Summary of Recent Advances
Single-molecule investigations promise to greatly advance our understanding of basic and regulated ribosome functions during the process of translation. Here, recent progress towards directly imaging the elemental translation elongation steps using fluorescence resonance energy transfer (FRET)-based imaging methods is discussed, which provide striking evidence of the highly dynamic nature of the ribosome. In this view, global rates and fidelities of protein synthesis reactions may be regulated by interactions of the ribosome with mRNA, tRNA, translation factors, and potentially many other cellular ligands, that modify intrinsic conformational equilibria in the translating particle. Future investigations probing this model must aim to visualize translation processes from multiple structural and kinetic perspectives simultaneously, to provide direct correlations between factor binding and conformational events.
Introduction
Here, recent single-molecule investigations of the complex biochemical system supporting protein synthesis are reviewed and discussed in the context of broader considerations in the translation and single-molecule imaging fields. To date, investigations have focused on the central component of the cell’s protein synthesis machinery, the ribosome, as it is the integration point for regulatory control of translation [1]. The ribosome is a structurally and functionally conserved two-subunit, ~3 megadalton (MDa) molecular assembly composed of ~70 distinct RNA and protein gene products. This RNA-rich enzyme works collaboratively with other cellular RNAs and proteins to convert mRNA-encoded genetic information into a specific polypeptide according to the messenger RNA sequence.
For the past two decades, beginning with atomic resolution structures of bacterial ribosomal subunits and more recently intact and functional ribosome complexes, characterizations of the physical architecture of the ribosome have taken center stage [2–4]. These landmark accomplishments provide the first views of the organization of components within the ribosome and unprecedented insights into the protein synthesis mechanism. Consequently, we now understand in molecular detail the determinants of each of the ribosome’s three transfer RNA (tRNA) binding sites –Aminoacyl (A), Peptidyl (P) and Exit (E) –as well as the interaction sites for specific translation factors, and distinct ribosome-targeting antibiotics. Yet even as breakthroughs on this front continue, questions remain as to how static views of the ribosome architecture are reconciled with the dynamic process of translation. Structures of the ribosome represent discrete local minima, or the set of microstates within local minima, on a complex free energy landscape [2,5,6]. At least five global conformational degrees of freedom have been experimentally identified in the ribosome –many more in translation factors –that are directly implicated in the translation mechanism •[7]. A deeper understanding of translation mechanism therefore hinges upon strategies enabling the delineation of the order and timing of conformational transitions in the translation apparatus and how sub-states visited along the reaction coordinate are rapidly and efficiently transited to achieve the faithful conversion of the genetic code into protein.
Single-molecule investigations of the bacterial translation apparatus were underway contemporaneous with reports of the first structures of ribosomal subunits [8–10]. These studies capitalized on emerging methods for room-temperature, aqueous investigations of conformational processes in enzymes pioneered in the context of relatively simple RNA, DNA and protein systems [11,12]. This work set the stage for recent investigations that have yielded remarkable new mechanistic insights into the basic and regulated aspects of ribosome and factor functions. In this review, specific focus is given to 4 papers published since 2007 that shed new light on a range of nanometer-scale motional and conformational processes underpinning the translation reaction coordinate. These studies are certain to shape many future experiments in this emerging field. Discussion will framed in the context of essential technological foundations ••[13–16] as well as outstanding questions raised by this research.
Why single-molecule imaging? Observing single ribosomes performing translation functions is a sharp departure from biochemical, genetic and biophysical methods where much of our understanding of the process of translation has been gained (Box 1). Through traditional techniques, translation activities are monitored by the rates of product (polypeptide) or by-product (GDP and Pi) formation. Motivated by cryo-electron microscopy (cryo-EM) evidence that dynamic processes in the ribosome mediate discrete translation steps [5] •[17], significant effort has been recently given to measuring inter- and intra-molecular motions between the ribosome, tRNAs, mRNA and translation factors during protein synthesis.
Box 1: The process of translation is replete with motion
The process of translation has been extensively investigated for the past half century [36]. The conversion of genetic information in the form of messenger RNA (mRNA) into protein begins with the assembly of both intact ribosomal subunits (30S and 50S in bacteria) into the functional (70S) particle at the start site of protein synthesis. Choice of the appropriate start site, a major determinant of translation fidelity, is facilitated by protein factors, initiator tRNA, and GTP hydrolysis. Localization of the ribosome to the mRNA start site is also mediated by direct, Shine-Dalgarno (S-D) type base pairing interactions between the small subunit ribosomal RNA as well as ribosomal protein S1.
