Abstract
Hepatic steatosis is present in insulin-resistant obese rodents and is concomitant with active lipogenesis. Hepatic lipogenesis depends on the insulin-induced activation of the transcription factor SREBP-1c. Despite prevailing insulin resistance, SREBP-1c is activated in the livers of genetically and diet-induced obese rodents. Recent studies have reported the presence of an ER stress response in the livers of obese ob/ob mice. To assess whether ER stress promotes SREBP-1c activation and thus contributes to lipogenesis, we overexpressed the chaperone glucose-regulated protein 78 (GRP78) in the livers of ob/ob mice using an adenoviral vector. GRP78 overexpression reduced ER stress markers and inhibited SREBP-1c cleavage and the expression of SREBP-1c and SREBP-2 target genes. Furthermore, hepatic triglyceride and cholesterol contents were reduced, and insulin sensitivity improved, in GRP78-injected mice. These metabolic improvements were likely mediated by restoration of IRS-2 expression and tyrosine phosphorylation. Interestingly, GRP78 overexpression also inhibited insulin-induced SREBP-1c cleavage in cultured primary hepatocytes. These findings demonstrate that GRP78 inhibits both insulin-dependent and ER stress–dependent SREBP-1c proteolytic cleavage and explain the role of ER stress in hepatic steatosis in obese rodents.
Introduction
Nonalcoholic fatty liver disease (NAFLD) is one of the most common chronic liver diseases. In Western societies, NAFLD is clearly associated with the features of the metabolic syndrome, including obesity, type 2 diabetes, hypertension, and dyslipidemia. Hepatic steatosis is considered the first stage leading to more severe complications, such as steatohepatitis, cirrhosis, and hepatocellular carcinoma. Moreover, hepatic fat accumulation is associated with the development of hepatic insulin resistance, characterized by increased hepatic glucose production leading to fasting hyperglycemia.
Recent studies in humans pointed out the important role of de novo lipogenesis in the excessive accumulation of triglycerides in the livers of patients with NAFLD, since one-third of total triglycerides might originate from this pathway (1). In addition, hepatic lipogenesis can also contribute to lipid accumulation by reducing fatty acid oxidation, as malonyl-CoA, generated in the lipogenic pathway, is a potent inhibitor of carnitine palmitoyl transferase I. Lipogenesis is highly dependent on nutritional conditions for its activation; it is induced by high-carbohydrate feeding and inhibited by fasting. SREBP-1c, a member of the SREBP family of transcription factors, and carbohydrate response element–binding protein (ChREBP) have emerged as transcription factors involved in the transcriptional effects of insulin and glucose, respectively, on lipogenic gene expression in the liver (2, 3). Activation of SREBP-1c by insulin during carbohydrate feeding involves 2 mechanisms: activation of SREBP-1c transcription and increase in proteolytic cleavage of the SREBP-1c precursor form embedded in the membranes of the ER (4–6). Within the ER membranes, the inactive SREBP proteins are associated with 2 essential proteins that take part in the control of the cleavage process: SREBP cleavage–activating protein (SCAP) and insulin-induced gene (Insig). SCAP interacts on the one hand with newly synthesized SREBP precursor and on the other hand with Insig, which functions as a retention protein of the SCAP/SREBP complex into the ER (7, 8). In the presence of specific signals such as insulin, SCAP dissociates from Insig and escorts SREBPs in coatomer protein II (COPII) vesicles from the ER to the Golgi apparatus, where SREBPs are proteolytically processed to yield the transcriptionally active form. Once released, the SREBP mature transcription factors can enter into the nucleus and, for the SREBP-1c mature isoform, activate the expression of lipogenic genes. The critical role of SREBP-1c in lipogenesis is emphasized in loss- and gain-of-function studies in vitro and in vivo. Hepatocytes or mice overexpressing an active form of SREBP-1c develop steatosis as a result of activation of the entire lipogenic program (9–11). In contrast, mice invalidated for hepatic SREBP-1c fail to induce lipogenic enzyme expression in response to fasting and refeeding (12).
Although highly dependent on insulin for its activation, lipogenesis is paradoxically very active in the livers of obese rodents such as ob/ob mice, which are characterized by severe hepatic insulin resistance resulting in glucose overproduction and hyperglycemia. Moreover, accumulation of nuclear SREBP-1c is detected in the livers of these animals (13). The failure of insulin to suppress gluconeogenesis while lipogenesis is still activated could reflect a differential sensitivity of these pathways to insulin. It was proposed that decreased IRS-2 content caused by hyperinsulinemia could account for the resistance to insulin-mediated repression of gluconeogenic genes (14) and that insulin could still activate lipogenic genes by IRS-1 or by another route (14). However, IRS tyrosine phosphorylation in the livers of ob/ob mice decreases for both IRS-1 and IRS-2 (15, 16). Thus the marked activation of SREBP-1c in the ob/ob mouse liver cannot be explained by stimulated activity of insulin signaling.
Recent studies have demonstrated the presence of an unfolded protein response (UPR) in the liver and adipose tissue of insulin-resistant rodents, which, when counteracted, improves the insulin resistance of these animals (17–20). Secreted and transmembrane proteins are folded in the ER, and the role of the UPR is to dynamically adjust the protein folding capacity to the cell requirements. Thus, any event that can disturb ER folding capacity, such as a load of unfolded proteins or altered redox state, calcium equilibrium, or glycosylation potential, will induce a UPR. The UPR is mediated by 3 transducer proteins that are integral membrane proteins of the ER: inositol-requiring kinase–1 (IRE1), an inositol-requiring kinase with endonuclease activity; activating transcription factor 6 (ATF6), a transcription factor that, like SREBP-1c and SREBP-2, is activated by proteolytic cleavage in the Golgi; and protein kinase RNA–like ER kinase (PERK). Each of these effectors activates specific pathways allowing for increased ER folding capacity, decreased protein folding load, induced degradation of misfolded proteins, and, ultimately, induced cell death (21, 22). It has been shown previously that a UPR induced by homocysteine is able to activate SREBP-1c and to induce lipogenic gene expression (23). We therefore reasoned that in the livers of obese insulin-resistant animals, an ER stress, by inducing SREBP-1c cleavage, could be an alternative explanation for the ongoing lipid synthesis.
In the present study, we showed that SREBP-1c cleavage was induced by the ER stress pathway independently of insulin. We also demonstrated that attenuation of ER stress by molecular chaperones in livers of ob/ob mice slowed down lipogenesis by inhibiting SREBP-1c proteolytic cleavage and thus improved steatosis and insulin sensitivity of these animals. Finally, we showed that insulin-induced SREBP cleavage was inhibited by overexpression of the chaperone glucose-regulated protein 78 (GRP78) and that the SREBP-1c complex was able to bind GRP78. Our results indicate that ER stress is a major component of the hepatic steatosis and insulin resistance observed in obese insulin-resistant rodents.
Results
The mature form of SREBP-1c and proteins of the UPR pathway are coinduced in the livers of insulin-resistant obese rodents.
We and others have previously shown that SREBP-1c is highly dependent on insulin for its expression and maturation in the liver. Insulin stimulates both SREBP-1c transcription and proteolytic cleavage in hepatocytes (4–6). Intriguingly, it was observed that the nuclear form of SREBP-1c is highly abundant in livers of ob/ob (lacking leptin secretion) and aP2–SREBP-1c lipodystrophic mice, models characterized by severe hepatic insulin resistance and steatosis (13). Here, we confirmed this finding in ob/ob mice (Figure 1A) and established that this was the case in obese insulin-resistant Zucker fa/fa rats, which lack the leptin receptor (Supplemental Figure 1; supplemental material available online with this article; doi:10.1172/JCI37007DS1). In these animals, the expression of SREBP-1c target genes, including fatty acid synthase (FAS; Figure 1A) and many glycolytic and lipogenic enzymes (data not shown), were robustly stimulated. The mRNA and the precursor form of SREBP-1c also increased in the livers of these animals, suggesting that the enhanced proteolytic cleavage of the precursor is compensated for by stimulated SREBP-1c transcription. This can be related to a feed-forward mechanism, because the SREBP-1c promoter is a target of active SREBP-1c itself (24, 25).
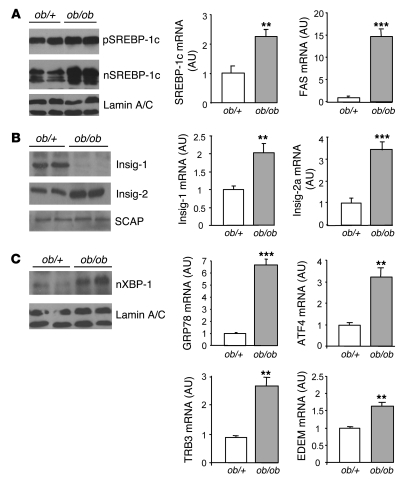
Figure 1. Expression of SREBP-1c and Insig proteins, SREBP-1c target genes, and ER stress markers in ob/+ and ob/ob mouse livers.
Livers from 8-wk-old mice were used to prepare nuclear extracts, microsomes, and total RNA. (A) Immunoblot analysis of the precursor form (p-) and nuclear (n-) SREBP-1c as well as lamin A/C from ob/+ and ob/ob mice. Quantification by qRT-PCR of SREBP-1c and FAS mRNA in the liver is shown at right. (B) Immunoblot analysis of Insig-1, Insig-2, and SCAP proteins. Quantification of Insig-1 and Insig-2a (liver specific) mRNA levels by qRT-PCR is shown at right. (C) Immunoblot analysis of nuclear XBP-1 in nuclear extracts. Quantification of GRP78, ATF4, TRB3, and EDEM mRNA levels by qRT-PCR is shown at right. Results are mean ± SEM (n = 4–5 per group). **P < 0.01, ***P < 0.001 versus ob/+.
Among the partners involved in SREBP processing, Insig-1 and Insig-2 proteins have been identified as proteins that promote SREBP retention in the ER through their interaction with SCAP, thus preventing SREBP-SCAP complex translocation to the Golgi apparatus for proteolytic processing (26). Insig-1 is transcriptionally induced by insulin and depends on nuclear SREBP-1c for its expression (27). Conversely, in the liver, Insig-2 is selectively downregulated by insulin and is independent of SREBP-1c for its expression (27). Insig mRNAs and protein contents were analyzed in the livers of lean ob/+ and ob/ob mice (Figure 1B). Insig-2 mRNA and protein increased in ob/ob compared with ob/+ mice, indicating that in these animals, insulin fails to inhibit Insig-2 expression. Insig-1 mRNA increased in the livers of ob/ob compared with ob/+ mice, in agreement with the accumulation of nuclear SREBP-1c. Surprisingly, despite this increase, Insig-1 proteins were undetectable in the livers of ob/ob mice, suggesting that Insig-1 is not translated or is degraded. In contrast to the changes observed for Insig proteins, the expression of SCAP, another partner of the SREBP processing complex, was not modified in the livers of ob/ob mice (Figure 1B).
