Abstract
Determining how neuronal networks encode memories is a key goal of neuroscience. Although neuronal circuit processes involved in encoding, storing and retrieving memory have attracted a great deal of attention, the processes that allocate individual memories to specific neurons within a network have remained elusive. Recent findings unraveled the first insights into the processes that modulate memory allocation in neuronetworks. They showed that neurons in the lateral amygdala compete to take part in auditory fear conditioned memory traces and that the levels of the transcription factor CREB (cAMP-response element binding protein) can affect the probability of a neuron to be recruited into a given memory representation. CREB-mediated transcriptional regulation involves several signaling pathways, known to mediate nuclear responses to diverse behavioral stimuli, along with coordinated interactions with multiple other transcription activators, co-activators and repressors. Moreover, activation of CREB triggers an autoinhibitory feedback loop, a metaplastic process that could be used to allocate memories away from cells that have been recently involved in memory. Beyond CREB, there may be a host of other processes that dynamically modulate memory allocation in neuronetworks by shaping cooperation and competition among neurons.
Keywords: Memory allocation, Competition, cAMP-response element binding protein (CREB), Activity-regulated cytoskeleton-associated protein (Arc), Metaplasticity
1. Memory allocation: a competitive process
Memory depends on specific sets of connected neurons which together support the ‘memory trace’ (McGaugh, 1972; Thompson, 2005). Electrophysiological and cellular imaging studies demonstrated that only a portion of neurons are involved in a given memory (Repa et al., 2001; Rumpel, LeDoux, Zador, & Malinow, 2005). Despite numerous studies on the nature and properties of memory traces, little is known about how memories are allocated into specific subsets of neurons in a given neuronetwork.
Activity-dependent competitive refinement of connections is a general feature of neural circuits in the central nervous system. Competition between bilateral monocular neural activities is critical for segregating projections from the two retinae into distinct laminae in the lateral geniculate nucleus and then into distinct columns in the visual cortex (Wiesel & Hubel, 1965; Cabelli, Hohn, & Shatz, 1995). Competition also sharpens the topographic mapping of retinal axons onto their central targets. In addition, competitive maintenance of long-term potentiation (LTP) of synaptic pathways has been described; when one of two previously potentiated synaptic pathways is stimulated again, further potentiation comes at the expense of the maintenance of potentiation in the other pathway (Miller, 1996; Fonseca, Nagerl, Morris, & Bonhoeffer, 2004).
Several studies have indicated that only a portion of eligible neurons participate in a given memory (see for example, Guzowski, McNaughton, Barnes, & Worley, 1999; Repa et al., 2001; Rumpel et al., 2005; Wilson & McNaughton, 1993). For example, plasticity within the lateral amygdala (LA) is essential for auditory conditioned fear memories (LeDoux, 2000; Fanselow & Gale, 2003), and although ∼70% of LA neurons receive the necessary sensory input, only one-quarter exhibit auditory fear conditioning-induced synaptic plasticity (Repa et al., 2001; Rumpel et al., 2005). Why are some neurons, rather than their neighbors, recruited in storing a given memory? A recent study from our laboratory suggests that neurons compete with each other to take part in fear memory traces and that the transcription factor cAMP-response element binding protein (CREB) plays a crucial role in determining which neurons will participate in a memory representation (Han et al., 2007).
2. Role of CREB in competitive memory allocation
CREB, a member of a family of structurally related transcription factors, is widely expressed in the brain and its activity is induced in response to calcium, neurotrophin, and cytokine signals as well as a variety of cellular stresses (Silva, Kogan, Frankland, & Kida, 1998; Shaywitz & Greenberg, 1999; Mayr & Montminy 2001; Lonze & Ginty, 2002; Carlezon, Duman, & Nestler, 2005). Membrane depolarization or/and an elevation of cAMP strongly induce the phosphorylation of CREB at serine 133, and thereby activate CREB-dependent transcription (Sheng, Thompson, & Greenberg, 1991; Gonzalez & Montminy, 1989). A large body of evidence indicates that CREB-dependent transcription is essential for both long-lasting forms of synaptic plasticity and long-term memory, but not short-term plasticity or short term memory (e.g., Silva et al., 1998; Shaywitz & Greenberg, 1999; Mayr & Montminy 2001; Lonze & Ginty, 2002; Carlezon, Duman, & Nestler, 2005). Genetic and pharmacological studies in several species demonstrate that CREB has a seemingly universal role in memory, tested in a wide range of tasks that span emotional, spatial and social memory (e.g., Yin, Del Vecchio, Zhou, & Tully, 1995; Bartsch, Casadio, Karl, Serodio, & Kandel, 1998; Josselyn et al., 2001; Wallace, Stellitano, Neve, & Duman, 2004; Jasnow, Shi, Israel, Davis, & Huhman, 2005). In addition, several lines of evidence have implicated CREB in the competition between neurons necessary for refining retinogeniculate axons and establishing ocular dominance within the visual cortex in the developing brain (Pham, Impey, Storm, & Stryker, 1999; Pham, Rubenstein, Silva, Storm, & Stryker, 2001; Pham et al., 2004; Mower, Liao, Nestler, Neve, & Ramoa, 2002).