The ribosome’s directional transit through the mRNA open reading frame (ORF), elongation, proceeds via sequential aminoacyl-tRNA (aa-tRNA) selection and translocation processes catalyzed by the GTPases Elongation Factors (EF-Tu and –G), respectively. EF-Tu and -G bind non-competitively to overlapping sites at the leading edge of the ribosome and hydrolyze GTP while bound to the particle. These reactions occur in rapid succession to drive amino acid polymerization according to the mRNA codon sequence. Both EF-Tu-catalyzed selection of specific aa-tRNA substrates at the A site and EF-G-catalyzed translocation are major fidelity determinants. aa-tRNA selection is a multistep, kinetically controlled reaction mediated by an interplay of EF-Tu, tRNA and ribosomal components and terminates with accommodation of the 3′-aminoacyl end of aa-tRNA at the Peptidyltransferase Center (PTC). Upon entering the PTC, peptide bond formation occurs rapidly, deacylating P-site tRNA and generating a nascent peptide on the newly incorporated tRNA. EF-G-catalyzed translocation, the movement of both A and P-site substrates ~30Å with respect to the ribosome, is also a multistep process. Faithful translocation ensures maintenance of the correct reading frame, vacates the A site for a new round of aa-tRNA selection and allows deacylated tRNA to dissociate from the E site. Despite much progress, the order and timing of structural events in the ribosome during elongation remains unclear.
Traditional, bulk biophysical investigations of such processes are often limited by the intrinsically dynamic nature of translation. Here, the interpretation of kinetic data may break down if conformational states of the ribosome interconvert in a highly asynchronous fashion or if the reaction endpoints are themselves highly dynamic or unstable. In such cases, ensemble-averaged information is obtained that may potentially mask important mechanistic information. These factors often thwart the reliable interpretation of bulk data and the generation of high-resolution structures, which both depend on the ribosome having a discrete number of conformationally distinct states separated by relatively large activation barriers. Crystallography efforts, in particular, demand both compositional and structural uniformity. Particle sorting algorithms implemented in cryo-EM investigations may at least partially circumnavigate these issues by separating particles in the ensemble with clear conformational or compositional distinctions [18–20].
By contrast, single-molecule investigations bypass the need compositionally and structurally uniform samples as well as synchronous behavior in the system. The optical detection of single molecules, which are smaller than the wavelength of visible light, requires that one or more components of the system be linked to fluorescent probes or particles large enough to scatter light. Single-molecule fluorescence resonance energy transfer (smFRET) imaging has proven a particularly powerful approach for directly observing dynamic processes in the ribosome. Here, excitation of a donor fluorophore attached directly to the biological sample may result in energy transfer to nearby acceptor fluorophores if spatially proximal (20–80Å). Given fast, isotropic rotational sampling of the fluorophores during the observation period, the efficiency of this dipole-dipole interaction reports on the distance between the two fluorophores according to the Förster relationship (Box 2). In the ideal case, FRET-based measurements yield the equivalent of a “molecular EKG” reporting on conformational processes in the system. Both stable and transient states are identified by unique structural and/or kinetic fingerprints[21]. Transitions between states, generally occurring faster (ca. >1ms) than the time scale of typical imaging capabilities, reveals the directionality of structural events. Mechanistic information regarding the underlying conformational event itself may therefore be lacking. Fast imaging systems have great potential to reveal insights in this area [22].
Box 2
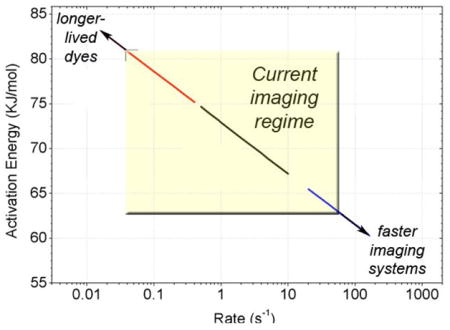
The Förster relationship defines the efficiency of energy transfer (E) between donor and acceptor fluorophore pairs where E = 1/(1 + (R/R0)6). Here, R is the distance between the fluorophores and R0 is the distance at which 50% energy transfer occurs. The R0 value, specific for each dye pair (typically ~50–60Å for single-molecule fluorophores such as Cy3/5 and Alexa555/647), is a function of the spectral overlap integral (J), dye mobility (κ ), and the refractive index (n) according to the equation: R0 = 0.211(κ2n−4QDJ (λ))1/6. FRET efficiency is experimentally determined from the ratio of acceptor fluorescence to total fluorescence, E = Iacceptor/(Iacceptor + Idonor) assuming similar photon collection efficiencies are achieved for both frequencies. The lifetimes of specific states (τ) prior to a conformation transition are defined computationally using thresholding or idealization procedures[14]. Inverse lifetime (1/τ), defines the rate leaving that state and reports on the activation energy for the underlying process observed according to the equation: Δ Gij‡ = − RTln(hkij/kBT), where kij is the rate of transition from state i to state j, h is Planck’s constant, kB is Boltzman’s constant, R is the gas constant, and T is the temperature[53]. The relationship between rates and activation energies (Figure insert) is such that tRNA motions and subunit ratcheting within the pre-translocation ribosome complex [33,35] are readily observed at time scales amenable present-day CCD technologies (~0.1–25 s−1) and typical fluorophore photobleaching rates (~10–30 sec). Higher and lower activation energy processes necessitate longer and shorter observation times, respectively as indicated by the arrows shown at the corners of the highlighted “current imaging regime” window.