Because activation of the ER stress pathway has previously been reported in the livers and adipose tissue of obese animals (17), we assessed whether ER stress is induced in our animal models. As shown in Figure 1C and Supplemental Figure 1, concomitantly to the activation of SREBP-1c, we observed activation of proteins of the UPR pathway in the livers of obese rodents. X box–binding protein–1 (XBP-1) nuclear protein content was highly increased in the livers of obese compared with lean animals, indicating that the IRE1 branch of the UPR pathway was activated (Figure 1C). Moreover, GRP78, ATF4, TRB3, and EDEM mRNA, induced in response to the activation of ATF6 and PERK, were increased in the livers of obese animals. Because it was previously shown that SREBP-1c is activated by homocysteine-induced ER stress in the liver (23), we hypothesized that activation of the UPR pathway in the livers of insulin-resistant rodents might account for the activation of SREBP-1c processing observed in these animals.
SREBP-1c proteolytic cleavage and SREBP-1c target genes are induced by ER stress in cultured hepatocytes.
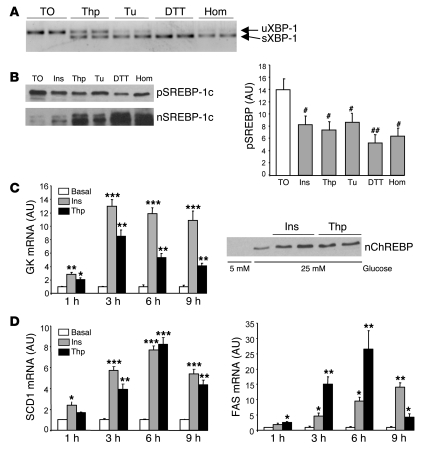
We sought to determine in cultured rat hepatocytes whether cleavage of the SREBP-1c precursor in the presence of ER stress is a generalized feature or whether it is restricted to homocysteine-induced ER stress. ER stress was thus induced by various compounds: thapsigargin, tunicamycin, DTT, and homocysteine. Thapsigargin modifies Ca2+ concentration in the ER lumen by inhibiting Ca2+ ATPase. Tunicamycin, DTT, and homocysteine lead to accumulation of proteins into the lumen of the ER by inhibiting protein glycosylation or the formation of disulfide bridges. We first verified that these compounds trigger an UPR activation in our culture conditions. As shown in Figure 2A, 1-h incubation in the presence of thapsigargin, tunicamycin, DTT, or homocysteine induced the splicing of XBP-1 mRNA, a classical index of the UPR. In order to analyze the effect of ER stress on SREBP-1c proteolytic cleavage, hepatocytes were incubated for 6 h in the presence of the LXR agonist TO-901317. This treatment led to accumulation of the precursor form of SREBP-1c in the ER membranes without inducing SREBP-1c proteolytic cleavage (Figure 2B and ref. 6). As previously observed (6), addition of insulin for 1 h decreased the precursor form of SREBP-1c and increased SREBP-1c in nuclear extracts, showing activation of the cleavage process. The treatment with ER stress inducers for the same time was followed by a decrease in the precursor form of SREBP-1c and a massive accumulation of nuclear SREBP-1c (Figure 2B). Although we cannot totally rule out stabilization of the mature form of SREBP-1c in the presence of ER stress inducers, these observations suggest that UPR activation induces proteolytic cleavage of SREBP-1c in vitro. We next analyzed the consequences of SREBP-1c activation on the expression of SREBP-1c target genes glucokinase (GK) and lipogenesis-related genes. Hepatocytes were cultured for 1, 3, 6, and 9 h in medium containing 25 mM glucose in the presence of thapsigargin or insulin. We found a high glucose concentration to be necessary to fully induce lipogenic transcripts through the activation of the glucose-responsive transcription factor ChREBP (see below). We studied expression of GK, a gene known to be induced by insulin through SREBP-1c action (6, 28). A marked increase in GK expression was observed in hepatocytes treated with thapsigargin (Figure 2C) or other ER stress inducers (data not shown), an effect close to that of insulin (Figure 2C). This increased GK expression is important, because an increase in glucose flux through GK will induce the generation of a metabolite necessary for the activation of the transcription factor ChREBP through its nuclear translocation (29). Thapsigargin in the presence of a high glucose concentration induced a marked increased in the nuclear concentration of ChREBP, an effect similar to that of insulin (Figure 2C). Lipogenic enzyme expression is dependent on the concomitant activation of SREBP-1c and ChREBP (30, 31). An increase in the expression of FAS and stearoyl-CoA desaturase 1 (SCD1) was thus logically observed in hepatocytes treated with thapsigargin (Figure 2D) or with other ER stress inducers (data not shown); the effect was comparable to that observed in the presence of insulin.
Figure 2. Effect of ER stress on SREBP-1c proteolytic cleavage and SREBP-1c target genes in cultured rat hepatocytes.
(A and B) Hepatocytes were incubated for 6 h in M199 medium supplemented with the LXR agonist TO-901317 (TO; 10 μM) and then treated for 1 h with 100 nM insulin (Ins), 300 nM thapsigargin (Thp), 1 μg/ml tunicamycin (Tu), 500 μM DTT, or 5 mM homocysteine (Hom). (A) Unspliced (u-) and spliced (s-) forms of XBP-1 mRNA, measured by RT-PCR. (B) Immunoblot analysis of SREBP-1c precursor and nuclear SREBP-1c. Quantification of SREBP-1c precursor is shown at right. Results are mean ± SEM of 3 independent cultures of hepatocytes in which 3 plates were collected for each condition. #P < 0.05, ##P < 0.01 versus TO-901317 alone. (C and D) Hepatocytes were treated for 1, 3, 6, and 9 h with 100 nM insulin or 300 nM thapsigargin in medium containing 25 mM glucose. (C) Total RNA from triplicate plates of hepatocytes was analyzed for the expression of GK. For the measurement of nuclear ChREBP, hepatocytes were cultured in the presence of 5 or 25 mM glucose and treated for 3 h in the presence of 100 nM insulin or 300 nM thapsigargin. (D) Total RNA from triplicate plates of hepatocytes was analyzed for expression of SCD1 and FAS mRNA. *P < 0.05, **P < 0.01, ***P < 0.001 versus basal value.
ER stress–induced SREBP-1c cleavage is reduced in vitro by overexpression of GRP78.
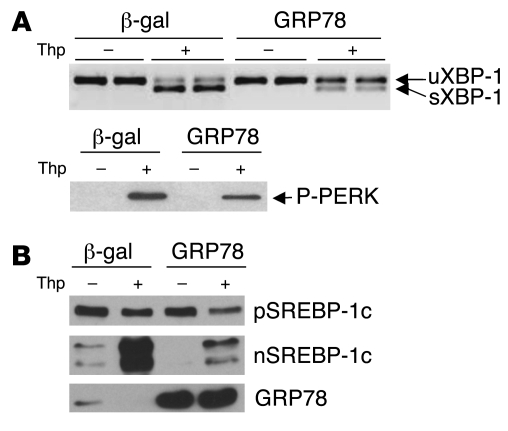
If ER stress is indeed responsible for the overexpression of nuclear SREBP-1c observed in the livers of insulin-resistant rodents, then a reduction of ER stress in the livers of ob/ob mice should reduce SREBP-1c processing, lipogenic gene expression, and hepatic steatosis. One of the key events in the activation of ER stress is the dissociation of the chaperone GRP78 from the luminal part of the ER integral membrane proteins: PERK, IRE1, and ATF6. It has previously been reported that supplementation with chaperones such as GRP78 or oxygen-related protein 150 (ORP150) is sufficient to prevent the activation of PERK, IRE1, and ATF6 and to limit ER stress (32). We thus counteracted ER stress in vivo by overexpressing GRP78 using an adenoviral vector. The efficiency of this procedure was first tested in cultured rat hepatocytes. Cultured hepatocytes were infected for 24 h with an adenovirus encoding human GRP78 and a control adenovirus encoding β-gal (referred to herein as Ad GRP78 and Ad β-gal, respectively), and ER stress was induced for 1 h by the addition of thapsigargin. As shown in Figure 3A, thapsigargin induced the splicing of XBP-1 mRNA and the phosphorylation of PERK, both of which were strongly reduced by overexpression of GRP78, demonstrating that GRP78 is effective to reduce ER stress in vitro. We next studied the consequences of GRP78 overexpression on SREBP-1c proteolytic cleavage induced by thapsigargin in cultured hepatocytes. As shown in Figure 3B and Figure 2, 1 h treatment with thapsigargin stimulated the proteolytic cleavage of SREBP-1c. Overexpression of GRP78 in hepatocytes strongly reduced the ER stress–induced release of nuclear SREBP-1c in response to thapsigargin (Figure 3B).
Figure 3. Effect of GRP78 overexpression on thapsigargin-induced ER stress and SREBP-1c proteolytic cleavage in cultured rat hepatocytes.
After a 24-h period of infection with Ad GRP78 or Ad β-gal, cultured hepatocytes were changed to a fresh M199 medium containing 10 μM TO-901317 for 6 h, then treated for 1 h with 300 nM thapsigargin or control DMSO. (A) Total RNA from triplicate plates of hepatocytes were extracted and analyzed for unspliced and spliced forms of XBP-1 mRNA by RT-PCR. The phosphorylation state of PERK in total lysates of hepatocytes was analyzed by Western blot. (B) Immunoblot of SREBP-1c in hepatic microsomal membranes and nuclear extracts and immunoblot of GRP78 measured in the microsomal fraction. Results are representative of 3 independent experiments with different preparations of hepatocytes.
In vivo overexpression of GRP78 reduces ER stress, SREBP-1c and SREBP-2 target genes, and steatosis in ob/ob mouse liver.
