Abstract
Imatinib mesylate (IM), a potent inhibitor of the BCR/ABL tyrosine kinase, has become standard first-line therapy for patients with chronic myeloid leukemia (CML), but the frequency of resistance increases in advancing stages of disease. Elimination of BCR/ABL-dependent intracellular signals triggers apoptosis, but it is unclear whether this activates additional cell survival and/or death pathways. We have shown here that IM induces autophagy in CML blast crisis cell lines, CML primary cells, and p210BCR/ABL-expressing myeloid precursor cells. IM-induced autophagy did not involve c-Abl or Bcl-2 activity but was associated with ER stress and was suppressed by depletion of intracellular Ca2+, suggesting it is mechanistically nonoverlapping with IM-induced apoptosis. We further demonstrated that suppression of autophagy using either pharmacological inhibitors or RNA interference of essential autophagy genes enhanced cell death induced by IM in cell lines and primary CML cells. Critically, the combination of a tyrosine kinase inhibitor (TKI), i.e., IM, nilotinib, or dasatinib, with inhibitors of autophagy resulted in near complete elimination of phenotypically and functionally defined CML stem cells. Together, these findings suggest that autophagy inhibitors may enhance the therapeutic effects of TKIs in the treatment of CML.
Introduction
Chronic myeloid leukemia (CML) is a malignancy arising from transformation of the hematopoietic stem cell, which typically evolves through 3 distinct disease stages: an indolent chronic phase (CP), characterized by the accumulation of mature granulocytes and myeloid precursors in the bone marrow and the peripheral blood; an accelerated phase, characterized by an increase in disease burden and in the frequency of progenitor/precursor cells; and an acute phase called blast crisis (BC), marked by increasing numbers of differentiation-arrested blast cells (1–3). The hallmark of all phases is the Philadelphia chromosome (Ph1), a reciprocal translocation of chromosomes 9 and 22, which generates the BCR/ABL fusion gene encoding a constitutively active tyrosine kinase (4). p210BCR/ABL exerts its oncogenic function by activating a cascade of intracellular signalling pathways, which leads to increased survival and proliferation and limited dependence on growth factors (5, 6). Two of the major pathways activated by BCR/ABL are the class I PI3K and the Ras pathways (7, 8), which are deregulated in most human cancers (9, 10). In normal hematopoietic cells, these 2 pathways are activated by stimulation of growth factor receptors with intrinsic or JAK-associated tyrosine kinase activity, suggesting that p210BCR/ABL effectively mimics growth factor–dependent signalling.
The generation of the BCR/ABL kinase, ATP-competitive inhibitor imatinib mesylate (IM) has revolutionized the therapy of CML, since this drug is highly effective in the CP of the disease (11). However, there are 3 major problems with IM-based therapy: (a) the limited response of CML-BC or Ph1 B cell acute lymphoblastic leukemia (ALL) patients to IM (11–13); (b) the development of resistance caused in approximately 40% of cases by mutations in the BCR/ABL kinase domain, which impair the ability of IM to interact with the protein (14–18); and (c) the relative insensitivity of Ph1 CML stem cells to IM (19). For these reasons, more potent BCR/ABL inhibitors, also targeting IM-resistant mutants, are being developed and tested (20, 21). However, at least one common BCR/ABL mutant (carrying the T315I mutation) is resistant to all tyrosine kinase inhibitors (TKIs) developed so far (22). A further limitation is that primitive Ph1 stem cells overexpress wild-type p210BCR/ABL and appear to be intrinsically resistant not only to treatment with IM but also to second generation (dasatinib [Das], nilotinib, and bosutinib) TKIs (19, 23–27). Therefore, there is the need to develop new therapeutic approaches that, in combination with TKIs, might be more effective in preventing the outgrowth of TKI-resistant CML/Ph1 ALL cells and target the stem cell population.
Macroautophagy (hereafter referred to as autophagy) is a degradative process in eukaryotic cells that results in the breakdown of intracellular material within lysosomes under homeostatic conditions or in response to stress signals (28, 29), allowing cells to adapt to environmental and/or developmental signals. Autophagy is a genetically controlled process, which progresses through definite steps, leading to the engulfment of long-lived proteins and whole organelles into multi-membraned vacuoles called autophagosomes (28, 29). Autophagosomes then fuse with lysosomes for final destruction and recycling (28, 29). While in certain cellular contexts autophagy can serve as an alternative cell death mechanism named type II cell death (30–32), it is becoming increasingly clear that this process can also act as a cell survival mechanism. In fact, autophagy is a process by which cells can adapt their metabolism to starvation caused by a decrease in metabolite concentrations or extracellular nutrients, a typical consequence of loss of growth factor signalling, allowing cells to evade programmed cell death (32, 33). Accordingly, inhibition of autophagy by knockdown of autophagy genes or by use of pharmacological inhibitors, such as chloroquine (CQ, an inhibitor of lysosomal acidification; ref. 34), results in cell death of growth factor–starved cells in which apoptosis has been genetically ablated (33, 35). In tumors displaying defective apoptosis, inhibition of autophagy causes caspase-independent necrotic cell death, which, in turn, augments inflammation, leading to enhanced tumor burden (36). In 2 recent studies, treatment of Myc-induced lymphomas with the autophagy inhibitor CQ, resulted in reduced tumor growth in vivo (34, 37), suggesting that induction of autophagy provides a protective mechanism in tumor cells. Thus, the consequences of autophagy inhibition are double-faced, as it can either promote or suppress tumorigenesis, depending on the tumor type or inflammation status.
Another interesting aspect of autophagy is its involvement in the response to chemotherapy. Autophagy inhibition has been shown to sensitize tumor cells to cell death induced by irradiation (38–40), alkylating agents (41), or arsenic trioxide (42), suggesting that cancer cells can react to chemotherapy by inducing autophagy as a self-defence mechanism. How an increase in intracellular lysosome-mediated catabolism results in reduced sensitivity to cell death is still unclear. Perhaps, clearance of mitochondria through autophagy (mitophagy) might cause a depletion of proapoptotic signals such as the release of cytochrome c (43, 44).
As mentioned above, p210BCR/ABL has the ability to activate several survival pathways, effectively mimicking growth factor signaling (5–8). This, in turn, causes transformed cells to acquire resistance to apoptosis induced by growth factor deprivation (5–8). Inhibition of BCR/ABL tyrosine kinase activity by IM results in induction of cell death, which is caused by a sudden decrease of intracellular survival signals and a relative increase in proapoptotic signals (45, 46). We show here that apoptosis is not the sole consequence of BCR/ABL inhibition, as IM treatment rapidly activates an autophagic process, which follows the induction of ER stress and relies on intracellular Ca2+. Of greater importance, inhibition of autophagy by pharmacological inhibitors potentiates IM-induced cell death in CML cell lines and primary CML cells, including those carrying partially IM-resistant BCR/ABL mutants. Colony formation of candidate CML stem cells, defined as CD34+CD38– and long-term culture–initiating cells (LTC-ICs), was also markedly suppressed by combination treatment. Knockdown of the autophagy genes ATG5 autophagy related 5 homolog (S. cerevisiae) (ATG5) and ATG7 autophagy related 7 homolog (S. cerevisiae) (ATG7) had comparable effects, supporting the specificity of the pharmacological inhibitors. These findings indicate that induction of autophagy provides a survival mechanism to IM-treated BCR/ABL-expressing cells, including the stem cell population, and strongly suggest that inhibition of autophagy may improve the therapeutic efficacy of TKIs in the treatment of CML.
Results
IM induces autophagy in p210BCR/ABL-expressing CML cell lines.
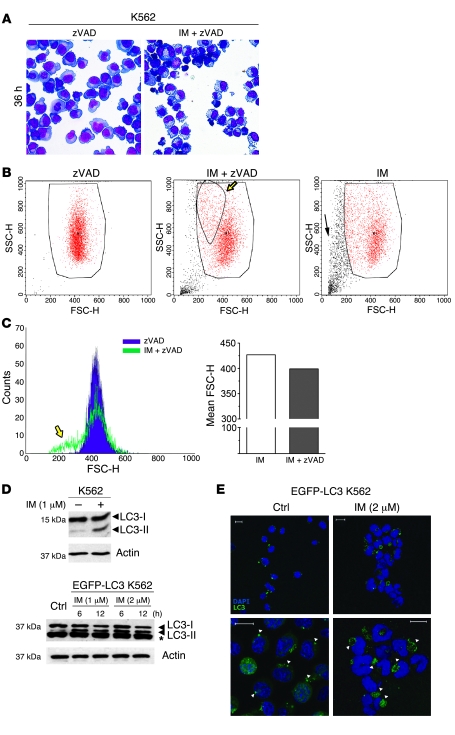
IM induces death of CML cells, but additional effects have not been studied in detail. Therefore, we designed experiments to study cellular changes induced by IM in CML cells. To this end, the effects of IM treatment were initially analyzed in the erythro-megakaryocytic CML-BC line K562. To exclude effects due to apoptosis-related changes, cells were treated with N-benzyloxycarbonyl-Val-Ala-Asp-fluoromethylketone (zVAD), a pan-caspase inhibitor. Morphological analysis of K562 cells cultured for 36 hours in the presence of 2 μM IM showed a consistent decrease in cell size as compared with cells treated with zVAD alone (Figure 1A). A closer examination showed that the cytoplasm of IM/zVAD-treated cells was severely reduced and almost entirely filled with several translucent vacuoles (Figure 1A). Flow cytometry of physical parameters of IM/zVAD-treated cells revealed the appearance of a distinct population of smaller and denser cells, compared with those treated with zVAD only (forward light scatter-height [FSC-H] and side light scatter-height [SSC-H] parameters; Figure 1B, yellow arrow). This population was clearly distinct from apoptotic cells, which are smaller and less dense, as shown in cultures treated with IM only (Figure 1B). Mean FSC-H was also decreased in IM/zVAD-treated cells (Figure 1C). Similar changes were observed at earlier time points in cells treated with IM alone (data not shown). These morphological changes were reminiscent of those induced by growth factor deprivation in hematopoietic cells (33). A previous study showed that growth factor withdrawal activated autophagy in cells lacking the intrinsic apoptotic pathway and that this process was critical for maintaining cell survival (35). Thus, we investigated whether inhibition of BCR/ABL-dependent survival signals by IM treatment could also trigger autophagy. Autophagy is invariably associated with conversion of the microtubule-associated protein 1 light chain 3 (LC3), also known as ATG8, from the cytosolic LC3-I to the autophagosome-associated LC3-II form (47). Increased levels of LC3-II were clearly detected in extracts of IM-treated K562 cells (Figure 1D, upper panel). Immunofluorescence/confocal microscopy (IF/confocal microscopy) showed an increase in LC3-positive vacuoles in IM-treated K562 cells (data not shown). In cells transduced with a retrovirus encoding LC3 fused to the EGFP (EGFP-LC3), Western blotting showed a clear accumulation of EGFP–LC3-II in protein extracts as early as 6 hours after treatment with 2 μM IM (Figure 1D, lower panel). Nevertheless, LC3-II accumulation could be due to inhibition, rather than induction, of autophagy. To exclude this possibility, we analyzed endogenous LC3-II levels in IM-treated K562 cells in the presence of CQ, an autophagy blocker. If IM blocked autophagy, one would expect equal levels of LC3-II in CQ- and CQ/IM-treated cells. Instead, LC3-II expression was higher in CQ/IM-treated cells (Supplemental Figure 1; supplemental material available online with this article; doi:10.1172/JCI35660DS1), supporting an autophagy-inducing function of IM.
Figure 1. Effects of IM treatment on morphology and autophagy in K562 cells.
