Abstract
The suppressors of cytokine signaling (SOCS) family of proteins act as intracellular inhibitors of several cytokine signal transduction pathways. Their expression is induced by cytokine activation of the Janus kinase/signal transducer and activator of transcription (JAK/STAT) pathway and they act as a negative feedback loop by subsequently inhibiting the JAK/STAT pathway either by direct interaction with activated JAKs or with the receptors. These interactions are mediated at least in part by the SH2 domain of SOCS proteins but these proteins also contain a highly conserved C-terminal homology domain termed the SOCS box. Here we show that the SOCS box mediates interactions with elongins B and C, which in turn may couple SOCS proteins and their substrates to the proteasomal protein degradation pathway. Analogous to the family of F-box-containing proteins, it appears that the SOCS proteins may act as adaptor molecules that target activated cell signaling proteins to the protein degradation pathway.
Members of the family of suppressors of cytokine signaling (SOCS) proteins contain a central SH2 domain and a C-terminal homology domain we have termed the SOCS box (1, 2). The first member of this family was called CIS (cytokine-inducible SH2-containing protein) (3) and was shown to inhibit erythropoietin and interleukin (IL) 3 receptor signaling. We cloned SOCS-1 from a retroviral expression library as a cDNA whose constitutive expression inhibited IL 6-induced differentiation of M1 cells (1) and it was simultaneously cloned by others as a protein that interacted with activated JAK kinases (JAK-binding protein, JAB) (4) and as a protein with antigenic similarity to signal transducers and activator of transcription (STATs) (STAT-inducible STAT inhibitor, SSI) (5). The sequence similarity of SOCS-1 and CIS led to the recognition of six additional members of this family (SOCS-2 through -7), each with an SH2 domain and a C-terminal SOCS box (2, 6, 7). An additional 12 proteins have been described that contain a C-terminal SOCS box but instead of an SH2 domain they contain different protein-protein interaction domains including WD40, ankyrin repeats, SP1a and ryanodine receptor, or small GTPase domains (2).
Following binding to their receptors, many cytokines activate receptor-associated cytoplasmic kinases called JAKs that in turn phosphorylate the receptor cytoplasmic domain and associated STATs. Phosphorylated STAT dimers translocate to the nucleus and activate transcription of specific genes including those of CIS and some of the SOCS. SOCS proteins recognize activated signaling molecules (including JAKs and cytokine receptors) through their SH2 and N-terminal domains and inhibit their activity (8, 9). Exactly how SOCS proteins inhibit JAK kinase activity and the role of the conserved SOCS box are currently unknown. In the present report we show that the SOCS box interacts with elongins B and C and through them potentially with the proteasome complex. Targeting of SOCS proteins and their bound activated signaling molecules to the protein degradation pathway may explain how SOCS proteins simultaneously terminate a cytokine stimulation cycle and their own inhibitory action so that cells may respond to a second round of stimulation.
MATERIALS AND METHODS
SOCS and Elongin Expression Vectors.
The cDNAs encoding mouse SOCS-1, SOCS-3, WSB-2 (WD-40 repeat-containing protein with a SOCS box), SSB-1 (SPRY domain-containing protein with a SOCS box), and ASB-1 (ankyrin repeat-containing protein with a SOCS box) have been described (1, 2, 9). Constructs in pEF-Flag1 encoding these proteins, with or without the SOCS box, with an N- terminal Flag epitope tag (DYKDDDDK) were generated by PCR essentially as described (1, 9) (found at http://www.wehi.edu.au/willson vectors). DNA fragments encoding mouse elongins B and C were amplified by using PCR from a 17-day embryo cDNA λ library (CLONTECH ML5014t) and were expressed with N-terminal Flag or Myc (DQKLISEEDL) epitope tags, respectively, by using the mammalian expression vector pEF-BOS.
Stable and Transient Transfection of Cell Lines, Immunoprecipitation, and Western Blot Analysis.
The murine monocytic leukemic cell line, M1, and the 293T human fibroblast cell line were maintained and transfected as described (9). Immunoprecipitations and Western blot analyses were performed as described (9), and goat anti-elongin antibodies were from Santa Cruz Biotechnology.
Preparation of Glutathione S-Transferase (GST) and GST-SOCS Box Affinity Resins.
DNA fragments encoding the SOCS boxes from mouse SOCS-1 (residues 172–212) and SOCS-3 (residues 186–225) with an N-terminal linker sequence (EGKSSGSGSESKVD) were generated by PCR and cloned into the bacterial expression vector pGEX-2T (10). The GST fusion proteins were purified by affinity chromatography on glutathione Sepharose 4B (Amersham Pharmacia), and affinity resins were prepared by covalently coupling 1 mg of purified proteins to 1 ml of NHS-activated Sepharose resin (Amersham Pharmacia). Before use, the affinity resins were washed with elution buffer (0.5% SDS/50 mM DTT/50 mM Tris⋅HCl, pH 8.0/150 mM NaCl) and equilibrated in lysis buffer (0.5% Nonidet P-40/10 mM Tris⋅HCl, pH 7.5/100 mM NaCl).