To determine whether CREB had a role in memory allocation, amygdala cells were transfected with a virus over-expressing either CREB or its dominant-negative form (Han et al., 2007) (Fig. 1). To visualize the memory trace, nuclear-localized transcripts of the neuronal activity-dependent gene Arc (activity-regulated cytoskeleton-associated protein; also termed Arg3.1) were detected with high-sensitivity fluorescence in situ hybridization (FISH). Neuronal activity induces a rapid and transient increase in Arc transcription, and thus nuclear-localized Arc RNA can serve as a molecular signature of a recently (5-15 min) active neuron (Guzowski et el., 1999). Only neurons active during the memory test have Arc RNA localized in the nucleus which can be detected with high-sensitivity FISH five minutes after the fear memory test (Guzowski et el., 1999). Arc is a particularly good marker for memory activation because not only is its expression associated with memory formation, but Arc expression is also needed for memory (Tzingounis & Nicoll, 2006).
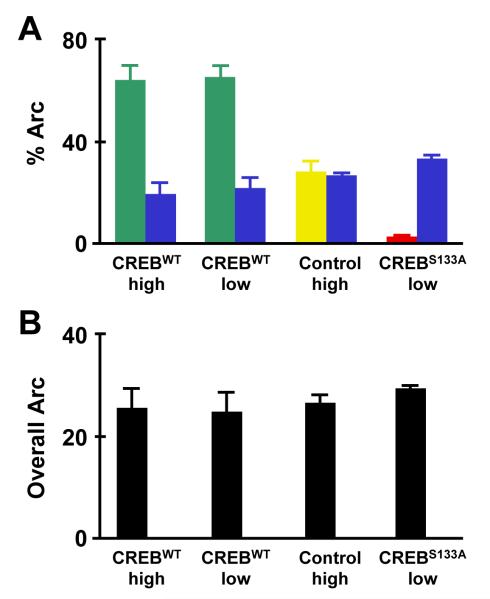
Fig. 1.
Relative CREB activity influences the competitive recruitment of neurons into a memory trace. (A) Distribution of Arc-positive neurons varied according to CREB manipulation. Arc-positive nuclei were more likely to be in neurons infected with a virus overexpressing wild-type CREB (CREBWT) (green bars) than in noninfected neighbors (paired blue bars), whether the mice were trained with high or low shock intensities. In contrast, Arc-positive nuclei were equally likely to be in neurons with (yellow bar) and without (paired blue bar) a control virus that does not express CREB. Arc-positive nuclei were less likely to be in neurons with decreased CREB function (neurons infected with a virus overexpressing CREBS133A; red bar) relative to noninfected neighbors (paired blue bar). (B) Irregardless of the conditions listed above, the percent of cells expressing Arc-positive neurons remained constant, regardless of virus used (control, CREBWT, CREBS133A) or training intensity (high or low) (modified from Han et al., 2007).
Neurons infected with a virus overexpressing CREB in the lateral amygdala preferentially expressed Arc after recall of a memory for auditory fear conditioning (Fig. 1A). Moreover, in comparison with their noninfected neighbors, neurons infected with a dominant-negative form of CREB (CREBS133A), in which serine 133 is replaced by alanine, have a much lower probability of having Arc-positive nuclei (Fig. 1A). Importantly, the overall proportion of Arc-positive neurons was constant regardless of CREB manipulation (Fig. 1B), suggesting that neuronal selection during memory formation is competitive rather than cell-autonomous. Importantly, a number of controls showed that the ability of CREB to bias memory allocation a) was not the result of a specific narrow set of training conditions, b) was not due to CREB function directly inducing Arc transcription, c) is dependent on training and learning, and d) is not due to changes in the threshold for Arc expression.
Taken together, these findings provide a novel approach to study memory allocation, and show that neuronal competition, which has previously been demonstrated to have an important role during brain development, is also an essential part of memory formation. Furthermore, the findings provide the first mechanistic insights into memory allocation: they show that CREB plays a crucial role in the selection of neurons to be recruited into a memory representation.
3. What are the mechanisms underlying CREB-mediated competitive memory allocation?
How do neurons with higher levels/activity of CREB gain a competitive edge during memory allocation? CREB regulates a diverse array of genes, and many CREB targets (e.g., c-fos, JunD, C/EBPβ, Egr1, Nurr1, etc.) are themselves transcription factors that regulate other genes. Multiple CREB target genes could contribute to the coordinate regulation of the memory allocation process. Much effort has been invested on identifying the CREB ‘transcriptome’ or ‘regulon’, a complex that includes all genes regulated by CREB (Cha-Molstad, Keller, Yochum, Impey, & Goodman, 2004; Impey et al., 2004; Zhang et al., 2005). Among this cohort of players, we will highlight a subset of CREB target genes and processes that could be involved in CREB-mediated competitive memory allocation.
Changes in neuronal excitability could directly affect memory allocation, since neurons with higher excitability would be more easily activated by learning and therefore would be more likely to be recruited into memory representations. Indeed, several lines of evidence indicate that CREB plays an important role in controlling the excitability of neurons (Marie, Morishita, Yu, Calakos, & Malenka, 2005; Dong et al., 2006; Han et al., 2006). Viral overexpression of CREB in the locus ceruleus (LC) of rats had no significant effect on neuronal firing at baseline, but enhanced the excitatory effect of forskolin (an activator of adenylate cyclase) on LC neurons, suggesting that the cAMP signaling pathway in these neurons was sensitized by CREB (Han et al., 2006); This is especially significant because this signaling pathway is known to be engaged during learning. Moreover, LC neurons expressing constitutively active CREB fired significantly faster and their resting membrane potential was more depolarized compared with control cells. Conversely, downregulating CREB activity in LC neurons decreased the firing rate and hyperpolarized the neurons. In addition, expression of active CREB in the rat nucleus accumbens (NAc) medium spiny neurons (MSNs) increases their excitability, whereas dominant-negative CREB has the opposite effect (Dong et al., 2006).
