Abstract
The fatal immune dysregulation that sometimes follows EBV infection in boys has been linked to mutations in two X chromosome–encoded genes, SLAM-associated protein (SAP) and X-linked inhibitor of apoptosis (XIAP). In this study we describe 2 girls from a consanguineous Turkish family who died after developing severe immune dysregulation and therapy-resistant EBV-positive B cell proliferation following EBV infection. SNP array–based genome-wide linkage analysis revealed IL-2–inducible T cell kinase (ITK) as a candidate gene for this immunodeficiency syndrome. Both girls harbored a homozygous missense mutation that led to substitution of a highly conserved residue (R335W) in the SH2 domain of ITK. Characteristics of ITK deficiency in mouse models, such as absence of NKT cells and high levels of eomesodermin in CD8+ cells, were seen in either one or both of the girls. Two lines of evidence suggested that R335W caused instability of the ITK protein. First, in silico modeling of the mutant protein predicted destabilization of the SH2 domain. Additionally, Western blot analysis revealed that, unlike wild-type ITK, the R335W mutant was nearly undetectable when expressed in 293 T cells. Our results suggest that ITK deficiency causes what we believe to be a novel immunodeficiency syndrome that leads to a fatal inadequate immune response to EBV. Because ITK deficiency resembles EBV-associated lymphoproliferative disorders in boys, we suggest that this molecular cause should be considered during diagnosis and treatment.
Introduction
Severe immune dysregulation in boys after primary infection with EBV, presenting as fatal mononucleosis, hemophagocytosis, hypogammaglobulinemia, and lymphoproliferation, is the hallmark of X-linked lymphoproliferative disease (XLP), a well-known inherited immunodeficiency disorder (1). Mutations in the SH2 domain protein 1A (SH2D1A), also referred to as signaling lymphocytic activation molecule family member 1–associated (SLAM-associated) protein (SAP), account for approximately 60% of XLP cases (2–4). In addition, mutations in the X-linked inhibitor of apoptosis (XIAP) protein have been shown to be causative in 3 XLP families (5). Recently, several studies have demonstrated an almost complete absence of NKT cells in lymph nodes, liver, spleen, and thymus of SAP-deficient mice as well as in the blood of patients with XLP carrying mutations in SAP and XIAP (5–8). As the lack of NKT cells is a shared feature of SAP and XIAP deficiency, a critical role has been postulated for NKT cells in the response to EBV infection.
In addition to SAP and XIAP, the SLAM receptor family member 1, the tyrosine kinase Fyn, and 2 members of the Tec family kinases, namely the resting lymphocyte kinase (RLK) and the IL-2–inducible T cell kinase (ITK), have been shown to be required for the development of NKT cells in mice (9–13).
We examined a consanguineous Turkish family in which 2 girls died after showing a fatal EBV-associated lymphoproliferative disorder clinically resembling XLP. In both girls we found homozygous mutations in the SH2 domain of ITK. The identified mutation R335W results in protein destabilization, lack of ITK in lymph node biopsies, and absence of NKT cells. Thus, we believe ITK deficiency to be a novel primary immunodeficiency characterized by fatal immune dysregulation following EBV infection.
Results
Patients
Patient 1.
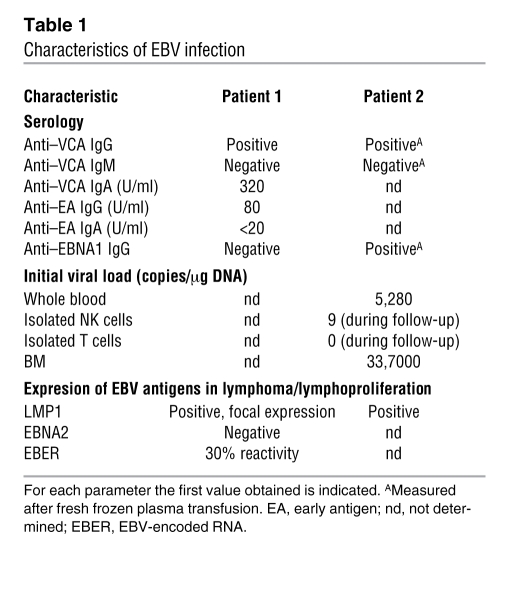
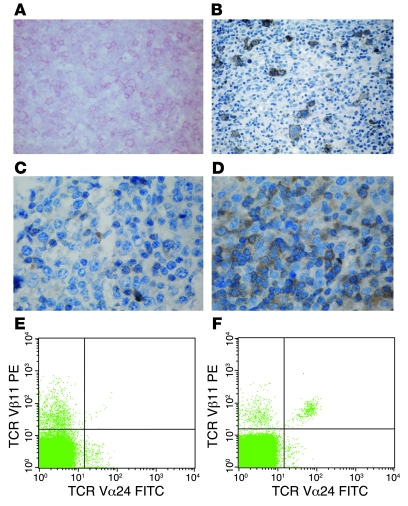
The female index patient was the first child of healthy consanguineous Turkish parents. At the age of 5 years 2 months, immunoglobulin levels, measured during an episode of aphthous stomatitis and subsequent candida stomatitis, were normal. From the age of 6 years, she successively developed recurrent episodes of fever and elevated C-reactive protein (CRP), nodular interstitial pulmonary infiltrates unresponsive to antibiotic treatment, generalized lymphadenopathy, hepatosplenomegaly with impaired liver function, pleural and pericardial effusion, anemia, thrombocytopenia, and hypogammaglobulinemia. EBV viral capsid antigen (VCA) IgG antibodies were positive in the absence of Epstein-Barr nuclear antigen 1 (EBNA1) and VCA IgM antibodies (Table 1). At the age of 6 years 10 months, she was transferred to our hospital. EBV PCR confirmed active EBV infection. The biopsy of an axillary lymph node showed polymorphic B cell proliferation. These cells were CD20+ (Figure 1A), CD22+, CD10–, and part of the population was CD30+. Focal expression of the EBV latent membrane protein 1 (LMP1) and 30% EBV-encoded RNA reactivity in in situ hybridization demonstrated the relation to EBV infection. Staining for EBNA2 was negative (Table 1). Clonality analysis revealed oligoclonal amplification of the CDRIII immunoglobulin heavy-chain region (data not shown). BM aspirate was normal, with no evidence for lymphoma infiltration, histiocytosis, or hemophagocytosis.