Purification of SOCS Box-Binding Proteins.
M1 cells (2 × 1010) were lysed on ice for 30 min in 100 ml of lysis buffer supplemented with protease inhibitors (Complete Mixture tablets; Boehringer Mannheim), 1 mM PMSF, 1 mM Na3VO4, and 1 mM NaF. The total cell lysate was centrifuged at 15,000 rpm (SS34 rotor) for 15 min at 4°C, and the clarified supernatant was preincubated with 1 ml of GST-Sepharose resin for 2 h at 4°C. Half of the GST-Sepharose-depleted M1 cell lysate was incubated with 1 ml of GST-SOCS-1 SOCS box and the other half with GST-SOCS-3 SOCS box-Sepharose resin for 2 h at 4°C. The affinity resins were washed with 40 ml of lysis buffer and then eluted with 8 × 0.5 ml of elution buffer. Eluates were concentrated to ≈40 μl, mixed with 15 μl of 4× SDS sample buffer containing 0.4 M DTT and resolved on a 14% polyacrylamide gel (Novex). The gel was stained for 5 min with 0.1% Coomassie blue in 50% methanol and destained in 12% methanol and 7% acetic acid.
Protein Identification by Peptide Sequence Analysis by Using Tandem MS.
Electrophoretically separated proteins were excised and digested in situ by using trypsin (11). Generated peptides were separated by using capillary chromatography (12) and sequenced by using an on-line electrospray ion-trap mass spectrometer (LCQ Finnigan–MAT, San Jose, CA) (13). The sequences of individual peptides were identified manually or by using the sequest algorithm to correlate the collision-induced dissociation spectra with amino acid sequences in the owl protein database (version 30.2) (14).
Peptide Synthesis and Biotinylation.
Peptide fragments of murine SOCS-1, WSB-2, and ASB-2 corresponding to the SOCS boxes and five upstream N-terminal residues (2) were synthesized according to the in situ neutralization/2-(1H-benzotriazol-1-yl)-1,1,3,3-tetramethyluronium hexafluorophosphate (HBTU) activation protocol for Boc solid phase chemistry (15), purified by using reverse-phase HPLC and the products characterized by electrospray MS. A sample of the SOCS-1 SOCS box peptide was postsynthetically biotinylated by treatment with sulfosuccinimidobiotin. Before biotinylation, the side chain of the unique cysteine residue was temporarily protected by oxidation to the peptide disulfide dimer and subsequently reduced with 5 mM DTT. Typically, peptide was bound to streptavidin-agarose resin (Pierce immunopure; 1–2 mg streptavidin/ml resin) by incubating equal volumes of resin and 1 mg/ml peptide for 1 h, followed by extensive washing.
Competition of SOCS 1 SOCS Box/Elongin C Interaction.
Streptavidin-agarose binding proteins were precleared from M1 cell lysate by treating overnight at 4°C with streptavidin-agarose resin. SOCS box peptides (SOCS-1, ASB-2, and WSB-2) were solubilized in water at 10 mg/ml, and aliquots of these, or water alone, were added to 350 μl fractions of cleared lysate, followed by incubation for 3 h at 4°C. These lysates were then added to 30 μl of SOCS-1 SOCS box peptide resin and incubated a further 2 h at 4°C. The resin was extensively washed with lysis buffer and bound proteins were eluted with 20 μl of 4× SDS sample buffer. Proteins were separated by SDS/PAGE on a 4–15% reducing gel.
Detection of SOCS-1 Interaction with Endogenous Elongins.
M1 cells stably expressing either full-length SOCS-1 or SOCS-1 lacking the SOCS box (with N-terminal Flag epitopes) were grown in DMEM containing 5% bovine calf serum, 10 μg/ml puromycin and 50 ng/ml murine IL-6. The cells were harvested and incubated in 20 ml of culture medium containing 10 μM proteasome-specific inhibitor, N-acetyl-l-leucinyl-l-leucinyl-norleucinal (LLnL; Sigma) for 30 min at 37°C. The cells were lysed in 14 ml of lysis buffer supplemented with protease inhibitors (Complete Mixture tablets), 1 mM PMSF, 1 mM Na3VO4, 1 mM NaF, and 10 μM LLnL. Total cell lysates were centrifuged at 15,000 rpm (SS34 rotor) for 15 min at 4°C, and the clarified supernatants were incubated with 0.3 ml of M2 anti-Flag antibody resin for 3 h at 4°C. Resin was then washed with 10 ml of lysis buffer and the bound proteins were eluted with 6 × 150 μl of 100 μg/ml Flag peptide in lysis buffer.