(A) Light microscopy images of May-Grünwald–stained untreated or IM-treated K562 cells. Cells were cultured for 36 hours in the absence or presence of 2 μM IM and processed for May-Grünwald staining. Original magnification, ×40. (B and C) Reduced size of IM-treated K562 cells. The size of K562 cells treated with zVAD, IM, or IM and zVAD was analyzed using a cytofluorimeter. Representative dot plots of physical parameters forward light scatter-height/side light scatter-height (FSC-H/SSC-H) are shown. The yellow arrows indicate a subpopulation of smaller and denser cells in IM/zVAD-treated cultures, while the black arrow shows dying cells after treatment with IM only. Large boxes indicate viable cells; small box indicates denser cells appearing upon IM/zVAD treatment. (C) Representative histograms of the forward light scatter-height parameter are shown. (D) Accumulation of autophagosome-associated LC3-II in IM-treated K562 cells. The Western blot shows endogenous LC3-I and LC3-II levels (upper panel) or ectopic EGFP–LC3-I and EGFP–LC3-II expression (lower panel) in extracts from untreated (control) or IM-treated cells (at 6 and 12 hours). The asterisk indicates that bands are not specific for LC3 (lower panel). Actin was measured as loading control. (E) Formation of LC3-positive vesicles in IM-treated EGFP-LC3–transduced K562 cells. EGFP-LC3 K562 cells were cultured in the presence or absence of IM and stained with anti-LC3 antibody (red). Nuclei were counterstained with DAPI. Scale bar: 10 μm.
IF/confocal microscopy confirmed the presence of large EGFP-positive cytoplasmic vacuoles in cells treated with 2 μM IM for 24 hours (Figure 1E, right panels). Conversely, untreated cells contained smaller EGFP-positive granules (Figure 1E, left panels), which are probably due to nonspecific aggregation of overexpressed LC3 (48).
EM was then performed to evaluate formation of autophagic structures. Examination of untreated K562 cells demonstrated the presence of multivesicular bodies (MVB) in the cytoplasm of approximately 40% of the cell profiles (Supplemental Figure 2). These organelles ranged from 1 to 4 μm in diameter and were characterized by the presence of numerous small vesicles, 25–100 nm in diameter, together with a few slightly larger, more electron-dense inclusions, containing a lipofuscin-like material. Treatment of K562 cells with 2 μM IM for 6 hours resulted in a large increase in the size and prevalence of cytoplasmic vacuoles, most of which contained many electron-dense inclusions (Supplemental Figure 3, A and B). Similar inclusions were present in small vacuoles distributed throughout the cytoplasm of the cells (Supplemental Figure 3A and Supplemental Figure 4). Many of these lysosome-like organelles appeared to be fusing, both together and with the larger vacuoles (Supplemental Figure 4). Untreated cells, transduced with EGFP-LC3 and processed for immunogold cytochemistry, exhibited a sparse and apparently nonspecific distribution of gold particles throughout the cytoplasm (data not shown). Conversely, cells treated with 2 μM IM for 6 hours showed a definite concentration of GFP-labelling over the dense inclusions present in the cytoplasm and in the large vacuoles (Supplemental Figure 3, C and D). Surprisingly, although the label was concentrated over the contents of these inclusions, their membrane was almost devoid of label. These results are consistent with the observations of Fader et al. (49), who reported the presence of constitutive MVBs in K562 cells. They also observed that the induction of autophagy led to increased interaction of MVBs with autophagosomes, yielding hybrid organelles containing vesicles, which were labelled for LC3. The presence of dense, lipofuscin-like material in the small cytoplasmic vacuoles, within 6 hours of treatment, probably indicates their rapid fusion with lysosomes and subsequent breakdown.
Cell shrinkage and induction of autophagy was confirmed also in the CML lymphoid BC cell line BV173, which was chosen because of the slow kinetics of cell death induction upon IM treatment. May-Grünwald staining and FACS analysis of BV173 cells cultured for 72 hours in the presence 4 μM IM revealed a decrease in cell size, compared with that of untreated cells (Supplemental Figure 5, A and B). The cytoplasm of treated cells was severely reduced and filled with translucent vacuoles (Supplemental Figure 5A). Because of the slow kinetics of cell death, it was not necessary to treat BV173 with zVAD, confirming that the reduction in cell size was not caused by caspase inhibition. Accumulation of LC3-II was clear only upon treatment with CQ, which blocks degradation of autophagosomes inside autolysosomes (Supplemental Figure 5C) (33, 34). Levels of LC3-II were higher in CQ/IM-treated cells than in CQ-treated cells, thus confirming the autophagy-inducing effect of IM. The same treatment demonstrated an accumulation of LC3-positive structures by IF/confocal microscopy (Supplemental Figure 5D) and EM (Supplemental Figure 6). This possibly reflects a very high rate of lysosome-mediated autophagosome degradation, resulting in very high turnover of LC3-II in these cells.
In control experiments a reduction in cell size was observed in IL-3–starved myeloid precursor 32Dcl3 (32D) cells, in which apoptosis was blocked by zVAD (Supplemental Figure 7, A and B). Moreover, a net increase in the conversion of LC3-I to LC3-II was also readily detected upon IL-3 withdrawal (Supplemental Figure 7C).
IM induces autophagy in p210BCR/ABL-expressing 32D cells.
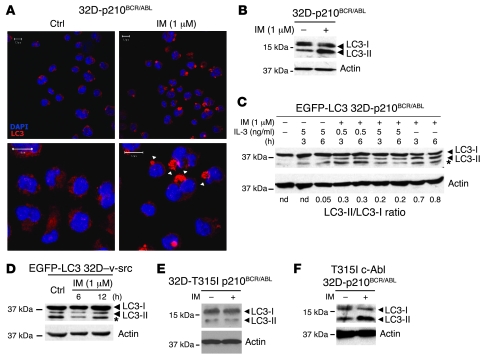
To confirm the link between inhibition of BCR/ABL tyrosine kinase and induction of autophagy, we analyzed endogenous levels of LC3 in IM-treated p210BCR/ABL–expressing 32D cells. Indeed, IM (1 μM) induced autophagy, as demonstrated by the presence of large LC3-positive vacuoles in the cytoplasm (Figure 2A). Moreover, Western blot analysis revealed the accumulation of LC3-II in protein extracts of treated cells (Figure 2B).
Figure 2. Autophagy in IM-treated 32D-p210BCR/ABL cells.
(A) Formation of LC3-positive vacuoles in IM-treated 32D-p210BCR/ABL cells. Cells were treated for 12 hours with 1 μM IM, and LC3 levels were analyzed by IF/confocal microscopy (anti-LC3, red). Arrowheads indicated autophagic vesicles. Scale bar: 10 μm. (B) Western blot detection of LC3-I and LC3-II in IM-treated 32D-p210BCR/ABL cells (6 hours). Actin was measured as loading control. (C) IL-3 inhibits IM-induced autophagy in EGFP-LC3–transduced 32D-p210BCR/ABL cells. Cells were cultured for 3 and 6 hours in the presence of different concentrations of IL-3 and IM, as indicated. The asterisk indicates that bands are not specific for LC3. The LC3-I to LC3-II ratio was calculated by densitometric analysis. Actin was measured as loading control. nd, not determined. (D) LC3-I/LC3-II levels in v-Src–transformed 32D cells stably expressing EGFP-LC3. Cells were treated for 6 or 12 hours with the indicated concentrations of IM, and LC3-I/LC3-II levels were analyzed by Western blot using an anti-LC3 antibody. The asterisk indicates that bands are not specific for LC3. Actin was detected as loading control. (E) IM treatment of 32D cells expressing the IM-resistant T315I p210BCR/ABL mutant (32D-T315Ip210BCR/ABL cells) does not induce autophagy. 32D cells expressing the IM-resistant T315I p210BCR/ABL mutant were treated with 1 μM IM and analyzed for LC3-I/LC3-II levels. Actin was detected as internal control. (F) Expression of T315I c-Abl in 32D-p210BCR/ABL cells does not block IM-induced LC3-II accumulation. p210BCR/ABL/T315I c-Abl coexpressing 32D cells were treated with IM and analyzed for LC3-I/LC3-II levels. Actin was detected as loading control.
To assess whether induction of autophagy was due to sudden lack of survival signals following BCR/ABL inhibition, IL-3–independent EGFP-LC3–transduced 32D-p210BCR/ABL cells were treated with 1 μM IM, alone or in the presence of 0.5–5 ng/ml IL-3 (Figure 2C). The presence of IL-3 in culture media markedly impaired the accumulation of LC3-II (Figure 2C), suggesting that the autophagic response is connected to the IM-dependent ablation of intracellular survival signals. Yet, autophagy induction could be a nonspecific consequence of IM treatment. To exclude this, we analyzed the effects of IM on EGFP-LC3–transduced 32D cells expressing the transforming oncogene of the Rous sarcoma virus (32D–v-Src) cells; in these cells, treatment with IM failed to induce accumulation of EGFP-LC3 (Figure 2D). Likewise, IM did not induce autophagy in EGFP-LC3–expressing HeLa cells (Supplemental Figure 8). Finally, we tested whether IM-dependent induction of autophagy was due only to inhibition of p210BCR/ABL. First, EGFP-LC3–transduced 32D cells expressing the IM-resistant T315I p210BCR/ABL mutant did not show accumulation of the LC3-II isoform upon IM treatment (Figure 2E). Second, ectopic expression of the IM-resistant T315I c-Abl mutant in 32D-p210BCR/ABL cells did not block IM-induced autophagy, demonstrating that the effect is BCR/ABL dependent (Figure 2F).
IM-induced autophagy is preceded by ER stress and relies on intracellular Ca2+.
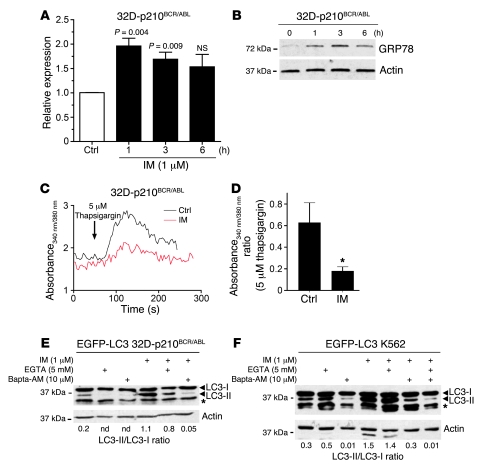
Several studies have implicated ER stress in the induction of autophagy through the activation of different downstream pathways (ref. 47 and references cited therein). Furthermore, inhibition of c-Abl by IM induces ER stress in cardiomyocytes (50). Therefore, we hypothesized that IM could cause ER stress also in BCR/ABL-expressing cells, and this could be involved in the induction of autophagy. To address this possibility, 32D-p210BCR/ABL cells were treated with or without 1 μM IM, and the mRNA levels of C/EBP homologous protein (CHOP), a marker of the ER stress response activated by transcriptional mechanisms, were measured by real-time quantitative PCR. We found a significant increase in CHOP mRNA levels (P = 0.004) as early as 1 hour after IM treatment (Figure 3A). Expression of Grp78, another marker of the ER stress response (51), was also rapidly induced in IM-treated cells (Figure 3B). Upregulation of both markers preceded the induction of autophagy, as LC3 conversion was detected only 3 hours after IM treatment (data not shown). Cellular Ca2+ homeostasis has been reported to be perturbed by ER stress and to be causally linked to the induction of autophagy (52, 53). Consistent with this, treatment of 32D-p210BCR/ABL cells with 2 μM IM produced a marked reduction in the ER Ca2+ pool as defined by thapsigargin-mediated Ca2+ release (Figure 3, C and D). To assess Ca2+ requirements in IM-induced autophagy, EGFP-LC3–transduced K562 and 32D-p210BCR/ABL cells were treated with 1 μM IM alone or in combination with 5 mM EGTA or 10 μM bis-(0-aminophenoxy)-ethane-N,N,N,N′-tetraacetic acid/tetra (acetoxymethyl)-ester (Bapta/AM) to chelate extracellular and intracellular Ca2+, respectively. Remarkably, IM-dependent conversion of LC3 was totally abrogated when cytosolic Ca2+ was depleted using Bapta/AM (Figure 3, E and F). By contrast, EGTA did not alter IM-dependent accumulation of LC3-II, implying that only Ca2+ derived from intracellular stores, such as ER, accounted for the induction of autophagy (Figure 3, E and F).
Figure 3. IM-induced autophagy is preceded by ER stress and relies on intracellular Ca2+.