The ribosome has proven an ideal biological system for obtaining FRET trajectories of this nature as the barriers for specific conformational transitions are generally large (ca. >60KJ.mol) and can be attenuated by increased magnesium ion concentrations [10,23]. Distinct conformational states of the ribosome can therefore be observed even at the finite time resolutions of present-day imaging technologies (Box 2). Using wide-field, charged coupled device (CCD) systems, data from large ensembles of single ribosome complexes (>1000) can be collected in minutes. Faster time scale, confocal imaging methods, achieved using avalanche photodiodes (APD), require much longer overall acquisition times for similar achievements [14]. In practice, both CCD and APD measurements are needed to understand fully the time scales of conformational events. The characterization of kinetic and/or structural behaviors of single-molecules is afforded by long-lived and stable fluorophore performance. Large observation numbers offer the potential to access the full distribution of kinetic and structural pathways in the biological system under investigation. Further insights into dispersed kinetic behaviors arising from compositional or functional heterogeneities or intrinsic dynamic disorder requires the extraction of rates for specific transitions for each single molecule in the ensemble ••[24] [25–28]. Here, a suite of computational tools is paramount to obtaining complete mechanistic information as individual molecules may traverse many distinct kinetic and structural pathways navigating through potentially complex reaction coordinates [28–30].
Technological considerations in single-molecule imaging of translation
Elemental protein synthesis reactions are rapid (ca 100ms) whereas synthesis of a complete protein requires minutes. Imaging translation processes on individual particles therefore requires sample immobilization strategies. Using wide-field imaging methods, hundreds of fluorescently-labeled translation components spatially-immobilized proximal to optically transparent surfaces can be illuminated simultaneously by the evanescent wave generated by total internal reflection (TIR) and observed over extended periods [6,16]. A number of practical solutions for biomolecule surface immobilization have now been achieved [14,16] and direct measurements of sub-nanometer scale conformational events on the ribosome have been obtained on rapid time scales (25–100ms) over the course of minutes.
Parlaying earlier breakthroughs [31,32], several groups have introduced fluorophores into the ribosome using tRNA molecules, chemically coupled to fluorophores via naturally occurring modified nucleotides. Bound to the ribosome, fluorescently-labeled tRNAs are sensitive amplifiers of ribosome conformational change, providing early insights into both the aa-tRNA selection process and tRNA motions within the ribosome preceding translocation [9,10]. New strategies for direct ribosome labeling have also been achieved, including the incorporation of single fluorescent proteins into the small ribosomal subunit via in vitro ribosome reconstitution strategies, genetic depletion of the ribosome of single proteins followed by biochemical reconstitution, and oligonucleotide hybridization to ribosomal RNA. Hybridization strategies may also be used for surface immobilization of the ribosome. To varying degrees, each of the FRET-based imaging studies discussed here have benefitted from enhanced conditions for fluorophore performance and the implementation of new data analysis procedures [14,16].
The ribosome has an intrinsic capacity to achieve functionally-relevant conformations
Employing distinct labeling methods, four groups using smFRET-based methods have investigated conformational processes in the ribosome from unique structural perspectives. Corroborating earlier reports [10,23], data from three of these groups [33–35] showed that conformational transitions related to the translocation reaction coordinate are thermally accessible. Sampling of functionally-relevant states in the absence of energy expenditure is consistent with early studies of template and factor-free protein synthesis [36] and suggestions that intrinsic conformational dynamics serves a central evolutionary pressure in the structure-function relationship of many, if not all, biomolecules ••[37].
Munro et al. ••[35] showed using fluorescently-labeled tRNA molecules bound at the A- and P-site that the pre-translocation ribosome dynamically samples three kinetically and structurally distinct FRET states at rates (ca. 2.5–10/sec) exceeding the rate of EF-G catalyzed translocation (1.5/sec) (Figure 1). Structural assignments of the observed FRET states were made using mutagenic strategies. This approach demonstrated that single base pair disruptions between the 3′-CCA ends of tRNA and the PTC dramatically and predictably reconfigured the relative positions of tRNA both with respect to the ribosome and each other. The 7–20Å motions observed revealed that the structurally-characterized “classical” tRNA configuration was in dynamic exchange with two distinct hybrid states previously implicated in the translocation reaction mechanism [38]. One important conclusion is that hybrid tRNA configurations may be achieved through independent and coupled motions of tRNA 3′CCA-ends with respect to the large subunit. Motion of P-site tRNA to a P/E hybrid state, rate determining in both transitions, arose from conformational events in the ribosome with activation energies (~70kJ/mol) similar in magnitude to the translocation reaction itself (~90KJ/mol)[6]. In agreement, bulk fluorescence and biochemical investigations showed that the independent motion of P-site tRNA to the P/E hybrid state precedes translocation [39,40]. Taken together, the implications for the translocation mechanism were that 1] P-site tRNA motions are mediated by a large-scale conformation events in the ribosome; 2] formation of the A/P tRNA hybrid state can occur through multiple kinetic pathways; 3] modest perturbations to the tRNA binding sites (single base pair disruptions; ca. 1–10kJ/mol) leads to large changes in the time-averaged distribution of states in the system. Thus, consistent with earlier hypotheses[41], the nature of tRNA-ribosome interactions biases substrate motions in the direction of translocation. smFRET methods demonstrated that such motions are thermally-activated, reversible processes. Although the contribution of these intrinsic, dynamic processes to the mechanism of translocation in not yet understood, thermal fluctuations have recently been implicated in the aa-tRNA selection process [42].