IL-6-Induced Expression of Endogenous SOCS-3 Protein.
Mouse macrophage-like J774 cells were grown continuously in DMEM containing 10% bovine calf serum. The cells were washed once in PBS, twice with DMEM, and starved for 1 h in DMEM containing 0.1% low-endotoxin BSA (Sigma). The proteasome inhibitor LLnL dissolved in DMSO or DMSO was added to the cells for 15 min and the cells then stimulated with 100 ng/ml of murine IL-6 for the indicated times.
RESULTS
The conserved SOCS box does not appear to be required for inhibition of the JAK/STAT signaling pathway when SOCS proteins are overexpressed (8, 9), suggesting that it might play a regulatory role in targeting proteins to particular cell compartments or in controlling the in vivo half-lives of associated proteins. The SOCS box domain is unlikely to be large enough to encode catalytic activity and is therefore likely to mediate such effects through protein–protein interactions. Consequently the ability of the SOCS box to interact with cellular target proteins was investigated.
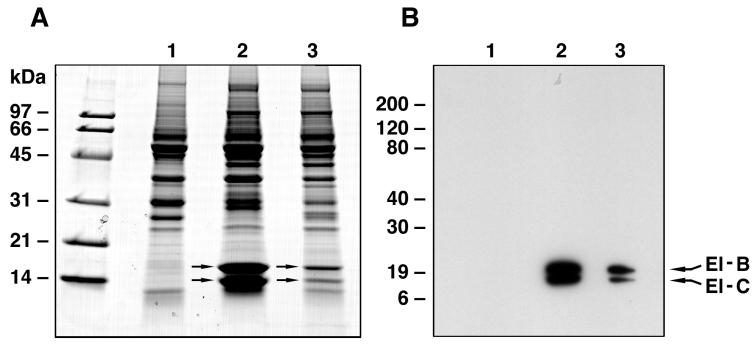
Isolated SOCS box sequences were used as affinity reagents to identify interacting proteins in cell lysates. GST fusion proteins containing the SOCS box sequences from SOCS-1 or SOCS-3 were coupled to Sepharose beads and used as affinity resins to bind proteins from M1 cell lysates. The two prominent bands seen coprecipitating with both GST–SOCS fusion proteins but not the GST control were proteins of 15 and 18 kDa (Fig. 1). These proteins were excised from the gel, digested with trypsin in situ, and the resulting peptides were analyzed by MS. The amino acid sequences of these peptides (Table 1) were determined either by manual interpretation of the collision-induced spectra of the major peptide ion or by computer-aided fragment matching algorithms (sequest). By these means the 18-kDa and the 15-kDa proteins were identified as elongins B and C, respectively. Western blotting of the gels of the same eluates with antibodies against elongins B and C confirmed that both elongins were present in eluates from beads containing SOCS-1 or SOCS-3-box fusion proteins but not from control GST bands (Fig. 1).
Figure 1.
Purification of SOCS box-binding proteins from murine myeloid M1 cells. (A) SDS/PAGE (14% gel) analysis of affinity column eluates from GST-Sepharose (lane 1), from GST–SOCS-1–SOCS-box–Sepharose column (lane 2), and from GST–SOCS-3–SOCS-box–Sepharose column (lane 3). The proteins were visualized by Coomassie blue staining. Arrows in lane 2 indicate the positions of the two protein bands excised for sequencing analysis by MS. (B) Western blot analysis of the three affinity column eluates shown in A by using (mouse cross-reactive) anti-rat elongin B and C antibodies.
Table 1.
Tandem mass spectrometric identification of elongin B and elongin C as proteins bound by SOCS box sequences
| Protein | Peptide no. | Experimental,* [M+H]+(Da) | Predicted,* [M+H+](Da) | Sequence† | Position in protein |
|---|---|---|---|---|---|
| Elongin B | 1 | 1161.6 | 1162.3 | HKTTIFTDAK | 10–19 |
| (18 kDa) | 2 | 771.5 | 772.0 | IVEGILK | 30–36 |
| 3 | 1196.3 | 1196.3 | ESSTVFELKR | 20–29 | |
| 4 | 927.7 | 928.2 | RIVEGILK | 29–36 | |
| 5 | 1664.9 | 1664.9 | IVEGILKRPPEEQR | 30–43 | |
| 6 | 2339.6 | 2339.7 | HKTTIFTDAKESSTVFELKR | 10–29 | |
| 7 | 1917.8 | 1918.2 | TTIFTDAKESSTVFELKR | 12–29 | |
| 8 | 3056.6 | 3056.3 | IEPFSSPPELPDVMKPQDSGGSANEQAVQ | 90–118 | |
| 9 | 4075.0 | 4075.4 | ADDTFEALRIEPFSSPPELPDVMKPQDSGGSANEQAVQ | 81–118 | |
| 10 | 1066.5 | 1067.4 | MDVFLMIR‡ | 1–8 | |
| Elongin C | 1 | 1213.2 | 1213.4 | REHALTSGTIK | 33–43 |
| (15 kDa) | 2 | 1009.6 | 1010.2 | EIPSHVLSK | 64–72 |
| 3 | 1596.5 | 1596.8 | TYGGCEGPDAMYVK§ | 7–20 | |
| 4 | 1501.5 | 1501.7 | LISSDGHEFIVKR | 21–33 | |
| 5 | 1159.5 | 1160.4 | VCMYFTYK§ | 73–80 | |
| 6 | 2213.1 | 2212.4 | AMLSGPGQFAENETNEVNFR | 44–63 |
Ten tryptic peptides analyzed from the 18-kDa protein correspond to the sequence of rat elongin B (Genbank accession no. L42855) and six tryptic peptides analysed from the 15-kDa protein corresponded to the sequence of rat elongin C (Genbank accession no. L29259) with mass errors of 0.004–0.080%. These peptides covered 68.6% and 66.1% of the elongins B and C sequence, respectively.
