Abstract
We identified a strong association (p=5.36×10−17) between rs492602 in FUT2 and plasma vitamin B12 levels in a genome-wide scan (n=1,658) and an independent replication sample (n=1,059) from the Nurses' Health Study. Women homozygous for the rs492602 G allele had higher B12 levels. This allele is in strong linkage disequilibrium with the FUT2 W143X nonsecretor variant, suggesting a plausible mechanism for altered B12 absorption and plasma levels.
Keywords: one-carbon metabolism, plasma vitamin B12, genome-wide association, FUT2, secretor locus
Plasma levels of vitamin B12 are modifiable quantitative traits associated with certain diseases1. Vitamin B12 found in meat (such as liver, shellfish, fish, poultry, and eggs) and milk products2 is composed of corrin and cobalt rings and is necessary for the formation of red blood cells, DNA synthesis during cell division, and maintenance of the myelin nerve sheath. Deficiency in vitamin B12, clinically associated with pernicious anemia, cardiovascular disease, cancer, and neurodegenerative disorders, is often related to poor intestinal B12 absorption2 rather than direct dietary deficiency (the recommended adult intake for vitamin B12 is 2.4 µg/day). In the Nurses’ Health Study (NHS), we previously observed that women in the lowest quartile of plasma vitamin B12 levels had marginally worse cognitive performance (based on a global score averaging 6 cognitive tests) than women in the highest quartile of plasma vitamin B12 and that combined folate and vitamin B12 deficiency was associated with the lowest cognitive performance3.
Epidemiologic studies provide evidence for the association of genes and metabolites in the B-vitamin-mediated plasma one-carbon metabolic pathway with chronic diseases2–4. Rare high penetrance mutations in genes in this pathway affect the ability to digest, absorb5, and utilize vitamin B122. However, common genetic variants in candidate genes have not been consistently associated with plasma vitamin B12 levels. Therefore, we conducted a genome-wide association study (GWAS) to identify novel loci that influence plasma vitamin B12 levels in 1,658 women genotyped with the HumanHap500 as part of the Cancer Genetic Markers of Susceptibility Project (CGEMS; Supplementary Methods). Participants were of self-reported European ancestry6. Detailed methods have been previously reported, including quality control assessment of genotypes with sample completion and SNP call rates, concordance rate, deviation from Hardy–Weinberg proportions in control DNA, and final sample selection for association analysis of the NHS CGEMS population6.
We tested association between each of the 528,134 SNP markers that passed quality control filters and log-transformed plasma vitamin B12 using linear regression, adjusted for age, total methyl intake (defined as total folate, dietary methionine, and alcohol intake), and assay batch. We saw no evidence for systematic bias in the distribution of p-values for analyses with and without further adjustment for residual population structure using the top four principal components of genetic variation (Supplementary Fig. 1), compatible with no confounding of SNP-metabolite associations due to population stratification.
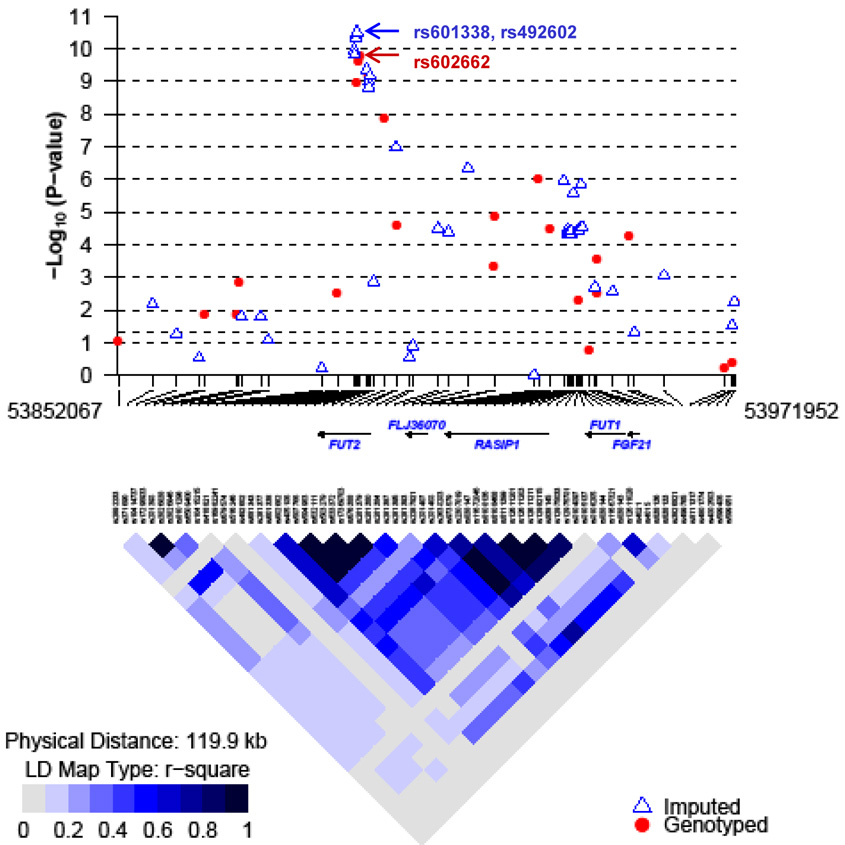