(A) Induction of CHOP expression in IM-treated 32D-p210BCR/ABL cells. CHOP mRNA levels were measured by quantitative real-time PCR in cells treated for 1, 3, or 6 hours with 1 μM IM. Values represent mean ± SEM. (B) Induction of Grp78 expression in IM-treated 32D-p210BCR/ABL cells. Cells were harvested and lysed at the indicated times after the addition of IM. Extracts were probed with anti-GRP78 and anti-actin as loading control. (C) ER Ca2+ content was estimated by adding 5 μM thapsigargin (a SERCA pump inhibitor) to Fura-2–loaded 32D-p210BCR/ABL cells. The experiment was conducted at 37°C in a Ca2+-free buffer. (D) Histogram shows mean ± SEM of Ca2+ rise (peak minus basal 340/380 fluorescence ratio) induced by 5 μM thapsigargin in 6 separate experiments. *P < 0.05, unpaired Student’s t test. (E) Western blot shows LC3-I/LC3-II levels in EGFP-LC3 32D-p210BCR/ABL cells treated (6 hours) with the indicated concentration of IM, EGTA, and Bapta/AM. The LC3-I/LC3-II ratio was determined by densitometric analysis and normalized according to the levels of β-actin in each sample (lower panel). (F) Western blot shows LC3-I/LC3-II levels in EGFP-LC3–transduced K562 cells after the indicated treatments (6 hours). The LC3-I/LC3-II ratio was determined as in E. The asterisks indicate that bands are not specific for LC3 (E and F).
Inhibition of autophagy potentiates IM-induced cell death in p210BCR/ABL-expressing 32D cells.
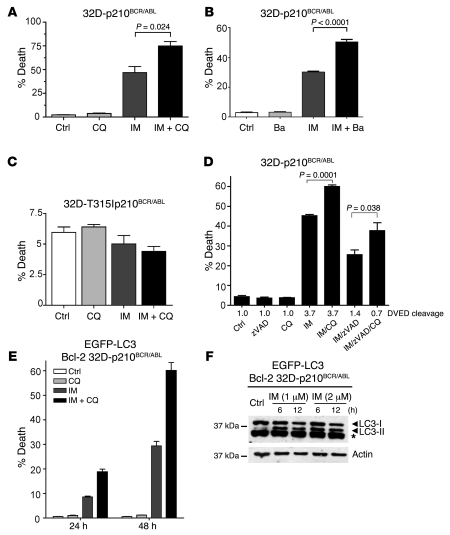
Several studies have suggested that autophagy may act as a protective mechanism in tumor cells (32, 33, 36, 37) and that therapy-induced cell death can be enhanced upon autophagy inhibition (38–42). To test whether autophagy acts as a survival mechanism in our system, we inhibited autophagy in 32D-p210BCR/ABL cells using CQ and analyzed the effects on IM-induced cell death. While treatment with CQ alone had no effect, IM-induced cell death was significantly increased (from 50% to over 75%; P = 0.024) when IM was combined with CQ (Figure 4A). Similar results were obtained using bafilomycin A1 (Ba), an inhibitor of autophagy that acts by blocking the activity of the H+-ATPase responsible for acidification of autophagolysosomes, or 3-methyladenine, an inhibitor which blocks the early stages of autophagy (28, 29) (Figure 4B and data not shown). In contrast, 32D cells expressing the IM-resistant T315I p210BCR/ABL mutant, which do not undergo autophagy upon IM treatment (Figure 2E), were not sensitive to CQ treatment (Figure 4C).
Figure 4. Inhibition of autophagy potentiates IM-induced cell death in 32D-p210BCR/ABL cells.
(A) 32D-p210BCR/ABL cells were cultured for 12 hours in the presence or absence of 1 μM IM, alone, or in combination with 5 μM CQ. Cell death was measured by annexin V staining. Values represent the mean ± SEM of 3 independent experiments. Data were analyzed by unpaired Student’s t test. (B) 32D-p210BCR/ABL cells were treated with 1 μM IM (12 hours), alone or in combination with 20 nM Ba. Cell death was measured as above. Values represent the mean ± SEM of 2 independent experiments. (C) CQ has no effects in 32D cells expressing the IM-resistant T315I p210BCR/ABL mutant. 32D cells expressing the IM-resistant T315I p210BCR/ABL mutant were treated with IM, with or without CQ, and analyzed for cell death induction as in A. (D) CQ-induced cell death is caspase independent. 32D-p210BCR/ABL cells were cultured for 6 hours in the presence or absence of different combination of 1 μM IM, 50 μM zVAD, and 5 μM CQ. The percentage of cell death was determined as above. Caspase activity was measured in extracts using a DVED fluorescent substrate. (E) Bcl-2 does not block CQ-mediated sensitization to IM. 32D-p210BCR/ABL cells ectopically expressing Bcl-2 (Bcl-2 32D-p210BCR/ABL) were cultured with 1 μM IM alone or in combination with 5 μM CQ. Cell death was measured as above. Values represent mean ± SEM (C–E). (F) Bcl-2 does not block IM-induced LC3-II accumulation. Western blot shows LC3-I/LC3-II levels in extracts of 32D-p210BCR/ABL cells ectopically expressing Bcl-2 treated for 6 or 12 hours with the indicated concentrations of IM. β-actin was detected as loading control.
We also assessed whether treatment with CQ would potentiate the effects of IM in mice inoculated with 32D-p210BCR/ABL cells. Thus, sublethally irradiated C3H/HeJ mice were injected intravenously with LC3-GFP 32D-p210BCR/ABL cells (2.5 × 105 per mouse) and divided in 4 groups: (a) untreated mice, (b) mice treated with CQ only, (c) mice treated with IM only, and (d) mice treated with IM and CQ.
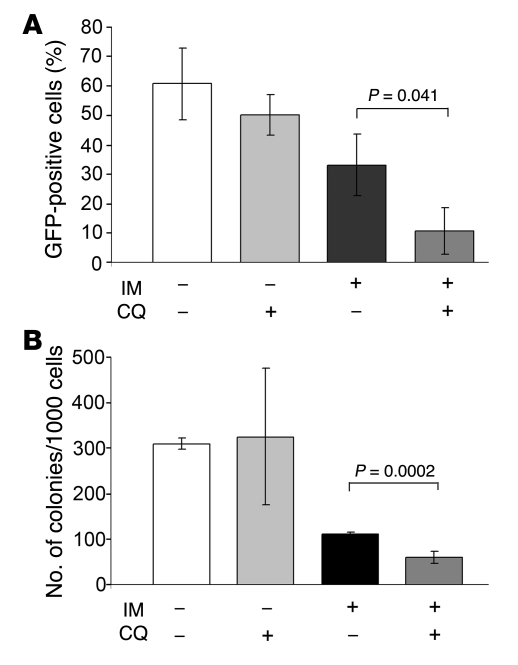
Leukemia was allowed to develop and 14 days after injection, when GFP-positive cells in the bone marrow were typically 30%–40%, mice were treated with IM (100 mg/kg for 5 consecutive days), CQ (60 mg/mouse, 7 intraperitoneal injections), or the IM/CQ combination. In the IM/CQ-treated group, mice were pretreated for 2 days with CQ before starting the therapy with IM. Twenty-four hours after the end of the treatment, mice (3 per group) were sacrificed and GFP-positive cells were assessed in the bone marrow. GFP-positive cells in untreated and CQ-treated mice were 60.0% ± 12.0% and 50.0% ± 6.8%, respectively. As expected, IM treatment induced a decrease in the number of bone marrow GFP–positive cells (33.0% ± 10.0%); this decrease was more pronounced in the mice treated with IM and CQ (10.6% ± 7.8% GFP positivity) (Figure 5A).
Figure 5. CQ treatment potentiates the effect of IM in mice inoculated with GFP-LC3 32D-p210BCR/ABL cells.
(A) GFP-positive cells in the bone marrow of untreated or treated (CQ, IM, or the CQ/IM combination) leukemic mice. (B) Cytokine-independent colony formation from bone marrow cells of untreated or treated (CQ, IM, or the CQ/IM combination) leukemic mice. Values represent SD.
The viability of GFP-positive cells in the bone marrow was evaluated by methylcellulose CFC assays in the absence of cytokines. On plating an equal number of cells (1,000 per plate), a similar number of colonies formed from bone marrow of untreated and CQ-treated mice, fewer colonies grew from marrow of mice treated with IM only, and even fewer colonies formed from marrow of mice treated with the IM/CQ combination (Figure 5B).
Next, we investigated the type of cell death induced by CQ-mediated sensitization and, specifically, whether caspase-dependent or -independent mechanisms were involved. To this end, 32D-p210BCR/ABL cells were treated with 1 μM IM along with 5 μM CQ and 50 μM zVAD, and the percentage of annexin V–positive cells was determined (Figure 4D). At the same time, caspase activity was measured in protein extracts using a DVED fluorescent substrate (Figure 4D). We found that CQ-mediated sensitization did not require caspase activation, as the combination of IM and CQ significantly augmented cell death in cells treated with zVAD to block caspase activity (Figure 4D). However, IM-induced cell death was reduced in cells treated with zVAD, consistent with the ability of BCR/ABL proteins to block caspase 3 activation (54). Next, we tested whether inhibition of the intrinsic apoptotic pathway would have any effect on CQ-mediated sensitization. 32D-p210BCR/ABL cells stably overexpressing Bcl-2 (55) and retrovirally transduced with MigRI–EGFP-LC3 were treated with 1 or 2 μM IM in the presence or absence of CQ and analyzed for induction of cell death at different time points (Figure 4E). While Bcl-2 overexpression delayed IM-induced cell death, it did not make cells refractory to CQ-mediated sensitization, which was even higher than in parental 32D-p210BCR/ABL cells (in Figure 4, compare A and E). GFP–LC3-II was readily detected as early as 6 hours after IM treatment of Bcl-2–expressing 32D-p210BCR/ABL cells (Figure 4F), indicating that in these cells, Bcl-2 overexpression does not block autophagy, which is at variance with what has been suggested by previous reports in other models (32, 56).
Inhibition of autophagy increases IM-induced cell death in cell lines and primary CML cells.
As CQ potentiates IM-induced cell death in 32D-p210BCR/ABL cells, we performed experiments to determine whether inhibition of autophagy leads to increased cell death in IM-treated CML cell lines. These studies were carried out by use of chemical inhibitors and by si/shRNA-dependent ablation of the expression of essential autophagy genes. The combination of IM and CQ treatment significantly augmented cell death in K562 cells compared with IM treatment alone (P = 0.015) (Figure 6A). Ba also enhanced IM-induced cell death, demonstrating that the potentiating effect is not limited to CQ (Figure 6B). To assess whether CQ affects survival in long-term assays, we performed CFC assays using K562 cells treated with 0.25 or 0.50 μM IM. CQ enhanced the effect of IM at 0.50 μM even more potently than in the short-term survival assay (in Figure 6, compare A and C). CQ treatment of a subclone of K562 cells resistant to IM as a consequence of gene amplification (57) also enhanced the survival-inhibitory effects of IM (Figure 6D). Finally, CQ treatment increased cell death in IM-treated BV173 cells, and the sensitizing role of autophagy inhibition was also confirmed using Ba (Supplemental Figure 9).
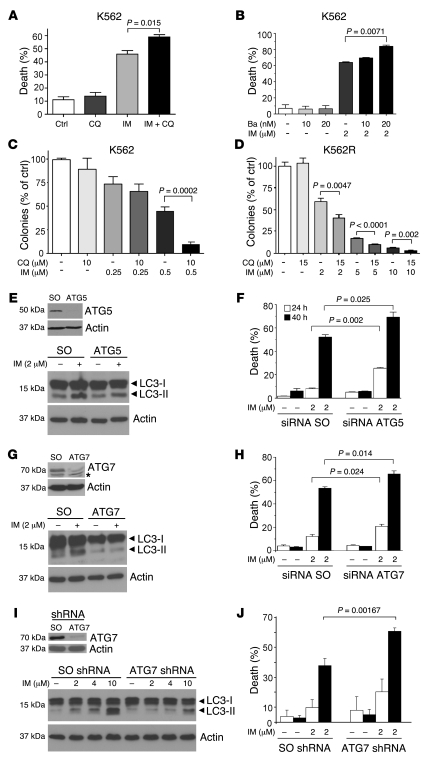
Figure 6. Inhibition of autophagy increases IM-induced cell death in K562 cells.