CREB could also affect the numbers of “silent” or “naïve” synapses (those expressing NMDA but not AMPA receptors) in each neuron, and thus affect where memories are more likely to be stored: neurons with higher CREB levels and therefore more naïve synapses would be more likely to store the memory than those with lower CREB levels and consequently fewer naive synapses. Neurons infected with a virus expressing constitutively active CREB showed an enhancement of N-methyl-D-aspartate (NMDA) receptor-mediated synaptic responses and LTP relative to their non-infected neighbors (Marie et al., 2005), a result consistent with the idea that CREB affects the number of silent synapses ready for synaptic changes, such as LTP. Importantly, additional electrophysiological and morphological studies provided compelling evidence for the idea that higher CREB levels lead to the generation of ‘silent synapses’, containing NMDA- but not AMPA- receptors, which are thought to provide an ideal substrate for the storage of memory traces (Marie et al., 2005).
The CREB target genes that are responsible for changes in either excitability or silent synapse numbers described above are not known. Likely candidates for CREB-mediated changes in neuronal excitability include voltage-dependent ion channels as well as second messenger systems that modulate these channels. Current-clamp recordings suggested that CREB-induced increases in neuronal excitability were mediated, at least in part, by an enhancement of Na+ conductances and an inhibition of K+ conductances (Dong et al., 2006). Consistent with these findings, a microarray analysis found that CREB expression in the NAc stimulated the transcription of a voltage-dependent sodium channel subunit, 1β, and inhibited the transcription of a voltage-dependent potassium channel subunit, Kv1.4 (McClung & Nestler, 2003). Additionally, adenylate cyclase VIII (ACVIII) appears to be a direct target for CREB (Lane-Ladd et al., 1997). Active CREB induces ACVIII promoter activity, whereas dominant-negative CREB inhibits it, both in vitro and in vivo in the brain (Chao et al., 2002). Since activation of the cAMP pathway increases neuronal excitability (Wang and Aghajanian, 1987; Alreja and Aghajanian, 1995; Ivanov and Aston-Jones, 2001), these observations support the hypothesis that increased CREB activity, through the consequent induction of ACVIII, increases neuronal excitability.
4. Potential molecular pathways regulating memory allocation
The finding that neurons with higher levels of CREB activity become memory attractors, while those with low levels are less likely to participate in a given memory trace, suggest that some or all of the cooperating and antagonizing signaling pathways known to regulate CREB activity (Fig. 2) might also affect the competitive memory allocation process.
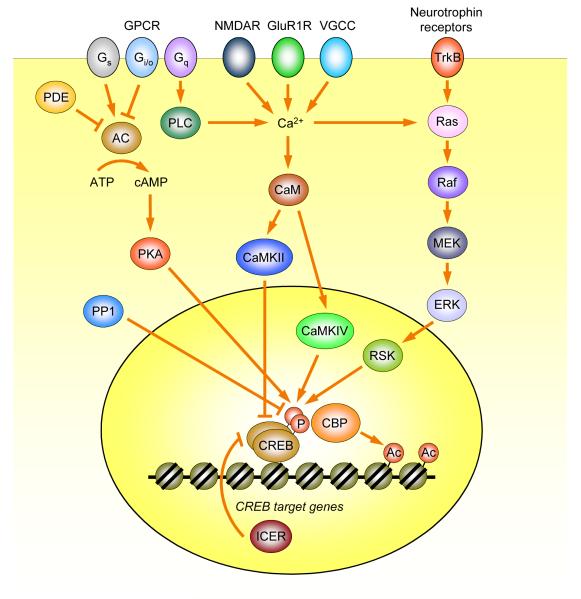
Fig. 2.
Signaling pathways regulating CREB-dependent transcription. Pathways and cross-interactions depicted here are grossly simplified. Arrows and barred lines indicate activation and suppression, respectively. In principle, each of the molecules in these pathways can contribute to the competitive memory allocation process. Abbreviations: Ac, acetylation of histone tails; AC, adenylate cyclase; CaM, calmodulin; CamKII, calcium/CaM-dependent protein kinase II; CaMKIV, calcium/CaM-dependent protein kinase IV; CBP, CREB-binding protein; CREB, cAMP-response element binding protein; ERK, extracellular signal-regulated kinase; GluR1R, glutamate receptor subunit GluR1 homomeric AMPA receptor; GPCR, G protein-coupled receptor; ICER, inducible cAMP early repressor; MEK, mitogen-activated protein kinase or extracellular signal-regulated kinase kinase; NMDAR, N-methyl-D-aspartate receptor; P, phosphorylation of CREB on Ser133; PDE, phosphodiesterase; PKA, protein kinase; PLC, phospholipase C; PP1, protein phosphatase 1; RSK, ribosomal S6 kinase; TrkB, tyrosine kinase receptor B; VGCC, voltage-gated calcium channel.
CREB is crucial for translating diverse behavioral stimuli into transcriptional responses in the nucleus. Several intracellular signaling pathways are involved in transmitting information initiated by activation of membrane receptors to CREB in the nucleus (Fig. 2). Multiple kinases, including protein kinase A (PKA), Ca2+/calmodulin-dependent kinase IV (CaMKIV), mitogen- and stress-activated protein kinase (MSK), and mitogen-activated protein kinase (MAPK)-activated ribosomal S6 kinases (RSKs), have been shown to phosphorylate CREB at serine 133 and thereby activate CREB-dependent transcription in response to a variety of stimuli. However, specific kinases and signaling pathways appear to respond primarily to subsets of these stimuli (Shaywitz & Greenberg, 1999; Mayr & Montminy 2001; West, Griffith, & Greenberg, 2002; Lonze & Ginty, 2002).