Table 1 .
Characteristics of EBV infection
Figure 1. Lymph node histology and NKT cells in patients 1 and 2.
(A) Immunohistochemical staining of CD20 antigen in an axillary lymph node of patient 1 with hyperimmune reaction and medium-sized blast reacting with CD20. Hodgkin-like cells were not detected (alkaline phosphatase anti–alkaline-phosphatase reaction; hematoxilin). Original magnification, ×200. (B) Immunohistochemical staining of CD20 antigen in an inguinal lymph node biopsy of patient 2 with positivity in Hodgkin- and Sternberg-Reed–like cells (avidin-biotin complex staining; hematoxilin). Original magnification, ×100. (C) Staining of the same lymph node specimen from patient 1 with an intracellular polyclonal antibody gave only very weak signals. Original magnification, ×500. (D) In contrast, a strong ITK expression was observed in control lymph node tissue. Original magnification, ×500. (E and F) Measurement of NKT cells by FACS. After gating on CD3+ T cells, NKT cells were determined as TCR Vβ11 and TCR Vα24 double-positive cells. In patient 2 (E) NKT cells were virtually absent, whereas the percentage of NKT cells was 0.35% in the healthy control subject (F).
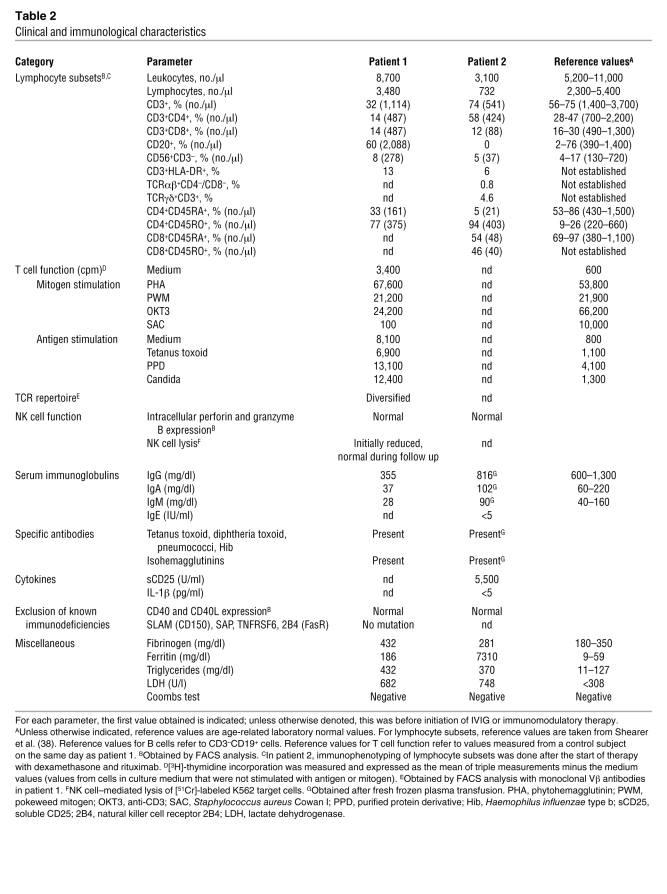
Because the disease phenotype resembled X-linked lymphoproliferative disease mutations in SLAM (CD150), SAP, natural killer cell receptor 2B4 (2B4), TNFRSF6 (also known as FAS), and EAT were excluded. An extensive immunological work-up, results of which are described in Table 2, did not contribute to elucidating the underlying defect. The patient received 4 courses of the monoclonal CD20 antibody rituximab at weekly intervals in addition to foscavir and steroids, which led to a marked reduction of EBV load and resolution of the clinical symptoms. A flare with increasing virus load and progressive intrapulmonary infiltrates at the age of 7 years 11 months was again responsive to rituximab. At the age of 8 years 2 months, a tumor of the right nasal concha was histologically identified as an EBV-associated Hodgkin lymphoma with strong expression of CD30 and LMP1, negative for CD15 and Ki-A10. CD20 negativity was attributed to the previous treatment with rituximab. The tumor was successfully treated according to a standard chemotherapy protocol using prednisone, procarbazine, vincristine, cyclophosphamide, and adriamycine followed by fractionated local and mediastinal irradiation. During the following years, the disease progressed with increasing EBV loads up to 3 × 107 copies/μg DNA and mainly cervical lymphadenopathy despite continued intensive treatment. We observed a continuous decline in lymphocytes, especially CD45RACD4+ T cells. At the age of 10 years 4 months, BK polyomavirus infection with high BK virus loads in urine and PB was diagnosed. At the same time, the girl acquired Pneumocystis jirovecii pneumonia and died from respiratory failure at the age of 10 years 5 months.
Table 2 .
Clinical and immunological characteristics
Patient 2.