Average mass values.
Amino acid sequence is given using the one-letter notation.
N-terminal methionine is acetylated (+42 Da).
Cysteine residue is alkylated with 4-vinyl pyridine during sample preparation (+105 Da).
Similar experiments by using a biotinylated SOCS-1 SOCS box peptide bound to streptavidin-agarose also resulted in the identification by MS of elongins B and C as interacting proteins in M1 cellular extracts. The specificity of this interaction was tested by preincubating extracts with unbiotinylated SOCS box peptides before addition of the immobilized SOCS-1 SOCS box peptide. As expected, unconjugated SOCS-1 SOCS box peptide competed for this interaction as did SOCS box peptides from WSB-2 and ASB-2, suggesting that interaction with elongins B and C is a general property of the conserved SOCS box (data not shown). Interestingly, identical results were obtained whether M1 cells were stimulated with cytokine (IL-6 or leukemia inhibitory factor) or not.
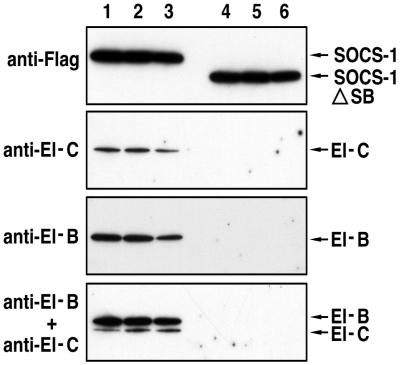
We next tested the ability of full-length or SOCS box-deleted SOCS proteins to interact with elongins B and C in M1 cells. IL-6-stimulated M1 cells stably transfected with vectors encoding N-terminally Flag-tagged full-length SOCS-1 or SOCS-1 lacking a SOCS box (SOCS-1/ΔSB) were lysed, the Flag-tagged proteins were immunoprecipitated with anti-Flag M2 antibody beads and the beads were eluted with Flag peptide. The eluates were electrophoresed on SDS/PAGE gels, transferred to poly(vinylidene difluoride) membranes, and Western blotted with anti-Flag antibodies or antibodies to elongins B and C (Fig. 2). Although full-length SOCS-1 and SOCS-1/ΔSB were expressed at similar levels, only the full-length SOCS-1 protein was associated with elongins B and C.
Figure 2.
Interaction of SOCS-1 with endogenous elongins B and C. Cellular extracts from M1 cells stably expressing either N-terminally Flag-tagged full-length SOCS-1 or SOCS-1 lacking the SOCS box (SOCS-1/ΔSB) were incubated with anti-Flag antibody M2 resin and bound cellular proteins eluted with Flag peptide. Lanes 1–3 correspond to column eluates 3–5 from M1 cells expressing full-length SOCS-1, and lanes 4–6 correspond to column eluates 3–5 from M1 cells expressing SOCS-1/ΔSB. The panels from top to bottom correspond to Western blot analyses with anti-Flag, anti-elongin C, anti-elongin B, and a mixture of anti-elongin B and anti-elongin C, respectively.