(A and B) Effects of CQ (A) and Ba (B) in IM-treated K562 cells. Cells were cultured for 48 hours in the presence or absence of 5 μM CQ and 2 μM IM (A) or Ba and IM as indicated (B), and cell death was measured by annexin V immunostaining. Values represent the mean ± SEM of 3 independent experiments. Data were analyzed by unpaired Student’s t test. (C and D) CQ potentiates the effect of IM in clonogenic assays of parental (C) and IM-resistant (D) K562 cells. Cells were plated in methylcellulose in the absence or in the presence of IM and CQ, and colonies were counted 10 days later. Values (expressed as percentage of control) represent the mean ± SEM of 3 (C) or 2 (D) independent experiments. (E–J) ATG5 or ATG7 downregulation enhances IM-induced cell death. K562 cells were transfected with control or ATG5 (E and F) or ATG7 (G and H) siRNAs and analyzed for ATG5 (E, upper panel) and ATG7 (G, upper panel) expression 48 hours after transfection. Levels of actin were measured as loading control. The asterisk indicates that bands are not specific for LC3. Forty-eight hours after transfection, cells were treated with IM and analyzed for LC3 and actin expression (E and G, lower panels). Cell death was measured by annexin V staining 24 (white bars) and 40 hours (black bars) after IM treatment (F and H). K562 cells were transduced with scrambled (SO) or ATG7 shRNA pGIPZ lentiviral vectors, and levels of ATG7, actin, and LC3 (I) and IM-induced cell death (annexin V staining) were measured at 24 (white bars) and 40 hours (black bars) (J). Values represent the mean ± SEM of 3 independent experiments.
Since CQ and Ba could enhance IM-induced cell death by mechanisms independent of their effects on autophagy (i.e., nonspecific effects), we knocked down expression of the essential autophagy genes ATG5 and ATG7 in K562 cells, using specific siRNAs, and analyzed whether autophagy was blocked and cells were sensitized to IM-induced cell death. Indeed, downregulation of ATG5 or ATG7 expression blocked the IM-induced accumulation of the autophagic LC3-II isoform and enhanced the cell death–inducing effect of IM (Figure 6, E–H). Finally, in K562 cells transduced with pGIPZ lentiviral vectors expressing a scrambled (SO) shRNA or an ATG7-specific shRNA, stable downregulation of ATG7 expression (Figure 6I, upper panel) blocked IM-induced LC3-II accumulation (Figure 6I, lower panel) and promoted an increase in IM-induced cell death (Figure 6K).
We next investigated whether IM treatment induced autophagy in primary CML cells and whether inhibition of autophagy enhanced the cell death–inducing effect of IM in these cells. Peripheral blood CD34+ cells from a partially IM-resistant (M351T) CML patient were treated with IM (2 μM) or left untreated for 24 hours and assessed for induction of autophagy by detection of the autophagic LC3-II isoform. Anti-LC3 Western blotting of protein extracts from CML cells showed a clear accumulation of LC3-II and an increase (an approximately 10-fold increase) of the LC3-II/LC3-I ratio in IM-treated cells (Figure 7A).
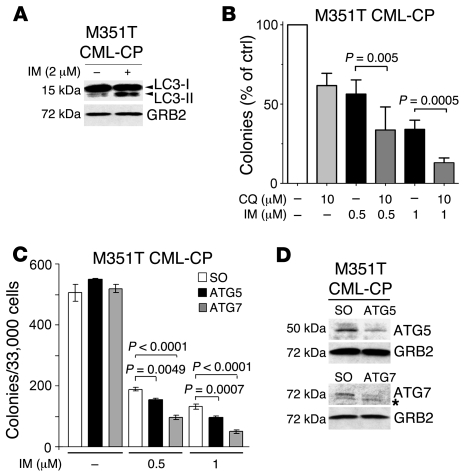
Figure 7. Autophagy is induced in IM-treated primary CML cells and acts as a survival mechanism.
(A) Accumulation of LC3-II in IM-treated CD34+ CML (M351T) cells. Cells were treated with IM (2 μM) or left untreated. Extracts were probed with anti-LC3 and anti-GRB2 antibodies. LC3-II/LC3-I ratio was determined by densitometric analysis. (B) CQ potentiates the effect of IM in clonogenic assays. CD34+ CML cells were plated in methylcellulose in the presence or in the absence of IM (0.5 or 1.0 μM) and CQ (10.0 μM). Colonies were counted 10 days later. Values (expressed as percentage of control) are representative of 3 independent experiments. (C) siRNA-mediated downregulation of ATG5 or ATG7 expression enhances IM-induced inhibition of colony formation. CD34+ CML cells were transfected with control or ATG5 or ATG7 siRNAs and, 24 hours later, left untreated or treated with IM (1 μM). Twenty-four hours later, cells were plated in methylcellulose in the presence or absence of IM. Colonies were counted 10 days later. Values are representative of 2 independent experiments. Values represent mean ± SEM (B and C). (D) ATG5 and ATG7 levels in siRNA transfected CML cells. Western blots show ATG5 and ATG7 expression in M351T CML-CP lysates 24 hours after transfection with scrambled or Atg5- or Atg7-specific siRNA. GRB2 levels were measured as loading control. The asterisk indicates that bands are not specific for LC3.
To assess whether IM-induced autophagy provided a survival advantage to these cells, CFC assays were performed after a 24 hour treatment with IM, CQ, or the IM/CQ combination. Treatment with CQ (10.0 μM) or IM (0.5 μM) alone had a modest effect on colony formation of these cells; by contrast, fewer colonies formed after treatment with 1.0 μM IM, and there was an additional 60% decrease upon treatment with the IM/CQ combination (Figure 7B). Finally, to determine whether genetic ablation of autophagy had similar effects on colony formation, we employed RNA interference of ATG5 or ATG7 to suppress autophagy in primary CML cells with the M351T mutation. Remarkably, knockdown of ATG5 or ATG7 expression (Figure 7D) enhanced the colony formation inhibitory effect of IM (Figure 7C).
CFC assays were also performed upon IM, CQ, and IM/CQ treatment of peripheral blood mononuclear cells from newly diagnosed CML-CP patients. Colony number was decreased in the presence of IM/CQ compared with the presence of IM alone (Supplemental Figure 10, A–D). Comparable results were obtained with 2 additional CML samples (data not shown). We next analyzed colony formation of mononuclear CML cells (70% CD34+) from a relapsed CML patient carrying the partially IM-resistant V299L p210BCR/ABL mutant (20). Like the CML sample with the M351T mutation (Figure 7B), CML cells became more sensitive to low doses of IM upon co-treatment with CQ (Supplemental Figure 10E). In contrast, normal marrow mononuclear (n = 3) or peripheral blood CD34+ cells (n = 2) did not show a further decrease in colony formation when treated with the IM/CQ combination (Supplemental Figure 10, F and G), demonstrating that the effect of the combination was specific for p210BCR/ABL-expressing cells.
Similar results were obtained using cells from a newly diagnosed CML-CP patient that had just started IM treatment (3 days at 800 mg/day; CML-CP-t-1; Supplemental Figure 11A) and a CML-CP patient undergoing IM treatment (10 days at 100 mg/day; CML-CP-t-2; Supplemental Figure 11B; low IM doses were used due to development of bone marrow aplasia). Interestingly, CQ alone markedly suppressed colony formation of CML cells from patient CML-CP-t-1, suggesting that high-dose IM treatment was particularly effective in sensitizing CML cells to CQ, probably because of induction of autophagy in vivo. Indeed, LC3-positive vacuoles were readily detected in peripheral blood mononuclear cells of both patients (data not shown).
TKI treatment in combination with inhibition of autophagy results in near complete eradication of CML stem cells.
Previous studies have shown that in vitro treatment with TKIs does not eradicate CML stem cells (23–27). To determine whether TKI-induced autophagy is a potential survival mechanism for this cell population, CFC and LTC-IC assays were performed. For CFC assays, CML-CP cells (n = 3) were first sorted into primitive (CD34+CD38–) and more mature progenitor (CD34+CD38+) populations and were left untreated or pretreated for 48 hours with TKIs (IM, nilotinib, or Das) or autophagy inhibitors (CQ or Ba) alone or the TKI/CQ/Ba combination in serum free medium (SFM), supplemented with a 5 growth factor (5GF) cocktail. The number of CFCs obtained for each sample was then compared with a baseline — cells which had no prior culture or treatment (100%). Notably, treatment with IM (2 μM) and Das (150 nM) induced accumulation of LC3-positive vesicles in primitive progenitor cells (Figure 8A). As shown in Figure 8B for the CD34+CD38+ population, the untreated cells showed a doubling of CFCs over the 48 hours “pretreatment”, consistent with stem/progenitor cell expansion in the culture medium. Neither CQ nor Ba alone had any effect on this expansion. However, as shown previously (19, 24), the TKIs IM, Das (10 nM), and nilotinib all exhibited an antiproliferative effect, which prevented any CFC expansion over baseline. Das, at the highest clinically achievable concentration (150 nM), was able to reduce the number of CFCs to 55% of baseline, suggesting some level of apoptosis. This effect was dramatically increased following TKI/autophagy inhibitor combination treatment, with 80%–95% reduction in colony formation versus baseline, and even more impressive compared with the no drug control (Figure 8B). For CD34+CD38– cells (Figure 8C), that are consistently resistant to TKIs, treatment with TKI/autophagy inhibitor was extremely effective, with only 2% CFCs remaining following Das/CQ exposure (150 nM and 10.0 μM, respectively).
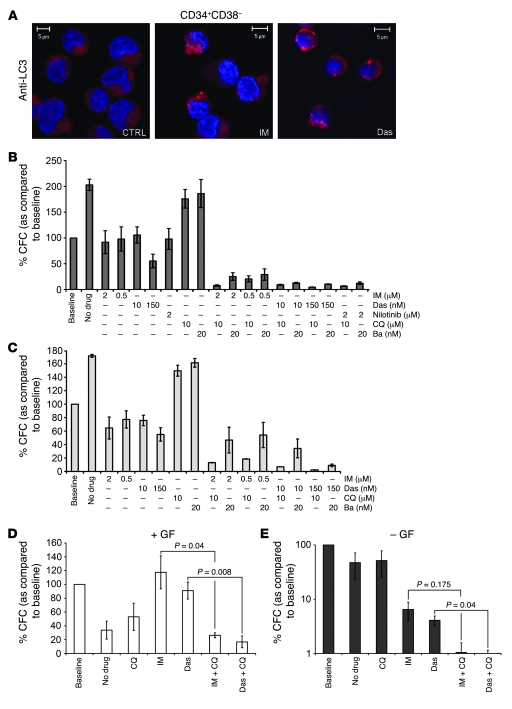
Figure 8. TKI treatment in combination with inhibition of autophagy results in near complete eradication of CML stem cells.
(A) Induction of LC3-positive autophagosomes in TKI-treated CML stem cells. CD34+CD38– cells were stained with an anti-LC3 antibody and analyzed by confocal microscopy. Nuclei were stained with DAPI; CML-CP CD34+ cells (n = 3) were sorted into progenitor (CD34+CD38+) (B) and stem (CD34+CD38–) (C) cell populations. Cells were left untreated or pretreated for 48 hours with IM (0.5 and 2.0 μM), Das (10.0 and 150.0 nM), or Nilotinib (2.0 μM) (CD34+CD38+ cells only), either alone or in combination with CQ (10.0 μM) or Ba (20.0 nM). Cells were then plated in methylcellulose in the presence or absence of indicated drug concentrations (as per pretreatment). Colonies were counted at 14 days and compared with those derived from cells at baseline (cells with no prior culture or treatment) taken as 100%. Values represent the mean ± SEM. (D and E) For long-term culture experiments, CML-CP CD34+ cells (n = 3) were cultured in SFM with (D) and without (E) growth factors (GFs) and left untreated or pretreated for 6 days with TKIs (IM, 2 μM, or Das, 150 nM) or CQ (10 μM) alone or the TKI/CQ combination, before being added to the LTC-IC feeders. Resulting colonies were compared with those derived from cells at baseline (cells with no prior culture or treatment) and taken as 100%. Values represent the mean ± SEM. E is presented on a logarithmic scale due to the great differences between baseline and TKI/combination-treated arms.