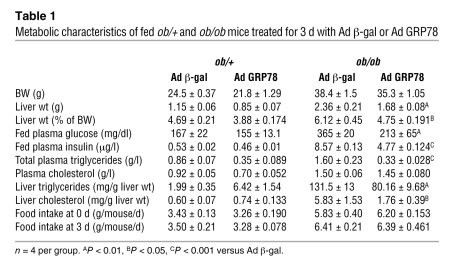
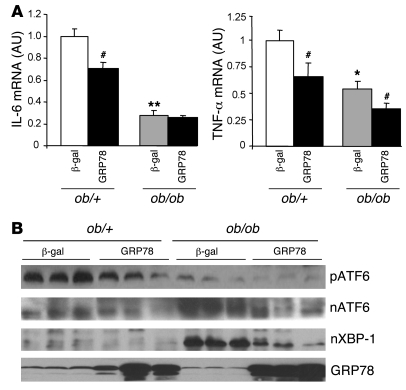
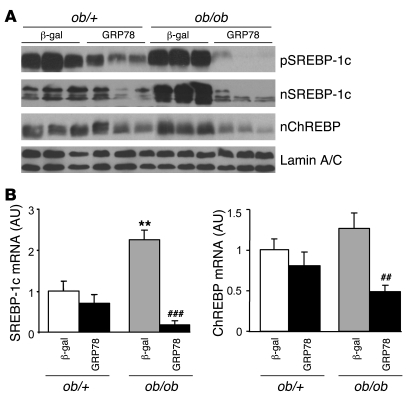
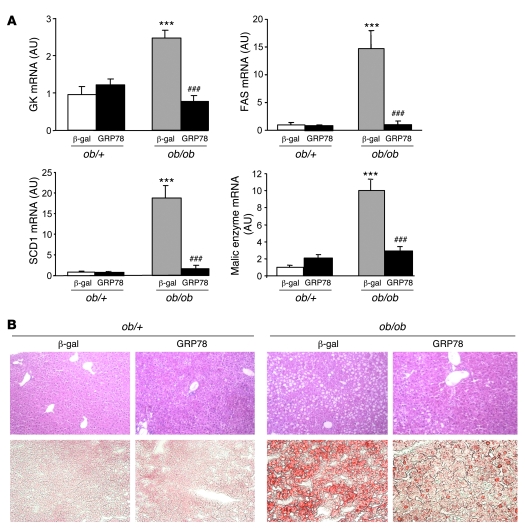
Having established in vitro the efficiency of GRP78 overexpression to reduce ER stress, we next determined whether GRP78 effectively reduces ER stress and SREBP-1c proteolytic cleavage in vivo in the ob/ob mouse liver. We injected ob/+ and ob/ob mice with Ad β-gal control or Ad GRP78 and analyzed them 72 h later in the fed state. Food intake and body weight were not altered in Ad GRP78–injected mice (Table 1). As shown previously (11), the delivery of adenovirus was liver specific, because the expression of β-gal was undetectable in tissues other than liver (data not shown). We also verified that GRP78 overexpression did not induce inflammation in the liver by measuring expression of the cytokines IL-6 and TNF-α, which in fact decreased (Figure 4A). Plasma alanine aminotransferase and aspartate aminotransferase were not modified in Ad GRP78–injected mice (data not shown). We next analyzed whether Ad GRP78 effectively reduces ER stress markers in the ob/ob mouse liver. As shown in Figure 4B, the mature nuclear forms of the UPR transcription factors ATF6 and XBP-1 accumulated in the livers of ob/ob compared with ob/+ mice. In contrast, the amount of ATF6 precursor was reduced in the livers of ob/ob compared with ob/+ mice, showing that ATF6 was processed in ob/ob mice. Adenovirally mediated expression of GRP78 caused a major increase in the protein content of GRP78 in ob/+ and ob/ob mice and a strong decrease in ATF6 and XBP nuclear forms in ob/ob mice (Figure 4B), demonstrating that ER stress was reduced in the livers of these animals. We also analyzed the consequences of ER stress inhibition on SREBP-1c mRNA and proteins levels in ob/+ and ob/ob mouse liver. The overexpression of GRP78 resulted in a near-complete disappearance of nuclear SREBP-1c in ob/ob mice (Figure 5A). A similar reduction was observed in ob/+ animals, although less marked than that in ob/ob mice. As a consequence of nuclear SREBP-1c decrease in the livers of Ad GRP78–injected mice, the SREBP-1c precursor (Figure 5A) and SREBP-1c mRNA (Figure 5B) were also strongly reduced. As previously stated, this might be explained by the fact that SREBP-1c participates in the control of its own transcription (24, 25). ChREBP nuclear content (Figure 5A) and ChREBP mRNA (Figure 5B) were also strongly diminished by GRP78 overexpression in the liver. This might result from the fact that the generation of a glucose metabolite necessary for the activation of ChREBP was reduced secondary to decreased expression of GK, a direct SREBP-1c target gene (Figure 6A). As a consequence of the fall in hepatic SREBP-1c and ChREBP nuclear forms in Ad GRP78–injected ob/ob mice, the levels of FAS, SCD1, and malic enzyme mRNA, target genes of SREBP-1c and ChREBP, were strongly reduced to levels observed in ob/+ mice (Figure 6A). The decrease in lipogenic gene expression in the livers of Ad GRP78–treated ob/ob mice prompted us to analyze the consequences on liver and serum triglycerides. Liver staining revealed large lipid droplets in ob/ob compared with ob/+ mice (Figure 6B). The overexpression of GRP78 considerably decreased the size and number of lipid droplets in ob/ob mouse livers (Figure 6B). This was associated with a decrease in hepatic triglyceride content, liver weight, and plasma triglyceride concentration (Table 1).
Table 1 .
Metabolic characteristics of fed ob/+ and ob/ob mice treated for 3 d with Ad β-gal or Ad GRP78
Figure 4. Expression of cytokines and ER stress markers in the livers of ob/+ and ob/ob mice injected with Ad GRP78 or Ad β-gal.
Mice were injected with the indicated adenoviruses and sacrificed in the fed state 72 h later. (A) Total RNA was extracted and analyzed for IL-6 and TNF-α mRNA by qRT-PCR. *P < 0.05, **P < 0.01 versus respective ob/+ value; #P < 0.05 versus respective Ad β-gal value. (B) Microsomal membranes and nuclear extracts were prepared and analyzed by Western blot for the expression of precursor and nuclear ATF6, nuclear XBP-1, and microsomal GRP78. Each lane represents a different animal. Results are representative of 3 independent experiments.
Figure 5. SREBP-1c and ChREBP protein levels and mRNA in the livers of ob/+ and ob/ob mice overexpressing GRP78.
Mice were injected with Ad β-gal or Ad GRP78, and livers were collected in the fed state 72 h later for the preparation of microsomal membranes, nuclear extracts, and isolation of total RNA. (A) Analysis of SREBP-1c precursor, nuclear SREBP-1c, nuclear ChREBP, and lamin A/C by Western blot. (B) Relative levels of SREBP-1c and ChREBP mRNA measured by qRT-PCR. Results are mean ± SEM (n = 5–6 per group). **P < 0.01 versus respective ob/+ value; ##P < 0.01, ###P < 0.001 versus respective Ad β-gal value.
Figure 6. SREBP-1c target gene expression and Oil Red O staining in the livers of ob/+ and ob/ob mice overexpressing GRP78.
Mice were injected with Ad β-gal or Ad GRP78, and livers were collected in the fed state 72 h later for the preparation of total RNA. (A) Relative levels of GK, FAS, SCD1, and malic enzyme mRNA measured by qRT-PCR. Results are mean ± SEM (n = 5–6 per group). ***P < 0.001 versus respective ob/+ value; ###P < 0.001 versus respective Ad β-gal value. (B) Liver sections were stained with H&E (top row) or Oil red O (bottom row). Original magnification, ×20.
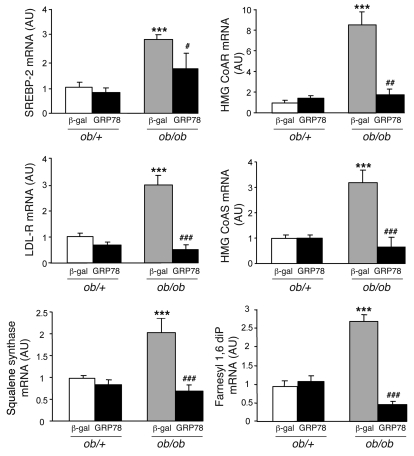
Because effects of ER stress have also been described for the SREBP-2 protein (33), a SREBP isoform activated by cholesterol depletion, we analyzed the consequences of GRP78 overexpression on SREBP-2 target genes. The expression of genes involved in cholesterol and isoprenoid metabolism (hydroxymethylglutaryl-CoA reductase, hydroxymethylglutaryl-CoA synthase, farnesyl diphosphatase, and squalene synthase) and cholesterol uptake (LDL receptor) were increased in livers of ob/ob compared with ob/+ mice (Figure 7), which indicates that SREBP-2 is activated. SREBP-2 expression was itself higher in ob/ob compared with ob/+ mice. Liver overexpression of GRP78 strongly reduced the mRNA levels of SREBP-2 and its target genes to values found in ob/+ mice, suggesting that SREBP-2 processing is also blunted in the livers of mice expressing GRP78 (Figure 7). Despite several attempts, we could not detect the SREBP-2 protein in these experiments. The liver content in total cholesterol was strongly reduced in GRP78 ob/ob mice, whereas no modification of plasma cholesterol concentration was observed (Table 1). Together, these results showed that overexpression of GRP78 leads to a clear improvement of hepatic steatosis and dyslipidemia in ob/ob mice.
Figure 7. SREBP-2 and SREBP-2 target gene expression in the livers of ob/+ and ob/ob mice overexpressing GRP78.
Mice were injected with Ad β-gal or Ad GRP78, and livers were collected in the fed state 72 h later for the preparation of total RNA. Shown are relative levels of SREBP-2, hydroxymethylglutaryl-CoA reductase (HMG CoAR), LDL receptor (LDL-R), hydroxymethylglutaryl-CoA synthase (HMG CoAS), squalene synthase, and farnesyl 1,6 diphosphatase (Farnesyl 1,6 diP) mRNA measured by qRT-PCR. Results are mean ± SEM (n = 5–6 per group). ***P < 0.001 versus respective ob/+ value; #P < 0.05, ##P < 0.01, ###P < 0.001 versus respective Ad β-gal value.
Hepatic overexpression of GRP78 improves liver insulin signaling and insulin sensitivity in ob/ob mice.