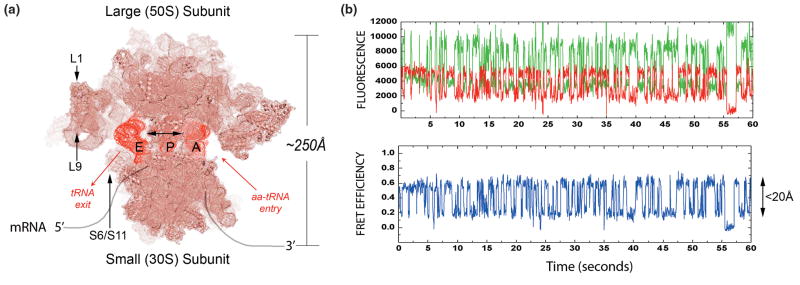
Figure 1. When placed in informative positions, FRET Pairs on the ribosome provide a “molecular EKG” reporting on transitions between discrete conformational states.
(A) Top-down view of an atomic representation of the two-subunit e.coli ribosome (derived from PDBIDs:1nkw;1ibm; and 486d), rendered in Pymol. The mRNA track (grey), A-, P- and E-site tRNA binding sites (red) as well as sites fluorescently labeled in the FRET studies discussed are highlighted. As diagrammed, aminoacyl-tRNA (aa-tRNA) enter the leading edge of the ribosome at the A site and transit the P and E sites before dissociating from the particle. Oligonucleotide tagging sites utilized by Marshall et al., cannot be seen here as both sites lie at the base of the ribosomal subunits opposite the illustrated vantage point. (B) A single-molecule fluorescence trajectory (green=donor; red=acceptor) and FRET trajectory (blue) obtained using a wide-field TIR imaging system at 40ms time resolution under equilibrium conditions from a surface-immobilized ribosome bearing site-specifically labeled A- and P-site tRNA molecules in the absence of energy input or translation factors[35]. The dynamic FRET data, reporting directly on thermally-accessible, nanometer-scale changes in tRNA position within the ribosome, persist until donor and/or acceptor fluorophore photobleaching. The recording shown is exemplary in that the duration of fluorescence extends for more than one minute.
Detailed structural characterizations of hybrid tRNA configurations were recently made by two cryo-EM groups employing particle sorting methods •[43,44]. Both groups reported sub-populations present in the ensemble of pre-translocation complexes investigated consistent with an intrinsic equilibrium between classical and hybrid states as proposed by earlier smFRET studies. Based on the relative stabilities of the two hybrid states and the intermolecular tRNA distances estimated in the work of Munro et al., both structures likely describe Hybrid state-2, the state wherein only P-site tRNA moves to its hybrid position. Interestingly, both groups reported that the hybrid state ribosome was characterized by ratchet-like motions at the subunit interface, a counterclockwise rotation of the small subunit with respect to the large and a direct interaction of the P/E hybrid tRNA with ribosomal protein L1, located at the tip of L1 stalk domain. Similar conformational rearrangements in the ribosome were previously observed with EF-G bound at the A site [45]. Thus, consistent with previous studies [46], the hybrid state tRNA configurations observed may represent intermediate states along the translocation reaction coordinate.
smFRET investigations also demonstrate that subunit ratcheting and L1 stalk closure are intrinsically dynamic processes that can occur spontaneously. Through the site-specific labeling of ribosomal proteins S6, S11 and L9, Cornish et al. [33], observed that subunit ratcheting and the reverse process, unratcheting, measured from two independent structural perspectives, occurred spontaneously (S6-L9 and S11-L9, Figure 1). The exchange rate between ratcheted and unratcheted conformations depended strongly on the nature of bound substrates. However, the rates of spontaneous ratcheting (ca. 0.1–1/sec) observed were significantly slower than the rates of P/E hybrid state formation reported by Munro et al. suggesting that subunit ratcheting and tRNA motions may be at least partially uncoupled. Consistent with an intimate relationship between ratcheting and translocation, EF-G binding at the A site in the presence of the non-hydrolyzable GTP analogue, GDPNP, dramatically increased ratcheting rates and decreased unratcheting rates.
