORFs show that the Sc-type sub-genome of the lager brewing yeast is clearly close to S. cerevisiae. Recently, Dunn and Sherlock,48 on the basis of results obtained using two species CGH array analysis, reported that the most probable ancestral parent of lager brewing yeasts Sc-type sub-genome is related to ale brewing yeasts. The contribution of ale brewing yeasts to the shaping of the lager brewing yeast Sc-type sub-genome, already proposed by de Barros Lopes et al.,49 is a very likely possibility.
Lager brewing yeast Sb-type sub-genome contributes to the most useful features typical of this yeast. However, the full identification of this sub-genome has never been achieved and this is probably due to the complexity of the S. bayanus species as well as the lack of a proper reference strain, as already described above. Our results show that the Sb-type sub-genome is clearly further apart from S. bayanus CBS 7001, which actually belongs to the S. uvarum group and is closely related to the enological environment. Additionally, although S. bayanus CBS 7001 genome has three reciprocal translocations (right arm of II and left arm of II, VI–X and II–XIV),26 the gene syntenies of the corresponding Sb-type lager brewing yeast chromosomal regions are the same as those of S. cerevisiae. We demonstrated in a previous study that the yeast that best represent the Sb-type sub-genome is strain NBRC 1948.50 This yeast, unlike S. bayanus CBS 7001, is currently classified as S. bayanus var. bayanus and was originally isolated from beer. The typing of 48 gene fragments in various lager brewing yeasts by RFLP analysis showed that the Sb-type sub-genome differed from the genome of strain NBRC 1948 only for a few genes. Saccharomyces uvarum sequences in lager brewing yeasts were limited to the few genes portions in which S. bayanus var. bayanus strains could not be distinguished from S. uvarum strains.50 This indicates a common origin of S. bayanus and S. uvarum yeasts and a subsequent differentiated evolution process in the brewing and in the enological environment, respectively.
The origin of the Sb-type sub-genome can also be confirmed by studying mtDNA inheritance. Several works have demonstrated that in lager brewing yeast the mtDNA genome is originating from the S. bayanus parental strain. In artificial interspecies, hybrid mtDNA is inherited by only one of the parental strains.51 Consequently, the sequence analysis of the mtDNA in lager brewing yeasts can be very useful to define the nature of the S. bayanus parental genome. According to our results, the nucleotide sequence of lager brewing yeast COXII, COXIII, ATP8, ATP6 and 21S rRNA showed a 2–4% sequence divergence with the corresponding S. uvarum genes (Supplementary Table S4). On the other hand, the nucleotide sequence of lager brewing yeast COXII gene was identical to S. bayanus var. bayanus NBRC 1948 COXII gene, and mtDNA restriction analysis carried out with three enzymes confirmed this result.52 Considering our present and previous results, the Sb-type sub-genome did not originate from S. uvarum, but from an S. bayanus var. bayanus yeast closely related to strain NBRC 1948.