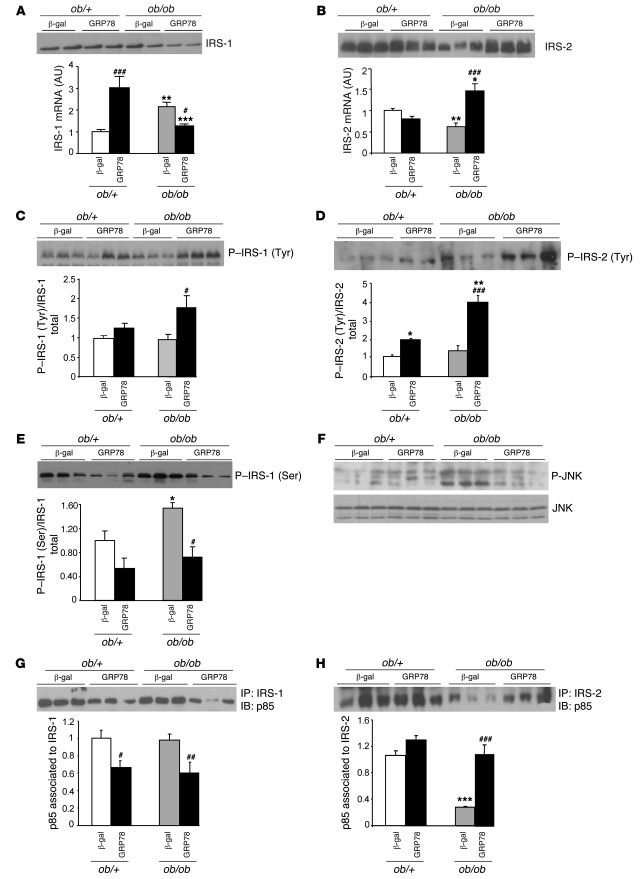
Because a reduction in hepatic steatosis has often been associated with improved hepatic insulin sensitivity (34, 35), we analyzed the effects of GRP78 overexpression on liver insulin signaling. Whereas we found the IRS-1 protein content to be mildly affected and its mRNA level even increased in ob/ob compared with ob/+ mice (Figure 8A), IRS-2 mRNA and protein contents were lower (Figure 8B), as previously described (14, 16). IRS-1 and IRS-2 phosphorylation on tyrosine residues was similar when expressed per unit of IRS protein in fed ob/ob and ob/+ mice (Figure 8, C and D). This occurred despite the huge increase in plasma insulin concentration in ob/ob compared with ob/+ mice (Table 1), indicating an impaired signaling process in the former. This could be secondary to increased serine phosphorylation on IRS proteins, as we observed here for IRS-1 (Figure 8E). In turn, this could be the consequence of increased JNK activity in ob/ob mice (Figure 8F), as shown previously by other groups (36, 37). Levels of the IRS-1–associated p85 subunit of PI3K were similar in fed ob/ob and ob/+ animals, but the IRS-2–associated p85 subunit of PI3K was strongly decreased in ob/ob compared with ob/+ mice (Figure 8, G and H). These results confirmed an overall impairment of insulin signaling in fed ob/ob mice that was more marked for the IRS-2 branch, a likely consequence of the decreased IRS-2 protein content.
Figure 8. Consequences of GRP78 overexpression on insulin signaling in the livers of ob/+ and ob/ob mice.
Mice were injected with Ad β-gal or Ad GRP78, and livers were collected in the fed state 72 h later. Total liver extracts and total RNA were prepared and analyzed by Western blotting and qRT-PCR, respectively. (A) Total IRS-1 protein content and relative expression of IRS-1 mRNA. (B) IRS-2 total protein content and relative mRNA expression. (C and D) Tyrosine phosphorylation of IRS-1 (C) and IRS-2 (D). Shown are a representative blot and quantification of 3 independent injection series (n = 4–5 per group). (E) Serine phosphorylation of IRS-1. Shown are a representative blot and quantification of 3 independent injection series (n = 4–5 per group). (F) Total liver extracts were analyzed for phosphorylated and total JNK. (G and H) Association of the PI3K p85 subunit with IRS-1 (G) and IRS-2 (H). Shown are a representative blot and quantification of 3 independent injection series (n = 4–5 per group). *P < 0.05, **P < 0.01, ***P < 0.001 versus respective ob/+ value; #P < 0.05, ##P < 0.01, ###P < 0.001 versus respective Ad β-gal value.
Counteracting ER stress in the livers of ob/ob mice leads to marked changes in the insulin signaling pathway. IRS-1 protein and mRNA content decreased in GRP78-treated ob/ob mice (Figure 8A), whereas IRS-2 protein and mRNA content markedly increased (Figure 8B) to a level comparable to that observed in ob/+ mice. IRS-1 and IRS-2 tyrosine phosphorylation per unit of IRS protein was stimulated (Figure 8, C and D), and IRS-1 serine phosphorylation was reduced (Figure 8E), concomitantly with decreased phosphorylation of JNK (Figure 8F). This led to a reduction in the IRS-1–associated p85 subunit of PI3K (Figure 8G), but a huge increase in the IRS-2–associated p85 subunit of PI3K (Figure 8H). In ob/+ animals, GRP78 overexpression had minor effects, except for a large increase in IRS-1 mRNA. In summary, IRS-2–mediated insulin signaling was the main pathway improved by counteracting ER stress in obese animals.
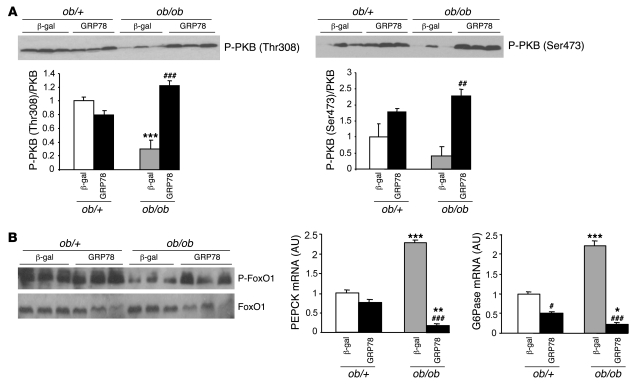
Finally, we analyzed more distal steps of insulin signaling. Akt/PKB phosphorylation on both Thr308 and Ser473, which was strongly reduced in ob/ob compared with ob/+ mice, was upregulated by ER stress inhibition in ob/ob mice, whereas there were no marked effects of GRP78 overexpression in ob/+ mice (Figure 9A). FoxO1 is considered a key transcription factor in the control of gluconeogenesis in the liver, because it can stimulate the expression of key gluconeogenic enzymes such as phosphoenolpyruvate carboxykinase (PEPCK) and glucose-6-phosphatase (G6Pase) (38). In the presence of insulin, FoxO1 is phosphorylated by PKB and then excluded from the nucleus (39). The increase of PKB activity in livers of ob/ob GRP78-treated animals prompted us to analyze the consequences of hepatic ER stress inhibition on FoxO1 and gluconeogenic enzyme gene expression. As a result of the state of insulin resistance described above, FoxO1 phosphorylation decreased, and gene expression of PEPCK and G6Pase was upregulated, in livers of ob/ob compared with ob/+ mice (Figure 9B). In ob/ob mice treated with Ad GRP78, phosphorylation of FoxO1 increased, and expression of PEPCK and G6Pase was strongly reduced, whereas the effects in ob/+ mice were minor (Figure 9B), corroborating the improvement in hepatic insulin sensitivity.
Figure 9. Consequences of GRP78 overexpression on PKB and FoxO phosphorylation and on gluconeogenic enzyme expression in livers of ob/+ and ob/ob mice.
Mice were injected with Ad β-gal or Ad GRP78, and livers were collected in the fed state 72 h later. Total liver extracts and mRNA were prepared and analyzed by Western blotting or qRT-PCR. (A) Phosphorylation of PKB on Thr308 and Ser473. Shown are representative blots and quantification of 3 independent injection series (n = 4–5 per group). (B) Total liver extracts were analyzed by Western blotting for phosphorylated and total FoxO1. Also shown is relative expression of PEPCK and G6Pase mRNA. *P < 0.05, **P < 0.01, ***P < 0.001 versus respective ob/+ value; #P < 0.05, ##P < 0.01, ###P < 0.001 versus respective Ad β-gal value.
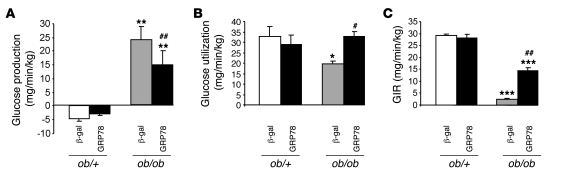
We next analyzed whether a liver-specific reduction in ER stress that improves hepatic insulin signaling has detectable effects on glucose homeostasis and overall insulin sensitivity. Plasma glucose and insulin were strongly diminished in fed ob/ob mice injected with GRP78 compared with their controls (Table 1), suggesting that their insulin sensitivity was improved. These differences were apparent 7 d after the Ad GRP78 injection (results not shown). In order to assess more precisely the improvement in insulin sensitivity, hyperinsulinemic-euglycemic clamp studies were performed. During clamp studies, hepatic glucose production was totally inhibited in ob/+ mice, whereas it was 22 mg/min/kg in ob/ob mice (Figure 10A). Glucose utilization was 40% lower in ob/ob mice (Figure 10B). We also observed in ob/ob mice a 15-fold lower rate of glucose infusion, a clear index of their insulin resistance. Inhibition of ER stress by overexpression of GRP78 caused a 6-fold increase in ob/ob mice of the glucose infusion rate necessary to maintain euglycemia (Figure 10C). This was secondary to a 40% decrease in hepatic glucose production and a 60% increase in glucose utilization rate, which suggests that both liver and peripheral tissues have improved insulin sensitivity. There was no effect of GRP78 overexpression during clamp studies in ob/+ mice (Figure 10).
Figure 10. Consequences of GRP78 overexpression on insulin sensitivity in awake ob/+ and ob/ob mice.
(A–C) Hepatic glucose production (A), glucose utilization (B), and glucose infusion rate (GIR; C) were measured in ob/+ and ob/ob mice treated with Ad β-gal or Ad GRP78 during hyperinsulinemic-euglycemic clamps. *P < 0.05, **P < 0.01, ***P < 0.001 versus respective ob/+ value; #P < 0.05, ##P < 0.01 versus respective Ad β-gal value.
GRP78 overexpression and inhibition of SREBP-1c cleavage.
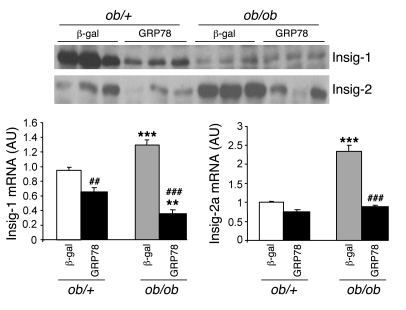
In order to analyze the mechanisms by which overexpression of GRP78 leads to reduced SREBP-1c cleavage, we studied the expression of Insig proteins. Both Insig-1 and Insig-2 mRNA contents were reduced by overexpression of GRP78 in ob/+ and ob/ob mice, leading to low protein concentrations (Figure 11). Thus, the reduced activation of SREBP-1c — and probably SREBP-2 — could not be explained by increased expression of Insig proteins.