Since the CFC assay measures a relatively mature, committed progenitor cell population, LTC-IC assays were also carried out to measure the effect of autophagy inhibition on the functionality of more primitive CML cells. As the presence of cytokines in the culture media may alter the requirement of autophagy for cell survival, the LTC-IC assays were carried out both in the presence and absence of a 5GF cocktail. CML-CP CD34+ cells (n = 3) were left untreated or pretreated for 6 days with TKIs (IM or Das) or CQ alone or the TKI/CQ combination. As with the CFC assay, the number of LTC-ICs obtained for each sample was then compared with a baseline. With 5GF, untreated and CQ-treated cells formed 67% and 47% fewer colonies than cells at baseline, respectively, suggesting that a large proportion of these cells has proceeded toward terminal differentiation (Figure 8D). Consistent with the CFC data and previous studies (24), treatment with IM or Das was antiproliferative, even in the presence of 5GF, and therefore resulted in a protective effect on the number of LTC-ICs over baseline. Combination treatment with TKIs and autophagy inhibitors counteracted this protective effect and resulted in an impressive decrease in the number of LTC-ICs, with a significant reduction for treatments with IM/CQ and Das/CQ combinations versus TKI alone (approximately 4.5- and 6.0-fold decrease, respectively) (Figure 8D). Untreated and CQ-treated cells cultured without supplemental growth factors formed reduced numbers of colonies, as compared with baseline cells (Figure 8E). The removal of growth factors combined with TKI treatment had a dramatic effect on the number of LTC-ICs (6.5% and 4.1% for IM and Das, respectively), suggesting that cytokines may play an important protective role for the survival of TKI-treated CML cells, consistent with the results of a previous study (58). Combination treatment with CQ resulted in an even greater decrease in LTC-IC numbers (1% and 0.6% for IM/CQ and Das/CQ, respectively), indicating that BCR/ABL inhibition via TKI treatment combined with autophagy inhibition of growth factor–starved CML cells promotes the most effective elimination of primitive CML cells.
Discussion
IM has revolutionized the therapy of CML, as most patients in the CP of the disease undergo durable remission when treated with this drug on a continuous basis. However, development of resistance and the limited efficacy of IM in CML-BC patients represent shortcomings of the therapy. In more than 40% of cases, resistance is the result of mutations inside and around the kinase domain of BCR/ABL that impair IM binding (14–18). Resistance also occurs through overexpression of BCR/ABL or gene amplification (59), involvement of glycoproteins (60), activation of Lyn kinase (61–63), and other mechanisms. To address this problem, new TKIs, such as the dual Src/BCR/ABL inhibitor Das, were developed (64, 65). However, Das treatment is also ineffective in CML patients carrying the BCR/ABL T315I mutation that prevents IM binding to the kinase domain (65). Although more potent and specific BCR/ABL inhibitors are being developed and tested in clinical trials, an ongoing problem with this approach is the selection of additional mutations, causing drug resistance in patients (66). In CML patients in CP, who are responding optimally to IM therapy, minimal residual disease is detectable using highly sensitive quantitative PCR, suggesting that the most primitive cells are not eradicated by TKIs. We have previously demonstrated that CML stem cells overexpress p210BCR/ABL and are inherently resistant to IM, Das, nilotinib, and bosutinib (24–27). For these reasons, alternative approaches that combine a TKI with a second agent exhibiting a different mechanism of action are required if we are to further improve CML therapy.
One of the most evident effects of IM on CML cells is induction of cell death due to insufficient survival signals following BCR/ABL inhibition. However, it is still unclear whether IM affects other cellular functions. We reasoned that it would be important to evaluate additional intracellular changes induced by IM treatment and to investigate whether some of the changes could be exploited for therapeutic intervention. We show here that cell shrinkage is one of the more evident effects of IM treatment in the CML cell lines K562 and BV173. This occurs in viable cells and is characterized by reduction of cytoplasmic content and accumulation of vacuoles, a phenomenon reminiscent of cell atrophy following growth factor deprivation–induced autophagy in myeloid cells (33). Indeed, IM treatment of BCR/ABL-expressing cell lines and primary cells induced morphological and biochemical hallmarks of autophagy such as generation of autophagosomes, LC3-positive autophagy-like vacuoles, and LC3-II accumulation. In the BV173 line, vacuoles were only evident upon treatment with CQ, which blocks degradation inside autophagolysosomes, suggesting that high lysosomal activity was responsible for the efficient turnover of LC3 and its associated vacuoles. By contrast, IM did not cause autophagy in v-Src–transformed 32D cells, IM-resistant 32D-p210BCR/ABL cells, and HeLa cells. Expression of an IM-resistant c-Abl mutant did not inhibit induction of autophagy in 32D-p210BCR/ABL cells, clearly demonstrating that the effect of the drug is specific and occurs predominantly through inhibition of BCR/ABL. Overall, these findings suggest that a sudden decrease of survival signals, triggered by inhibition of the p210BCR/ABL tyrosine kinase, induces the rapid activation of an autophagy program.
We next focused on the potential pathways involved in IM-dependent induction of autophagy. Recent studies have shown that ER stress can trigger autophagy by several mechanisms (52, 53). Interestingly, BCR/ABL has been shown to regulate ER Ca2+ homeostasis and inhibit ER stress–induced cell death (67). Therefore, it is conceivable that inhibition of p210BCR/ABL could affect ER function. Indeed, IM rapidly induced expression of the ER stress markers CHOP and Grp78, and this preceded the accumulation of LC3-II, suggesting that ER stress is implicated in IM-triggered autophagy. Interestingly, Ca2+ was released from the ER following IM treatment and was required for autophagy induction. Together, these data suggest that induction of ER stress upon BCR/ABL inhibition may cause autophagy. ER stress has been reported to trigger autophagy through inhibition of mTOR (52, 53). Consistent with this, treatment of K562 cells with the mTOR inhibitor, rapamycin, induced the conversion of LC3-I into the autophagosome-associated LC3-II (data not shown).
What would the role of autophagy be in IM-treated p210BCR/ABL-expressing cells? We hypothesize that autophagy represents a condition upon which BCR/ABL-transformed cells rely to reach a state of metabolic inertia following cessation of antiapoptotic signals. Therefore, autophagy would represent a survival mechanism. To directly test this, we assessed whether IM-induced cell death would be enhanced by chemical and genetic inhibition of autophagy using CQ to inhibit the last step of the autophagic process (i.e., the degradation of autophagosomes following fusion with lysosomes; ref. 34) and RNA interference of essential autophagy genes. Remarkably, CQ treatment potentiates IM-induced cell death. However, this effect is abrogated in cells expressing the IM-resistant T315I p210BCR/ABL mutant but is not affected by expression of the IM-resistant T315I c-Abl mutant (data not shown). This confirms that inhibition of BCR/ABL is the main trigger of autophagy. CQ also has a sensitizing effect on IL-3–deprived parental 32D cells undergoing autophagy (data not shown), again supporting the notion that BCR/ABL inhibition mimics growth factor deprivation and that, in this context, autophagy is a protective mechanism. CQ-dependent potentiation of IM-induced cell death (by annexin V staining and clonogenic assays) is also evident in BV173 and K562 cells, including a partially IM-resistant K562 subline. Inhibition of autophagy by RNA interference–mediated knockdown of either ATG5 or ATG7 resulted in comparable sensitizing effects, demonstrating that the effects of CQ and Ba depend, to a large extent, on suppression of autophagy. A recent study suggested that the cell death–promoting effect of CQ in a model of Myc-induced lymphomagenesis is p53 dependent (34); this mechanism clearly does not explain the effect of CQ (or Ba) in the p53-null K562 cells (68). Since p53 is mutated in approximately 25% of CML-BC patients (69, 70), it would be attractive to be able to enhance the therapeutic effects of TKIs via p53-independent mechanisms. Remarkably, the IM/CQ combination is effective in primary CML cells and also in cells carrying partially IM-resistant p210BCR/ABL mutants, while normal mononuclear and CD34+ cells were much less affected. Treatment with CQ alone had some inhibitory effect on colony formation of CML mononuclear cells and, to a lesser degree, of the normal mononuclear cells. Whether this reflects a baseline level of autophagy in normal and CML late myeloid progenitor/precursors is unclear; however, early progenitors from healthy donors and CML patients were almost completely insensitive to treatment with CQ alone, suggesting that CQ toxicity to normal marrow progenitors may not be a concern in CML patients treated with the TKI/CQ combination.
To date TKIs used as single agents have been ineffective in killing primitive CML cells (22–27). Recently, we demonstrated that the combination of a TKI with the farnesyl transferase inhibitor, BMS-214662, resulted in potent, selective eradication of CML stem cells, including functionally defined CFCs and LTC-ICs (24). Importantly, we show here that these cells undergo autophagy upon TKI treatment and are highly susceptible to killing by the combination of TKIs and inhibition of autophagy. These data are extremely encouraging and suggest it will be possible to target the cancer stem cell population in a selective manner without compromising the survival of normal stem cells. Finally, bone marrow cells isolated from a CML-CP patient responsive to IM treatment appear to be particularly sensitive to treatment with CQ alone, suggesting that IM treatment can induce autophagy in vivo.
Overall, our findings clearly demonstrate that (a) autophagy acts as a survival signal in p210BCR/ABL-expressing cells treated with TKIs; (b) its inhibition can be exploited to potentiate TKI-induced cell death, raising the possibility that the combination of IM and CQ, or more clinically applicable inhibitors of autophagy, may be beneficial in the treatment of CML (Figure 9); and (c) that such a combination can target the TKI-resistant CML stem cell population. Consistent with the concept that induction of autophagy provides a survival mechanism to IM-treated CML cells, a recent report has proposed that resistance of CML cells to histone deacetylase inhibitors can be reverted by using autophagy inhibitors (71).
Figure 9. Inhibition of autophagy potentiates IM-induced cell death in CML cells.
In addition to programmed cell death, inhibition of BCR/ABL causes autophagy, which acts as a survival mechanism in CML cells. Treatment with autophagy inhibitors such as CQ results in potentiation of IM-induced cell death.
More generally, our work reinforces the notion that cancer cells, including cancer stem cells, can survive in a stressful environment, following inhibition of critical oncogenic pathways, by inducing autophagy. These tumor cells are primed to resume proliferation under more favorable conditions, e.g., if drug concentrations drop below a certain threshold, due perhaps to drug withdrawal related to toxicity or noncompliance, or if mutations that promote drug resistance develop.
Thus, future therapeutic strategies that aim to inhibit autophagy in cancer cells treated with conventional chemotherapy or novel small molecules, including TKIs, represent a promising approach that will target both bulk tumor and cancer stem cells.
Methods
Reagents.
IM and nilotinib were obtained from Novartis Pharma. A 100 mM stock solution of IM in sterile distilled water was stored at 4°C. A 10 mM stock solution of nilotinib in dimethyl sulfoxide (Sigma-Aldrich) was stored at –20°C. Das was purchased from LC Laboratories. Stock solutions of 10 mg/ml were stored at –20°C. Dilutions of these drugs in PBS were freshly prepared for each experiment.
Cell cultures.
BV173 and K562 cells were cultured in Iscove’s modified Dulbecco’s medium (IMDM), supplemented with l-glutamine, 10% FBS (Hyclone), and penicillin/streptomycin. All other hematopoietic cell lines (32D and derivatives) used in this study were cultured in IMDM with l-glutamine, supplemented with 10% FBS and penicillin/streptomycin. CML cells were obtained from newly diagnosed or relapsed patients after receiving written informed consent, and approval was obtained from the North Glasgow University Hospital Division of NHS Greater Glasgow Institutional Review Board or the Bioethical Committee of the University of Catania. Mononuclear cells were obtained by centrifugation on a Ficoll-Hypaque gradient (Amersham Biosciences), while CD34+ cells were FACS sorted with the PE-conjugated anti-human CD34 antibody (clone 563; BD Biosciences — Pharmingen). Presence of mutations in the kinase domain was assessed by sequencing. Normal marrow mononuclear cells were obtained from healthy donors with written informed consent and approval of the Bioethical Committee of the University of Catania. Peripheral blood (G-CSF–mobilized) normal CD34+ cells were purchased from StemCell Technologies. Cells were cultured in StemSpan SFEM medium (StemCell Technologies), supplemented with a cytokine cocktail (StemSpan CC100; StemCell Technologies), containing Flt-3 ligand (100 ng/ml), Kit ligand (100 ng/ml), IL-3 (20 ng/ml), and IL-6 (20 ng/ml). HeLa cells were cultured in Dulbecco’s modified Eagle’s medium, supplemented with 10% FBS and penicillin/streptomycin. Cells were maintained in a Heraeus CO2 Auto-Zero incubator at 37°C with 5% CO2. All reagents were purchased from GIBCO unless otherwise stated.