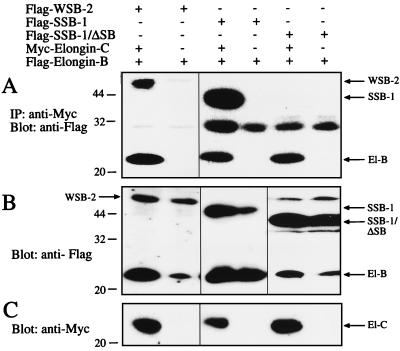
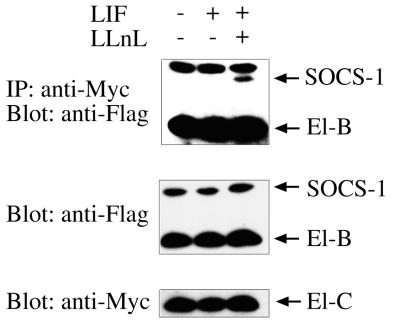
To further confirm the generality of this interaction for other proteins containing a SOCS box, 293T fibroblasts were transfected with N-terminally Flag-tagged WSB-2 or SSB-1 (with or without a SOCS box) along with elongin B containing a Flag epitope and elongin C containing a Myc epitope. When elongin C was immunoprecipitated with anti-Myc antibodies and the eluates Western blotted with anti-Flag antibodies, both WSB-2 and SSB-1 were found to coimmunoprecipitate (along with elongin B) with elongin C (see Fig. 3). As with SOCS-1, the interaction of elongins B and C with SSB-1 depended on the SOCS box because a truncated form lacking only the SOCS box failed to coimmunoprecipitate with the elongins (Fig. 3). In these experiments elongins B and C did not coimmunoprecipitate with full-length SOCS-1 or SOCS-3 in unstimulated 293T cells. However, when these cells were stimulated with leukemia inhibitory factor, especially in the presence of the proteasomal inhibitor LLnL, SOCS-1 could be coimmunoprecipitated with elongin C (Fig. 4).
Figure 3.
Coimmunoprecipitation of SOCS box-containing proteins and elongin B (El-B) with elongin C (El-C) in 293T cells. Cells were cotransfected with Flag-elongin B with or without Myc-elongin C and Flag-WSB-2, -SSB-1 or -SSB-1/ΔSB. Elongin C and associated proteins were immunoprecipitated with anti-Myc antibodies and immunoprecipitates Western blotted with anti-Flag antibodies (A). Whole-cell extracts were Western blotted with anti-Flag (B) or anti-Myc (C).
Figure 4.
Interaction of full-length SOCS-1 with elongins B and C in 293T cells depends on cytokine stimulation and proteasomal inhibition to be visualized. Lysates from 293T cells cotransfected with Myc-elongin C and Flag-tagged elongin B and full-length SOCS-1 were immunoprecipitated with anti-Myc antibodies and Western blotted with anti-Flag antibodies (Top). Whole-cell lysate was Western blotted with anti-Flag or anti-Myc antibodies as loading controls (Middle and Bottom).
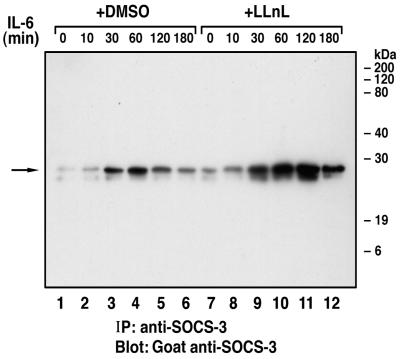
Because elongins B and C have been proposed to target proteins to proteasomal destruction (16, 17), we tested whether endogenous SOCS proteins are degraded through the proteasomal complex. When the J774 macrophage cell line was stimulated with IL-6, SOCS-3 protein expression was elevated by 30 min, peaked at 60 min, and was significantly depleted by 120 and 180 min. In contrast, cells incubated with the proteasomal inhibitor LLnL and stimulated with IL-6 showed a continual increase in SOCS-3 protein levels from 30 to 180 min (Fig. 5), suggesting that the proteasomal complex plays a major role in rapidly degrading SOCS-3 after its induction.
Figure 5.
Effect of the proteasomal inhibitor LLnL on the endogenous expression of SOCS-3 protein. J774 cells (4 × 107) were treated with either dimethyl sulfoxide (0.1%) or LLnL (50 μM) for 15 min and then stimulated with 100 ng/ml of murine IL-6 for the indicated times. The cellular extracts were immunoprecipitated with 1 μl of a rabbit anti-SOCS-3 polyclonal antiserum and immune complexes eluted from protein G-Sepharose beads were resolved by SDS/PAGE (13%) under reducing conditions and analyzed by Western blot by using goat anti-SOCS-3.
DISCUSSION
The present report has shown that a common role of SOCS boxes from several different classes of proteins is to bind to elongins B and C. The elongin B and C complex has previously been shown to bind to elongin A to form an active transcriptional elongation complex or to the von Hippel Lindau (VHL) tumor suppressor protein (16, 17). The sites on elongin A and VHL that interact with elongin C have been mapped and the consensus binding sequence (T,S,P)LXXX(C,S)XXX(LIV) is also conserved in the N-terminal half of all SOCS boxes (17), suggesting that the primary interaction is between the SOCS box and elongin C.