Figure 11. Insig-1 and Insig-2 gene expression and protein content in the livers of ob/+ and ob/ob mice overexpressing GRP78.
Mice were injected with Ad β-gal or Ad GRP78, and livers were collected in the fed state 72 h later. Total liver extracts were prepared and analyzed by Western blotting for Insig-1 and Insig-2 protein content. The graph shows relative expression of Insig-1 and Insig-2 mRNA quantified by qRT-PCR. **P < 0.01, ***P < 0.001 versus respective ob/+ value; ##P < 0.01, ###P < 0.001 versus respective Ad β-gal value.
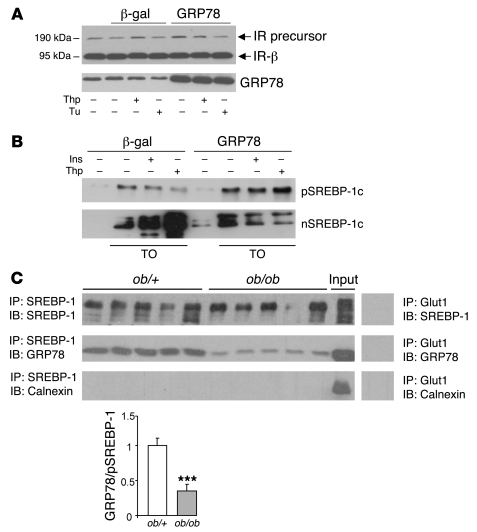
We were puzzled by the fact that in Ad GRP78–injected ob/ob mice, despite the improvement of insulin sensitivity and the still-high insulin concentrations compared with ob/+ mice (see Table 1), the expression of SREBP-1c and its usual target genes such as GK, FAS, and Insig-1 were lower than in ob/+ mice. This suggests that in mice overexpressing GRP78, the insulin-dependent SREBP-1c cleavage is blunted. One obvious explanation would be that insulin-induced SREBP-1c processing is inhibited by GRP78 overexpression. We thus tested this possibility in primary cultured hepatocytes. A crucial step in SREBP-1c processing is its transfer from the ER to the Golgi apparatus. In order to demonstrate that GRP78 does not induce a general inhibition of ER to Golgi transport, we measured the expression of the IR in cultured hepatocytes overexpressing GRP78. The IR is synthesized as a 190-kDa precursor in the ER and processed in the Golgi to yield 2 subunits of 135 and 95 kDa (the IR-α and IR-β subunits, respectively; ref. 40), and its half-life has been estimated to be about 2 h. We found that the IR precursor did not accumulate in Ad GRP78–treated hepatocytes and that the concentration of the β subunit did not decrease, indicating that there is no general impairment of the ER to Golgi transport (Figure 12A).
Figure 12. In vitro effects of GRP78 overexpression on insulin-induced SREBP-1c cleavage in rat hepatocytes and analysis of SREBP-1c complex and GRP78 interaction in mouse livers.
(A) Immunoblot of the IR precursor and IR-β in total lysates of hepatocytes infected for 24 h with Ad GRP78 or Ad β-gal. The blot was hybridized with a GRP78 antibody to verify the overexpression of the transgene. (B) After a 24-h period of infection with Ad GRP78 or Ad β-gal, cultured hepatocytes were changed to a fresh M199 medium containing 10 μM TO-901317 for 6 h, then treated for 1 h with 300 nM thapsigargin or 100 nM insulin. Shown is immunoblot analysis of microsomal SREBP-1c precursor and nuclear SREBP-1c. (C) Microsomal membranes were isolated from the livers of fed ob/+ and ob/ob mice. Detergent-solubilized membranes were subjected to immunoprecipitation with polyclonal H160 anti–SREBP-1 antibody. Immunoprecipitated proteins were probed by Western blot with anti-GRP78 and anti–SREBP-1 or anti-calnexin as a nonrelevant microsomal antibody. An aliquot of microsomal proteins before immunoprecipitation from a pooled preparation of ob/+ mice livers was run on the gel in parallel (Input). Control immunoprecipitation with an irrelevant antibody, Glut1, was made on microsomal proteins from a pooled preparation of ob/+ mouse livers. The quantified ratio of GRP78 to SREBP-1 precursor is shown below. ***P < 0.001 versus ob/+.
Overexpression of GRP78 in cultured hepatocytes reduced both thapsigargin- and insulin-induced SREBP-1c cleavage, suggesting that this chaperone could be a common actor of ER stress and insulin-induced SREBP-1c cleavage (Figure 12B). An interesting possibility could be that GRP78 interacts with the SREBP-1c complex, as has been described for other membrane effectors (PERK, IRE1, and ATF6) of the UPR (21), keeping them in an inactivated state. As preliminary evidence for this possibility, we determined the ability of the SREBP-1c precursor complex to associate with GRP78 in the ob/+ mouse liver. Membranes of ob/+ livers were immunoprecipitated with an antibody against SREBP-1c, and immunoprecipitates were tested for the presence of GRP78. GRP78 was indeed coimmunoprecipitated (Figure 12C). Appropriate controls demonstrated that this association between SREBP-1c precursor complex and GRP78 was specific (Figure 12C). Interestingly, in liver membranes of ob/ob mice expressing amounts of SREBP-1c precursor roughly similar to those of ob/+ mice, coimmunoprecipitated GRP78 was much lower (Figure 12C), which suggests that in these mice GRP78 is dissociated from the SREBP complex, a feature that fits with active SREBP-1c cleavage.
Discussion
The results of the present studies point to a potential role for GRP78 in the regulation of SREBP-1c cleavage induced by insulin or ER stress. They also demonstrate that the hepatic steatosis and lipid disorders present in obese insulin-resistant rodents are strongly related to the activation of hepatic ER stress through the processing of SREBP-1c and probably SREBP-2. In addition, ER stress could contribute to hepatic insulin resistance through both increased IRS serine phosphorylation and decreased IRS-2 expression.
ER stress and insulin resistance.
The ob/ob mice are characterized by a strong decrease in the expression and insulin-induced tyrosine phosphorylation of IRS-2 and a lower decrease of IRS-1 expression and phosphorylation (ref. 16 and the present study). The decreased tyrosine phosphorylation status of IRS-1 and IRS-2 seems to be the consequence of the activation of JNK1 (36), a phenomenon that can be attributed to concomitant ER stress (ref. 17 and the present study). JNK1 in turn phosphorylates IRS proteins on serine residues, which reduces their tyrosine phosphorylation by the IR. In the livers of steatotic animals, PKC-e has also been associated with JNK1 in explaining hepatic insulin resistance (41). It is now well established, as reinforced in the present study, that in addition to decreased tyrosine phosphorylation per IRS unit, IRS-2 expression is markedly decreased in the livers of obese insulin-resistant rodents. Previously, it was convincingly demonstrated that hyperinsulinemia per se could mimic the decreased IRS-2 expression (14). We show here that in a situation of hyperinsulinemia, inhibition of ER stress reversed the decreased IRS-2 expression. This raises the interesting possibility that ER stress is a mediator of hyperinsulinemia for decreased IRS-2 expression; in other words, that insulin can induce ER stress. This could explain the prior observation that in refed rodents, markers of ER stress are increased (42).
Distinct roles for IRS-1 and IRS-2 in hepatic metabolism have been described previously: IRS-2 is involved in the inhibitory effects of insulin on hepatic glucose production, particularly PEPCK and G6Pase expression, whereas IRS-1 transduces the stimulatory effects of insulin on glucose-utilizing processes and particularly SREBP-1c and its target genes, GK and lipogenic enzymes (14, 43). Thus, in ob/ob mice, the high expression of PEPCK and G6Pase can be explained by the reduced expression and phosphorylation of IRS-2. However, because signaling through IRS-1 is not increased in ob/ob mice, it cannot explain the overactivation of glycolytic and lipogenic genes.
ER stress and hepatic steatosis.
A number of recent reports have emphasized the link between the ER stress response and hepatic lipid metabolism, mainly through genetic ablation studies. Rutkowski et al. showed that unresolved ER stress — obtained by injecting tunicamycin in mice deficient in specific genes encoding ER stress pathways — induces hepatic lipid accumulation (44), whereas Lee et al. showed that conditional liver XBP-1 disruption is concomitant with decreased hepatic lipogenesis (45). Oyadomari et al. showed that compromised signaling through translation initiation factor 2α (eIF2α) leads to diminished hepatosteatosis in animals fed a high-fat diet (42). We propose here that hepatic ER stress is the missing link in explaining the high rate of lipogenesis in obese insulin-resistant rodents. We demonstrated in vitro that ER stress indeed activated the lipogenic pathway by increasing SREBP-1c cleavage and ChREBP nuclear concentrations. A likely explanation for ChREBP activation is that activation of SREBP-1c induces expression of the gene encoding GK. This is a compulsory step for ChREBP activation, since it allows a high flux of glycolysis and generation of the ChREBP-activating metabolite. Inhibition of hepatic ER stress reduced in vivo SREBP-1c and ChREBP activation, which in turn markedly reduced the concomitant steatosis through decreased fatty acid synthesis. Increased fatty acid oxidation could also be a contributing factor by deinhibition of carnitine palmitoyl transferase I through a decrease in malonyl-CoA concentration.
ER stress and cholesterol metabolism.
In ob/ob mice, hepatic cholesterol concentration was markedly elevated and was related to increased expression of enzymes involved in cholesterol synthesis and cholesterol uptake via the LDL receptor. This increased expression demonstrated that the SREBP-2 transcription factor is activated. However, it occurred despite cholesterol abundance, which suggests that the usual control of SREBP-2 processing was bypassed. Expression of SREBP-2 and target genes was strongly reduced in vivo by overexpression of GRP78, leading to a large decrease in hepatic cholesterol concentration. Plasma cholesterol was not affected, but this may be explained by decreased cholesterol removal by the liver linked to the lower expression of the LDL receptor. This strongly suggests that ER stress is responsible for SREBP-2 processing and high hepatic cholesterol concentration in the ob/ob mouse liver.
A role for GRP78 in potential mechanisms relating insulin, ER stress, and SREBP-1c processing.