Using native ribosome complexes bearing fluorescently labeled P-site tRNA and L1 protein (Figure 1), Fie et al. reported that subunit ratcheting, P/E hybrid state formation and L1 stalk closure are tightly-coupled processes that occur spontaneously and reversibly during the process of translocation[34]. While clearly demonstrating that relative motions between the L1 stalk and P-site tRNA occur in a highly-reversible and EF-G-induced manner, conclusions that all three conformational processes occur in lock-step remains speculative as neither subunit ratcheting nor tRNA motions were directly measured and data were obtained from just a single structural perspective.
Contrasting the conclusions of both Cornish et al. and Fie et al., using native ribosomes, site-specifically hybridized with fluorescently labeled oligonucleotides on both the small and large subunit, Marshall et al. [47] observed that subunit ratcheting only occurred after aa-tRNA accommodation at the A site. Here, accommodation was monitored by the co-localization of dye-labeled aa-tRNA to the ribosome under observation. Unratcheting was only observed upon EF-G catalyzed translocation. The authors argue that some aspect of the tRNA selection process, or subsequent peptide-bond formation, provides the energy necessary to promote ratcheting. The energy for unratcheting is provided by EF-G-catalyzed translocation, which is coupled with GTP hydrolysis. In this view, subunit ratcheting must be specifically induced by substrate and factor binding events [45,48]. However, even a slightly increased activation barriers for ratcheting and unratcheting processes in their system, just 5KJ/mol (1–2 base pairs), would result in ratcheting rates ~10-fold slower than observed by Cornish et al. (Box 2). In this case, ratcheting and unratcheting events would be significantly slower than the apparent photobleaching rate (ca. 10 seconds).
Conclusions: An integrated perspective on the protein synthesis mechanism
In order to examine the many unanswered questions that remain in the translation field, in particular the coupling of tRNA motions, subunit ratcheting and L1 stalk closure during elongation processes, the translating ribosome must be monitored from multiple structural perspectives. Here, platform improvements increasing photon collection efficiencies through enhanced optical components, extending fluorescence lifetimes, and/or multicolor detection systems may contribute substantially to progress in these pursuits. In combination with robust chemical strategies to label the ribosome and translation components, detailed studies comparing and contrasting translation mechanisms from other domains of life may also be possible. Future efforts towards integrated optical trapping and FRET platforms may also yield many new insights into this complex and highly regulated biological system by shedding light on the relationship between conformational events in the ribosome and directional motion in translation. Given the tremendous intellectual and capital foundation required for such studies, collaborative groups may be ideally positioned for these next-generation experiments.
The metastable energy landscape model of the protein synthesis mechanism [6,17] is an attractive foundation for future studies given that it helps explain longstanding observations of allostery in translation including mutations distal to the functional centers of the ribosome that affect numerous protein synthesis reactions [36]. It also adequately explains new observations of distinct ribosome sub-populations in recent cryo-EM studies [43,44] and the known sensitivity of ribosome functions to solution conditions. In this view, modifications to the ribosome and/or substrates of translation may alter the conformational dynamics observed and/or change coupling efficiencies of specific events. In this view, even small changes in microscopic parameters of discrete reactions become amplified at the systems level offering a plausible regulation paradigm. Here, the presence of unique molecular signatures in ribosomes from distinct domains ••[49] and conserved and divergent features of ribosome architecture neighboring regions of known degrees of freedom [49–51], foreshadow mechanistic distinctions between bacterial, archaeal and mammalian translation systems. Such differences, in addition to sequence variations may contribute to the known specificities of particular antibiotics for ribosomes of distinct domains [52]. Here, to the examination of mechanistic disparities will hinge on robust dynamics measurements and high experimental throughput in order to ensure a meaningfully diverse range of samples and conditions are examined.
Towards the goal of interpreting these data in a rigorous physical framework, future efforts must also be made to recapitulate smFRET data at the atomic level through molecular dynamics simulations. This pursuit will demand significant computational investments in order to simulate experimentally-observed FRET changes in stereochemically feasible terms. Progress in this area will also facilitate labeling strategies that may be used to detect functionally-relevant conformational events that have yet to be observed by structural means. The successful integration of data obtained from single-molecule imaging and computational platforms in the area of translation will doubtless contribute meaningfully towards ongoing and future efforts aiming to understand the nature and role of conformational events underpinning many other biological processes. A robust suite of tools enabling these emerging and integrated technologies will be critical to bringing single-molecule science into laboratories around the world in the short-term future.