Phosphorylation of CREB at serine 133 triggers the recruitment of the transcriptional coactivator CREB-binding protein (CBP), which induces transcription via its intrinsic and associated histone acetylase activities and/or by interacting with the core transcriptional machinery (Vo & Goodman, 2001; Lonze & Ginty, 2002). In contrast, CaMKII phosphorylates CREB at serine 142, which promotes the dissociation of CREB dimers and thus reduces CREB-mediated gene transcription (Matthews et al., 1994; Wu & McMurray, 2001). Calcium-dependent activation of protein phosphatases PP1 and PP2A leads to the dephosphorylation of CREB at serine 133 (Shaywitz & Greenberg, 1999; Lonze & Ginty, 2002). Phosphodiesterase type IV (PDE4), which degrades cAMP, can also regulate CREB-dependent transcription. Dynamic regulation of these signaling pathways, stimulating and antagonizing CREB activity, might fine tune the process that allocates memories in neuronetworks.
The CREB family of transcription factors comprise CREB, CREM (cAMP response element modulatory protein) and ATF-1 (activating transcription factor 1), which can form both homo- and heterodimers to bind to the same cis-regulatory element, cAMP response element (CRE), a sequence identified in the promoters of many inducible genes (De Cesare et al. 1999; Mayr and Montminy 2001; Shaywitz and Greenberg 1999). CREB and CREM genes can be alternatively spliced to encode both transcriptional activators and repressors (Foulkes, Borrelli, & Sassone-Corsi, 1991; Walker, Girardet, & Habener, 1996). In addition, ATF-4 (CREB-2), an unconventional member of the CREB family, has been reported to negatively regulate CRE-mediated transcription and long-term memory (Bartsch et al., 1995; Chen et al., 2003). Thus, CREB-mediated memory allocation could be regulated at the level of alternative splicing of CREB family members as well as by their physical interactions and competition for binding sites on target promoters.
Multiple lines of evidence indicate that epigenetic alterations, including DNA methylation and histone modifications are actively engaged in neural plasticity, learning, and memory via regulation of gene expression critical for these processes (Levenson & Sweatt, 2005; Feng, Fouse, & Fan, 2007). In resting neurons, neural plasticity genes, many of which are direct targets of CREB (e.g., BDNF), are associated with more inactive chromatin structures, in which histones are deacetylated or methylated on certain lysine residues (e.g., lysine 9 of histone H3) and/or DNA is more methylated (Martinowich et al., 2003; Chen et al., 2003). Upon induction of neural plasticity, calcium signaling activates the CREB kinase RSK2, CREB, and CBP. These events lead to chromatin remodeling and to a more open chromatin structure that allows for long-lasting expression of plasticity genes and consequently to long-term memory storage. Recent studies have demonstrated that increased histone acetylation, caused by environmental enrichment or by inhibitors of histone deacetylases (HDACs), induce sprouting of dendrites, an increase in the number of synapses and increased access to long-term memories (Fischer, Sananbenesi, Wang, Dobbin, & Tsai, 2007). Moreover, a recent study has shown that HDAC inhibitors enhance memory processes by the activation of key genes regulated by the CREB-CBP transcriptional complex (Alarcon et al., 2004; Korzus et al., 2004; Vecsey et al., 2007). HDAC inhibitors seem to potentiate CREB activity by prolonging serine 133 phosphorylation in response to cAMP stimuli, suggesting a potential role for HDAC complexes in silencing CREB activity (Canettieri et al., 2003).
Promoters harboring CRE sites are subject to combinatorial regulation by CREB and other transcription factors and coactivators, which themselves are under control of various signaling pathways. Moreover, their transcriptional activities are influenced by nearby chromatin structure. Therefore, integration of multiple signals can occur in the context of CREB target genes, which themselves could control memory allocation. This perspective illustrates a novel mechanism by which diverse signaling and chromatin-modifying activities act coordinately to dynamically allocate memories in neuronetworks.
5. Metaplasticity in memory allocation
It is possible that the acquisition of a memory changes the activity of CREB (activation followed by repression due to the transcription of CREB repressors such as inducible cAMP early repressor, ICER), which then decreases the probability that the cells engaged in the first memory participate in a second memory some time later. Memories created very close in time are a special challenge because of the high likelihood that there will be common attributes and overlapping contexts. Dynamic memory allocation to different sets of neurons may increase capacity for, and decrease interference between, the encoding of these multiple distinct attributes that together constitute an epoch. These considerations support the existence of a form of ‘metaplasticity’, by which predisposition of neurons to participate in a memory trace can be dynamically adjusted according to the history of neuronal activity (Abraham & Bear, 1996), thus resulting in the effective separation of distinct memories.
Besides CREB and related transcriptional processes that could serve to separate memories by transcribing inhibitors (requiring tens of minutes), there may be other processes that could affect memory allocation more quickly. For example, feedback inhibition in neuronal circuits could affect the allocation of two subsequent memories within a given episode by immediately decreasing the probability that cells engaged by one aspect of an episode, are again recruited into encoding a closely related aspect of the same episode seconds/minutes later.
Recently, Guzowski et al. demonstrated that the coupling between cell firing and Arc transcription, which is required for memory consolidation (Guzowski et al., 2000), is plastic, not static, because it is influenced strongly by recent behavioral history (Guzowski et al., 2006). They showed that the number of Arc-positive CA1 neurons in the hippocampus decreased dramatically in rats exposed repeatedly to an environment (25 min between exposures in a single day), although the firing properties of CA1 neurons did not change across these repeated sessions. Intriguingly, if after repeated exposures to the same environment rats were exposed to a novel environment, the percentage of Arc-positive CA1 neurons was that predicted if the reduction of Arc transcriptional responsiveness was limited to the cell population repeatedly activated in a repeatedly exposed environment (Guzowski et al., 2006). These results indicate that the altered association and Arc transcription observed with repeated exposures is cell and experience specific and not a generalized inhibition of Arc transcription in all CA1 neurons. But, what could be the underlying mechanisms for this inhibition?