Several studies have been carried out to explain the origin and the evolutionary processes that lager brewing yeasts has undergone. Either a simple hybridization event between an S. cerevisiae and S. bayanus yeast, or more likely, a multiple hybridization event as proposed by de Barros Lopes et al.,49 followed by chromosomal loss and rearrangements are the most likely processes to have occurred, since they are common mechanisms for yeast genome evolution.53 The hybridization between haploid S. bayanus and S. cerevisiae spores proposed by Dunn and Sherlock48 seems a less probable mechanism, given the fact that interspecies hybrid constructed in vitro by conjugating haploid spores are generally stable and do not undergo genome rearrangements.54
According to our results, following the hybridization event, lager brewing yeast genome was shaped as follows: eight chromosomal translocations between the Sc- and the Sb-type sub-genomes occurred and due to chromosomal translocations, the number of chromosomes increased with respect to the parental species. We have demonstrated that the lager brewing yeast possess 36 chromosomes, whereas its putative parental species, S. cerevisiae and S. bayanus, possess 16. The translocations between diverged sequences are rare in Saccharomyces yeast as described above.33 The stressful condition typical of alcoholic fermentation might have driven the evolution of lager brewing yeast and the translocation that occurred to shape its genome. An experimental study of S. cerevisiae in a continuous culture under glucose limitation, for example, revealed that the selection pressure imposed by the environmental conditions can result in the selection of strains having translocations, being aneuploid or increasing the copy number of the high-affinity hexose transporter genes.55 Another study proposed that gross chromosomal translocations were enhanced by ethanol and acetaldehyde.56 A chromosomal translocation mediated by short homology has also been found in wine strains which are also exposed to multiple stress factors during alcoholic fermentation.57 Thus, it seems that the multiple stress factors that lager brewing yeast have been exposed to (such as low fermentation temperature, low pH, sugar depletion and high ethanol conditions) could have determined or facilitated chromosomal translocations, as a process of evolutionary adaptation. Most translocations after the hybridization event occurred by non-reciprocal recombination. It has been demonstrated that in S. cerevisiae, chromosomal translocations seem to increase as diploid yeast cells age. While in young cells, translocations occur by reciprocal recombination, in the old mother’s progeny, they occur by non-reciprocal recombination.58 This phenomenon could explain the translocation that occurred in lager brewing yeast by non-reciprocal recombination. Lager brewing fermentation is traditionally carried out by transferring the yeast culture from one fermentation batch to the following; thus, it is possible that non-reciprocal translocations between the Sc- and the Sb-type chromosomes occurred in an aged lager brewing yeast cell that adapted to lager brewing conditions. Non-reciprocal translocations usually cause the change of gene dosage. In the lager brewing yeast we sequenced, one large region of the Sc-type chromosomes and five large regions of the Sb-type chromosomes have been deleted and unilateral chromosomal regions have been increased by these non-reciprocal translocations. The increased number of loci caused by the translocations of one gene associated to an efficient maltose and maltotriose uptakes and of genes associated to sulfite production might have been the result not only of adaptation to the lager brewing conditions, but also and especially to human-driven selection. More than any other yeast the evolution of lager brewing yeasts has been strongly influenced by human-imposed selection, which has favored strains with optimal fermentation characteristics and good organoleptic traits. The lager brewing yeast sequenced in this study, possess a non-functional Sc-type AGT1 gene; however, it still showed an increase in sugar assimilation rate when compared with lager brewing yeast with functional AGT1, suggesting that not only chromosomal translocation but also gene deletion and truncation after the hybridization event have been beneficial for lager brewing yeast.
Dunn and Sherlock48 classified lager brewing yeasts into two distinct groups based on their shared sets of chromosomes; group 1 included strains that had lost a significant portion of the S. cerevisiae genome, and group 2 included strains that had preserved all genomic contents of both S. cerevisiae and S. bayanus. The set of 17 lager brewing strains these authors tested did not include any current production strains. The strain sequenced in this work is currently used in lager beer production and could be considered as a representative of lager brewing production strains. According to this grouping, our representative production strain is part of Group 2, in fact it possess almost the two entire sets of sub-genomes. Furthermore, most of the breakpoints of non-reciprocal translocations that we found seem to be consistent with the CGH analysis these authors performed, although they have not been confirmed by sequencing analysis.
3.11. Implications
Lager brewing yeast is economically the most important yeast for beer brewing, but its genome sequence had not yet been determined, partly due to its complexity and partly to the lack of knowledge of its genome structure. In this study, the genome sequence was determined by the whole genome shotgun sequencing, by the additional cosmid sequencing and subsequently confirmed by opmap. This work represents the first example of the whole genome sequencing of a lager brewing yeast strain and demonstrates that lager brewing yeasts comprises two sub-genomes, one originating from S. cerevisiae and the other from S. bayanus. Sequence comparison between Sb-type sub-genome and mtDNA of lager brewing yeast and those of S. bayanus CBS 7001 (S. uvarum) supports the notion that the Sb-type sub-genome in the current lager brewing strain is not originating from S. uvarum, but rather from S. bayanus var. bayanus.