Isolation of CD34+CD38+ and CD34+CD38– cells.
Fresh leukapheresis or peripheral blood samples were obtained with written informed consent from patients with newly diagnosed CML in CP (n = 6). Samples were enriched for CD34+ cells using CliniMACS (Miltenyi Biotec) according to the manufacturer’s instructions and were cryopreserved in 10% (v/v) dimethyl sulfoxide in ALBA (4% [w/v] human albumin solution; Scottish National Blood Transfusion Service). Purity of CML CD34+ cells was more than 95% and more than 95% were Ph1 as assessed by dual-color dual-fusion fluorescence in situ hybridization. CD34+ CML cells were simultaneously stained with allophycocyanin-conjugated CD34 and FITC-conjugated CD38 (BD Biosciences) and sorted to isolate the CD34+ and the CD34+CD38– subpopulations using a FACSAria (BD Biosciences). Cells were then cultured in IMDM (Sigma-Aldrich), supplemented with a serum substitute (BIT; StemCell Technologies), 10 mg/ml LDL, 0.1 μM 2-mercaptoethanol (Sigma-Aldrich), 1% glutamine (100 mM; Invitrogen), and 1% penicillin-streptomycin (100 mM; Invitrogen) (SFM). SFM was further supplemented with a 5GF cocktail, composed of 100 ng/ml Flt-3 ligand and 100 ng/ml Kit ligand and 20 ng/ml each of IL-3 and IL-6 (StemCell Technologies) and G-CSF (Chugai Pharma Europe Ltd.) as indicated.
Colony forming assays.
K562 cells, primary CML cells, normal marrow mononuclear cells, and peripheral blood CD34+ cells were treated for 24 hours with the indicated concentrations of CQ or IM, individually or in combination. Drug-treated cells were seeded in Methocult H4230 methylcellulose medium (StemCell Technologies) in the absence of cytokines (K562 cells) or in the presence of a cytokine cocktail (StemSpan CC100; StemCell Technologies), containing Flt-3 ligand (100 ng/ml), Kit ligand (100 ng/ml), IL-3 (20 ng/ml), and IL-6 (20 ng/ml) (CML, normal marrow mononuclear, and CD34+ cells) with the indicated concentrations of CQ or IM. Formation of colonies was assessed 7 days after plating 1,000 (K562), 2,500 (normal CD34+), or 25,000–50,000 (normal mononuclear and primary CML) cells per plate. CD34+CD38+ and CD34+CD38– CML-CP cells were pretreated for 48 hours with the indicated concentrations of IM, Das, nilotinib, CQ, and Ba, either alone or in combination. Untreated and drug-treated cells and cells that had no prior culture or treatment (baseline) were seeded in Methocult methylcellulose medium (StemCell Technologies), in the presence of indicated drug concentrations (from pretreatment). Formation of colonies was assessed 14 days after plating 10,000 cells per plate.
LTC-IC assays.
M2-10B4 cells and S1/S1 fibroblasts were first established as feeder layers and then irradiated at 80 Gy. CML-CP CD34+ were pretreated with or without 5GF for 6 days with IM (2 μM), Das (150 nM), and CQ (10 μM), alone or in combination. All CML cells remaining after culture were washed twice to remove residual drugs, and then untreated and drug-treated cells and cells that had no prior culture or treatment (baseline) were plated in duplicated on the irradiated feeders in Myelocult medium (StemCell Technologies). Cultures were maintained for 5 weeks with weekly half medium changes. After 5 weeks, cell counts were performed, and 1 × 105/ml cells were transferred to CFC assays and maintained in culture for an additional 2 weeks in Methocult medium (StemCell Technologies) before colonies were scored.
Treatment of leukemic mice with IM, CQ, or the IM/CQ combination.
Sublethally irradiated C3H/HeJ mice were injected intravenously with MigRI GFP-LC3–transduced 32D-p210BCR/ABL cells (2.5 × 105 cells per mouse) and divided into 4 groups (untreated, treated with IM only, treated with CQ only, or treated with the IM/CQ combination). For IM-only treatment, mice were treated with 100 mg/kg/day for 5 consecutive days (by gavage), starting 14 days after injection of leukemic cells. For CQ only treatment, mice were treated with 60 mg/kg/day for 7 consecutive days (by intraperitoneal injection), starting 12 days after injection of leukemic cells. For the IM/CQ combination treatment, mice were pretreated with CQ (60 mg/kg/mouse) for 2 days (starting at day 12 after injection of leukemic cells) and, on day 14, were treated with IM and CQ at the dose and schedule indicated above for single treatments. Untreated and treated mice were sacrificed 20 days after injection of LC3-GFP 32D-p210BCR/ABL cells.
Fluorimetric assay of caspase activity.
Activity of the effector caspases 3 and 7 in cell lysates was assessed by the cleavage of carbobenzoxy-Asp-Glu-Val-Asp-7-amino-trifluoromethyl coumarin (Z-DEVD.AFC) (72) and measured (excitation/emission = 405/510 nm) at 37°C in 96-well plates in 200 μl of assay buffer (20 μM Z-DEVD.AFC, 0.1% for 3-[(3-cholamidopropyl)dimethylammonio]-propanesulfonic acid, 10 mM dithiothreitol, 100 mM HEPES, and 10% sucrose, pH 7.0) using a Wallac Victor2 1420 Multilabel counter. The 96-well plates were assayed for 10 cycles, and cleavage rates were determined by linear regression. The protease activities were expressed as pmol/min/mg protein.
Quantification of Ca2+ content in the ER.
p210BCR/ABL 32D cells were incubated with 2 μM IM in cell culture medium (see above). For the last 45 minutes, cells were additionally incubated with the fluorescent Ca2+ indicator Fura-2 AM. At the end of this time period, cells were pelleted and then resuspended in a Ca2+-free Krebs-Henseleit buffer (118 mM NaCl, 4.7 mM KCl, 1.2 mM MgSO4, 1.2 mM KH2PO4, 4.2 mM NaHCO3, 10 mM HEPES, 11.7 mM glucose, pH 7.4). The cell suspension was placed in a UV grade cuvette, and intracellular Ca2+ was measured in a PerkinElmer LS55 fluorimeter. Cells were stirred continuously and maintained at 37°C. The content of the ER Ca2+ pool was estimated from the magnitude of Ca2+ rise evoked by addition of the SERCA pump inhibitor thapsigargin (5 μM).
Retroviral infection.
For retroviral infection, Phoenix cells (gift of G.P. Nolan, Stanford University School of Medicine, Stanford, California, USA) were transiently transfected with the indicated plasmids. The infectious supernatant was collected 24 hours later and used for infection. Twenty-four hours later, infected cells were sorted (EPICS Profile Analyzer, Coulter) for GFP expression.
Determination of autophagy.
Autophagy was assessed in cells cultured in the presence or absence of IM by measuring levels of cytosolic LC3-I and autophagosomes-associated LC3-II. Briefly, cells were plated at a density of 1 × 105 cells/ml unless otherwise stated. Cells were cultured in the presence of 1 to 4 μM IM and harvested 6 or 12 hours later. The effect of Ca2+ on IM-dependent autophagy was assessed by culturing the cells in the presence or absence of 5 mM EGTA (Sigma-Aldrich) and/or 10 μM Bapta/AM (Calbiochem) for 6 or 12 hours. Then, 3.5 × 105 cells were harvested, rinsed twice with cold PBS (Sigma-Aldrich), and lysed directly in Laemmli’s sample buffer, supplemented with β-mercaptoethanol (Sigma-Aldrich). Finally, protein extracts were subjected to SDS-PAGE and analyzed by standard Western blotting. HeLa cells stably expressing EGFP-LC3 were seeded at 1.5 × 105/ml in a 6-well plate and left adhering overnight. The following day, media were replaced and cells were cultured in the presence or absence of 1 and 5 μM IM for 6 or 12 hours. Cells were then harvested, and protein levels were measured. Primary antibodies used were anti-LC3 (Sigma-Aldrich) and anti-actin (Sigma-Aldrich). To assess expression of Grp78, cells were cultured in the presence of 2 μM IM and harvested 1, 3, and 6 hours later. Grp78 was detected using anti-BiP/Grp78 monoclonal antibody (BD Transduction Laboratories). Anti-rabbit and -mouse horseradish peroxidase-conjugated secondary antibodies were purchased from Amersham and ECL solution was purchased from Pierce.
Immunohistochemistry.
Morphological analysis was performed by standard May-Grünwald/Giemsa staining as described (73). In brief, cells were cytospun on top on poly-l-lysine coated microscope slides (Menzel) at 6.72 g for 5 minutes using a cytocentrifuge (Shandon). Slides were dried for 30 minutes at room temperature, stained for 3 minutes with a May-Grünwald solution (Sigma-Aldrich), and rinsed with an excess of bidistilled H2O for 1 minutes. Then, nuclei were counterstained for 30 minutes using a Giemsa solution (Sigma-Aldrich) and rinsed as described above. Slides were analyzed using an Axiostar direct light microscope (Zeiss) and pictures of cells were acquired using a ProgRes C-14 RGB camera.
IF.
IF was performed as described (74) with some modifications. Briefly, 5 × 104 cells were cytospun on poly-l-lysine coated microscope slides (Menzel) at 6.72 g for 5 minutes by using a cytocentrifuge (Shandon). Slides were dried for 30 minutes, fixed with 4% paraformaldehyde (Sigma-Aldrich), permeabilized with 0.1% Triton-X 100 (Sigma-Aldrich) for 3 minutes, and incubated with a primary antibody for 1 hour. FITC- or TRITC-conjugated secondary anti-mouse antibodies used in this study were purchased from Molecular Probes. Finally, nuclei were counterstained by Hoechst (Sigma-Aldrich), and slides were analyzed using a laser scanning confocal microscope (LSM 510 Zeiss system). Primary antibody used in this study was anti-LC3 (Nanotools).
Quantification of cell death.
Apoptosis was quantified by phosphatidylserine externalization in the presence of propidium iodide as described (75). In brief, cells were cultured in the presence of 1 or 2 μM IM, alone or in combination with 5 μM CQ for 12 and 48 hours. Cells were stained with allophycocyanin-conjugated annexin V (Molecular Probes, Invitrogen) and propidium iodide (Sigma-Aldrich) and analyzed using a FACSCalibur system (Becton Dickinson). Data were expressed as mean ± SEM of 3 independent experiments and analyzed by unpaired Student’s t test (Graphpad Software Inc.), and P values less than 0.05 were considered statistically significant.
EM and immunogold cytochemistry.
Cells were fixed with 2% glutaraldehyde in 0.1 M sodium cacodylate buffer, pH 7.4, postfixed with 1% osmium tetroxide/1% potassium ferrocyanide, en bloc stained with 5% uranyl acetate, and embedded in Taab epoxy resin (Taab Ltd.). Duplicate pellets were fixed with 4% formaldehyde, freshly made from paraformaldehyde in Dulbecco’s PBS, pH 7.4, for 1 hour at room temperature. These cells were embedded in LR-White Resin (Agar Scientific); ultrathin sections were incubated with the anti-GFP antibody and labeled with immunogold (British Biocell International). Control incubations involved the replacement of primary antibody with an equivalent concentration of an isotype-specific IgG. All sections were examined unstained or after staining with lead citrate and/or uranyl acetate.
RNA interference.