It is possible that cell-intrinsic oscillating feedback loops control the intracellular levels of CREB and thereby modulate dynamically memory allocation in neuronetworks. Studies of the CREB transcriptome suggest the existence of a negative feedback loop under transcriptional control (Fass, Butler, & Goodman, 2003; Impey et al., 2004). For example, one of the genes most highly induced by activation of CREB is ICER, which is a potent inhibitor of CREB function (De Cesare & Sassone-Corsi, 2000; Fass, Butler, & Goodman, 2003). In addition, calcium-dependent activation of protein phosphatases PP1 and PP2A, which leads to the dephosphorylation of CREB at serine 133, might also contribute to this intracellular negative feedback loop (Shaywitz & Greenberg, 1999; Lonze & Ginty, 2002).
6. Reconsolidating the allocation of stored memories
Transgenic studies with inducible CREB mice showed that CREB plays a key role in the reconsolidation as well as consolidation of memory (Kida et al., 2002). It would be exciting to examine whether reconsolidation, just as consolidation, involves the re-allocation of memories, and whether CREB plays a role in this process. The levels and activities of CREB in each neuron might differ dramatically during acquisition and retrieval. Therefore, reactivation of memory circuits during retrieval and subsequent reconsolidation could alter the set of neurons dedicated to the storage of a particular memory. Putative memory reallocation processes could have an important role in the slow reorganization of cortical-dependent remote memories, where fine-tuning storage sites may underlie the emergence of statistical regularities underlying semantic memory (Frankland & Bontempi, 2005). It is conceivable that memory relocation processes play a role during the prolonged periods required to consolidate memories in the neocortex, when these memories are thought to be interleaved with previous related memories into integrated semantic-like representations. New memories may force the relocation of previous related memories so that the two are seamlessly integrated within neocortical networks. In another words, memory allocation and memory reconsolidation processes may work together to generate semantic-like integrated knowledge structures in neocortical networks.
7. Concluding remarks
Recent findings show that competition between neurons, which has been demonstrated to be necessary for refining neural circuits during development, may be important for selecting the neurons that participate in encoding memories in the adult brain. They also suggest that CREB mediates the competition between neuronal cells that leads to the formation of memory traces. Yet, there are both competing as well as cooperating pathways regulating CREB activity in neurons (Shaywitz & Greenberg, 1999; Mayr & Montminy 2001; Lonze & Ginty, 2002; Carlezon, Duman, & Nestler, 2005) and both of these could also affect memory allocation. Thus, the many dynamic signaling processes that converge on CREB could play a role in modulating and fine-tuning where memories are stored in neuronetworks.
Much remains to be done regarding the molecular and cellular basis of memory allocation processes. Identification of critical CREB target genes and the mechanism(s) by which their expressed products control competitive memory allocation is a key goal for future studies. It also remains to be determined whether CREB plays a role in the allocation of memory in brain regions other than the amygdala. For example, it was shown that ∼40% of CA1 hippocampal neurons are recruited during spatial learning (Guzowski et al., 2006) and it would be of interest to examine whether CREB-mediated competition also affects which CA1 neurons encode a given spatial memory. In addition, the mechanism(s) by which neurons with higher CREB activity keep other neurons from participating in a given memory trace will be undoubtedly the target of future studies.
The studies reviewed above are the first of what will definitely be an exciting new line of research probing the molecular and cellular mechanisms that determine the addresses of memories in neuronetworks. The combination of approaches that made this study possible represent a new trend in the study of memory where powerful new tools are allowing us to probe deeper into the mechanisms that process and store information in the brain.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abraham WC, Bear MF. Metaplasticity: the plasticity of synaptic plasticity. Trends in Neurosciences. 1996;19:126–130. doi: 10.1016/s0166-2236(96)80018-x. [DOI] [PubMed] [Google Scholar]
- Alarcon JM, Malleret G, Touzani K, Vronskaya S, Ishii S, Kandel ER, Barco A. Chromatin acetylation, memory, and LTP are impaired in CBP+/- mice: a model for the cognitive deficit in Rubinstein-Taybi syndrome and its amelioration. Neuron. 2004;42:947–959. doi: 10.1016/j.neuron.2004.05.021. [DOI] [PubMed] [Google Scholar]