There are several specific biological features of lager brewing yeast such as the production of a significant amount of sulfite and the ability to ferment at low temperatures (ranging from 8 to 15°C). The Sb-type sub-genome is possibly responsible for these features since S. cerevisiae produces a relatively small amount sulfite18 and ferments at higher temperatures, whereas the hybrid strains between S. cerevisiae and S. bayanus ferment fast and efficiently at low-temperature.59 However, a clear understanding of these characteristics has been limited because of the lack of a complete genome sequence of this yeast. We have made the lager brewing yeast genome a freely accessible resource, and this represent a significant step towards further analysis of the lager brewing yeast. This may benefit research programs of genetic engineering or breeding designed to improve brewing yeast strains. New technologies for comprehensive analyses (gene expression analysis, polymorphism analysis and proteome analysis) can also be developed. These technologies promise further advances in the science of lager brewing yeast, such as understanding why lager brewing yeast is good for lager beer brewing, and the detection of sequence polymorphisms among lager strains for reference of the strain-specific characteristics. Indeed, comprehensive gene expression analysis of lager brewing yeasts under lager beer production using lager brewing yeast DNA microarray has revealed that the expression level of the Sb-type SSU1 gene, encoding a sulfite transporter, was causing the elevated sulfite production of lager brewing yeast.60
To our knowledge, this work is the first example of a genome sequencing project of a hybrid species. In principle, whole genome duplication may allow for large-scale adaptation to new environments, and subsequent gene-loss events then led to current genome by a large number of rearrangement events and individual gene deletions, which persists until the genome returns to functionally normal ploidy through the mutation, gene loss and genomic rearrangements.29 The same is the case in the lager brewing yeast, in which larger chromosomal regions have been deleted, the rDNA cluster of the Sb-type chromosome XII has been drastically reduced, and many truncated genes were found. This indicates that such rearrangements are subsequent to genome hybridization.
The genome information and the new technology involved will be useful not only for their possible industrial applications but also for academic basic research. In particular, lager brewing yeast provides an excellent case to understand whole genome duplication and multiple genomes evolution and organization.
Supplementary data
Supplementary Material is available at www.dnaresearch.oxfordjournals.org.
Acknowledgements
We thank Y. Kaneko and H. Yoshikawa for their valuable participations in scientific discussions and H. Hatanaka for her valuable discussions on α-glucoside transporters. We are deeply appreciative of K. Olesen, M. C. Kielland-Brandt and F. Omura for critically reading the manuscript. We are also grateful to K. Togami for her assistance with this study and to R. Komatsu for his analysis of the opmap. The expert technical assistance of K. Yasui and H. Toyonaga is gratefully acknowledged.