Acknowledgments
The author is grateful to members of the laboratory for helpful discussions and careful reviews of the manuscript during its preparation. This work was supported by the National Institutes of Health (GM079238), the National Science Foundation (0747230), the Human Frontiers in Science Program and the New York State Foundation for Science, Technology and Innovation (C060021).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Wilson DN, Nierhaus KH. The weird and wonderful world of bacterial ribosome regulation. Crit Rev Biochem Mol Biol. 2007;42:187–219. doi: 10.1080/10409230701360843. [DOI] [PubMed] [Google Scholar]
- 2.Korostelev A, Noller HF. The ribosome in focus: new structures bring new insights. Trends Biochem Sci. 2007;32:434–441. doi: 10.1016/j.tibs.2007.08.002. [DOI] [PubMed] [Google Scholar]
- 3.Ramakrishnan V. What we have learned from ribosome structures. Biochem Soc Trans. 2008;36:567–574. doi: 10.1042/BST0360567. [DOI] [PubMed] [Google Scholar]
- 4.Steitz TA. A structural understanding of the dynamic ribosome machine. Nat Rev Mol Cell Biol. 2008;9 :242–253. doi: 10.1038/nrm2352. [DOI] [PubMed] [Google Scholar]
- 5.Mitra K, Frank J. Ribosome dynamics: insights from atomic structure modeling into cryo-electron microscopy maps. Annu Rev Biophys Biomol Struct. 2006;35:299–317. doi: 10.1146/annurev.biophys.35.040405.101950. [DOI] [PubMed] [Google Scholar]
- 6.Munro JB, Vaiana A, Sanbonmatsu KY, Blanchard SC. A New View of Protein Synthesis: Mapping the Free Energy Landscape of the Ribosome Using Single-Molecule FRET. Biopolymers. 2008;89 :565–577. doi: 10.1002/bip.20961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7•.Berk V, Cate JH. Insights into protein biosynthesis from structures of bacterial ribosomes. Curr Opin Struct Biol. 2007;17:302–309. doi: 10.1016/j.sbi.2007.05.009. An excellent review summarizing many of the key degrees of freedom observed in the ribosome through structural investigations. [DOI] [PubMed] [Google Scholar]
- 8.Sytnik A, Vladimirov S, Jia Y, Li L, Cooperman BS, Hochstrasser RM. Peptidyl transferase center activity observed in single ribosomes. J Mol Biol. 1999;285:49–54. doi: 10.1006/jmbi.1998.2312. [DOI] [PubMed] [Google Scholar]
- 9.Blanchard SC, Gonzalez RL, Kim HD, Chu S, Puglisi JD. tRNA selection and kinetic proofreading in translation. Nat Struct Mol Biol. 2004;11:1008–1014. doi: 10.1038/nsmb831. [DOI] [PubMed] [Google Scholar]
- 10.Blanchard SC, Kim HD, Gonzalez RL, Jr, Puglisi JD, Chu S. tRNA dynamics on the ribosome during translation. Proc Natl Acad Sci USA. 2004;101:12893–12898. doi: 10.1073/pnas.0403884101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sugawa M, Arai Y, Iwane AH, Ishii Y, Yanagida T. Single molecule FRET for the study on structural dynamics of biomolecules. Biosystems. 2007;88:243–250. doi: 10.1016/j.biosystems.2006.09.041. [DOI] [PubMed] [Google Scholar]
- 12.Zhuang X. Single-molecule RNA science. Annu Rev Biophys Biomol Struct. 2005;34:399–414. doi: 10.1146/annurev.biophys.34.040204.144641. [DOI] [PubMed] [Google Scholar]
- 13••.Roy R, Hohng S, Ha T. A practical guide to single-molecule FRET. Nat Methods. 2008;5:507–516. doi: 10.1038/nmeth.1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14••.Joo C, Balci H, Ishitsuka Y, Buranachai C, Ha T. Advances in Single-Molecule Fluorescence Methods for Molecular Biology. Annu Rev Biochem. 2008 doi: 10.1146/annurev.biochem.77.070606.101543. [DOI] [PubMed] [Google Scholar]
- 15••.Neuman KC, Nagy A. Single-molecule force spectroscopy: optical tweezers, magnetic tweezers and atomic force microscopy. Nat Methods. 2008;5:491–505. doi: 10.1038/nmeth.1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16••.Marshall RA, Aitken CE, Dorywalska M, Puglisi JD. Translation at the Single-Molecule Level. Annu Rev Biochem. 2008;77:177–203. doi: 10.1146/annurev.biochem.77.070606.101431. Excellent discussions of the basic precepts and foundations of single-molecule science and detailed descriptions of its application to specific biological systems. The article of Marshall et al. thoroughly covers the history and findings of single-molecule and optical trapping methods applied to investigations of translation. [DOI] [PubMed] [Google Scholar]
- 17•.Frank J, Gao H, Sengupta J, Gao N, Taylor DJ. The process of mRNA-tRNA translocation. Proc Natl Acad Sci USA. 2007;104:19671–19678. doi: 10.1073/pnas.0708517104. An excellent review of the process of translocation on the ribosome and the changing view of dynamic processes in the ribosome during protein synthesis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Elad N, Clare DK, Saibil HR, Orlova EV. Detection and separation of heterogeneity in molecular complexes by statistical analysis of their two-dimensional projections. J Struct Biol. 2008;162:108–120. doi: 10.1016/j.jsb.2007.11.007. [DOI] [PubMed] [Google Scholar]