For siRNA transfection, 1 × 106 cells were resuspended in 100 μl of nucleofector V solution and cell suspensions were mixed with 5 μg of siRNA. The solution was added to the Amaxa electrode cuvettes and electroporated in Amaxa Electroporator II, using program T-16 for K562 and U-08 for CD34+ CML cells, respectively. Immediately afterward, cells were diluted in 2 ml IMDM (supplemented with 10% FBS, 100 U/ml penicillin, 0.1 mg/ml streptomycin, and 2 mM l-glutamine) at 37°C (5 × 105 cells/ml). The siRNA corresponding to the human cDNA sequence for ATG5 (5′-GCAACTCTGGATGGGATTG-3′) and ATG7 (5′-CAGTGGATCTAAATCTCAAACTGAT-3′) were synthesized by Sigma-Aldrich and Ambion/Applied Biosystems. Control siRNAs were purchased from Ambion. Downregulation of ATG5 and ATG7 expression was verified by Western blotting with the anti-ATG5 rabbit polyclonal (catalog no. 2630; Cell Signalling Technology) and anti-ATG7 rabbit polyclonal (catalog no. 2631; Cell Signalling Technology) antibodies.
For stable ATG7 downregulation, scrambled and ATG7 shRNA pGIPZ vectors were purchased from Open Biosystems. Lentivirus supernatants were prepared following the manufacturer’s instructions and lentiviral infections carried out accordingly.
Statistics.
Means were compared using the unpaired, 2-tailed Student’s t test. A P value of less than 0.05 was considered statistically significant in all calculations.
Supplementary Material
Acknowledgments
We thank Craig Thompson for the gift of plasmid MigRI–EGFP-LC3 and Elisabeth Buchdunger (Novartis) for the gift of IM. We also thank Marja Jaattela (Danish Cancer Society, Copenhagen, Denmark); Gerry Cohen, Ed Bampton, Mario Rossi, Gerry Melino, and Melania Capasso (Medical Research Council Toxicology Unit, Leicester, United Kingdom); and Tommaso Zanocco (University of Modena and Reggio Emilia, Modena, Italy) for reagents and critical discussion. P. Salomoni, A. Hamilton, and G.V. Helgason were supported by the Medical Research Council, United Kingdom. A. Hamilton was partly funded by the Leukaemia Research Fund, United Kingdom. This work was supported, in part, by NCI grant PO1 CA78890 to B. Calabretta. M.R. Lidonnici and A.R. Soliera were supported, in part, by a fellowship of the American-Italian Foundation for Cancer Research (AIFCR). C. Bellodi was the recipient of a PhD studentship at the University of Leicester, United Kingdom. We thank all the patients and United Kingdom haematologists who have provided normal and CML samples for our Biobank.
Footnotes
Authorship note: Cristian Bellodi, Maria Rosa Lidonnici, and Ashley Hamilton contributed equally to this work.
Conflict of interest: The authors have declared that no conflict of interest exists.
Nonstandard abbreviations used: Ba, bafilomycin A1; Bapta/AM, bis-(0-aminophenoxy)-ethane-N,N,N,N′-tetraacetic acid/tetra (acetoxymethyl)-ester; BC, blast crisis; CML, chronic myeloid leukemia; CP, chronic phase; CQ, chloroquine; 32D cell, 32Dcl3 cell; Das, dasatinib; EGFP-LC3, LC3 fused to the EGFP; IF, immunofluorescence; IM, imatinib mesylate; LC3, microtubule-associated protein 1 light chain 3; LTC-IC, long-term culture–initiating cell; Ph1, Philadelphia chromosome; SFM, serum free medium; TKI, tyrosine kinase inhibitor; zVAD, N-benzyloxycarbonyl-Val-Ala-Asp-fluoromethylketone.
Citation for this article: J. Clin. Invest. 119:1109–1123 (2009). doi:10.1172/JCI35660
References
- 1.Kantarjian H.M., et al. Chronic myelogenous leukemia in blast crisis. Analysis of 242 patients. Am. J. Med. 1987;83:445–454. doi: 10.1016/0002-9343(87)90754-6. [DOI] [PubMed] [Google Scholar]
- 2.Savage D.G., Szydlo R.M., Goldman J.M. Clinical features at diagnosis in 430 patients with chronic myeloid leukaemia seen at a referral centre over a 16-year period. Br. J. Haematol. 1997;96:111–116. doi: 10.1046/j.1365-2141.1997.d01-1982.x. [DOI] [PubMed] [Google Scholar]
- 3.Spiers A.S. Clinical manifestations of chronic granulocytic leukemia. Semin. Oncol. 1995;22:380–395. [PubMed] [Google Scholar]
- 4.Ben-Neriah Y., Daley G.Q., Mes-Masson A.M., Witte O.N., Baltimore D. The chronic myelogenous leukemia-specific P210 protein is the product of the bcr/abl hybrid gene. Science. 1986;233:212–214. doi: 10.1126/science.3460176. [DOI] [PubMed] [Google Scholar]
- 5.Cortez D., Kadlec L., Pendergast A.M. Structural and signaling requirements for BCR-ABL-mediated transformation and inhibition of apoptosis. Mol. Cell. Biol. 1995;15:5531–5541. doi: 10.1128/mcb.15.10.5531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gordon M.Y. Biological consequences of the BCR/ABL fusion gene in humans and mice. J. Clin. Pathol. 1999;52:719–722. doi: 10.1136/jcp.52.10.719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goga A., McLaughlin J., Afar D.E., Saffran D.C., Witte O.N. Alternative signals to RAS for hematopoietic transformation by the BCR-ABL oncogene. Cell. 1995;82:981–988. doi: 10.1016/0092-8674(95)90277-5. [DOI] [PubMed] [Google Scholar]
- 8.Skorski T., et al. Phosphatidylinositol-3 kinase activity is regulated by BCR/ABL and is required for the growth of Philadelphia chromosome-positive cells. Blood. 1995;86:726–736. [PubMed] [Google Scholar]
- 9.Vivanco I., Sawyers C.L. The phosphatidylinositol 3-Kinase AKT pathway in human cancer. Nat. Rev. Cancer. 2002;2:489–501. doi: 10.1038/nrc839. [DOI] [PubMed] [Google Scholar]
- 10.Sebolt-Leopold J.S., Herrera R. Targeting the mitogen-activated protein kinase cascade to treat cancer. Nat. Rev. Cancer. 2004;4:937–947. doi: 10.1038/nrc1503. [DOI] [PubMed] [Google Scholar]
- 11.Van Etten R.A. Oncogenic signaling: new insights and controversies from chronic myeloid leukemia. J. Exp. Med. 2007;204:461–465. doi: 10.1084/jem.20062335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ottmann O.G., et al. A phase 2 study of imatinib in patients with relapsed or refractory Philadelphia chromosome-positive acute lymphoid leukemias. Blood. 2002;100:1965–1971. doi: 10.1182/blood-2001-12-0181. [DOI] [PubMed] [Google Scholar]
- 13.Sawyers C.L., et al. Imatinib induces hematologic and cytogenetic responses in patients with chronic myelogenous leukemia in myeloid blast crisis: results of a phase II study. Blood. 2002;99:3530–3539. doi: 10.1182/blood.v99.10.3530. [DOI] [PubMed] [Google Scholar]
- 14.Hofmann W.K., et al. Ph(+) acute lymphoblastic leukemia resistant to the tyrosine kinase inhibitor STI571 has a unique BCR-ABL gene mutation. Blood. 2002;99:1860–1862. doi: 10.1182/blood.v99.5.1860. [DOI] [PubMed] [Google Scholar]
- 15.Branford S., et al. Detection of BCR-ABL mutations in patients with CML treated with imatinib is virtually always accompanied by clinical resistance, and mutations in the ATP phosphate-binding loop (P-loop) are associated with a poor prognosis. Blood. 2003;102:276–283. doi: 10.1182/blood-2002-09-2896. [DOI] [PubMed] [Google Scholar]
- 16.Nardi V., Azam M., Daley G.Q. Mechanisms and implications of imatinib resistance mutations in BCR-ABL. Curr. Opin. Hematol. 2004;11:35–43. doi: 10.1097/00062752-200401000-00006. [DOI] [PubMed] [Google Scholar]
- 17.Deininger M., Buchdunger E., Druker B.J. The development of imatinib as a therapeutic agent for chronic myeloid leukemia. Blood. 2005;105:2640–2653. doi: 10.1182/blood-2004-08-3097. [DOI] [PubMed] [Google Scholar]
- 18.Soverini S., et al. ABL mutations in late chronic phase chronic myeloid leukemia patients with up-front cytogenetic resistance to imatinib are associated with a greater likelihood of progression to blast crisis and shorter survival: a study by the GIMEMA Working Party on Chronic Myeloid Leukemia. J. Clin. Oncol. 2005;23:4100–4109. doi: 10.1200/JCO.2005.05.531. [DOI] [PubMed] [Google Scholar]
- 19.Graham S.M., et al. Primitive, quiescent, Philadelphia-positive stem cells from patients with chronic myeloid leukemia are insensitive to STI571 in vitro. Blood. 2002;99:319–325. doi: 10.1182/blood.v99.1.319. [DOI] [PubMed] [Google Scholar]
- 20.Shah N.P., et al. Overriding imatinib resistance with a novel ABL kinase inhibitor. Science. 2004;305:399–401. doi: 10.1126/science.1099480. [DOI] [PubMed] [Google Scholar]
- 21.Weisberg E., et al. Characterization of AMN107, a selective inhibitor of native and mutant Bcr-Abl. Cancer Cell. 2005;7:129–141. doi: 10.1016/j.ccr.2005.01.007. [DOI] [PubMed] [Google Scholar]
- 22.O’Hare T., Eide C.A., Deininger M.W. Bcr-Abl kinase domain mutations, drug resistance, and the road to a cure for chronic myeloid leukemia. Blood. 2007;110:2242–2249. doi: 10.1182/blood-2007-03-066936. [DOI] [PubMed] [Google Scholar]
- 23.Jiang X., et al. Chronic myeloid leukemia stem cells possess multiple unique features of resistance to BCR-ABL targeted therapies. Leukemia. 2007;21:926–935. doi: 10.1038/sj.leu.2404609. [DOI] [PubMed] [Google Scholar]
- 24.Copland M., et al. Dasatinib (BMS-354825) targets an earlier progenitor population than imatinib in primary CML but does not eliminate the quiescent fraction. Blood. 2006;107:4532–4539. doi: 10.1182/blood-2005-07-2947. [DOI] [PubMed] [Google Scholar]
- 25.Jorgensen H.G., Allan E.K., Jordanides N.E., Mountford J.C., Holyoake T.L. Nilotinib exerts equipotent antiproliferative effects to imatinib and does not induce apoptosis in CD34+ CML cells. Blood. 2007;109:4016–4019. doi: 10.1182/blood-2006-11-057521. [DOI] [PubMed] [Google Scholar]
- 26.Konig H., et al. Enhanced BCR-ABL kinase inhibition does not result in increased inhibition of downstream signaling pathways or increased growth suppression in CML progenitors. Leukemia. 2008;22:748–755. doi: 10.1038/sj.leu.2405086. [DOI] [PubMed] [Google Scholar]
- 27.Konig H., Holyoake T.L., Bhatia R. Effective and selective inhibition of chronic myeloid leukemia primitive hematopoietic progenitors by the dual Src/Abl kinase inhibitor SKI-606. Blood. 2008;111:2329–2338. doi: 10.1182/blood-2007-05-092056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kroemer G., Jaattela M. Lysosomes and autophagy in cell death control. Nat. Rev. Cancer. 2005;5:886–897. doi: 10.1038/nrc1738. [DOI] [PubMed] [Google Scholar]
- 29.Klionsky D.J. Autophagy: from phenomenology to molecular understanding in less than a decade. Nat. Rev. Mol. Cell Biol. 2007;8:931–937. doi: 10.1038/nrm2245. [DOI] [PubMed] [Google Scholar]
- 30.Baehrecke E.H. Autophagy: dual roles in life and death? Nat. Rev. Mol. Cell Biol. 2005;6:505–510. doi: 10.1038/nrm1666. [DOI] [PubMed] [Google Scholar]
- 31.Kondo Y., Kanzawa T., Sawaya R., Kondo S. The role of autophagy in cancer development and response to therapy. Nat. Rev. Cancer. 2005;5:726–734. doi: 10.1038/nrc1692. [DOI] [PubMed] [Google Scholar]