Footnotes
Edited by Katsumi Isono
References
- 1.Barth S. J. 2007. The barth report hops 2006/2007. http://www.barthhaasgroup.com/cmsdk/content/bhg/barth_report.htm . [Google Scholar]
- 2.Hansen E. C. Investigations on the physiology and morphology of budding yeast. XIII. New studies of bottom fermenting brewers yeasts. C. R. Trav. Lab. Carlsberg. 1908;7:179–217. [Google Scholar]
- 3.Martini A. V., Martini A. In: The Yeasts, a Taxonomic Study: Saccharomyces Myen ex Reessm. Kurtzman C. P., Fell J. W., editors. Amsterdam: Elsevier; 1998. pp. 358–371. Chapter 44. [Google Scholar]
- 4.Martini A. V., Martini A. Three newly delimited species of Saccharomyces sensu stricto. Antonie Van Leeuwenhoek. 1987;53:77–84. doi: 10.1007/BF00419503. [DOI] [PubMed] [Google Scholar]
- 5.Martini A. V., Kurtzman C. P. Deoxyribonucleic acid relatedness among species of the genus Saccharomyces sensu stricto. Int. J. Syst. Bacteriol. 1985;35:508–511. [Google Scholar]
- 6.Tamai Y., Momma T., Yoshimoto H., Kaneko Y. Co-existence of two types of chromosome in the bottom fermenting yeast, Saccharomyces pastorianus. Yeast. 1998;14:923–933. doi: 10.1002/(SICI)1097-0061(199807)14:10<923::AID-YEA298>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 7.Yamagishi H., Ogata T. Chromosomal structures of bottom fermenting yeasts. Syst. Appl. Microbiol. 1999;22:341–353. doi: 10.1016/S0723-2020(99)80041-1. [DOI] [PubMed] [Google Scholar]
- 8.Nilsson-Tillgren T., Petersen J. G. L., Hohnberg S., Kielland-Brandt M. C. Transfer of chromosome III during kar mediated cytoduction in yeast. Carlsberg Res. Commun. 1980;45:113–117. [Google Scholar]
- 9.Nilsson-TiIlgren T., Gjermansen C., Holmberg S., Petersen J. G. L., Kielland-Brandt M. C. Analysis of chromosome V and the ILV1 gene from Saccharomyces carlsbergensis. Carlsberg Res. Commun. 1986;51:309–326. [Google Scholar]
- 10.Kielland-Brandt M. C., Nilsson-Tillgren T., Gjermansen C., Holmberg S., Pedersen M. B. Genetics of brewing yeasts. In: Rose A. H., Wheals A. E., Harrison J. S., editors. The Yeasts. 2nd Ed. Vol. 6. London: Academic Press; 1995. pp. 223–254. [Google Scholar]
- 11.Fujii T., Yoshimoto H., Nagasawa N., Bogaki T., Tamai Y., Hamachi M. Nucleotide sequences of alcohol acetyltransferase genes from lager brewing yeast Saccharomyces carlsbergensis. Yeast. 1996;12:593–598. doi: 10.1002/(SICI)1097-0061(199605)12:6%3C593::AID-YEA593%3E3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 12.Tamai Y., Tanaka K., Umemoto N., Tomizuka K., Kaneko Y. Diversity of the HO gene encoding an endonuclease for mating-type conversion in the bottom fermenting yeast Saccharomyces pastorianus. Yeast. 2000;16:1335–1343. doi: 10.1002/1097-0061(200010)16:14<1335::AID-YEA623>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 13.Kodama Y., Omura F., Ashikari T. Isolation and characterization of a gene specific to lager brewing yeast that encodes a branched-chain amino acid permease. Appl. Environ. Microbiol. 2001;67:3455–3462. doi: 10.1128/AEM.67.8.3455-3462.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hansen J., Kielland-Brandt M. C. Saccharomyces carlsbergensis contains two functional MET2 alleles similar to homologues from S. cerevisiae and S. monacensis. Gene. 1994;140:33–40. doi: 10.1016/0378-1119(94)90727-7. [DOI] [PubMed] [Google Scholar]
- 15.Groth C., Petersen R. F., Piskur J. Diversity in organization and the origin of gene orders in the mitochondrial DNA molecules of the genus Saccharomyces. Mol. Biol. Evol. 2000;17:1833–1841. doi: 10.1093/oxfordjournals.molbev.a026284. [DOI] [PubMed] [Google Scholar]