Patient 2 was the index patient’s younger sister. At the age of 16 months, immunological evaluation at our institution showed normal blood count, CRP, IgG and IgG subclasses, specific vaccination antibodies, hemagglutinins, and immunophenotyping of lymphocyte subsets. She was EBV VCA IgG positive and VCA IgM negative at that time. However, at the age of 16 months, the presence of maternal VCA IgG antibodies could not be excluded. At the age of 5 years, she developed recurrent episodes of fever and elevated CRP and erythrocyte sedimentation rate unresponsive to antibiotic treatment. Within 4 months the symptoms progressed to a life-threatening condition with profound pancytopenia, retroperitoneal and abdominal lymphadenopathy, hepatosplenomegaly, severely impaired liver function, ascites, and pleural effusion. At that time, aged 5 years 4 months, she was transferred to our hospital. PCR showed an active EBV infection with a virus load of 3,920 copies/μg DNA in PB and 33,700 copies/μg in BM. EBV VCA IgG and anti-EBNA1 antibodies were positive, whereas VCA IgM antibodies were absent. In isolated T and NK cells, no relevant EBV load could be detected (Table 1). The biopsy specimen of an inguinal lymph node showed a proliferation of Hodgkin- and Sternberg-Reed–like giant cells positive for CD20 (Figure 1B) and LMP1 and negative for CD30 and CD15. BM aspirate was normal. Further laboratory results are summarized in Table 2. Within the first day after admission to our hospital, the girl developed multiple organ failure requiring mechanical ventilation and hemodialysis for several days. Treatment with ganciclovir, steroids, and rituximab at weekly intervals led to an improvement of symptoms and a stable virus load not exceeding 5,000 copies/μg DNA but did not induce a complete remission as initially seen in the sister. At the age of 5 years 9 months, she received haploidentical PB stem cell transplantation. During aplasia severe nasopharyngeal hemorrhage led to an acute airway obstruction with respiratory insufficiency and cardiovascular arrest. Unfortunately, she died 7 days after resuscitation from the consequences of ischemic brain injury.
Molecular findings
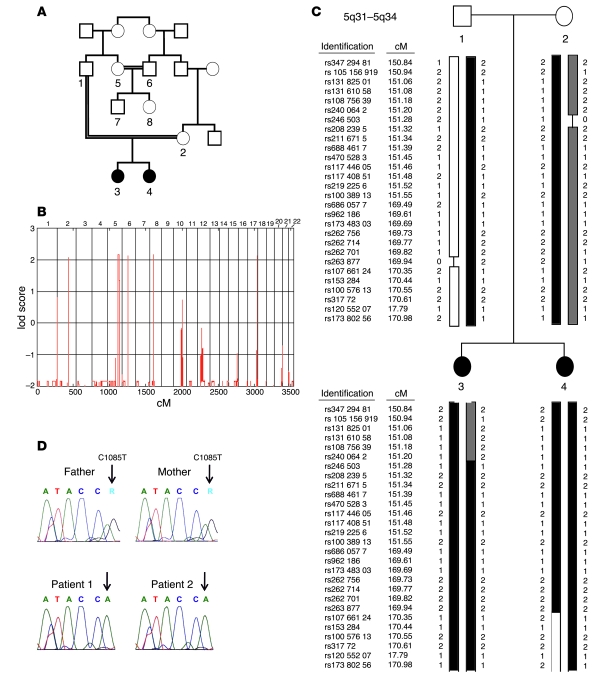
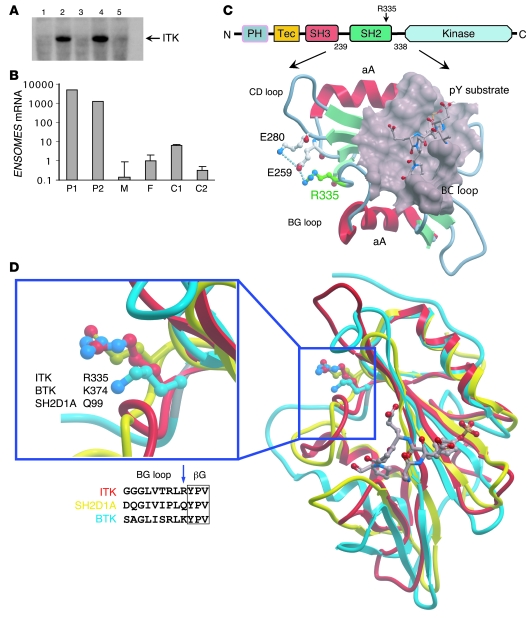
Consanguinity of the parents and the similar clinical course in both daughters were suggestive for a genetic basis of the disease. We therefore initiated a genome-wide linkage analysis with 8 family members including patient 1 (individual 3), patient 2 (individual 4), father (individual 1), mother (individual 2), an aunt (individual 5), an uncle (individual 6), a male cousin (individual 7), and a female cousin (individual 8) (Figure 2A). Parametric linkage analysis with a recessive model revealed genome-wide 5 regions on chromosomes 2, 5, 6, 8, and 17 where the maximal possible lod score of 2.15 was reached (Figure 2B). The largest segment was on chromosome 5q31–5q34, where the homozygous region between recombinant markers rs2400642 and rs17066124 spanned 17.2 Mb (Figure 2C). Within this region we identified 78 positional candidate genes including ITK, which appeared the most likely candidate for the development of the disease. Sequencing of ITK revealed the homozygous missense mutation C1085T in both affected individuals (Figure 2D). The parents, aunt, and male cousin (individuals 1, 2, 5, and 7) were identified as heterozygous carriers, whereas individuals 6 and 8 showed the wild-type sequence. To our knowledge, C1085T has not been published as a SNP, and the C1085T mutation was not present in 100 screened children of Turkish descent and 100 children of German descent (data not shown). Comparison of ITK sequences between different species revealed complete conservation of this residue from human to frog Xenopus (data not shown). The discovered mutation leads to the substitution of an arginine by tryptophan residue at position 335 (R335W) in ITK. R335 is located in the BG loop of the ITK SH2 domain (Figure 3C). Due to the distant location of the mutated residue to the phosphotyrosine binding pocket, it is not likely that this mutation impairs recruitment of the SH2 domain substrate. Attempts to functionally express the R335W mutant SH2 domain in bacteria failed due to aggregation of the recombinant protein testing 8 different expression constructs, whereas wild-type protein was stable and expressed functionally at high levels (data not shown). In silico prediction of protein stability revealed a severe destabilization of the SH2 domain by the mutation. We therefore expressed the R335W mutant and wild-type ITK along with different controls in 293 T cells. By Western blot analysis, the R335W mutant protein was nearly undetectable (Figure 3A). To control that similar amounts of ITK mRNA were expressed in R335W and wild-type transfected cells, we performed quantitative RT-PCR after DNase digestion of the transfected plasmid. We found equal mRNA expression levels for R335W mutated and wild-type ITK (data not shown), which underlines the protein destabilizing effect of the R335W mutant. As a result of this protein instability, using a polyclonal ITK antibody, we found extremely low staining levels in the lymph node biopsy from patient 1 as compared with normal lymph node tissue (Figure 1, C and D).