- 19.Penczek PA, Frank J, Spahn CM. A method of focused classification, based on the bootstrap 3D variance analysis, and its application to EF-G-dependent translocation. J Struct Biol. 2006;154:184–194. doi: 10.1016/j.jsb.2005.12.013. [DOI] [PubMed] [Google Scholar]
- 20.Scheres SH, Nunez-Ramirez R, Gomez-Llorente Y, San Martin C, Eggermont PP, Carazo JM. Modeling experimental image formation for likelihood-based classification of electron microscopy data. Structure. 2007;15:1167–1177. doi: 10.1016/j.str.2007.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu S, Bokinsky G, Walter NG, Zhuang X. Dissecting the multistep reaction pathway of an RNA enzyme by single-molecule kinetic “fingerprinting”. Proc Natl Acad Sci U S A. 2007;104:12634–12639. doi: 10.1073/pnas.0610597104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee TH, Lapidus LJ, Zhao W, Travers KJ, Herschlag D, Chu S. Measuring the folding transition time of single RNA molecules. Biophys J. 2007;92:3275–3283. doi: 10.1529/biophysj.106.094623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim HD, Puglisi JD, Chu S. Fluctuations of Transfer RNAs between Classical and Hybrid States. Biophys J. 2007;93:3575–3582. doi: 10.1529/biophysj.107.109884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24••.Xie XS. Single-molecule approach to dispersed kinetics and dynamic disorder: Probing conformational fluctuation and enzymatic dynamics. J Chem Phys. 2002;117:11024–11032. A seminal article addressing intrinsic heterogeneities present in biological systems. [Google Scholar]
- 25.Xie Z, Srividya N, Sosnick TR, Pan T, Scherer NF. Single-molecule studies highlight conformational heterogeneity in the early folding steps of a large ribozyme. Proceedings of the National Academy of Sciences. 2004;101:534–539. doi: 10.1073/pnas.2636333100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kuzmenkina EV, Heyes CD, Nienhaus GU. Single-molecule FRET Study of Denaturant Induced Unfolding of RNase H. J Mol Biol. 2006;357:313–324. doi: 10.1016/j.jmb.2005.12.061. [DOI] [PubMed] [Google Scholar]
- 27.Coban O, Lamb DC, Zaychikov E, Heumann H, Nienhaus GU. Conformational Heterogeneity in RNA Polymerase Observed by Single-Pair FRET Microscopy. Biophys J. 2006;90:4605–4617. doi: 10.1529/biophysj.105.078840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McKinney SA, Joo C, Ha T. Analysis of Single-Molecule FRET Trajectories Using Hidden Markov Modeling. Biophys J. 2006;91:1941–1951. doi: 10.1529/biophysj.106.082487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Michalet X, Weiss S, Jager M. Single-Molecule Fluorescence Studies of Protein Folding and Conformational Dynamics. Chem Rev. 2006;106:1785–1813. doi: 10.1021/cr0404343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ritort F. Single-molecule experiments in biological physics: methods and applications. J Phys: Condens Matter. 2006;18:R531. doi: 10.1088/0953-8984/18/32/R01. [DOI] [PubMed] [Google Scholar]
- 31.Plumbridge JA, Baumert HG, Ehrenberg M, Rigler R. Characterisation of a new, fully active fluorescent derivative of E. coli tRNA Phe Nucleic Acids Res. 1980;8:827–843. [PMC free article] [PubMed] [Google Scholar]
- 32.Watson BS, Hazlett TL, Eccleston JF, Davis C, Jameson DM, Johnson AE. Macromolecular arrangement in the aminoacyl-tRNA.elongation factor Tu.GTP ternary complex. A fluorescence energy transfer study. Biochemistry. 1995;34:7904–7912. doi: 10.1021/bi00024a015. [DOI] [PubMed] [Google Scholar]
- 33.Cornish PV, Ermolenko DN, Noller HF, Ha T. Spontaneous intersubunit rotation in single ribosomes. Mol Cell. 2008;30:578–588. doi: 10.1016/j.molcel.2008.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fei J, Kosuri P, MacDougall DD, Gonzalez RL., Jr Coupling of ribosomal L1 stalk and tRNA dynamics during translation elongation. Mol Cell. 2008;30:348–359. doi: 10.1016/j.molcel.2008.03.012. [DOI] [PubMed] [Google Scholar]
- 35••.Munro JB, Altman RB, O’Connor N, Blanchard SC. Identification of two distinct hybrid state intermediates on the ribosome. Mol Cell. 2007;25:505–517. doi: 10.1016/j.molcel.2007.01.022. smFRET investigations of tRNA dynamics on the ribosome and the identification of two distinct hybrid states dynamically sampled in pre-translocation complexes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nierhaus KH, Wilson DN. Protein synthesis and ribosome structure : translating the genome. Weinheim: Wiley-VCH; 2004. [Google Scholar]