- 16.Boulton C., Quain D. In: Brewing Yeast and Fermentation: Brewing yeast. Boulton C., Quain D., editors. Oxford: Blackwell Science Ltd; 2001. pp. 143–259. Chapter 4. [Google Scholar]
- 17.Zheng X. T., D’Amore T., Russell I., Stewart G. G. Factors influence maltotriose utilization during brewery wort fermentations. J. Am. Soc. Brew. Chem. 1994;52:41–47. [Google Scholar]
- 18.Crumplen R., D’Amore T., Slaughter C., Stewart G. G. Novel differences between ale and lager brewing yeasts. Proc. Congr. Eur. Brew. Conv. 1993;24:267–274. [Google Scholar]
- 19.Ewing B., Hillier L., Wendl M. C., Green P. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 1998;8:175–185. doi: 10.1101/gr.8.3.175. [DOI] [PubMed] [Google Scholar]
- 20.Huang X., Wang J., Aluru S., Yang S. P., Hillier L. PCAP: a whole-genome assembly program. Genome Res. 2003;13:2164–2170. doi: 10.1101/gr.1390403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reslewic S., Zhou S., Place M., et al. Whole-genome shotgun optical mapping of Rhodospirillum rubrum. Appl. Environ. Microbiol. 2005;71:5511–5522. doi: 10.1128/AEM.71.9.5511-5522.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Montrocher R., Verner M. C., Briolay J., Gautier C., Marmeisse R. Phylogenetic analysis of the Saccharomyces cerevisiae group based on polymorphisms of rDNA spacer sequences. Int. J. Syst. Bacteriol. 1998;48(Pt 1):295–303. doi: 10.1099/00207713-48-1-295. [DOI] [PubMed] [Google Scholar]
- 23.Rokas A., Williams B. L., King N., Carroll S. B. Genome-scale approaches to resolving incongruence in molecular phylogenies. Nature. 2003;425:798–804. doi: 10.1038/nature02053. [DOI] [PubMed] [Google Scholar]
- 24.Nguen H. V., Gaillardin C. Two subgroups within the Saccharomyces bayanus species evidenced by PCR amplification and restriction polymorphism of the non-transcribed spacer2 in the ribosomal DNA unit. System. Appl. Microbiol. 1997;20:286–294. [Google Scholar]
- 25.Nguyen H. V., Gaillardin C. Two subgroups within the Saccharomyces bayanus species evidenced by PCR amplification and restriction polymorphism of the non-transcribed spacer2 in the ribosomal DNA unit. System. Appl. Microbiol. 1997;20:286–294. [Google Scholar]
- 26.Kellis M., Patterson N., Endrizzi M., Birren B., Lander E. S. Sequencing and comparison of yeast species to identify genes and regulatory elements. Nature. 2003;423:241–254. doi: 10.1038/nature01644. [DOI] [PubMed] [Google Scholar]
- 27.Török T., Mortimer R. K., Romano P., Suzzi G., Polsinelli M. Quest for wine yeasts—An old story revisited. J. Ind. Microbiol. Biotechnol. 1996;17:303–313. [Google Scholar]
- 28.Wei W., McCusker J. H., Hyman R. W., et al. Genome sequencing and comparative analysis of Saccharomyces cerevisiae strain YJM789. Proc. Natl Acad. Sci. USA. 2007;104:12825–12830. doi: 10.1073/pnas.0701291104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kellis M., Birren B. W., Lander E. S. Proof and evolutionary analysis of ancient genome duplication in the yeast Saccharomyces cerevisiae. Nature. 2004;428:617–624. doi: 10.1038/nature02424. [DOI] [PubMed] [Google Scholar]
- 30.Klein H. L. Different types of recombination events are controlled by the RAD1 and RAD52 genes of Saccharomyces cerevisiae. Genetics. 1988;120:367–377. doi: 10.1093/genetics/120.2.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Petes T. D., Hill C. W. Recombination between repeated genes in microorganisms. Annu. Rev. Genet. 1988;22:147–168. doi: 10.1146/annurev.ge.22.120188.001051. [DOI] [PubMed] [Google Scholar]