In the initial GWAS, the SNPs on chromosome 19p13.3 accounted for the excess of p-values <10−7 (Supplementary Fig. 1: Quantile-Quantile plot). The strongest association with plasma vitamin B12 was rs602662 (Ptrend = 6.54×10−10; Fig. 1, Table 1, Supplementary Fig. 2), a non-synonymous SNP in FUT2 on chromosome 19p13.3, which has a minor allele frequency of 0.49 (Table 1). The association with plasma vitamin B12 and rs602662 was independently replicated using Taqman allelic discrimination assays in 1,059 women from two nested case control studies drawn from the same cohort (Table 1, Supplementary Table 2) with ptrend=1.13 × 10 −06. The joint analysis of the CGEMS scan data and the replication dataset supports the association of rs602662 with plasma vitamin B12 (ptrend = 3.51×10−15). Results were similar using untransformed plasma vitamin B12 values. A similar association was observed using the non-parametric Kruskal-Wallis test (joint analysis Kruskal-Wallis p = 1.48×10−21). Plasma vitamin B12 has an inverse correlation in this data set with plasma homocysteine, an integrated marker of the one-carbon metabolism pathway, (Spearman correlation coefficient −0.26, p<0.0001). FUT2 rs602662 also has a modest association with plasma homocysteine, (ptrend=0.0085 in the GWAS data and ptrend=0.0081 in the replication data). The pattern of mean log-transformed plasma vitamin B12 levels by rs602662 genotype suggest a dominant genetic effect, with lower B12 levels among variant carriers (test comparing mean log-transformed vitamin B12 levels between variant carriers and non-carriers p = 1.35 × 10−39).
Figure 1. LD Structure of Chromosome 19.
This panel shows the trend P values for association testing with plasma vitamin B12 from 25 observed ( ) and 44 imputed (
) and 44 imputed ( ) SNPs from the GWA study displaying SNPs on chromosome 19p13.3. All known genes are shown (National Center for Biotechnology Information build 36.1)
) SNPs from the GWA study displaying SNPs on chromosome 19p13.3. All known genes are shown (National Center for Biotechnology Information build 36.1)
Table 1.
Association of plasma vitamin B12 levels with FUT2 genotypes in women participants from GWAS and replication studies
| SNP Association by Study | Age | N | Allele Freq | Estimate (S.E.)c | P-valued | |||
|---|---|---|---|---|---|---|---|---|
| Geometric Mean (95% CI), pg/ml |
||||||||
| rs602662 GWAS | G (Gly)=0.49 | Ser/Ser | Ser/Gly | Gly/Gly | ||||
| NHS CGEMS | 59 | 1,658 | 489.82 | 418.67 | 417.05 | −0.08 (0.01) | 6.54 × 10−10 | |
| (472.24–508.06) | (407.74–429.90) | (401.29–433.43) | ||||||
| Replication | ||||||||
| Combined NHS | 63 | 1,056 | 483.68 | 406.96 | 409.60 | −0.08 (0.02) | 1.13 × 10−06 | |
| Colorectal (CR e) | (461.78–506.63) | (393.69–420.67) | (390.22–429.93) | |||||
| Adenoma and CR Cancer | ||||||||
| Case-control Datasets | ||||||||
| TOTAL (GWAS and Replication) e | 487.72 | 413.35 | 413.52 | −0.08 (0.01) | 3.52 × 10−15 | |||
| (473.80–502.06) | (404.83–422.06) | (401.14–426.27) | ||||||
| rs601338b | aA (Trp)=0.51 | Nonsecretor | Secretor | Secretor | ||||
| (X/X) | (X/Trp) | (Trp/Trp) | ||||||
| GWAS | ||||||||
| NHS CGEMS | 59 | 1,658 | 496.60 | 418.69 | 418.32 | −0.08 (0.01) | 4.11 × 10−10 | |
| (477.89–516.04) | (407.76–429.92) | (403.36–433.84) | ||||||
| rs492602 GWAS | a A=0.51 | GG/GG | AG/AG | AA/AA | ||||
| NHS CGEMS | 59 | 1,637 | 496.02 | 419.95 | 416.97 | −0.08 (0.01) | 2.68 × 10−10 | |
| (477.32–515.45) | (408.88–431.31) | (402.14–432.35) | ||||||
| Replication | ||||||||
| Combined NHS CR | 63 | 1,059 | 491.13 | 407.67 | 406.61 | −0.10 (0.02) | 5.60 × 10−09 | |
| Adenoma and CR Cancer | (469.03–514.26) | (394.34–421.45) | (389.46–424.53) | |||||
| Case-control Datasets | ||||||||
| TOTAL (GWAS and Replication)f | 493.89 | 414.27 | 412.70 | −0.09 (0.01) | 5.36 × 10−17 | |||
| (479.42–508.81) | (405.69–423.04) | (401.33–424.40) | ||||||
Major allele according to HapMap CEU data