- 37••.Bahar I, Chennubhotla C, Tobi D. Intrinsic dynamics of enzymes in the unbound state and relation to allosteric regulation. Curr Opin Struct Biol. 2007;17:633–640. doi: 10.1016/j.sbi.2007.09.011. A concise summary of recent experimental data shedding light on the intrinsically dynamic nature of biological molecules and the implications of these insights to enzyme function. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sharma D, Southworth DR, Green R. EF-G-independent reactivity of a pre-translocation-state ribosome complex with the aminoacyl tRNA substrate puromycin supports an intermediate (hybrid) state of tRNA binding. RNA. 2004;10:102–113. doi: 10.1261/rna.5148704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pan D, Kirillov S, Cooperman BS. Kinetically competent intermediates in the translocation step of protein synthesis. Mol Cell. 2007;25:519–529. doi: 10.1016/j.molcel.2007.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Walker SE, Shoji S, Pan D, Cooperman BS, Fredrick K. Role of hybrid tRNA-binding states in ribosomal translocation. Proc Natl Acad Sci USA. 2008 doi: 10.1073/pnas.0710146105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moazed D, Noller HF. Intermediate states in the movement of transfer RNA in the ribosome. Nature. 1989;342:142–148. doi: 10.1038/342142a0. [DOI] [PubMed] [Google Scholar]
- 42.Lee TH, Blanchard SC, Kim HD, Puglisi JD, Chu S. The role of fluctuations in tRNA selection by the ribosome. Proc Natl Acad Sci U S A. 2007;104:13661–13665. doi: 10.1073/pnas.0705988104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43•.Agirrezabala X, Lei J, Brunelle JL, Ortiz-Meoz RF, Green R, Frank J. Visualization of the hybrid state of tRNA binding promoted by spontaneous ratcheting of the ribosome. Mol Cell. 2008;32:190–197. doi: 10.1016/j.molcel.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44•.Julian P, Konevega AL, Scheres SHW, Lazaro M, Gil D, Wintermeyer W, Rodnina M, Valle M. Structure of ratcheted ribosomes with tRNAs in hybrid states. PNAS. 2008;105:16924–16927. doi: 10.1073/pnas.0809587105. Articles describing the first structural characterization of tRNA hybrid states on the pre-translocation ribosome using cryo-EM and particle sorting methods. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Frank J, Agrawal RK. A ratchet-like inter-subunit reorganization of the ribosome during translocation. Nature. 2000;406:318–322. doi: 10.1038/35018597. [DOI] [PubMed] [Google Scholar]
- 46.Dorner S, Brunelle JL, Sharma D, Green R. The hybrid state of tRNA binding is an authentic translation elongation intermediate. Nat Struct Mol Biol. 2006;13:234–241. doi: 10.1038/nsmb1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Marshall RA, Dorywalska M, Puglisi JD. Irreversible chemical steps control intersubunit dynamics during translation. Proc Natl Acad Sci U S A. 2008;105:15364–15369. doi: 10.1073/pnas.0805299105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Valle M, Zavialov AV, Sengupta J, Rawat U, Ehrenberg M, Frank J. Locking and unlocking of ribosomal motions. Cell. 2003;114:123–134. doi: 10.1016/s0092-8674(03)00476-8. [DOI] [PubMed] [Google Scholar]
- 49.Roberts E, Sethi A, Montoya J, Woese CR, Luthey-Schulten Z. Molecular signatures of ribosomal evolution. Proc Natl Acad Sci U S A. 2008;105:13953–13958. doi: 10.1073/pnas.0804861105. A first-of-its-kind article presenting comparative analyses of ribosomal RNA and ribosomal protein sequences from diverse species representing the three domains of life, discussing these findings in the context of potential convergent and divergent ribosome functions. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Spahn CM, Gomez-Lorenzo MG, Grassucci R, Jorgensen R, Andersen GR, Beckmann R, Penczek P, Ballesta JP, Frank J. Domain movements of elongation factor eEF2 and the eukaryotic 80S ribosome facilitate tRNA translocation. EMBO J. 2004;23:1008–1019. doi: 10.1038/sj.emboj.7600102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chandramouli P, Topf M, Menetret JF, Eswar N, Cannone JJ, Gutell RR, Sali A, Akey CW. Structure of the mammalian 80S ribosome at 8.7 A resolution. Structure. 2008;16:535–548. doi: 10.1016/j.str.2008.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tenson T, Mankin A. Antibiotics and the ribosome. Mol Microbiol. 2006;59:1664–1677. doi: 10.1111/j.1365-2958.2006.05063.x. [DOI] [PubMed] [Google Scholar]
- 53.Fersht A. Structure and Mechanism in Protein Science. New York: W.H. Freeman and Company; 1999. [Google Scholar]