- 32.Kupiec M., Petes T. D. Allelic and ectopic recombination between Ty elements in yeast. Genetics. 1988;119:549–559. doi: 10.1093/genetics/119.3.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Datta A., Hendrix M., Lipsitch M., Jinks-Robertson S. Dual roles for DNA sequence identity and the mismatch repair system in the regulation of mitotic crossing-over in yeast. Proc. Natl Acad. Sci. USA. 1997;94:9757–9762. doi: 10.1073/pnas.94.18.9757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Argueso J. L., Westmoreland J., Mieczkowski P. A., Gawel M., Petes T. D., Resnick M. A. Double-strand breaks associated with repetitive DNA can reshape the genome. Proc. Natl Acad. Sci. USA. 2008;105:11845–11850. doi: 10.1073/pnas.0804529105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kodama Y., Kielland-Brandt M. C., Hansen J. In: Top. Curr. Genet.: Lager brewing yeast. Sunnerhagen P., Piskur J., editors. Vol. 15. Heidelberg: Springer-Verlag; 2006. pp. 145–164. [Google Scholar]
- 36.Kotseruba V., Gernand D., Meister A., Houben A. Uniparental loss of ribosomal DNA in the allotetraploid grass Zingeria trichopoda (2n equals 8) Genome. 2003;46:156–163. doi: 10.1139/g02-104. [DOI] [PubMed] [Google Scholar]
- 37.Cardazzo B., Minuzzo S., Sartori G., Grapputo A., Carignani G. Evolution of mitochondrial DNA in yeast: gene order and structural organization of the mitochondrial genome of Saccharomyces uvarum. Curr. Genet. 1998;33:52–59. doi: 10.1007/s002940050308. [DOI] [PubMed] [Google Scholar]
- 38.Giaever G., Chu A. M., Ni L., et al. Functional profiling of the Saccharomyces cerevisiae genome. Nature. 2002;418:387–391. doi: 10.1038/nature00935. [DOI] [PubMed] [Google Scholar]
- 39.Kodama Y., Fukui N., Ashikari T., Shibano Y. Improvement of maltose fermentation efficiency: constitutive expression of MAL genes in brewing yeasts. J. Am. Soc. Brew. Chem. 1995;53:24–29. [Google Scholar]
- 40.Dietvorst J., Londesborough J., Steensma H. Y. Maltotriose utilization in lager yeast strains: MTT1 encodes a maltotriose transporter. Yeast. 2005;22:775–788. doi: 10.1002/yea.1279. [DOI] [PubMed] [Google Scholar]
- 41.Jespersen L., Cesar L. B., Meaden P. G., Jakobsen M. Multiple alpha-glucoside transporter genes in brewer’s yeast. Appl. Environ. Microbiol. 1999;65:450–456. doi: 10.1128/aem.65.2.450-456.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vidgren V., Ruohonen L., Londesborough J. Characterization and functional analysis of the MAL and MPH Loci for maltose utilization in some ale and lager yeast strains. Appl. Environ. Microbiol. 2005;71:7846–7857. doi: 10.1128/AEM.71.12.7846-7857.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Turakainen H., Korhola M., Aho S. Cloning, sequence and chromosomal location of a MEL gene from Saccharomyces carlsbergensis NCYC396. Gene. 1991;101:97–104. doi: 10.1016/0378-1119(91)90229-5. [DOI] [PubMed] [Google Scholar]
- 44.Goncalves P., Rodrigues de Sousa H., Spencer-Martins I. FSY1, a novel gene encoding a specific fructose/H(+) symporter in the type strain of Saccharomyces carlsbergensis. J. Bacteriol. 2000;182:5628–5630. doi: 10.1128/jb.182.19.5628-5630.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Omura F., Hatanaka H., Nakao Y. Characterization of a novel tyrosine permease of lager brewing yeast shared by Saccharomyces cerevisiae strain RM11-1a. FEMS Yeast Res. 2007;7:1350–1361. doi: 10.1111/j.1567-1364.2007.00310.x. [DOI] [PubMed] [Google Scholar]