Imputed distribution for nonsense polymorphism, rs601338 (also known as W143X)
Estimates (regression coefficients) calculated from linear regression adjusted for age using log-transformed plasma vitamin B12 (same estimates obtained when adjusting for age, assay batch and total dietary methyl status)
P-values: GWAS and replication p-values are calculated from linear regression adjusted for age
colorectal is abbreviated as CR
Dominant model p-value for the joint analysis: rs602662 = 1.35 × 10−39; rs492602= 8.26 × 10−45
The SNP rs602662 is in strong linkage disequilibrium (D’=1, r2=0.76) with the FUT2 nonsense SNP W143X (rs601338; Fig. 1). We imputed the nonsense SNP in the initial GWAS samples using the observed chromosome 19 genotyping data augmented by data from the densely-genotyped CEPH European HapMap samples and found that it was strongly associated with plasma vitamin B12 levels (ptrend=4.11×10−10; Fig. 1, Table 1). Since the nonsense SNP rs601338 could not be genotyped by Taqman because of a neighboring SNP (rs1800459) located one nucleotide upstream, we genotyped a proxy SNP, rs492602 (which is in perfect LD with rs601338 in HapMap data; Supplementary Table 1, Supplementary Fig. 3), in both the initial GWAS and the replication data set. The association between rs492602 and plasma vitamin B12 was stronger than the association for rs602662 and plasma vitamin B12 (Table 1: rs492602 joint ptrend=5.36×10−17; joint dominant model p =8.26×10−45). The rs492602 SNP explains 2.5% of the variance in log-transformed plasma vitamin B12 levels using the log-additive genetic model and 3.5% of variance using the dominant genetic model.
Fucosylated carbohydrate structures are involved in a range of biological processes7,8 including tissue development, angiogenesis, fertilization, cell adhesion, inflammation, and tumor metastasis. The classic human secretor locus (Se) FUT2 encodes α(1,2)fucosyltransferase, which regulates expression of the Lewis ABO(H) histo-blood group antigens on the surface of epithelial cells and in body fluids and determines the secretion status of the ABO antigens7. Secretor status of this polymorphic protein was used by Mohr to provide the first autosomal linkage in humans between secretor factor and the Lutheran blood group; subsequently secretor linkage was established with the APOE and myotonic dystrophy locus7. The family of α-1,2-fucosyltransferases catalyze the addition of fucose in α-1,2-linkage to the galactose of type 1(Galβ1,3GlcNAc-R) and type 2 Galβ1,4GlcNAc- R) disaccharide to form H type 1 and H type 2 antigens7, respectively.
In the highly polymorphic FUT2 gene7, three SNPs, rs602662, rs492602 and rs601338 (W143X; nucleotide position 428), are in strong LD, we note that W143X is a nonsense mutation7 and is plausibly the causal variant for the association with plasma B12 levels. The 143X variant is characteristic for the nonsecretor allele in Europeans and has an allele frequency of 0.46 in populations of European ancestry7. In Europeans, Africans, and Iranians, the FUT2 W143X nonsense mutation is the primary nonsecretor allele, with a frequency of approximately 50 percent. In contrast, in Asian populations, the FUT2 I129F (nucleotide position 385) missense mutation is the primary nonsecretor allele8. In non-Asian populations, nonsecretors are frequently homozygous for the FUT2 W143X polymorphism, resulting in an inactive FUT2. Individuals homozygous for the FUT2 nonsecretor genotype appear to be resistant to infection with Norovirus9, suggesting that individuals homozygous for nonsecretor status may be unable to mediate host-microbe interactions9.