Association of ER stress with the activation of SREBPs has been found in several situations: in obese insulin-resistant rodents (the present study); in homocysteine-induced ER stress (23); in cellular stress (46); in alcohol-fed animals (47); and in several cell types, including hepatocytes, adipocytes, and mammary cells (48) as well as Hela and CHO cells. The cellular mechanisms linking ER stress to SREBP activation are still not fully understood, but some observations, including our present findings, allow us to raise the hypothesis that the disappearance of Insig-1 protein could be a key step in this process. In CHO-7 cells, ER stress inhibits Insig-1 synthesis, leading to rapid disappearance of the protein linked to its high turnover rate despite high Insig-1 mRNA content (49). Overexpression of Insig-1 reverses the effect of ER stress on SREBP processing (49). In murine embryonic fibroblasts, SREBP maturation during ER stress is concomitant with Insig-1 depletion, a process that seems to depend on PERK activity (48). We found very similar results in the livers of ob/ob mice: absence of Insig-1 protein despite high levels of its mRNA. Interestingly, in these situations, Insig-2 protein was unaffected or even increased, which suggests that Insig-1 could be the major Insig isoform involved in the ER stress–induced cleavage process. However, this is not the sole mechanism involved in the regulation of SREBP processing by ER stress, because overexpression of GRP78 — which strongly reduces SREBP cleavage — is not concomitant with increased expression of Insig proteins. We propose another explanation, initially based on the fact that overexpression of GRP78 both decreases in vitro ER stress– and insulin-induced SREBP-1c cleavage and decreases in vivo SREBP-1c nuclear content in lean mice. Thus, a provocative but logical hypothesis would be that the SREBP-SCAP complex is like other key integral membrane proteins of the UPR associated with GRP78, and, as with the transcription factor ATF6, the export of the complex toward the Golgi requires its dissociation from GRP78 (50). As shown in Figure 12, we did observe interaction between the SREBP-1c complex and GRP78. We cannot from this sole experiment distinguish between association of GRP78 with SREBP-1c itself or association with other proteins, such as SCAP. The proposed mechanism for SREBP-SCAP complex sorting into COPII-coated vesicles is that a conformational change of SCAP exposes a specific MELADL sequence on the cytosolic side that then binds to COPII proteins (51, 52). Cholesterol and oxysterols favor the binding of SCAP to Insig, and this interaction renders the MELADL sequence inaccessible to COPII proteins. It is presently not known how insulin acts on the complex, but it is likely that it also induces a change in the accessibility of the MELADL sequence. One potential scenario is that GRP78 associates with a protein of the SREBP complex and that this association blocks the accessibility of the MELADL sequence. Modification of the conformation of regulatory proteins such as SCAP by insulin, by altered cholesterol concentration, or by ER stress would induce GRP78 dissociation and activate the SREBP complex sorting into COPII vesicles. This would explain why ER stress is able to bypass the usual regulations for SREBP processing and why overexpression of GRP78 inhibited both ER stress and insulin-induced SREBP-1c proteolytic cleavage. GRP78 may therefore be a new regulatory input in the general process of SREBP-1c cleavage, although further studies are required to address this process in detail.
Potential sequence of events leading to triglyceride accumulation.
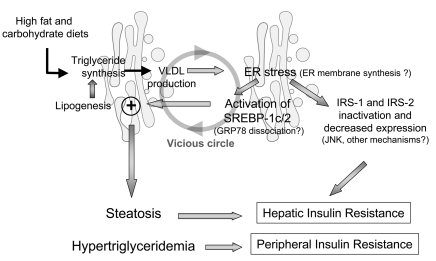
A potential succession of events in obese rodent models (Figure 13) could consist of first an abnormally high synthesis of triglycerides in the liver, caused by either excessive insulin-mediated lipogenic rate (e.g., intake of a high-carbohydrate diet in leptin-deficient ob/ob mice) or by excessive hepatic fatty acid uptake (e.g., consumption of a high-fat diet). Because triglyceride synthesis and packaging take place initially in the ER and involve de novo membrane formation, this could be interpreted as a stress situation by this organelle, which then reacts by synthesizing more membranes. Indeed, increased triglyceride synthesis in hepatic cells induces ER stress (53). Activation of the UPR would then rapidly affect the functional activity or quantity of a key protein (e.g., Insig-1 or GRP78) in the SREBP complex, leading to the release of SREBP-SCAP and proteolytic cleavage in the Golgi apparatus of the mature forms of these transcription factors independent of insulin and cholesterol status. SREBP factors in the liver — but probably also in other cell types — could then be considered as a branch of the UPR, allowing the synthesis of lipids for extra membranes. However, activation of these factors by exacerbating lipogenesis and cholesterol synthesis would perpetuate the initial disequilibrium, leading to a vicious circle. In addition, ER stress would contribute to hepatic insulin resistance by activating the process of IRS serine phosphorylation and decreased IRS-2 expression.
Figure 13. Model illustrating how an initial ER stress linked to a high triglyceride synthesis rate in the liver could induce SREBP-1c cleavage and insulin resistance and initiate a vicious circle leading to steatosis and dyslipidemia.
These findings are highly relevant to steatotic syndromes in humans. Activation of lipogenesis in humans with NAFLD has been shown previously (1), and recently, the UPR was found to be activated to varying degrees in the livers of individuals with NAFLD (54). Thus, in the livers of obese insulin-resistant rodents, ER stress was responsible for the steatosis and cholesterol anomalies by activating the processing of SREBPs and bypassing their usual regulation. In addition, ER stress modifies IRS expression in a way that contributes to the prevailing insulin resistance. We now have several arguments to consider in support of the fact that, at least in the liver, SREBPs can be considered full partners of the UPR response: (a) they are transmembrane proteins of the ER, activated by various ER stresses in a way similar to ATF6; (b) this activation is repressed, as is done for PERK, IRE1, and ATF6, by GRP78 overexpression; and (c) SREBPs deliver a message to the nucleus that can be considered essential for the expansion of the ER membranes and thus for the functional capacity of the ER.
In conclusion, the results of the present study demonstrated that SREBP-1c proteolytic cleavage induced by insulin and ER stress is inhibited by GRP78 and strongly suggest that GRP78 dissociation from the SREBP-1c–SCAP complex is a mechanism involved in insulin action in lean animals. In obese rodents, the GRP78 dissociation induced by ER stress would lead to SREBP-1c maturation and result in hepatic steatosis.
Methods
Animals.
Male ob/+ and ob/ob mice as well as Wistar rats were purchased from Charles River Laboratories and adapted to the environment for 1 wk prior to the study. Lean (fa/+) and obese (fa/fa) Zucker rats were bred in our laboratory. All animals were housed with a 12-h light/12-h dark cycle in a temperature-controlled environment and had free access to water and regular diet (65% carbohydrate, 11% fat, 24% protein). All procedures were approved by the Regional Ethics Committee in animal experiments no. 3 of Ile-de-France (agreement no. p3/2008/029).
Injection of Ad GRP78.
Male ob/+ and ob/ob mice (8 wk old) were anesthetized with Aerane (Baxter) and injected through the penis vein with 108 PFU/g BW of Ad β-gal or Ad GRP78 in a final volume of 250 μl sterile physiological serum. Ad GRP78 was constructed as described previously by Satoh et al. (55). Adenoviruses were amplified in HEK 293 cells and purified on a cesium chloride gradient.
Hyperinsulinemic-euglycemic clamps.
At 8 d before the hyperinsulinemic-euglycemic clamp studies, indwelling catheters were placed into the jugular vein. The mice were allowed to recover for 3–4 d and were injected into the catheter with 108 PFU/g BW of Ad β-gal or Ad GRP78. Clamp studies were performed on 5-h fasted mice as previously described (56) 4 d after injection. Briefly, a bolus of insulin (16 mU/kg) and D-(3H)3-glucose (5 μCi/kg) were injected, and thereafter insulin was infused at a constant rate of 3.3 mU/min/kg. D-(3H)3-glucose was concomitantly infused with insulin at a rate of 30 μCi/kg•min. Blood was sampled by caudal vessels every 15 min to determine blood glucose concentration and to adjust the rate of glucose infusion to maintain blood glucose at its initial basal level in each group. The euglycemic clamp was attained within 60 min and maintained for 30–40 min thereafter. At the end of the clamp, study blood samples were taken in order to determine blood glucose–specific activity, thus allowing calculation of glucose turnover rate and hepatic glucose production by standard equations at equilibrium, as previously described (56).
Rat hepatocyte isolation, culture, and infection with recombinant adenoviruses.
Rat hepatocytes were isolated and cultured as described previously (57). Hepatocytes were cultured in M199 with Earle’s salts (Invitrogen) supplemented with antibiotics and dexamethasone. For the induction of ER stress, hepatocytes were incubated in the presence of thapsigargin (Merck), tunicamycin (Sigma-Aldrich), or DTT (Invitrogen) as detailed in the figure legends. For adenovirus infection, hepatocytes were incubated for 2 h at 37°C in medium M199 with 25 mM glucose and containing Ad GRP78 or Ad β-gal at a titer of 10 PFU/cell.
Isolation of total RNA and quantitative RT-PCR.
Total RNA was isolated as described previously (57). We retro transcribed 1 μg RNA using Superscript II (Invitrogen). Real-time quantitative RT-PCR (qRT-PCR) analyses were performed with 25 ng cDNA and 250 nM sense and antisense primers (Eurogentec) in a final reaction volume of 25 μl by using the qPCR Core Kit (Eurogentec) and the MyiQ real-time PCR detection system (Bio-Rad). Specific primers were designed using Primer Express software (version 1.0; Applied Biosystems) and are shown in Supplemental Table 1. Relative quantification of each gene was calculated after normalization to 18S ribosomal RNA by using the comparative Ct method. For analysis of XBP-1 mRNA splicing, 200 ng cDNA was amplified with specific primers for the rat XBP-1 gene (forward, 5′-CCATGGGAAGATGTTCTGGG-3′; reverse, 5′-ACACGCTTGGGGATGAATGC-3′). The conditions used for the PCR were described previously (20). PCR products were separated by electrophoresis on 2.5% agarose gels and visualized by ethidium bromide staining.
Preparation of liver extracts, Western blot analysis, and immunoprecipitation.
Microsomal membranes and nuclear extracts from mouse livers and hepatocytes were isolated as previously described (6, 58). Total lysates from mouse livers were prepared by homogenizing 20–50 mg tissue in 300 μl lysis buffer (50 mM Tris, pH 7.4; 0.27 M sucrose; 1 mM Na3VO4; 1 mM EDTA; 1 mM EGTA; 10 mM β-glycerophosphate; 50 mM NaF; 5 mM tetrasodium pyrophosphate; 0.1% [v/v] β-mercaptoethanol; 1% [v/v] Triton X-100; and 1 mM phenylmethylsulfonylfluoride) supplemented with a cocktail of protease inhibitors (Roche).