Figure 2. Identification of a homozygous missense mutation in ITK.
(A) Pedigree of the affected sisters (individuals 3 and 4) with 2 consanguineous loops. Circles indicate females, squares indicate males. Individuals 1–8 were genotyped with a SNP array for linkage analysis. (B) lod score (ordinate) yielded by parametric linkage analysis with a recessive model. The maximum lod score of 2.15 was reached for 5 regions on chromosomes 2, 5, 6, 8, and 17, with the largest region detected on chromosome 5q31–5q34. (C) Haplotype of individuals 1–4 on chromosome 5q31–5q34. Reference SNP (rs) identification, genetic position (in cM), and genotype variant (1 or 2) are indicated for each SNP. “0” and a black line indicate an uninformative result for the particular marker. Both affected sisters showed a large homozygous region between rs2400642 and rs17066124 containing the candidate gene ITK. To simplify the graphic representation, between rs10038913 (151.55 cM) and rs6860577 (169.49 cM), 518 markers were skipped. (D) Relevant section of the 3′→5′ ITK sequencing data for individuals 1–4. Both affected siblings carried a homozygous G→A single nucleotide mutation in exon 11 (ATA/CCA; 5′→3′, c.1,085 C→T). The father and mother were heterozygous (ATA/CCA/G). The affected position is marked with an arrow. The wild-type sequence is ATA/CCG (not shown). c.1,085 C→T leads to the amino acid substitution R335W.
Figure 3. Evidence of severe destabilization of the mutant ITK protein.
(A) Western blot with transiently transfected 293 T cells using a monoclonal ITK antibody. Cyclophilin A was used as negative control (lane 1). No expression was detected for the R335W mutant (lane 5) and an ITK variant with complete deletion of the SH2 domain (lane 3), whereas wild-type ITK (lane 4) and the kinase-inactivating mutant (F511Y) (lane 2) showed strong ITK expression. (B) Results of quantitative real-time PCR for EOMES mRNA in CD8+ single-positive T cells on a logarithmic scale. We found 1,000- to 6,000-fold higher mRNA levels of EOMES in both affected children compared with their heterozygous parents or healthy controls. Error bars denote the standard deviation. P1, patient 1; P2, patient 2; F, father; M, mother; C1, control 1; C2, control 2. (C) Domain architecture of ITK and the location of the mutated residue (R335) (highlighted by a red label) in the SH2 domain. Shown is the NMR minimized average structure (protein data bank code 2etz). Protein instability probably results from a compromised salt bridge network (dotted lines) as well as surface exposure of the introduced tryptophan side chain. A phosphotyrosine substrate peptide and the corresponding binding pocket are located on the opposite side of the molecule. The main structural elements are labeled. (D) Site of the BG loop mutations in ITK (R335W, red), BTK (K374N, blue), and SAP (Q99P, yellow).
Studies in Itk–/– mice suggest that ITK is required for repressing the T box transcription factor eomesodermin (Eomes) in CD8+ single-positive T cells (24). In agreement with the low ITK levels detected in the biopsy, quantitative RT-PCR revealed a 1,000- to 6,000-fold overexpression of Eomes in both affected children as compared with their heterozygous parents and 2 healthy male controls (Figure 3B). Finally, functional ITK has been shown to be essential for the development of NKT cells. After gating on CD3+ T cells, NKT cells were determined as TCR Vβ11 and TCR Vα24 double-positive cells. In patient 2, NKT cells were virtually absent (Figure 1E), whereas the percentage of NKT cells in the healthy control subject shown in Figure 1F was 0.35%. Unfortunately we were not able to determine in retrospect the level of NKT cells in patient 1 due to the lack of appropriate material. However, the heterozygous parents showed a low percentage of NKT cells, which was 0.15% in the mother and 0.08% in the father, but both were healthy throughout their lives.
Discussion
We report what we believe is a novel primary immunodeficiency, which leads to an XLP-like phenotype in girls, caused by a homozygous mutation in ITK on chromosome 5q31–5q32. This is the first molecular cause of an autosomal recessively inherited lymphoproliferative disease.
Both ITK-deficient individuals suffered from uncontrolled EBV infection meeting several criteria for secondary hemophagocytic lymphohistiocytosis, but without evidence of hemophagocytosis in BM and lymph nodes. In contrast to SAP deficiency, in which the development of preferably Burkitt lymphoma is not restricted to EBV-positive individuals, and XIAP deficiency, in which no case of lymphoma occurred so far, the leading symptom was an atypical, generalized EBV-positive B cell proliferation with progression to Hodgkin lymphoma in patient 1 (5, 14). It is not clear whether the progressive decline of lymphocytes in patient 1, which led to Pneumocystis jirovecii and BK virus infection, was due to ITK deficiency itself, chronic EBV infection, or the long-lasting virostatic therapy.