Absorption of B12 requires the secretion of the glycoprotein intrinsic factor (IF) from the gastric cells, binding of IF to vitamin B12 and a functional gastrointestinal absorption system2. The H-antigen synthesized by FUT2, Lewis ABO antigens, and FUT2 genotypes have all been reported to mediate H. pylori attachment to human gastric mucosa10. Atrophic gastritis is a consequence of H. pylori infection11 and leads to reduced secretion of IF12,14. The FUT2 secretor status has been associated with both H. pylori infection and gastritis10; patients with vitamin B12 malabsorption and low levels of serum vitamin B12 have higher seroprevalence of H. pylori infection10. These data suggest a potential mechanism by which vitamin B12 absorption12–15 may be reduced in carriers of the secretor genotype due to the sequelae of susceptibility to H. pylori infection compared to individuals with the nonsecretor genotype.
In summary, among generally well-nourished women, we found that common variation in the FUT2 secretor gene is associated with plasma vitamin B12 at high levels of statistical confidence. Although the participants included in these analyses are a selected subset of women who had plasma vitamin B12 levels measured, numerous demographic and lifestyle factors are comparable between this sample and the overall NHS cohort. Insights gained from the study of plasma vitamin B12 are likely to have implications for the study of complex diseases such as cognitive decline, cancer, and cardiovascular disease. Further study is required to investigate the biological basis of our reported association findings.
Acknowledgements
We thank Hardeep Ranu, Constance Chen, and the staff at the Core Genotyping Facility at the National Cancer Institute for their expertise.
This research is supported by the National Institutes of Health Research Grants U54 CA100971, P01 CA87969, P01 CA55075, U01 CA098233, R01 CA 065725 and CA070817.
A.H. is supported in part by training grant NIH T-32 CA 09001-30.
The commonly used abbreviations are
- FUT2
Fucosyltransferase
- GWAS
genome-wide association study
- H. pylori
Helicobacter pylori
- NHS
Nurses’ Health Study
- CI
confidence interval
- SNPs
single nucleotide polymorphisms
References
- 1.Siva A, et al. Qjm. 2007;100:495–499. doi: 10.1093/qjmed/hcm054. [DOI] [PubMed] [Google Scholar]
- 2.Watanabe F. Exp Biol Med (Maywood) 2007;232:1266–1274. doi: 10.3181/0703-MR-67. [DOI] [PubMed] [Google Scholar]
- 3.Kang JH, et al. Epidemiology. 2006;17:650–657. doi: 10.1097/01.ede.0000239727.59575.da. [DOI] [PubMed] [Google Scholar]
- 4.Dahlin AM, et al. Int J Cancer. 2008;122:2057–2061. doi: 10.1002/ijc.23299. [DOI] [PubMed] [Google Scholar]
- 5.Tanner SM, et al. Hum Mutat. 2004;23:327–333. doi: 10.1002/humu.20014. [DOI] [PubMed] [Google Scholar]
- 6.Hunter DJ, et al. Nat Genet. 2007;39:870–874. doi: 10.1038/ng2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kelly RJ, et al. J Biol Chem. 1995;270:4640–4649. doi: 10.1074/jbc.270.9.4640. [DOI] [PubMed] [Google Scholar]
- 8.Koda Y, et al. Genetics. 2001;158:747–756. doi: 10.1093/genetics/158.2.747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lindesmith LC, et al. PLoS Med. 2008;5:e31. doi: 10.1371/journal.pmed.0050031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carmel R, et al. J Lab Clin Med. 1987;109:454–463. [PubMed] [Google Scholar]
- 11.Azevedo M, et al. J Pathol. 2008;215:308–316. doi: 10.1002/path.2363. [DOI] [PubMed] [Google Scholar]
- 12.van Oijen MG, et al. J Nutr Sci Vitaminol (Tokyo) 2004;50:305–308. doi: 10.3177/jnsv.50.305. [DOI] [PubMed] [Google Scholar]
- 13.Dholakia KR, et al. World J Gastroenterol. 2005;11:7078–7083. doi: 10.3748/wjg.v11.i45.7078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Annibale B, et al. Dig Liver Dis. 2002;34 Suppl 2:S72–S77. doi: 10.1016/s1590-8658(02)80170-0. [DOI] [PubMed] [Google Scholar]
- 15.Tamura A, et al. Am J Gastroenterol. 2002;97:861–866. doi: 10.1111/j.1572-0241.2002.05601.x. [DOI] [PubMed] [Google Scholar]