Proteins were separated electrophoretically by SDS-PAGE and transferred to a nitrocellulose membrane (GE Healthcare). The membranes were hybridized with the following antibodies: SREBP-1 (clone 2A4; Thermo Scientific), ATF6 (Imgenex), ChREBP (Novus Biological), XBP-1 (Prosci), and GRP78 and Insig-1 (Santa Cruz Biotechnology Inc.). The antibody against Insig-2 was developed as described previously (6). Phospho-PKB (Thr308/Ser473), PKB, phospho-FoxO1, FoxO1, phospho-JNK (Thr183/Tyr185), and JNK were purchased from Cell Signaling Technology. Phospho-Tyr-HRP antibody was a kind gift of J. Cherfils (INSERM, UMR-S 872, Centre de Recherche des Cordeliers, Paris, France). IRS-2, phospho–IRS-1 (Tyr612), phospho–IRS-1 (Ser307), IRS-1, and anti-PI3K p85 were obtained from Upstate. Mouse lamin A/C antibody was used as a loading control for nuclear proteins, and calnexin was used as a nonrelevant microsomal antibody (BD Biosciences). The IR-β antibody and the anti–phosphotyrosine HRP were obtained from Santa Cruz Biotechnology Inc. Immunoprecipitating anti–SREBP-1 polyclonal antibody (H-160) and anti-Glut1 polyclonal antibody (H-43) were obtained from Santa Cruz Biotechnology Inc.
For immunoprecipitation of IRS-1 and IRS-2, 500 μg of liver extracts were incubated with specific antibodies against IRS-1 or IRS-2 overnight at 4°C. Protein A sepharose beads (Sigma-Aldrich) were added, followed by incubation for 4 h at 4°C. After washing 3 times with lysis buffer, the immunocomplexes were resolved on 7% SDS-PAGE. Phosphorylated or total protein was analyzed by immunoblotting with specific antibodies against phosphotyrosine, IRS-1, IRS-2, and anti-PI3K p85.
SREBP-1 and GRP78 coimmunoprecipitation.
Microsomal membrane proteins (1 mg) were solubilized for 2 h at 4°C on a rotating wheel in 50 mM HEPES buffer (pH 7.6), 100 mM NaCl, 1.5 mM MgCl2, 1% (w/v) 3-[(3-Cholamidopropyl)dimethylammonio]-1-propanesulfonate, and protease inhibitors (phenylmethylsulfonylfluoride, aprotinin, and leupeptin). Aggregates were removed by centrifugation at 16,000 g for 10 min. The supernatant was precleared by rotating incubation for 1 h with 40 μl protein G sepharose beads (Sigma-Aldrich). Soluble proteins were immunoprecipitated for 16 h at 4°C with 10 μl of the polyclonal anti–SREBP-1 or anti-Glut1 antibody (irrelevant antibody) bound to 40 μl protein G sepharose and washed 3 times in solubilization buffer. Bound proteins were resolved by 10% SDS-PAGE under reducing conditions and transferred to nitrocellulose membranes. Blots were incubated with anti–SREBP-1, anti-GRP78, or anti-calnexin.
Histological analysis.
For H&E (Sigma-Aldrich), liver tissues were fixed in 10% neutral-buffered formalin, embedded in paraffin, and cut into 7-μm sections. For Oil red O (Sigma-Aldrich), liver was frozen in liquid nitrogen and cut into 10-μm sections. Sections were stained and analyzed at ×20 magnification using a Leica DMRB microscope.
Analytical procedures.
Serum concentrations of aspartate aminotransferase, alanine aminotransferase, triglycerides, and cholesterol were determined using commercial kits (Randox Laboratories).
Statistics.
Results are expressed as mean ± SEM. Statistical significance was assessed using 2-way ANOVA. A P value less than 0.05 was considered significant.
Supplementary Material
Acknowledgments
This work was supported by Agence Nationale de la Recherche grant ANR-2005-PCOD-035; by Alfediam-Takeda; and by EXGENESIS Integrated Project Grant LSHM-CT-2004-005272, funded by the European Commission. H.L. Kammoun is the recipient of a doctoral fellowship from the Ministère de la Recherche et de l’Enseignement supérieur. We thank Bertrand Blondeau for helpful advice for the staining experiments, Delphine Dorchenne for performing liver sections, and Julien Castel for surgical procedures.
Footnotes
Authorship note: Hélène L. Kammoun, Hervé Chabanon, and Isabelle Hainault contributed equally to this work.
Conflict of interest: The authors have declared that no conflict of interest exists.
Nonstandard abbreviations used: ATF, activating transcription factor; ChREBP, carbohydrate response element–binding protein; COPII, coatomer protein II; FAS, fatty acid synthase; GK, glucokinase; G6Pase, glucose-6-phosphatase; GRP78, glucose-regulated protein 78; Insig, insulin-induced gene; IRE1, inositol-requiring kinase–1; NAFLD, nonalcoholic fatty liver disease; PEPCK, phosphoenolpyruvate carboxykinase; PERK, protein kinase RNA–like ER kinase; qRT-PCR, quantitative RT-PCR; SCAP, SREBP cleavage–activating protein; SCD, stearoyl-CoA desaturase; UPR, unfolded protein response; XBP, X box–binding protein.
Citation for this article: J. Clin. Invest. 119:1201–1215 (2009). doi:10.1172/JCI37007
References
- 1.Donnelly K.L., et al. Sources of fatty acids stored in liver and secreted via lipoproteins in patients with nonalcoholic fatty liver disease. . J. Clin. Invest. 2005;115:1343–1351. doi: 10.1172/JCI23621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Foufelle F., Ferre P. New perspectives in the regulation of hepatic glycolytic and lipogenic genes by insulin and glucose: a role for the transcription factor sterol regulatory element binding protein-1c. Biochem. J. 2002;366:377–391. doi: 10.1042/BJ20020430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Postic C., Dentin R., Denechaud P.D., Girard J. ChREBP, a transcriptional regulator of glucose and lipid metabolism. Annu. Rev. Nutr. 2007;27:179–192. doi: 10.1146/annurev.nutr.27.061406.093618. [DOI] [PubMed] [Google Scholar]
- 4.Foretz M., et al. ADD1/SREBP-1c is required in the activation of hepatic lipogenic gene expression by glucose. Mol. Cell. Biol. 1999;19:3760–3768. doi: 10.1128/mcb.19.5.3760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shimomura I., et al. Insulin selectively increases SREBP-1c mRNA in the livers of rats with streptozotocin-induced diabetes. Proc. Natl. Acad. Sci. U. S. A. 1999;96:13656–13661. doi: 10.1073/pnas.96.24.13656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hegarty B.D., et al. Distinct roles of insulin and liver X receptor in the induction and cleavage of sterol regulatory element-binding protein-1c. Proc. Natl. Acad. Sci. U. S. A. 2005;102:791–796. doi: 10.1073/pnas.0405067102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brown M.S., Goldstein J.L. A proteolytic pathway that controls the cholesterol content of membranes, cells, and blood. Proc. Natl. Acad. Sci. U. S. A. 1999;96:11041–11048. doi: 10.1073/pnas.96.20.11041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goldstein J.L., DeBose-Boyd R.A., Brown M.S. Protein sensors for membrane sterols. Cell. 2006;124:35–46. doi: 10.1016/j.cell.2005.12.022. [DOI] [PubMed] [Google Scholar]
- 9.Shimano H., et al. Isoform 1c of sterol regulatory element binding protein is less active than isoform 1a in livers of transgenic mice and in cultured cells. J. Clin. Invest. 1997;99:846–854. doi: 10.1172/JCI119248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shimano H., et al. Sterol regulatory element-binding protein-1 as a key transcription factor for nutritional induction of lipogenic enzyme genes. . J. Biol. Chem. 1999;274:35832–35839. doi: 10.1074/jbc.274.50.35832. [DOI] [PubMed] [Google Scholar]
- 11.Becard D., et al. Adenovirus-mediated overexpression of sterol regulatory element binding protein-1c mimics insulin effects on hepatic gene expression and glucose homeostasis in diabetic mice. Diabetes. 2001;50:2425–2430. doi: 10.2337/diabetes.50.11.2425. [DOI] [PubMed] [Google Scholar]
- 12.Liang G., et al. Diminished hepatic response to fasting/refeeding and liver X receptor agonists in mice with selective deficiency of sterol regulatory element-binding protein-1c. J. Biol. Chem. 2002;277:9520–9528. doi: 10.1074/jbc.M111421200. [DOI] [PubMed] [Google Scholar]
- 13.Shimomura I., Bashmakov Y., Horton J.D. Increased levels of nuclear SREBP-1c associated with fatty livers in two mouse models of diabetes mellitus. J. Biol. Chem. 1999;274:30028–30032. doi: 10.1074/jbc.274.42.30028. [DOI] [PubMed] [Google Scholar]
- 14.Shimomura I., et al. Decreased IRS-2 and increased SREBP-1c lead to mixed insulin resistance and sensitivity in livers of lipodystrophic and ob/ob mice. Mol. Cell. 2000;6:77–86. doi: 10.1016/S1097-2765(00)00009-5. [DOI] [PubMed] [Google Scholar]
- 15.Saad M.J., et al. Regulation of insulin receptor substrate-1 in liver and muscle of animal models of insulin resistance. J. Clin. Invest. 1992;90:1839–1849. doi: 10.1172/JCI116060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kerouz N.J., Horsch D., Pons S., Kahn C.R. Differential regulation of insulin receptor substrates-1 and -2 (IRS-1 and IRS-2) and phosphatidylinositol 3-kinase isoforms in liver and muscle of the obese diabetic (ob/ob) mouse. J. Clin. Invest. 1997;100:3164–3172. doi: 10.1172/JCI119872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ozcan U., et al. Endoplasmic reticulum stress links obesity, insulin action, and type 2 diabetes. Science. 2004;306:457–461. doi: 10.1126/science.1103160. [DOI] [PubMed] [Google Scholar]
- 18.Ozawa K., et al. The endoplasmic reticulum chaperone improves insulin resistance in type 2 diabetes. Diabetes. 2005;54:657–663. doi: 10.2337/diabetes.54.3.657. [DOI] [PubMed] [Google Scholar]
- 19.Nakatani Y., et al. Involvement of endoplasmic reticulum stress in insulin resistance and diabetes. J. Biol. Chem. 2005;280:847–851. doi: 10.1074/jbc.M411860200. [DOI] [PubMed] [Google Scholar]
- 20.Ozcan U., et al. Chemical chaperones reduce ER stress and restore glucose homeostasis in a mouse model of type 2 diabetes. Science. 2006;313:1137–1140. doi: 10.1126/science.1128294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu J., Kaufman R.J. From acute ER stress to physiological roles of the Unfolded Protein Response. Cell Death Differ. 2006;13:374–384. doi: 10.1038/sj.cdd.4401840. [DOI] [PubMed] [Google Scholar]