The non-receptor tyrosine kinase ITK is a member of the Tec kinase family, which also includes Bruton’s tyrosine kinase (BTK), Tec protein tyrosine kinase (TEC), RLK, and BM kinase X-linked (BMX) (15). Tec kinases contain a conserved SH3, SH2 and catalytic (kinase) domain. The SH2 domain of non-receptor tyrosine kinases is a non-catalytic protein interaction module, which by engaging with a phosphotyrosine-containing signaling partner regulates activity of the catalytic domain (16). Our results suggest that R335W mutant ITK causes a profound instability of the ITK protein rather than an impairment of ITK activation. Interestingly, mutations at exactly analogous positions within the SH2 domain have been described in BTK (Y361C) (17) and SAP (Q99P) (18), both leading to protein instability and the clinical phenotype of atypical X-linked agammaglobulinemia (XLA) and XLP, respectively (Figure 3D).
Given the pivotal role of ITK in a number of T cell signaling pathways, absence of Itk has been studied in great detail in mice, but no human disease with ITK deficiency has been described yet. The previously published experimental data in Itk–/– mouse cells greatly facilitated the choice of ITK among our candidate gene list of 78 genes after linkage analysis. Loss of Itk in mice leads to major immunological abnormalities, e.g., decreased responses to TCR stimulation, T cell proliferation, production of IL-2 and other effector cytokines, as well as altered thymic selection of CD4+ and CD8+ cells by modulation of TCR signal strength (19–23). In a set of elegant experiments, different research groups demonstrated that Tec kinase–deficient mice develop a large population of abnormal CD8+ T cells exhibiting innate immune properties but with high expression of the memory markers CD44 and CD122 (24–26).
We also found a clearly increased memory phenotype in our patients pointing to a profoundly altered differentiation of the conventional T cell lineage (Table 2). Perhaps the most important paralleling feature between Itk-deficient mice and humans is the absence or reduced numbers of NKT cells. Itk–/– mice show a block of NKT cell maturation and reduced peripheral survival of NKT cells (8, 10, 11).
In mice, NKT cells have been shown to provide a protective innate-type immune response to several microbes (27) including viruses, e.g., HSV1 and HSV2 (28, 29) and hepatitis B virus (30). Strong evidence for a paramount biological relevance of NKT cells in humans comes from the observation that boys with XLP due to SAP or XIAP deficiency show a severely reduced number, or even a complete absence, of NKT cells (5, 6). Thus, a critical role for NKT cells in the immune response to EBV infection in humans has been postulated (5, 6). The absence of NKT cells in patient 2 and the concomitant inability to clear EBV infection in the 2 ITK-deficient sisters strengthens the hypothesis of a role for NKT cells in the control of EBV infection. The fact that both heterozygous parents have low but still detectable numbers of NKT cells also suggests that ITK is required for the selection and/or survival of NKT cells not only in mice, but also in humans. Rigaud et al. found normal numbers of NKT cells in 8 patients with acute infectious mononucleosis, showing that the absence of NKT cells is not an immediate consequence of acute EBV infection (5), whereas to our knowledge there are no data on NKT cell numbers in patients with chronic active EBV infection. EBV affects more than 90% of the world’s population, but why the adequate immunological control of this successful member of the herpes family seems to be critically dependent on this numerically rather tiny cell population remains obscure.
However, as there are autoimmune conditions in which patients with a severely reduced number of NKT cells are not especially susceptible to EBV infection (31), the lack of NKT cells is likely to contribute to the impaired immune response to EBV but may not be the single underlying factor. Other evidence for a protective role of invariant NKT cells in human infection is rather limited. In 2003 Levy et al. reported a girl with severe varicella infection after varicella vaccination, in whom an absolute deficiency of invariant NKT cells was the only detectable immunologic abnormality (32).
The immunophenotypical as well as functional analyses in our cases were performed following several months of active EBV infection. Thus, it is not possible to clearly distinguish between the impact of EBV infection and ITK deficiency itself. In the future, the identification and examination of further patients with ITK deficiency will certainly contribute to elucidating the functional implications of ITK deficiency in different lymphocyte lineages on host defense. We recommend that in all children with EBV-associated lymphoproliferative disorders, ITK deficiency should be considered because it may show striking clinical and immunological similarities to SAP or XIAP deficiency in boys.
Methods
The studies were reviewed and approved by the institutional review board (ethics committee) of Heinrich Heine University. The parents gave informed consent to carry out the investigations described below.
Linkage analysis.
Samples were genotyped with the Affymetrix Human Mapping 250K Sty I array according to the manufacturer’s guidelines (Affymetrix). Genotype calling was performed with the Dynamic Model algorithm implemented in the Affymetrix software GTYPE version 4.1. The Call rate was above 94% for all samples; the average call rate was 97.15%.
Quality control and data conversion were managed by ALOHOMORA (http://gmc.mdc-berlin.de/alohomora/) (33). The correct relationship of individuals within the families was verified with the Graphical Representation of Relationships software (http://www.sph.umich.edu/csg/abecasis/GRR/index.html) using 14,339 autosomal markers (34). Mendelian errors were detected by PedCheck (http://watson.hgen.pitt.edu/register/soft_doc.html) and genotypes deleted in all individuals, while unlikely genotypes (double recombinants), identified with Merlin (http://www.sph.umich.edu/csg/abecasis/Merlin/), were deleted in the individuals in which they appeared (35, 36). Parametric multipoint lod score analysis and haplotyping were performed using Merlin. We used a recessive model with complete penetrance and a trait locus allele frequency of 0.0001. Marker allele frequencies for a European population as well as genetic positions were extracted from the Affymetrix SNP annotation file (version na23) (http://www.affymetrix.com/support/technical/byproduct.affx?product=500k).