The elongin B/C complex appears to have two distinct roles. When bound to elongin A it acts as a positive transcriptional regulator by increasing the activity of the RNA polymerase II elongation complex (18) but when bound to VHL it acts to suppress the accumulation of hypoxia-inducible mRNAs (19). Initially it was thought that VHL might act as a transcriptional suppressor by sequestering elongins B and C and making them unavailable to interact with elongin A but more recent studies have suggested an alternate mechanism of action. The VHL/elongin B–C complex also contains a putative E3 ubiquitin ligase (Cullin-2) that may target VHL-binding proteins to destruction by the proteasome. Cullin-2 appears to interact with elongin C (directly or indirectly) independently of subsequent association with VHL (19). Analysis of the VHL gene in individuals with VHL disease has revealed that the interaction domain with elongin C is commonly mutated and that most affected individuals show a reduced ability of VHL to interact with elongins B and C (20–23).
Interestingly, Elongin B also contains a ubiquitin-like (UBL) sequence at its N terminus (19) in common with several other proteins. One of these (RAD23) has recently been shown to interact directly with proteasomal subunit proteins (Cim3 and Cim5) through its UBL domain, leading to an increase of protease activity associated with RAD23 (24). These observations suggest that coupling of VHL or RAD23 proteins to the proteasome is essential for the correct functioning of these proteins.
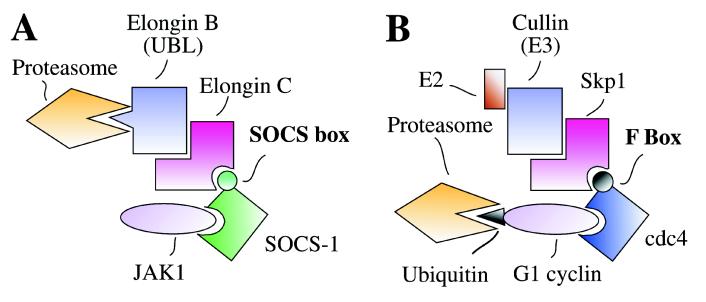
Together with our data on the binding of elongin B/C to the SOCS box, these observations suggest a model for the action of SOCS proteins (Fig. 6). As shown previously (9) the N-terminal and SH2 domains of SOCS-1 and SOCS-3, at least, are required for recognition and binding to activated (tyrosine phosphorylated) signal transduction molecules (e.g., JAKs). The SOCS box brings into this complex elongins B and C and either through direct interactions of the elongin B UBL domain with the proteasome or through associated Cullin-2-induced ubiquitination of substrates and subsequent proteosomal association, the substrate and associated SOCS protein may be destroyed. In this scheme both activated signal transduction molecules and their negative regulators (SOCS proteins) would be destroyed after a cytokine activation cycle and the cell would be ready to respond again if cytokine is still present.
Figure 6.
Model of the interaction of SOCS box-containing proteins with elongins C and B (A) and comparison with the PULC assembled by F box–containing proteins (B).
In overexpression studies, inhibition of cytokine signaling by SOCS proteins did not require the SOCS box (8, 9). This implies that specific interaction of SOCS proteins with components of the JAK/STAT pathway is sufficient to inhibit signaling and that the role of the SOCS box interaction with elongins B and C may be primarily to terminate the inhibitory signal by destroying the SOCS protein. The data in Fig. 5 indeed suggest that SOCS-3 is degraded in a proteasome-dependent manner. In situations where SOCS proteins are expressed at physiological levels, the ability to degrade SOCS-associated signaling molecules may become important to achieve maximal inhibition of cytokine-generated signals.
It was also noted in the present study that intact SOCS proteins bound less well to elongins B and C than did isolated SOCS box peptides (at least for SOCS-1 and SOCS-3), but this could be improved by cytokine stimulation in the presence of proteasomal inhibitor. This finding may suggest that SOCS box availability for interaction with elongins depends on changes associated with SOCS protein binding to its activated targets (e.g., JAKs). Given the efficiency of the proteosomal protein degradation system, it may be appropriate for SOCS proteins and signal transduction molecules to be destroyed only after they have interacted with their targets.
The model proposed above for the function of the SOCS box has a very strong parallel with components of the phosphoprotein-ubiquitin ligase complex (PULC) that is utilized to control various aspects of the cell cycle (25) (Fig. 6). In the yeast PULC system, serine phosphorylation of the cyclin-dependent kinase (cdk) inhibitor sicI or G1 cyclins Cln1 and Cln2 leads to their recognition by adaptor proteins such as Cdc4 or Grr1, which contain conserved N-terminal domains called F-boxes. The F-box mediates interaction with Skp1, an elongin C homologue, which in turn interacts with E2 and E3 (Cullin homologue) ubiquitin ligases. This results in ubiquitination of the phosphorylated substrates and targeting for proteasomal degradation so that the cell cycle can progress from G1 to S.