- 22.Ron D., Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat. Rev. Mol. Cell Biol. 2007;8:519–529. doi: 10.1038/nrm2199. [DOI] [PubMed] [Google Scholar]
- 23.Werstuck G.H., et al. Homocysteine-induced endoplasmic reticulum stress causes dysregulation of the cholesterol and triglyceride biosynthetic pathways. J. Clin. Invest. 2001;107:1263–1273. doi: 10.1172/JCI11596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen G., Liang G., Ou J., Goldstein J.L., Brown M.S. Central role for liver X receptor in insulin-mediated activation of Srebp-1c transcription and stimulation of fatty acid synthesis in liver. Proc. Natl. Acad. Sci. U. S. A. 2004;101:11245–11250. doi: 10.1073/pnas.0404297101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cagen L.M., et al. Insulin activates the rat sterol-regulatory-element-binding protein 1c (SREBP-1c) promoter through the combinatorial actions of SREBP, LXR, Sp-1 and NF-Y cis-acting elements. Biochem. J. 2005;385:207–216. doi: 10.1042/BJ20040162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang T., et al. Crucial step in cholesterol homeostasis: sterols promote binding of SCAP to INSIG-1, a membrane protein that facilitates retention of SREBPs in ER. Cell. 2002;110:489–500. doi: 10.1016/S0092-8674(02)00872-3. [DOI] [PubMed] [Google Scholar]
- 27.Yabe D., Komuro R., Liang G., Goldstein J.L., Brown M.S. Liver-specific mRNA for Insig-2 down-regulated by insulin: implications for fatty acid synthesis. Proc. Natl. Acad. Sci. U. S. A. 2003;100:3155–3160. doi: 10.1073/pnas.0130116100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim S.Y., et al. SREBP-1c mediates the insulin-dependent hepatic glucokinase expression. . J. Biol. Chem. 2004;279:30823–30829. doi: 10.1074/jbc.M313223200. [DOI] [PubMed] [Google Scholar]
- 29.Yamashita H., et al. A glucose-responsive transcription factor that regulates carbohydrate metabolism in the liver. Proc. Natl. Acad. Sci. U. S. A. 2001;98:9116–9121. doi: 10.1073/pnas.161284298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Koo S.H., Dutcher A.K., Towle H.C. Glucose and insulin function through two distinct transcription factors to stimulate expression of lipogenic enzyme genes in liver. J. Biol. Chem. 2001;276:9437–9445. doi: 10.1074/jbc.M010029200. [DOI] [PubMed] [Google Scholar]
- 31.Dentin R., et al. Hepatic glucokinase is required for the synergistic action of ChREBP and SREBP-1c on glycolytic and lipogenic gene expression. J. Biol. Chem. 2004;279:20314–20326. doi: 10.1074/jbc.M312475200. [DOI] [PubMed] [Google Scholar]
- 32.Reddy R.K., et al. Endoplasmic reticulum chaperone protein GRP78 protects cells from apoptosis induced by topoisomerase inhibitors: role of ATP binding site in suppression of caspase-7 activation. J. Biol. Chem. 2003;278:20915–20924. doi: 10.1074/jbc.M212328200. [DOI] [PubMed] [Google Scholar]
- 33.Colgan S.M., Tang D., Werstuck G.H., Austin R.C. Endoplasmic reticulum stress causes the activation of sterol regulatory element binding protein-2. Int. J. Biochem. Cell Biol. 2007;39:1843–1851. doi: 10.1016/j.biocel.2007.05.002. [DOI] [PubMed] [Google Scholar]
- 34.Neschen S., et al. Prevention of hepatic steatosis and hepatic insulin resistance in mitochondrial acyl-CoA:glycerol-sn-3-phosphate acyltransferase 1 knockout mice. Cell Metab. 2005;2:55–65. doi: 10.1016/j.cmet.2005.06.006. [DOI] [PubMed] [Google Scholar]
- 35.Postic C., Girard J. Contribution of de novo fatty acid synthesis to hepatic steatosis and insulin resistance: lessons from genetically engineered mice. J. Clin. Invest. 2008;118:829–838. doi: 10.1172/JCI34275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hirosumi J., et al. A central role for JNK in obesity and insulin resistance. Nature. 2002;420:333–336. doi: 10.1038/nature01137. [DOI] [PubMed] [Google Scholar]
- 37.Nakatani Y., et al. Modulation of the JNK pathway in liver affects insulin resistance status. . J. Biol. Chem. 2004;279:45803–45809. doi: 10.1074/jbc.M406963200. [DOI] [PubMed] [Google Scholar]
- 38.Matsumoto M., Pocai A., Rossetti L., Depinho R.A., Accili D. Impaired regulation of hepatic glucose production in mice lacking the forkhead transcription factor Foxo1 in liver. Cell Metab. 2007;6:208–216. doi: 10.1016/j.cmet.2007.08.006. [DOI] [PubMed] [Google Scholar]
- 39.Puigserver P., et al. Insulin-regulated hepatic gluconeogenesis through FOXO1-PGC-1alpha interaction. Nature. 2003;423:550–555. doi: 10.1038/nature01667. [DOI] [PubMed] [Google Scholar]
- 40.Hedo J.A., Simpson I.A. Biosynthesis of the insulin receptor in rat adipose cells. Intracellular processing of the Mr-190 000 pro-receptor. Biochem. J. 1985;232:71–78. doi: 10.1042/bj2320071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Samuel V.T., et al. Inhibition of protein kinase Cepsilon prevents hepatic insulin resistance in nonalcoholic fatty liver disease. J. Clin. Invest. 2007;117:739–745. doi: 10.1172/JCI30400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Oyadomari S., Harding H.P., Zhang Y., Oyadomari M., Ron D. Dephosphorylation of translation initiation factor 2alpha enhances glucose tolerance and attenuates hepatosteatosis in mice. Cell Metab. 2008;7:520–532. doi: 10.1016/j.cmet.2008.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kubota N., et al. Dynamic functional relay between insulin receptor substrate 1 and 2 in hepatic insulin signaling during fasting and feeding. Cell Metab. 2008;8:49–64. doi: 10.1016/j.cmet.2008.05.007. [DOI] [PubMed] [Google Scholar]
- 44.Rutkowski D.T., et al. UPR pathways combine to prevent hepatic steatosis caused by ER stress-mediated suppression of transcriptional master regulators. Dev. Cell. 2008;15:829–840. doi: 10.1016/j.devcel.2008.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee A.H., Scapa E.F., Cohen D.E., Glimcher L.H. Regulation of hepatic lipogenesis by the transcription factor XBP1. Science. 2008;320:1492–1496. doi: 10.1126/science.1158042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ye J., et al. ER stress induces cleavage of membrane-bound ATF6 by the same proteases that process SREBPs. Mol. Cell. 2000;6:1355–1364. doi: 10.1016/S1097-2765(00)00133-7. [DOI] [PubMed] [Google Scholar]
- 47.Ji C., Chan C., Kaplowitz N. Predominant role of sterol response element binding proteins (SREBP) lipogenic pathways in hepatic steatosis in the murine intragastric ethanol feeding model. J. Hepatol. 2006;45:717–724. doi: 10.1016/j.jhep.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 48.Bobrovnikova-Marjon E., et al. PERK-dependent regulation of lipogenesis during mouse mammary gland development and adipocyte differentiation. Proc. Natl. Acad. Sci. U. S. A. 2008;105:16314–16319. doi: 10.1073/pnas.0808517105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lee J.N., Ye J. Proteolytic activation of sterol regulatory element-binding protein induced by cellular stress through depletion of Insig-1. . J. Biol. Chem. 2004;279:45257–45265. doi: 10.1074/jbc.M408235200. [DOI] [PubMed] [Google Scholar]
- 50.Shen J., Chen X., Hendershot L., Prywes R. ER stress regulation of ATF6 localization by dissociation of BiP/GRP78 binding and unmasking of Golgi localization signals. Dev. Cell. 2002;3:99–111. doi: 10.1016/S1534-5807(02)00203-4. [DOI] [PubMed] [Google Scholar]
- 51.Sun L.P., Li L., Goldstein J.L., Brown M.S. Insig required for sterol-mediated inhibition of Scap/SREBP binding to COPII proteins in vitro. J. Biol. Chem. 2005;280:26483–26490. doi: 10.1074/jbc.M504041200. [DOI] [PubMed] [Google Scholar]
- 52.Sun L.P., Seemann J., Goldstein J.L., Brown M.S. Sterol-regulated transport of SREBPs from endoplasmic reticulum to Golgi: Insig renders sorting signal in Scap inaccessible to COPII proteins. Proc. Natl. Acad. Sci. U. S. A. 2007;104:6519–6526. doi: 10.1073/pnas.0700907104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ota T., Gayet C., Ginsberg H.N. Inhibition of apolipoprotein B100 secretion by lipid-induced hepatic endoplasmic reticulum stress in rodents. J. Clin. Invest. 2008;118:316–332. doi: 10.1172/JCI32752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Puri P., et al. Activation and dysregulation of the unfolded protein response in nonalcoholic fatty liver disease. Gastroenterology. 2008;134:568–576. doi: 10.1053/j.gastro.2007.10.039. [DOI] [PubMed] [Google Scholar]
- 55.Satoh T., et al. Facilitatory roles of novel compounds designed from cyclopentenone prostaglandins on neurite outgrowth-promoting activities of nerve growth factor. J. Neurochem. 2000;75:1092–1102. doi: 10.1046/j.1471-4159.2000.0751092.x. [DOI] [PubMed] [Google Scholar]
- 56.Troy S., et al. Intestinal gluconeogenesis is a key factor for early metabolic changes after gastric bypass but not after gastric lap-band in mice. Cell Metab. 2008;8:201–211. doi: 10.1016/j.cmet.2008.08.008. [DOI] [PubMed] [Google Scholar]
- 57.Foretz M., Guichard C., Ferre P., Foufelle F. Sterol regulatory element binding protein-1c is a major mediator of insulin action on the hepatic expression of glucokinase and lipogenesis-related genes. Proc. Natl. Acad. Sci. U. S. A. 1999;96:12737–12742. doi: 10.1073/pnas.96.22.12737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bobard A., Hainault I., Ferre P., Foufelle F., Bossard P. Differential regulation of sterol regulatory element-binding protein 1c transcriptional activity by insulin and liver X receptor during liver development. J. Biol. Chem. 2005;280:199–206. doi: 10.1074/jbc.M406522200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.