Of the 238,304 SNPs on the array, 238,230 had a physical and genetic position. To reduce the impact of linkage disequilibrium between closely linked markers on the lod score analysis, the pedigree was recalculated with smaller sets of markers (86,162 and 67,334 SNPs) with a minimal distance of 10,000 bp and minimal minor allele frequencies of 0.05 and 0.1. Graphical plots of the Merlin haplotypes were created using HaploPainter (http://haplopainter.sourceforge.net) (37).
ITK staining in lymph node tissue.
The polyclonal ITK antibody ab32113 (Abcam) was used for staining of ITK in lymph node tissue.
ITK sequencing and expression of ITK constructs in 293T cells.
Genomic DNA was extracted from PB leukocytes. Mutation analysis was performed by direct sequencing of PCR fragments obtained after nested amplification of the exonic and flanking intron region coding sequences of ITK with 17 exons. Primers to amplify the genomic DNA samples were designed according to GenBank sequences (Table 3). Direct cycle sequencing of all PCR fragments was performed with BigDye Terminator v3.1 cycle sequencing kit (Applied Biosystems) and analyzed by capillary electrophoresis on an ABI Prism 3130 Genetic Analyzer (Applied Biosystems). Analyzed sequences were compared with the cDNA and genomic DNA sequences in GenBank accession numbers NM_005546 (human ITK mRNA). The mutation nomenclature used follows the recommendation of the Human Genome Variation Society (http://www.hgvs.org/mutnomen).
Table 3 .
5′→3′ primer sequences for nested amplification of the coding regions of ITK
Confluent 293T cells were transfected with either full-length mutated ITK or wild-type ITK plasmids, which were purchased from GenScript. Transient transfections were performed by polyethylenimine transfection using standard parameters, and Western blotting was performed 24 hours thereafter.
ITK Western blot.
Postnuclear supernatant equivalents of 1 × 106 (or 0.5 × 106 stimulated) cells were separated by 12% SDS-PAGE and blotted onto PVDF membranes (1 h, 100 V, Hybond-P; GE Healthcare) and blocked with 5% nonfat dry milk in PBS-Tween (0.05% Tween-20 in PBS) for 1 hour. After washing with PBS-Tween, the blots were incubated overnight with the primary antibody ITK 2F12 (catalog 2380; Cell Signaling Technology) at 4°C. Blots were washed again with PBS-Tween, incubated with the HRP-coupled secondary antibodies (1:20,000) goat anti-rabbit (catalog RPN4301) and sheep anti-mouse IgG (catalog RPN4201V) horseradish peroxide (GE Healthcare) for 1 hour at room temperature, washed again, and then developed with a chemiluminescence reagent (Super Signal West Pico Chemiluminescent Substrate; Thermo Scientific). Western blots were stripped with Re-Blot Plus (Millipore) and reprobed with ITK (Y402; catalog 1595-1; Epitomics) (for protocol, see above, beginning with blocking) to ensure the specificity of the antibody used first.
Quantitative real-time PCR analysis of Eomes expression.
CD8+ T cells were isolated from PB using Miltenyi Biotec beads and columns. Total RNA was prepared from sorted cells using RNase Mini kit (Qiagen) and reverse transcribed into cDNA using Quantitect Reverse Transcription Kit (Qiagen) according to the manufacturer’s instructions. Quantitative PCR was performed using the primer and probe set for Eomes (Applied Biosystems), with S18 as a housekeeping gene. Data were analyzed using the ΔΔCT method and normalized to S18. The relative gene expression levels were then determined by comparing them with the expression found in the CD8+ population of the father, which were set as 1.
Immunophenotyping by flow cytometry.
Whole blood was anticoagulated with EDTA and processed within 24 hours. A differential white blood cell count was done automatically. Four-color fluorescence-activated cell sorting (FACS) analysis was performed with a FACS Calibur flow cytometer with CellQuest version 3.2 software (BD). Lymphocyte subpopulations were analyzed by FACS staining using FITC-, PE-, peridinin-chlorophyll protein– (PerCP-), and allophycocyanin-labeled monoclonal antibodies (BD or Immunotech) following the manufacturer’s instructions. Antibodies (clone) against the following epitopes were used: CD3 (SK7; BD), CD4 (SK3; BD), CD8 (SK1; BD), TCRab (WT31; BD), TCRgd (11F2; BD), TCR Vα24 (C15; Immunotech), TCR Vβ11 (C21; Immunotech), CD45RA (L48; BD), CD45RO (UCHL-1; BD), CD56 (MY31; BD), HLA-DR (L243; BD), and CD20 (L27; BD). Conserved thawed cells of both patients and healthy controls were CD3 positively selected using whole-blood CD3 MicroBeads (Miltenyi Biotec) following the manufacturer’s instructions. After gating on 1 × 105 CD3+ T cell lymphocytes, the percentage of NKT cells (CD3+ TCR Vα24+ TCR Vβ11+) was determined by FACS.
Prediction of protein stability.
For in silico prediction of protein stability, the Eris program was used (http://troll.med.unc.edu/eris).