Recently, Verdier et al. (26) showed that CIS, a member of the SOCS family of proteins, is monoubiquinated and subject to proteasomal degradation. Moreover, they also showed that inhibitors of the proteasome lead to sustained expression of activated forms of the erythropoietin receptor and STAT5 following erythropoietin stimulation of UT-7 cells. Similarly, Yu and Burakoff (27) showed that inhibition of the proteasome resulted in sustained activation of the JAK/STAT pathway following IL-2 stimulation although neither appeared to be ubiquitinated. The present results provide a mechanism for targeting such proteins to proteasomal degradation via association of signaling molecules with the SOCS/CIS proteins followed by SOCS-box-mediated interaction with elongins B and C. It is possible that this interaction results in ubiquitination of SOCS/CIS and associated molecules (mediated by cullins) or that nonubiquitinated proteins in the complex are delivered to the proteasome via the UBL sequence in elongin B.
The present report has demonstrated that the single conserved domain in 20 structurally diverse proteins (the SOCS box) serves to couple bound proteins to the ubiquitination or proteasomal compartments through interaction with elongins B and C. The SOCS-box-containing proteins thus form a family of adapter proteins, analogous to the F-box-containing proteins, which potentially terminate cell signaling by targeting critical molecules for intracellular degradation. It will be necessary to define the molecules interacting with the various protein interaction domains of the SOCS family of proteins to understand better the regulatory roles subserved by this interesting family of proteins.
Acknowledgments
We thank Lorraine O’Reilly and Andreas Strasser (The Walter and Eliza Hall Institute) for rat-anti-FLAG antibody and Jim Johnson (DNAX, Palo Alto, CA) for rabbit anti-SOCS-3 polyclonal antiserum. We also thank Bronwyn Roberts and Marta Brysha for their technical advice and Jason Corbin for his technical assistance. This work was supported by the Anti-Cancer Council of Victoria (Melbourne, Australia), AMRAD Corp. (Melbourne), the National Health and Medical Research Council (Canberra, Australia), the J. D. and L. Harris Trust, the National Institutes of Health (Grant CA-22556), and the Australia Federal Government Cooperative Research Centres Program.
ABBREVIATIONS
- ASB
ankyrin-repeat-containing protein with a SOCS box
- CIS
cytokine-inducible SH2-containing protein
- IL
interleukin
- Jak
Janus kinase
- LLnL
N-acetyl-l-leucinyl-l-leucinyl-norleucinal
- PULC
phosphoprotein-ubiquitin-ligase complex
- SOCS
suppressors of cytokine signaling
- SSB
SPRY domain-containing protein with a SOCS box
- UBL
ubiquitin-like
- VHL
von Hippel Lindau
- WSB
WD-40-repeat-containing protein with a SOCS box
- GST
glutathione S-transferase
Note Added in Proof
After submission of this manuscript, a paper was published by Kamura et al. (28) that also demonstrates interaction of the elongin BC complex with the SOCS box motif of several proteins. However, in contrast to us, they suggest that this interaction inhibits degradation of SOCS box-containing proteins.
References
- 1.Starr R, Willson T A, Viney E M, Murray L J, Rayner J R, Jenkins B J, Gonda T J, Alexander W S, Metcalf D, Nicola N A, Hilton D J. Nature (London) 1997;387:917–921. doi: 10.1038/43206. [DOI] [PubMed] [Google Scholar]
- 2.Hilton D J, Richardson R T, Alexander W S, Viney E M, Willson T A, Sprigg N S, Starr R, Nicholson S E, Metcalf D, Nicola N A. Proc Natl Acad Sci USA. 1998;95:114–119. doi: 10.1073/pnas.95.1.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yoshimura A, Ohkubo T, Kiguchi T, Jenkins N A, Gilbert D J, Copeland N G, Hara T, Miyajima A. EMBO J. 1995;14:2816–2826. doi: 10.1002/j.1460-2075.1995.tb07281.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Endo T A, Masuhara M, Yokouchi M, Suzuki R, Mitsui K, Sakamoto H, Ohtsubo M, Misawa H, Kanekura Y, Yoshimura A. Nature (London) 1997;387:921–924. doi: 10.1038/43213. [DOI] [PubMed] [Google Scholar]