- Bartsch D, Casadio A, Karl KA, Serodio P, Kandel ER. CREB1 encodes a nuclear activator, a repressor, and a cytoplasmic modulator that form a regulatory unit critical for long-term facilitation. Cell. 1998;95:211–223. doi: 10.1016/s0092-8674(00)81752-3. [DOI] [PubMed] [Google Scholar]
- Bartsch D, Ghirardi M, Skehel PA, Karl KA, Herder SP, Chen M, Bailey CH, Kandel ER. Aplysia CREB2 represses long-term facilitation: relief of repression converts transient facilitation into long-term functional and structural change. Cell. 1995;83:979–992. doi: 10.1016/0092-8674(95)90213-9. [DOI] [PubMed] [Google Scholar]
- Cabelli RJ, Hohn A, Shatz CJ. Inhibition of ocular dominance column formation by infusion of NT-4/5 or BDNF. Science. 1995;67:1662–1666. doi: 10.1126/science.7886458. [DOI] [PubMed] [Google Scholar]
- Carlezon WA, Jr., Duman RS, Nestler EJ. The many faces of CREB. Trends in Neurosciences. 2005;28:436–445. doi: 10.1016/j.tins.2005.06.005. [DOI] [PubMed] [Google Scholar]
- Canettieri G, Morantte I, Guzman E, Asahara H, Herzig S, Anderson SD, Yates JR, 3rd., Montminy M. Attenuation of a phosphorylation-dependent activator by an HDAC-PP1 complex. Nature Structural Biology. 2003;10:175–181. doi: 10.1038/nsb895. [DOI] [PubMed] [Google Scholar]
- Cha-Molstad H, Keller DM, Yochum GS, Impey S, Goodman RH. Cell-type-specific binding of the transcription factor CREB to the cAMP-response element. Proceedings of the National Academy of Sciences United States of America. 2004;101:13572–13577. doi: 10.1073/pnas.0405587101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen A, Muzzio IA, Malleret G, Bartsch D, Verbitsky M, Pavlidis P, Yonan AL, Vronskaya S, Grody MB, Cepeda I, Gilliam TC, Kandel ER. Inducible enhancement of memory storage and synaptic plasticity in transgenic mice expressing an inhibitor of ATF4 (CREB-2) and C/EBP proteins. Neuron. 2003;39:655–669. doi: 10.1016/s0896-6273(03)00501-4. [DOI] [PubMed] [Google Scholar]
- Chen WG, Chang Q, Lin Y, Meissner A, West AE, Griffith EC, Jaenisch R, Greenberg ME. Derepression of BDNF transcription involves calcium-dependent phosphorylation of MeCP2. Science. 2003;302:885–889. doi: 10.1126/science.1086446. [DOI] [PubMed] [Google Scholar]
- De Cesare D, Sassone-Corsi P. Transcriptional regulation by cyclic AMP-responsive factors. Progress in Nucleic Acid Research and Molecular Biology. 2000;64:343–369. doi: 10.1016/s0079-6603(00)64009-6. [DOI] [PubMed] [Google Scholar]
- Dong Y, Green T, Saal D, Marie H, Neve R, Nestler EJ, Malenka RC. CREB modulates excitability of nucleus accumbens neurons. Nature Neuroscience. 2006;9:475–477. doi: 10.1038/nn1661. [DOI] [PubMed] [Google Scholar]
- Egger G, Liang G, Aparicio A, Jones PA. Epigenetics in human disease and prospects for epigenetic therapy. Nature. 2004;429:457–463. doi: 10.1038/nature02625. [DOI] [PubMed] [Google Scholar]
- Fanselow MS, Gale GD. The amygdala, fear, and memory. Annals of the New York Academy of Sciences. 2003;985:125–134. doi: 10.1111/j.1749-6632.2003.tb07077.x. [DOI] [PubMed] [Google Scholar]
- Fass DM, Butler JE, Goodman RH. Deacetylase activity is required for cAMP activation of a subset of CREB target genes. Journal of Biological Chemistry. 2003;278:43014–43019. doi: 10.1074/jbc.M305905200. [DOI] [PubMed] [Google Scholar]
- Feng J, Fouse S, Fan G. Epigenetic regulation of neural gene expression and neuronal function. Pediatric Research. 2007;61:58R–63R. doi: 10.1203/pdr.0b013e3180457635. [DOI] [PubMed] [Google Scholar]
- Fischer A, Sananbenesi F, Wang X, Dobbin M, Tsai LH. Recovery of learning and memory is associated with chromatin remodelling. Nature. 2007;447:178–182. doi: 10.1038/nature05772. [DOI] [PubMed] [Google Scholar]
- Fonseca R, Nagerl UV, Morris RG, Bonhoeffer T. Competing for memory: hippocampal LTP under regimes of reduced protein synthesis. Neuron. 2004;44:1011–1020. doi: 10.1016/j.neuron.2004.10.033. [DOI] [PubMed] [Google Scholar]
- Foulkes NS, Borrelli E, Sassone-Corsi P. CREM gene: use of alternative DNA-binding domains generates multiple antagonists of cAMP-induced transcription. Cell. 1991;64:739–749. doi: 10.1016/0092-8674(91)90503-q. [DOI] [PubMed] [Google Scholar]
- Frankland PW, Bontempi B. The organization of recent and remote memories. Nature Reviews Neuroscience. 2005;6:119–130. doi: 10.1038/nrn1607. [DOI] [PubMed] [Google Scholar]
- Gonzalez GA, Montminy MR. Cyclic AMP stimulates somatostatin gene transcription by phosphorylation of CREB at serine 133. Cell. 1989;59:675–680. doi: 10.1016/0092-8674(89)90013-5. [DOI] [PubMed] [Google Scholar]
- Guzowski JF, McNaughton BL, Barnes CA, Worley PF. Environment-specific expression of the immediate-early gene Arc in hippocampal neuronal ensembles. Nature Neuroscience. 1999;2:1120–1124. doi: 10.1038/16046. [DOI] [PubMed] [Google Scholar]