Acknowledgments
Supported by grants from the European Community (LSHB-CT-2004-005276, to A. Borkhardt) and from the German Ministry for Science and Education (NGFN2, to N. Hübner). The Structural Genomics Consortium is a registered charity that receives funds from the Canadian Institutes for Health Research, Canadian Foundation for Innovation, Genome Canada through the Ontario Genomics Institute, GlaxoSmithKline, Karolinska Institutet, Knut and Alice Wallenberg Foundation, Ontario Innovation Trust, Ontario Ministry for Research and Innovation, Merck, Novartis Research Foundation, Swedish Agency for Innovation Systems, Swedish Foundation for Strategic Research, and Wellcome Trust. We thank S. Bellert in Düsseldorf for excellent technical assistance, M. Gombert in Düsseldorf for assistance with real-time PCR, H. Hanenberg in Düsseldorf for providing stored patient material, H.K. Müller-Hermelink in Würzburg for evaluation of histological samples, M. Schneider in Ulm and S. Ehl in Freiburg for NK cell cytotoxicity assays, and D. Dilloo and all other members of the BM transplantation team in Düsseldorf involved in patient care.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Nonstandard abbreviations used: BTK, Bruton’s tyrosine kinase; CRP, C-reactive protein; EBNA, Epstein-Barr nuclear antigen; Eomes, eomesodermin; FACS, fluorescence-activated cell sorting; ITK, IL-2–inducible T cell kinase; LMP1, latent membrane protein 1; SAP, SLAM-associated protein; SLAM, signaling lymphocytic activation molecule family member 1; VCA, viral capsid antigen; XIAP, X-linked inhibitor of apoptosis; XLP, X-linked lymphoproliferative disease.
Citation for this article: J. Clin. Invest. 119:1350–1358 (2009). doi:10.1172/JCI37901
References
- 1.Purtilo D.T., Grierson H.L., Davis J.R., Okano M. The X-linked lymphoproliferative disease: from autopsy toward cloning the gene 1975-1990. Pediatr. Pathol. 1991;11:685–710. doi: 10.3109/15513819109065466. [DOI] [PubMed] [Google Scholar]
- 2.Coffey A.J., et al. Host response to EBV infection in X-linked lymphoproliferative disease results from mutations in an SH2-domain encoding gene. Nat. Genet. 1998;20:129–135. doi: 10.1038/2424. [DOI] [PubMed] [Google Scholar]
- 3.Nichols K.E., et al. Inactivating mutations in an SH2 domain-encoding gene in X-linked lymphoproliferative syndrome. Proc. Natl. Acad. Sci. U. S. A. 1998;95:13765–13770. doi: 10.1073/pnas.95.23.13765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sayos J., et al. The X-linked lymphoproliferative-disease gene product SAP regulates signals induced through the co-receptor SLAM. Nature. 1998;395:462–469. doi: 10.1038/26683. [DOI] [PubMed] [Google Scholar]
- 5.Rigaud S., et al. XIAP deficiency in humans causes an X-linked lymphoproliferative syndrome. Nature. 2006;444:110–114. doi: 10.1038/nature05257. [DOI] [PubMed] [Google Scholar]
- 6.Pasquier B., et al. Defective NKT cell development in mice and humans lacking the adapter SAP, the X-linked lymphoproliferative syndrome gene product. J. Exp. Med. 2005;201:695–701. doi: 10.1084/jem.20042432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chung B., Aoukaty A., Dutz J., Terhorst C., Tan R. Signaling lymphocytic activation molecule-associated protein controls NKT cell functions. J. Immunol. 2005;174:3153–3157. doi: 10.4049/jimmunol.174.6.3153. [DOI] [PubMed] [Google Scholar]
- 8.Nichols K.E., et al. Regulation of NKT cell development by SAP, the protein defective in XLP. Nat. Med. 2005;11:340–345. doi: 10.1038/nm1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Borowski C., Bendelac A. Signaling for NKT cell development: the SAP-FynT connection. J. Exp. Med. 2005;201:833–836. doi: 10.1084/jem.20050339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gadue P., Morton N., Stein P.L. The Src family tyrosine kinase Fyn regulates natural killer T cell development. J. Exp. Med. 1999;190:1189–1196. doi: 10.1084/jem.190.8.1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Felices M., Berg L.J. The Tec kinases Itk and Rlk regulate NKT cell maturation, cytokine production, and survival. J. Immunol. 2008;180:3007–3018. doi: 10.4049/jimmunol.180.5.3007. [DOI] [PubMed] [Google Scholar]
- 12.Au-Yeung B.B., Fowell D.J. A key role for Itk in both IFN gamma and IL-4 production by NKT cells. J. Immunol. 2007;179:111–119. doi: 10.4049/jimmunol.179.1.111. [DOI] [PubMed] [Google Scholar]
- 13.Gadue P., Stein P.L. NK T cell precursors exhibit differential cytokine regulation and require Itk for efficient maturation. J. Immunol. 2002;169:2397–2406. doi: 10.4049/jimmunol.169.5.2397. [DOI] [PubMed] [Google Scholar]
- 14.Sumegi J., et al. Correlation of mutations of the SH2D1A gene and epstein-barr virus infection with clinical phenotype and outcome in X-linked lymphoproliferative disease. Blood. 2000;96:3118–3125. [PubMed] [Google Scholar]
- 15.Gibson S., et al. Identification, cloning, and characterization of a novel human T-cell-specific tyrosine kinase located at the hematopoietin complex on chromosome 5q. Blood. 1993;82:1561–1572. [PubMed] [Google Scholar]
- 16.Pletneva E.V., Sundd M., Fulton D.B., Andreotti A.H. Molecular details of Itk activation by prolyl isomerization and phospholigand binding: the NMR structure of the Itk SH2 domain bound to a phosphopeptide. J. Mol. Biol. 2006;357:550–561. doi: 10.1016/j.jmb.2005.12.073. [DOI] [PubMed] [Google Scholar]