- 46.Yoshida S., Hashimoto K., Tanaka-Kanai K., Yoshimoto H., Kobayashi O. Identification and characterization of amidase- homologous AMI1 genes of bottom-fermenting yeast. Yeast. 2007;24:1075–1084. doi: 10.1002/yea.1551. [DOI] [PubMed] [Google Scholar]
- 47.Halestrap A. P., Price N. T. The proton-linked monocarboxylate transporter (MCT) family: structure, function and regulation. Biochem. J. 1999;343(Pt 2):281–299. [PMC free article] [PubMed] [Google Scholar]
- 48.Dunn B., Sherlock G. Reconstruction of the genome origins and evolution of the hybrid lager yeast Saccharomyces pastorianus. Genome Res. 2008;18:1610–1623. doi: 10.1101/gr.076075.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.de Barros Lopes M., Bellon J. R., Shirley N. J., Ganter P. F. Evidence for multiple interspecific hybridization in Saccharomyces sensu stricto species. FEMS Yeast Res. 2002;1:323–331. doi: 10.1111/j.1567-1364.2002.tb00051.x. [DOI] [PubMed] [Google Scholar]
- 50.Rainieri S., Kodama Y., Kaneko Y., Mikata K., Nakao Y., Ashikari T. Pure and mixed genetic lines of Saccharomyces bayanus and Saccharomyces pastorianus and their contribution to the lager brewing strain genome. Appl. Environ. Microbiol. 2006;72:3968–3974. doi: 10.1128/AEM.02769-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pulvirenti A., Caggia C., Restuccia C., Giudici P., Zambonelli C. Inheritance of mitochondrial DNA in interspecific Saccharomyces hybrids. Ann. Microbiol. 2000;50:61–64. [Google Scholar]
- 52.Rainieri S., Kodama Y., Nakao Y., Pulvirenti A., Giudici P. The inheritance of mtDNA in lager brewing strains. FEMS Yeast Res. 2008;8:586–596. doi: 10.1111/j.1567-1364.2008.00363.x. [DOI] [PubMed] [Google Scholar]
- 53.Wolfe K. H., Shields D. C. Molecular evidence for an ancient duplication of the entire yeast genome. Nature. 1997;387:708–713. doi: 10.1038/42711. [DOI] [PubMed] [Google Scholar]
- 54.Rainieri S., Pulvirenti A., Giudici P. Construction of model lager brewing strains by interspecific hybridization. Yeast. 2007;24:01–41. [Google Scholar]
- 55.Dunham M. J., Badrane H., Ferea T., et al. Characteristic genome rearrangements in experimental evolution of Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA. 2002;99:16144–16149. doi: 10.1073/pnas.242624799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Infante J. J., Dombek K. M., Rebordinos L., Cantoral J. M., Young E. T. Genome-wide amplifications caused by chromosomal rearrangements play a major role in the adaptive evolution of natural yeast. Genetics. 2003;165:1745–1759. doi: 10.1093/genetics/165.4.1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Perez-Ortin J. E., Querol A., Puig S., Barrio E. Molecular characterization of a chromosomal rearrangement involved in the adaptive evolution of yeast strains. Genome Res. 2002;12:1533–1539. doi: 10.1101/gr.436602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.McMurray M. A., Gottschling D. E. An age-induced switch to a hyper-recombinational state. Science. 2003;301:1908–1911. doi: 10.1126/science.1087706. [DOI] [PubMed] [Google Scholar]
- 59.Sato M., Kishimoto M., Watari J., Takashio M. Breeding of brewer’s yeast by hybridization between a top-fermenting yeast Saccharomyces cerevisiae and a cryophilic yeast Saccharomyces bayanus. J. Biosci. Bioeng. 2002;93:509–511. doi: 10.1016/s1389-1723(02)80101-3. [DOI] [PubMed] [Google Scholar]
- 60.Nakao Y., Kodama Y., Shimonaga T., et al. Gene expression analysis of lager brewing yeast under different oxygenation condition using newly developed DNA microarray; Proc. Eur. Conv. Congr.; 2007. pp. 406–419. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.