- Guzowski JF, Lyford GL, Stevenson GD, Houston FP, McGaugh JL, Worley PF, Barnes CA. Inhibition of activity-dependent arc protein expression in the rat hippocampus impairs the maintenance of long-term potentiation and the consolidation of long-term memory. Journal of Neuroscience. 2000;20:3993–4001. doi: 10.1523/JNEUROSCI.20-11-03993.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzowski JF, Miyashita T, Chawla MK, Sanderson J, Maes LI, Houston FP, Lipa P, McNaughton BL, Worley PF, Barnes CA. Recent behavioral history modifies coupling between cell activity and Arc gene transcription in hippocampal CA1 neurons. Proceedings of the National Academy of Sciences United States of America. 2006;103:1077–1082. doi: 10.1073/pnas.0505519103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han JH, Kushner SA, Yiu AP, Cole CJ, Matynia A, Brown RA, Neve RL, Guzowski JF, Silva AJ, Josselyn SA. Neuronal competition and selection during memory formation. Science. 2007;316:457–460. doi: 10.1126/science.1139438. [DOI] [PubMed] [Google Scholar]
- Han MH, Bolanos CA, Green TA, Olson VG, Neve RL, Liu RJ, Aghajanian GK, Nestler EJ. Role of cAMP response element-binding protein in the rat locus ceruleus: regulation of neuronal activity and opiate withdrawal behaviors. Journal of Neuroscience. 2006;26:4624–4629. doi: 10.1523/JNEUROSCI.4701-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Impey S, McCorkle SR, Cha-Molstad H, Dwyer JM, Yochum GS, Boss JM, McWeeney S, Dunn JJ, Mandel G, Goodman RH. Defining the CREB regulon: a genome-wide analysis of transcription factor regulatory regions. Cell. 2004;119:1041–1054. doi: 10.1016/j.cell.2004.10.032. [DOI] [PubMed] [Google Scholar]
- Jasnow AM, Shi C, Israel JE, Davis M, Huhman KL. Memory of social defeat is facilitated by cAMP response element-binding protein overexpression in the amygdala. Behavioral Neuroscience. 2005;119:1125–1130. doi: 10.1037/0735-7044.119.4.1125. [DOI] [PubMed] [Google Scholar]
- Josselyn SA, Shi C, Carlezon WA, Jr., Neve RL, Nestler EJ, Davis M. Long-term memory is facilitated by cAMP response element-binding protein overexpression in the amygdala. Journal of Neuroscience. 2001;21:2404–2012. doi: 10.1523/JNEUROSCI.21-07-02404.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kida S, Josselyn SA, de Ortiz SP, Kogan JH, Chevere I, Masushige S, Silva AJ. CREB required for the stability of new and reactivated fear memories. Nature Neuroscience. 2002;5:348–355. doi: 10.1038/nn819. [DOI] [PubMed] [Google Scholar]
- LeDoux JE. Emotion circuits in the brain. Annual Review of Neuroscience. 2000;23:155–184. doi: 10.1146/annurev.neuro.23.1.155. [DOI] [PubMed] [Google Scholar]
- Levenson JM, Sweatt JD. Epigenetic mechanisms in memory formation. Nature Reviews Neuroscience. 2005;6:108–118. doi: 10.1038/nrn1604. [DOI] [PubMed] [Google Scholar]
- Lonze BE, Ginty DD. Function and regulation of CREB family transcription factors in the nervous system. Neuron. 2002;35:605–623. doi: 10.1016/s0896-6273(02)00828-0. [DOI] [PubMed] [Google Scholar]
- Marie H, Morishita W, Yu X, Calakos N, Malenka RC. Generation of silent synapses by acute in vivo expression of CaMKIV and CREB. Neuron. 2005;45:741–752. doi: 10.1016/j.neuron.2005.01.039. [DOI] [PubMed] [Google Scholar]
- Mayr B, Montminy M. Transcriptional regulation by the phosphorylation-dependent factor CREB. Nature Reviews Molecular Cell Biology. 2001;2:599–609. doi: 10.1038/35085068. [DOI] [PubMed] [Google Scholar]
- Malenka RC, Bear MF. LTP and LTD: an embarrassment of riches. Neuron. 2004;44:5–21. doi: 10.1016/j.neuron.2004.09.012. [DOI] [PubMed] [Google Scholar]
- Martinowich K, Hattori D, Wu H, Fouse S, He F, Hu Y, Fan G, Sun YE. DNA methylation-related chromatin remodeling in activity-dependent BDNF gene regulation. Science. 2003;302:890–893. doi: 10.1126/science.1090842. [DOI] [PubMed] [Google Scholar]
- Matthews RP, Guthrie CR, Wailes LM, Zhao X, Means AR, McKnight GS. Calcium/calmodulin-dependent protein kinase types II and IV differentially regulate CREB-dependent gene expression. Molecular and Cellular Biology. 1994;14:6107–6116. doi: 10.1128/mcb.14.9.6107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClung CA, Nestler EJ. Regulation of gene expression and cocaine reward by CREB and DeltaFosB. Nature Neuroscience. 2003;6:1208–1215. doi: 10.1038/nn1143. [DOI] [PubMed] [Google Scholar]
- McGaugh JL. The search for the memory trace. Annual Review of Neuroscience. 1972;193:112–123. doi: 10.1111/j.1749-6632.1972.tb27828.x. [DOI] [PubMed] [Google Scholar]
- Miller KD. Synaptic economics: competition and cooperation in synaptic plasticity. Neuron. 1996;17:371–374. doi: 10.1016/s0896-6273(00)80169-5. [DOI] [PubMed] [Google Scholar]
- Mower AF, Liao DS, Nestler EJ, Neve RL, Ramoa AS. cAMP/Ca2+ response element-binding protein function is essential for ocular dominance plasticity. Journal of Neuroscience. 2002;22:2237–2245. doi: 10.1523/JNEUROSCI.22-06-02237.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham TA, Impey S, Storm DR, Stryker MP. CRE-mediated gene transcription in neocortical neuronal plasticity during the developmental critical period. Neuron. 1999;22:63–72. doi: 10.1016/s0896-6273(00)80679-0. [DOI] [PubMed] [Google Scholar]