- 17.Saffran D.C., et al. Brief report: a point mutation in the SH2 domain of Bruton’s tyrosine kinase in atypical X-linked agammaglobulinemia. N. Engl. J. Med. 1994;330:1488–1491. doi: 10.1056/NEJM199405263302104. [DOI] [PubMed] [Google Scholar]
- 18.Morra M., et al. Characterization of SH2D1A missense mutations identified in X-linked lymphoproliferative disease patients. J. Biol. Chem. 2001;276:36809–36816. doi: 10.1074/jbc.M101305200. [DOI] [PubMed] [Google Scholar]
- 19.Schwartzberg P.L., Finkelstein L.D., Readinger J.A. TEC-family kinases: regulators of T-helper-cell differentiation. Nat. Rev. Immunol. 2005;5:284–295. doi: 10.1038/nri1591. [DOI] [PubMed] [Google Scholar]
- 20.Liu K.Q., Bunnell S.C., Gurniak C.B., Berg L.J. T cell receptor-initiated calcium release is uncoupled from capacitative calcium entry in Itk-deficient T cells. J. Exp. Med. 1998;187:1721–1727. doi: 10.1084/jem.187.10.1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Berg L.J., Finkelstein L.D., Lucas J.A., Schwartzberg P.L. Tec family kinases in T lymphocyte development and function. Annu. Rev. Immunol. 2005;23:549–600. doi: 10.1146/annurev.immunol.22.012703.104743. [DOI] [PubMed] [Google Scholar]
- 22.Liao X.C., Littman D.R. Altered T cell receptor signaling and disrupted T cell development in mice lacking Itk. Immunity. 1995;3:757–769. doi: 10.1016/1074-7613(95)90065-9. [DOI] [PubMed] [Google Scholar]
- 23.Lucas J.A., Miller A.T., Atherly L.O., Berg L.J. The role of Tec family kinases in T cell development and function. Immunol. Rev. 2003;191:119–138. doi: 10.1034/j.1600-065X.2003.00029.x. [DOI] [PubMed] [Google Scholar]
- 24.Atherly L.O., et al. The Tec family tyrosine kinases Itk and Rlk regulate the development of conventional CD8+ T cells. Immunity. 2006;25:79–91. doi: 10.1016/j.immuni.2006.05.012. [DOI] [PubMed] [Google Scholar]
- 25.Broussard C., et al. Altered development of CD8+ T cell lineages in mice deficient for the Tec kinases Itk and Rlk. Immunity. 2006;25:93–104. doi: 10.1016/j.immuni.2006.05.011. [DOI] [PubMed] [Google Scholar]
- 26.Dubois S., Waldmann T.A., Muller J.R. ITK and IL-15 support two distinct subsets of CD8+ T cells. Proc. Natl. Acad. Sci. U. S. A. 2006;103:12075–12080. doi: 10.1073/pnas.0605212103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kinjo Y., Kronenberg M. Valpha14i NKT cells are innate lymphocytes that participate in the immune response to diverse microbes. J. Clin. Immunol. 2005;25:522–533. doi: 10.1007/s10875-005-8064-5. [DOI] [PubMed] [Google Scholar]
- 28.Ashkar A.A., Rosenthal K.L. Interleukin-15 and natural killer and NKT cells play a critical role in innate protection against genital herpes simplex virus type 2 infection. J. Virol. 2003;77:10168–10171. doi: 10.1128/JVI.77.18.10168-10171.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grubor-Bauk B., Simmons A., Mayrhofer G., Speck P.G. Impaired clearance of herpes simplex virus type 1 from mice lacking CD1d or NKT cells expressing the semivariant V alpha 14-J alpha 281 TCR. J. Immunol. 2003;170:1430–1434. doi: 10.4049/jimmunol.170.3.1430. [DOI] [PubMed] [Google Scholar]
- 30.Kakimi K., Guidotti L.G., Koezuka Y., Chisari F.V. Natural killer T cell activation inhibits hepatitis B virus replication in vivo. J. Exp. Med. 2000;192:921–930. doi: 10.1084/jem.192.7.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miyake S., Yamamura T. NKT cells and autoimmune diseases: unraveling the complexity. Curr. Top. Microbiol. Immunol. 2007;314:251–267. doi: 10.1007/978-3-540-69511-0_10. [DOI] [PubMed] [Google Scholar]
- 32.Levy O., et al. Disseminated varicella infection due to the vaccine strain of varicella-zoster virus, in a patient with a novel deficiency in natural killer T cells. J. Infect. Dis. 2003;188:948–953. doi: 10.1086/378503. [DOI] [PubMed] [Google Scholar]
- 33.Ruschendorf F., Nurnberg P. ALOHOMORA: a tool for linkage analysis using 10K SNP array data. Bioinformatics. 2005;21:2123–2125. doi: 10.1093/bioinformatics/bti264. [DOI] [PubMed] [Google Scholar]
- 34.Abecasis G.R., Cherny S.S., Cookson W.O., Cardon L.R. GRR: graphical representation of relationship errors. Bioinformatics. 2001;17:742–743. doi: 10.1093/bioinformatics/17.8.742. [DOI] [PubMed] [Google Scholar]
- 35.O’Connell J.R., Weeks D.E. PedCheck: a program for identification of genotype incompatibilities in linkage analysis. Am. J. Hum. Genet. 1998;63:259–266. doi: 10.1086/301904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Abecasis G.R., Cherny S.S., Cookson W.O., Cardon L.R. Merlin--rapid analysis of dense genetic maps using sparse gene flow trees. Nat. Genet. 2002;30:97–101. doi: 10.1038/ng786. [DOI] [PubMed] [Google Scholar]
- 37.Thiele H., Nurnberg P. HaploPainter: a tool for drawing pedigrees with complex haplotypes. Bioinformatics. 2005;21:1730–1732. doi: 10.1093/bioinformatics/bth488. [DOI] [PubMed] [Google Scholar]
- 38.Shearer W.T., et al. Lymphocyte subsets in healthy children from birth through 18 years of age: the Pediatric AIDS Clinical Trials Group P1009 study. J. Allergy Clin. Immunol. 2003;112:973–980. doi: 10.1016/j.jaci.2003.07.003. [DOI] [PubMed] [Google Scholar]