- 5.Naka T, Narazaki M, Hirata M, Matsumoto T, Minamoto S, Aono A, Nishimoto N, Kajita T, Taga T, Yoshizaki K, Akira S, Kishimoto T. Nature (London) 1997;387:924–929. doi: 10.1038/43219. [DOI] [PubMed] [Google Scholar]
- 6.Masuhara M, Sakamoto H, Matsumoto A, Suzuki R, Yasukawa H, Mitsui K, Wakioka T, Tanimura S, Sasaki A, Misawa H, Yokouchi M, Ohtsubo M, Yoshimura A. Biochem Biophys Res Commun. 1997;239:429–446. doi: 10.1006/bbrc.1997.7484. [DOI] [PubMed] [Google Scholar]
- 7.Minamoto S, Ikegame K, Ueno K, Narazaki M, Naka T, Yamamoto H, Matsumoto T, Saito M, Hosoe S, Kishimoto T. Biochem Biophys Res Commun. 1997;237:79–83. doi: 10.1006/bbrc.1997.7080. [DOI] [PubMed] [Google Scholar]
- 8.Narazaki M, Fujimoto M, Matsumoto T, Morita T, Saito H, Kajita T, Yoshizaki K, Naka T, Kishimoto T. Proc Natl Acad Sci USA. 1998;95:13130–13134. doi: 10.1073/pnas.95.22.13130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nicholson SE, Willson T A, Farley A, Starr R, Zhang J-G, Baca M, Alexander W S, Metcalf D, Hilton D J, Nicola N A. EMBO J. 1999;18:375–385. doi: 10.1093/emboj/18.2.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Smith D B, Johnson K S. Gene. 1988;67:31–40. doi: 10.1016/0378-1119(88)90005-4. [DOI] [PubMed] [Google Scholar]
- 11.Moritz R L, Eddes J S, Reid G E, Simpson R J. Electrophoresis. 1996;17:907–917. doi: 10.1002/elps.1150170512. [DOI] [PubMed] [Google Scholar]
- 12.Moritz R L, Simpson R J. J Chromatogr. 1992;599:119–130. doi: 10.1016/0021-9673(92)85464-5. [DOI] [PubMed] [Google Scholar]
- 13.Zugaro L M, Reid G E, Ji H, Eddes J S, Murphy A A, Burgess A W, Simpson R J. Electrophoresis. 1998;19:867–876. doi: 10.1002/elps.1150190544. [DOI] [PubMed] [Google Scholar]
- 14.Eng J K, McCormack A L, Yates J R. J Am Soc Mass Spectrom. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 15.Schnölzer M, Alewood P, Jones A, Alewood D, Kent S B H. Int J Peptide Protein Res. 1992;40:180–193. doi: 10.1111/j.1399-3011.1992.tb00291.x. [DOI] [PubMed] [Google Scholar]
- 16.Kaelin W G, Jr, Maher E R. Trends Genet. 1998;14:423–426. doi: 10.1016/s0168-9525(98)01558-3. [DOI] [PubMed] [Google Scholar]
- 17.Conaway J W, Kamura T, Conaway R C. Biochim Biophys Acta. 1998;1377:M49–M54. doi: 10.1016/s0304-419x(97)00035-8. [DOI] [PubMed] [Google Scholar]
- 18.Garrett K P, Aso T, Bradsher J N, Foundling S I, Lane W S, Conaway R C, Conaway J W. Proc Natl Acad Sci USA. 1995;92:7172–7176. doi: 10.1073/pnas.92.16.7172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lonergan K M, Iliopoulos O, Ohh M, Kamura T, Conaway R C, Conaway J W, Kaelin W G. Mol Cell Biol. 1998;18:732–741. doi: 10.1128/mcb.18.2.732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Duan D R, Pause A, Burgess W H, Aso T, Chen D Y T, Garrett K P, Conaway R C, Conaway J W, Linehan W M, Klausner R D. Science. 1995;269:1402–1406. doi: 10.1126/science.7660122. [DOI] [PubMed] [Google Scholar]
- 21.Kibel A, Iliopoulos O, DeCaprio J D, Kaelin W G. Science. 1995;269:1444–1446. doi: 10.1126/science.7660130. [DOI] [PubMed] [Google Scholar]
- 22.Pause A, Lee S, Worrell R A, Chen D Y T, Burgess W H, Linehan W M, Klausner R D. Proc Natl Acad Sci USA. 1997;94:2156–2161. doi: 10.1073/pnas.94.6.2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kishida T, Stackhouse T M, Chen F, Lerman M I, Zbar B. Cancer Res. 1995;55:4544–4548. [PubMed] [Google Scholar]
- 24.Schauber C, Chen L, Tongaaokar P, Vega I, Lambertson D, Potts W, Madura K. Nature (London) 1998;391:715–718. doi: 10.1038/35661. [DOI] [PubMed] [Google Scholar]
- 25.Hershko A, Ciechanover A. Annu Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 26.Verdier F, Chretien S, Muller O, Varlet P, Yoshimura A, Gisselbrecht S, Lacombe C, Mayeux P. J Biol Chem. 1998;273:28185–28190. doi: 10.1074/jbc.273.43.28185. [DOI] [PubMed] [Google Scholar]
- 27.Yu C-L, Burakoff S J. J Biol Chem. 1997;272:14017–14020. doi: 10.1074/jbc.272.22.14017. [DOI] [PubMed] [Google Scholar]
- 28.Kamura T, Sato S, Haque D, Liu L, Kaelin W G, Jr, Conaway R C, Conaway J W. Genes Dev. 1998;12:3872–3881. doi: 10.1101/gad.12.24.3872. [DOI] [PMC free article] [PubMed] [Google Scholar]