- Pham TA, Rubenstein JL, Silva AJ, Storm DR, Stryker MP. The CRE/CREB pathway is transiently expressed in thalamic circuit development and contributes to refinement of retinogeniculate axons. Neuron. 2001;31:409–420. doi: 10.1016/s0896-6273(01)00381-6. [DOI] [PubMed] [Google Scholar]
- Pham TA, Graham SJ, Suzuki S, Barco A, Kandel ER, Gordon B, Lickey ME. A semi-persistent adult ocular dominance plasticity in visual cortex is stabilized by activated CREB. Learning & Memory. 2004;11:738–747. doi: 10.1101/lm.75304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Repa JC, Muller J, Apergis J, Desrochers TM, Zhou Y, LeDoux JE. Two different lateral amygdala cell populations contribute to the initiation and storage of memory. Nature Neuroscience. 2001;4:724–731. doi: 10.1038/89512. [DOI] [PubMed] [Google Scholar]
- Rumpel S, LeDoux J, Zador A, Malinow R. Postsynaptic receptor trafficking underlying a form of associative learning. Science. 2005;308:83–88. doi: 10.1126/science.1103944. [DOI] [PubMed] [Google Scholar]
- Shaywitz AJ, Greenberg ME. CREB: a stimulus-induced transcription factor activated by a diverse array of extracellular signals. Annual Review of Biochemistry. 1999;68:821–861. doi: 10.1146/annurev.biochem.68.1.821. [DOI] [PubMed] [Google Scholar]
- Sheng M, Thompson MA, Greenberg ME. CREB: a Ca(2+)-regulated transcription factor phosphorylated by calmodulin-dependent kinases. Science. 1991;252:1427–1430. doi: 10.1126/science.1646483. [DOI] [PubMed] [Google Scholar]
- Silva AJ, Kogan JH, Frankland PW, Kida S. CREB and memory. Annual Review of Neuroscience. 1998;21:127–148. doi: 10.1146/annurev.neuro.21.1.127. [DOI] [PubMed] [Google Scholar]
- Thompson RF. In search of memory traces. Annual Review of Psychology. 2005;56:1–23. doi: 10.1146/annurev.psych.56.091103.070239. [DOI] [PubMed] [Google Scholar]
- Tzingounis AV, Nicoll RA. Arc/Arg3.1: linking gene expression to synaptic plasticity and memory. Neuron. 2006;52:403–407. doi: 10.1016/j.neuron.2006.10.016. [DOI] [PubMed] [Google Scholar]
- Vecsey CG, Hawk JD, Lattal KM, Stein JM, Fabian SA, Attner MA, Cabrera SM, McDonough CB, Brindle PK, Abel T, Wood MA. Histone deacetylase inhibitors enhance memory and synaptic plasticity via CREB: CBP-dependent transcriptional activation. Journal of Neuroscience. 2007;27:6128–6140. doi: 10.1523/JNEUROSCI.0296-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vo N, Goodman RH. CREB-binding protein and p300 in transcriptional regulation. Journal of Biological Chemistry. 2001;276:13505–13508. doi: 10.1074/jbc.R000025200. [DOI] [PubMed] [Google Scholar]
- Walker WH, Girardet C, Habener JF. Alternative exon splicing controls a translational switch from activator to repressor isoforms of transcription factor CREB during spermatogenesis. Journal of Biological Chemistry. 1996;271:20145–21050. [PubMed] [Google Scholar]
- Wallace TL, Stellitano KE, Neve RL, Duman RS. Effects of cyclic adenosine monophosphate response element binding protein overexpression in the basolateral amygdala on behavioral models of depression and anxiety. Biological Psychiatry. 2004;56:151–160. doi: 10.1016/j.biopsych.2004.04.010. [DOI] [PubMed] [Google Scholar]
- West AE, Griffith EC, Greenberg ME. Regulation of transcription factors by neuronal activity. Nature Reviews Neuroscience. 2002;3:921–931. doi: 10.1038/nrn987. [DOI] [PubMed] [Google Scholar]
- Wiesel TN, Hubel DH. Comparison of the effects of unilateral and bilateral eye closure on cortical unit responses in kittens. Journal of Neurophysiology. 1965;28:1029–1040. doi: 10.1152/jn.1965.28.6.1029. [DOI] [PubMed] [Google Scholar]
- Wilson MA, McNaughton BL. Dynamics of the hippocampal ensemble code for space. Science. 1993;261:1055–1058. doi: 10.1126/science.8351520. [DOI] [PubMed] [Google Scholar]
- Wu X, McMurray CT. Calmodulin kinase II attenuation of gene transcription by preventing cAMP response element-binding protein (CREB) dimerization and binding of the CREB-binding protein. Journal of Biological Chemistry. 2001;276:1735–1741. doi: 10.1074/jbc.M006727200. [DOI] [PubMed] [Google Scholar]
- Yin JC, Del Vecchio M, Zhou H, Tully T. CREB as a memory modulator: induced expression of a dCREB2 activator isoform enhances long-term memory in Drosophila. Cell. 1995;81:107–115. doi: 10.1016/0092-8674(95)90375-5. [DOI] [PubMed] [Google Scholar]
- Zhang X, Odom DT, Koo SH, Conkright MD, Canettieri G, Best J, Chen H, Jenner R, Herbolsheimer E, Jacobsen E, Kadam S, Ecker JR, Emerson B, Hogenesch JB, Unterman T, Young RA, Montminy M. Genome-wide analysis of cAMP-response element binding protein occupancy, phosphorylation, and target gene activation in human tissues. Proceedings of the National Academy of Sciences United States of America. 2005;102:4459–4464. doi: 10.1073/pnas.0501076102. [DOI] [PMC free article] [PubMed] [Google Scholar]