- 32.Maiuri M.C., Zalckvar E., Kimchi A., Kroemer G. Self-eating and self-killing: crosstalk between autophagy and apoptosis. Nat. Rev. Mol. Cell Biol. 2007;8:741–752. doi: 10.1038/nrm2239. [DOI] [PubMed] [Google Scholar]
- 33.Lum J.J., et al. Growth factor regulation of autophagy and cell survival in the absence of apoptosis. Cell. 2005;120:237–248. doi: 10.1016/j.cell.2004.11.046. [DOI] [PubMed] [Google Scholar]
- 34.Maclean K.H., Dorsey F.C., Cleveland J.L., Kastan M.B. Targeting lysosomal degradation induces p53-dependent cell death and prevents cancer in mouse models of lymphomagenesis. J. Clin. Invest. 2008;118:79–88. doi: 10.1172/JCI33700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lum J.J., DeBerardinis R.J., Thompson C.B. Autophagy in metazoans: cell survival in the land of plenty. Nat. Rev. Mol. Cell Biol. 2005;6:439–448. doi: 10.1038/nrm1660. [DOI] [PubMed] [Google Scholar]
- 36.Degenhardt K., et al. Autophagy promotes tumor cell survival and restricts necrosis, inflammation, and tumorigenesis. Cancer Cell. 2006;10:51–64. doi: 10.1016/j.ccr.2006.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Amaravadi R.K., et al. Autophagy inhibition enhances therapy-induced apoptosis in a Myc-induced model of lymphoma. J. Clin. Invest. 2007;117:326–336. doi: 10.1172/JCI28833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Paglin S., et al. A novel response of cancer cells to radiation involves autophagy and formation of acidic vesicles. Cancer Res. 2001;61:439–444. [PubMed] [Google Scholar]
- 39.Ito H., Daido S., Kanzawa T., Kondo S., Kondo Y. Radiation-induced autophagy is associated with LC3 and its inhibition sensitizes malignant glioma cells. Int. J. Oncol. 2005;26:1401–1410. [PubMed] [Google Scholar]
- 40.Abedin M.J., Wang D., McDonnell M.A., Lehmann U., Kelekar A. Autophagy delays apoptotic death in breast cancer cells following DNA damage. Cell Death Differ. 2007;14:500–510. doi: 10.1038/sj.cdd.4402039. [DOI] [PubMed] [Google Scholar]
- 41.Kanzawa T., et al. Role of autophagy in temozolomide-induced cytotoxicity for malignant glioma cells. Cell Death Differ. 2004;11:448–457. doi: 10.1038/sj.cdd.4401359. [DOI] [PubMed] [Google Scholar]
- 42.Kanzawa T., Kondo Y., Ito H., Kondo S., Germano I. Induction of autophagic cell death in malignant glioma cells by arsenic trioxide. Cancer Res. 2003;63:2103–2108. [PubMed] [Google Scholar]
- 43.Berger Z., et al. Rapamycin alleviates toxicity of different aggregate-prone proteins. Hum. Mol. Genet. 2006;15:433–442. doi: 10.1093/hmg/ddi458. [DOI] [PubMed] [Google Scholar]
- 44.Colell A., et al. GAPDH and autophagy preserve survival after apoptotic cytochrome c release in the absence of caspase activation. Cell. 2007;129:983–997. doi: 10.1016/j.cell.2007.03.045. [DOI] [PubMed] [Google Scholar]
- 45.Calabretta B., Perrotti D. The biology of CML blast crisis. Blood. 2004;103:4010–4022. doi: 10.1182/blood-2003-12-4111. [DOI] [PubMed] [Google Scholar]
- 46.Sharma S.V., et al. A common signaling cascade may underlie “addiction” to the Src, BCR-ABL, and EGF receptor oncogenes. Cancer Cell. 2006;10:425–435. doi: 10.1016/j.ccr.2006.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kabeya Y., et al. LC3, GABARAP and GATE16 localize to autophagosomal membrane depending on form-II formation. J. Cell Sci. 2004;117:2805–2812. doi: 10.1242/jcs.01131. [DOI] [PubMed] [Google Scholar]
- 48.Kuma A., Matsui M., Mizushima N. LC3, an autophagosome marker, can be incorporated into protein aggregates independent of autophagy: caution in the interpretation of LC3 localization. Autophagy. 2007;3:323–328. doi: 10.4161/auto.4012. [DOI] [PubMed] [Google Scholar]
- 49.Fader C.M., Sanchez D., Furlan M., Colombo M.I. Induction of autophagy promotes fusion of multivesicular bodies with autophagic vacuoles in K562 cells. Traffic. 2008;9:230–250. doi: 10.1111/j.1600-0854.2007.00677.x. [DOI] [PubMed] [Google Scholar]
- 50.Kerkela R., et al. Cardiotoxicity of the cancer therapeutic agent imatinib mesylate. Nat. Med. 2006;12:908–916. doi: 10.1038/nm1446. [DOI] [PubMed] [Google Scholar]
- 51.Tirasophon W., Welihinda A.A., Kaufman R.J. A stress response pathway from the endoplasmic reticulum to the nucleus requires a novel bifunctional protein kinase/endoribonuclease (Ire1p) in mammalian cells. Genes Dev. 1998;12:1812–1824. doi: 10.1101/gad.12.12.1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hoyer-Hansen M., et al. Control of macroautophagy by calcium, calmodulin-dependent kinase kinase-beta, and Bcl-2. Mol. Cell. 2007;25:193–205. doi: 10.1016/j.molcel.2006.12.009. [DOI] [PubMed] [Google Scholar]
- 53.Hoyer-Hansen M., Jaattela M. Connecting endoplasmic reticulum stress to autophagy by unfolded protein response and calcium. Cell Death Differ. 2007;14:1576–1582. doi: 10.1038/sj.cdd.4402200. [DOI] [PubMed] [Google Scholar]
- 54.Deming P.B., et al. Bcr-Abl-mediated protection from apoptosis downstream of mitochondrial cytochrome c release. Mol. Cell. Biol. . 2004;24:10289–10299. doi: 10.1128/MCB.24.23.10289-10299.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cirinna M., et al. Bcl-2 expression restores the leukemogenic potential of a BCR/ABL mutant defective in transformation. Blood. 2000;96:3915–3921. [PubMed] [Google Scholar]
- 56.Pattingre S., Levine B. Bcl-2 inhibition of autophagy: a new route to cancer? Cancer Res. 2006;66:2885–2888. doi: 10.1158/0008-5472.CAN-05-4412. [DOI] [PubMed] [Google Scholar]
- 57.Puttini M., et al. In vitro and in vivo activity of SKI-606, a novel Src-Abl inhibitor, against imatinib-resistant Bcr-Abl+ neoplastic cells. Cancer Res. 2006;66:11314–11322. doi: 10.1158/0008-5472.CAN-06-1199. [DOI] [PubMed] [Google Scholar]
- 58.Wang Y., et al. Adaptive secretion of granulocyte-macrophage colony stimulating factor (GM-CSF) mediates imatinib and nilotinib resistance in BCR/ABL+ progenitors via JAK-2/STAT-5 pathway activation. Blood. 2007;109:2147–2155. doi: 10.1182/blood-2006-08-040022. [DOI] [PubMed] [Google Scholar]
- 59.Gorre M.E., et al. Clinical resistance to STI-571 cancer therapy caused by BCR-ABL gene mutation or amplification. Science. 2001;293:876–880. doi: 10.1126/science.1062538. [DOI] [PubMed] [Google Scholar]
- 60.Maekawa T., Ashihara E., Kimura S. The Bcr-Abl tyrosine kinase inhibitor imatinib and promising new agents against Philadelphia chromosome-positive leukemias. Int. J. Clin. Oncol. 2007;12:327–340. doi: 10.1007/s10147-007-0699-1. [DOI] [PubMed] [Google Scholar]
- 61.Donato N.J., et al. BCR-ABL independence and LYN kinase overexpression in chronic myelogenous leukemia cells selected for resistance to STI571. Blood. 2003;101:690–698. doi: 10.1182/blood.V101.2.690. [DOI] [PubMed] [Google Scholar]
- 62.Dai Y., Rahmani M., Corey S.J., Dent P., Grant S. A Bcr/Abl-independent, Lyn-dependent form of imatinib mesylate (STI-571) resistance is associated with altered expression of Bcl-2. J. Biol. Chem. 2004;279:34227–34239. doi: 10.1074/jbc.M402290200. [DOI] [PubMed] [Google Scholar]
- 63.Ptasznik A., Nakata Y., Kalota A., Emerson S.G., Gewirtz A.M. Short interfering RNA (siRNA) targeting the Lyn kinase induces apoptosis in primary, and drug-resistant, BCR-ABL1(+) leukemia cells. Nat. Med. 2004;10:1187–1189. doi: 10.1038/nm1127. [DOI] [PubMed] [Google Scholar]
- 64.Tokarski J.S., et al. The structure of Dasatinib (BMS-354825) bound to activated ABL kinase domain elucidates its inhibitory activity against imatinib-resistant ABL mutants. Cancer Res. 2006;66:5790–5797. doi: 10.1158/0008-5472.CAN-05-4187. [DOI] [PubMed] [Google Scholar]
- 65.Soverini S., et al. Second-line treatment with dasatinib in patients resistant to imatinib can select novel inhibitor-specific BCR-ABL mutants in Ph+ ALL. Lancet Oncol. 2007;8:273–274. doi: 10.1016/S1470-2045(07)70078-5. [DOI] [PubMed] [Google Scholar]
- 66.Shah N.P., et al. Sequential ABL kinase inhibitor therapy selects for compound drug-resistant BCR-ABL mutations with altered oncogenic potency. J. Clin. Invest. 2007;117:2562–2569. doi: 10.1172/JCI30890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Piwocka K., Vejda S., Cotter T.G., O’Sullivan G.C., McKenna S.L. Bcr-Abl reduces endoplasmic reticulum releasable calcium levels by a Bcl-2-independent mechanism and inhibits calcium-dependent apoptotic signaling. Blood. 2006;107:4003–4010. doi: 10.1182/blood-2005-04-1523. [DOI] [PubMed] [Google Scholar]
- 68.Law J.C., Ritke M.K., Yalowich J.C., Leder G.H., Ferrell R.E. Mutational inactivation of the p53 gene in the human erythroid leukemic K562 cell line. Leuk. Res. 1993;17:1045–1050. doi: 10.1016/0145-2126(93)90161-d. [DOI] [PubMed] [Google Scholar]
- 69.Ahuja H., Bar-Eli M., Advani S.H., Benchimol S., Cline M.J. Alterations in the p53 gene and the clonal evolution of the blast crisis of chronic myelocytic leukemia. Proc. Natl. Acad. Sci. U. S. A. 1989;86:6783–6787. doi: 10.1073/pnas.86.17.6783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Feinstein E., et al. p53 in chronic myelogenous leukemia in acute phase. Proc. Natl. Acad. Sci. U. S. A. 1991;88:6293–6297. doi: 10.1073/pnas.88.14.6293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Carew J.S., et al. Targeting autophagy augments the anticancer activity of the histone deacetylase inhibitor SAHA to overcome Bcr-Abl-mediated drug resistance. Blood. 2007;110:313–322. doi: 10.1182/blood-2006-10-050260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cain K., Brown D.G., Langlais C., Cohen G.M. Caspase activation involves the formation of the aposome, a large (approximately 700 kDa) caspase-activating complex. J. Biol. Chem. 1999;274:22686–22692. doi: 10.1074/jbc.274.32.22686. [DOI] [PubMed] [Google Scholar]
- 73.Ferrari-Amorotti G., et al. Leukemogenesis induced by wild-type and STI571-resistant BCR/ABL is potently suppressed by C/EBPalpha. Blood. 2006;108:1353–1362. doi: 10.1182/blood-2006-01-011833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bellodi C., et al. Cytoplasmic function of mutant promyelocytic leukemia (PML) and PML-retinoic acid receptor-alpha. J. Biol. Chem. 2006;281:14465–14473. doi: 10.1074/jbc.M600457200. [DOI] [PubMed] [Google Scholar]
- 75.Inoue S., et al. Histone deacetylase inhibitors potentiate TNF-related apoptosis-inducing ligand (TRAIL)-induced apoptosis in lymphoid malignancies. Cell Death Differ. 2004;11(Suppl. 2):S193–S206. doi: 10.1038/sj.cdd.4401535. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.